Abstract
Rhodopsins possess retinal chromophore surrounded by seven transmembrane α-helices, are widespread in prokaryotes and in eukaryotes, and can be utilized as optogenetic tools. Although rhodopsins work as distinctly different photoreceptors in various organisms, they can be roughly divided according to their two basic functions, light-energy conversion and light-signal transduction. In microbes, light-driven proton transporters functioning as light-energy converters have been modified by evolution to produce sensory receptors that relay signals to transducer proteins to control motility. In this study, we cloned and characterized two newly identified microbial rhodopsins from Haloquadratum walsbyi. One of them has photochemical properties and a proton pumping activity similar to the well known proton pump bacteriorhodopsin (BR). The other, named middle rhodopsin (MR), is evolutionarily transitional between BR and the phototactic sensory rhodopsin II (SRII), having an SRII-like absorption maximum, a BR-like photocycle, and a unique retinal composition. The wild-type MR does not have a light-induced proton pumping activity. On the other hand, a mutant MR with two key hydrogen-bonding residues located at the interaction surface with the transducer protein HtrII shows robust phototaxis responses similar to SRII, indicating that MR is potentially capable of the signaling. These results demonstrate that color tuning and insertion of the critical threonine residue occurred early in the evolution of sensory rhodopsins. MR may be a missing link in the evolution from type 1 rhodopsins (microorganisms) to type 2 rhodopsins (animals), because it is the first microbial rhodopsin known to have 11-cis-retinal similar to type 2 rhodopsins.
Keywords: Membrane Biogenesis, Membrane Biophysics, Membrane Proteins, Photoreceptors, Rhodopsin
Introduction
Rhodopsin molecules are photochemically active membrane-embedded proteins having seven transmembrane α-helices and retinal chromophore (vitamin A aldehyde) (1, 2). Rhodopsins are classified into two groups, microbial (type 1) and animal (type 2) rhodopsins (3). Type 1 rhodopsins are widespread in the microbial world in prokaryotes (bacteria and archaea) and in eukaryotes (fungi and algae) (1, 4–7), and type 2 rhodopsins, such as visual pigments, are G-protein-coupled receptors, widespread in vertebrates and in invertebrates (3, 8). An interesting feature of this photoactive protein family is a wide range of seemingly dissimilar functions performed by its members. Some rhodopsins are light-driven transporters, such as proton pumps bacteriorhodopsin (BR)2 in haloarchaea, xanthorhodopsin in halophilic bacteria, proteorhodopsin in marine bacteria, and Leptosphaeria rhodopsin in fungi (1, 9–13). Those proteins generate an electrochemical membrane potential upon light activation, which is utilized by ATP synthase to produce ATP (14). Other rhodopsins are light sensors, such as the phototaxis receptors sensory rhodopsins I (SRI) and II (SRII) in haloarchaea (1, 5) and mammalian rod rhodopsin and color visual pigments both in vertebrates and in invertebrates (3, 8). These sensory receptors relay signals through protein-protein interactions to integral membrane transducer proteins I (HtrI) and II (HtrII) for SRI and SRII, respectively (15), and to a trimeric G-protein transducin for rhodopsin (3, 8). SRI mediates attractant motility responses to green/orange wavelengths used by ion pumps BR and halorhodopsin (HR) (16), whereas SRII mediates blue light avoidance responses (15). SRI also mediates repellent responses to damaging near-UV light by a color-sensitive two-photon mechanism, so that cells migrate toward green/orange regions but not into full spectrum solar radiation containing damaging short wavelength photons (16, 17). The SRI-HtrI and SRII-HtrII complexes control a cytoplasmic phosphorylation pathway composed of a histidine kinase CheA and a response regulator CheY that modulates the motility of the cells (18). Light-activated rhodopsin binds to the transducin molecule and mediates the GDP-GTP exchange reaction (3, 8), which is common among G-protein-coupled receptors. The signal relay mechanisms by SR-Htr and rhodopsin-transducin complexes have become a focus of research efforts of many groups, in part because of their importance for general understanding of communication between proteins and signal transduction mediated by membrane proteins.
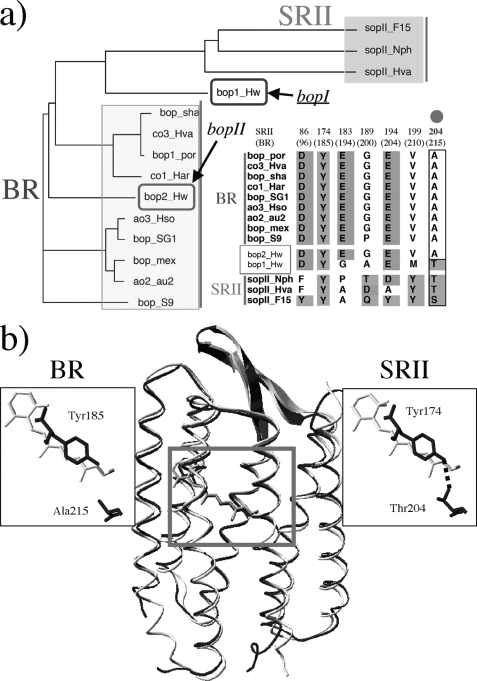
But how did the signaling rhodopsins evolve? In the case of type 1 rhodopsins, phylogenetic analysis strongly suggests that rhodopsin photosensors like SRI and SRII evolved from light-driven proton pumps such as BR (Fig. 1a) (5). In fact, when SRI from Halobacterium salinarum and SRII from Natronomonas pharaonis are separated from their tightly bound transducers HtrI and HtrII, they exhibit light-driven electrogenic proton transport across the membrane (19–21). This suggests that the essential features of the proton transport mechanism of BR are conserved in the signal transduction mechanism of SRs, although primary structures of BR and SRII differ by 74% (supplemental Fig. S1). Moreover, the replacement of only three residues converts BR into an SRII-like phototaxis receptor (22), indicating that the essential features of the signal transduction mechanism of SRII are conserved in the proton transport mechanism of BR as well. In fact, the atomic structure of BR from H. salinarum (23) aligns with that of SRII from the related haloarchaeon N. pharaonis fairly well (Fig. 1b) (24, 25). The studies of the signal-competent BR mutant tell us what changes would be required to convert BR into SRII. A single replacement (A215T) corresponding to Thr-204 in SRII, which bridges the retinal and the membrane-embedded surface (Fig. 1b), confers weak phototaxis signaling activity. Two additional changes (surface substitutions P200T and V210Y), which are expected to align BR and HtrII similar to SRII and HtrII, greatly enhance the signaling (22). Combining this with other findings (26–30), we proposed a model for signaling in which the initial storage of energy of photoisomerization in hydrogen bond between Tyr-174, which is in contact with retinal, and Thr-204 is followed by the transfer of that chemical energy to drive structural transitions in HtrII (Fig. 1b) (31). However, questions on when and how the molecular evolution from BR to SRII occurred remain unsolved, because no evolutionary intermediate between BR and SRII has been previously discovered. In other words, it is unclear when the SRII-like properties (blue-shifted absorption maximum, slow photocycle, interaction with HtrII, etc. (32)) appeared.
FIGURE 1.
a, phylogenetic tree of microbial rhodopsins and alignment of critical amino acids of SRII- and BR-like proteins, including the two newly identified microbial rhodopsins from H. walsbyi (bopI and bopII), created using ClustalW. b, x-ray crystallographic structures of BR from H. salinarum (Protein Data Bank code 1C3W) (23) and SRII from N. pharaonis (Protein Data Bank code 1JGJ) (24), and magnified view of the critical functional site of SRII, the hydrogen bond between Tyr-174 and Thr-204.
In 2006, Oesterhelt and co-worker s (33) reported the genomic sequence of the archaeon halophile Haloquadratum walsbyi, which encodes two microbial rhodopsins as shown in Fig. 1a. Interestingly, one of them is located between BR and SRII phylogenetically and has a Thr residue critical for signaling (Fig. 1a) (22, 28). In this study, we characterized properties of these microbial rhodopsins and discussed their implications on the evolutionary pathway from BR to SRII.
EXPERIMENTAL PROCEDURES
Plasmids and Strains
H. walsbyi (a kind gift from Dr. Dyall-Smith) was grown aerobically at 40 °C, pH 7.0, in a medium with the following composition (g/liter): NaCl, 195; MgSO4·7H2O, 25; MgCl2·6H2O, 16.3; CaCl2·2H2O, 1.25; KCl, 5.0; NaHCO3, 0.25; NaBr, 0.625; and yeast extract (Difco), 1.0. Cells were harvested by centrifugation and stored at −80 °C. Genomic DNA was prepared using the method of Marmur (34). Escherichia coli DH5α cells were used as a host for DNA manipulation, and BL21(DE3) cells were used to express the genes. For DNA manipulation, the rhodopsin genes (bopI and bopII, Fig. 1a) were amplified using PCR from the genomic DNA of H. walsbyi. For bopI, the forward primer 5′-CATATGGCGACACCAGGCTCAGAAGCG-3′ and the reverse primer 5′-CTCGAGGTGTGATTTTATATCGTTCGTCGA-3′ were designed (underlined parts indicate the restriction sites for NdeI and XhoI). The stop codon was deleted during amplification. The NdeI and XhoI fragment was ligated to the NdeI and XhoI sites of the pET21c(+) vector (Novagen). Consequently, the plasmid encodes rhodopsin with six histidines at the C terminus, and it was named pET-sqbopI. This cloning strategy resulted in the following N- and C-terminal peptide sequences: 1MATP(X)5DIKSH246LEHHHHHH. For bopII, the forward primer 5′-GCTAGCATGTCTCAGCTGGCGCTGCAAATG-3′ and the reverse primer 5′-CTCGAGGTCCGCAGCAGTTTCTTGTGCTGA-3′ were designed (underlined parts indicate the restriction sites for NheI and XhoI). The stop codon was deleted during amplification. Because the bopII gene contains an NheI site, the amplified fragment was digested into two pieces (large and small fragments) by NheI and XhoI. Therefore, the complete bopII gene cloning was composed of two steps as follows: large fragment cloning into the NheI and XhoI sites of the expression vector and the following small fragment cloning at the NheI site. The expression vector was constructed by introducing multicloning sites of pET21a into the pBAD vector. The direction of the inserted small fragment was confirmed by sequencing. Consequently, the plasmid encodes rhodopsin with six histidines at the C terminus, and it was named pBAD-sqbopII. This cloning strategy resulted in the following N- and C-terminal peptide sequences: 1MASMSQLA(X)5TAAD254LEHHHHHH. All constructed plasmids were analyzed using an automated sequencer (ABI 3100) to confirm the expected nucleotide sequences.
Protein Expression, Purification, and Reconstitution into Phosphatidylcholine Liposomes
Cells were grown in LB medium supplemented with ampicillin (final concentration 50 μg/ml). E. coli BL21(DE3) cells harboring the plasmid were grown to an OD660 of 0.3–0.5 in a 30 °C incubator, followed by the addition of 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for MR or 0.1% l-arabinose for HwBR and 5 μm all-trans-retinal (35). Cells were harvested 10 h post-induction at 18 °C by centrifugation at 4 °C, resuspended in buffer (50 mm MES, pH 6.5) containing 1 m NaCl, and disrupted by sonication or a French press. Cell debris was removed by low speed centrifugation (5000 × g, 10 min, 4 °C). Crude membranes were collected by centrifugation (100,000 × g for 30 min at 4 °C) and washed with buffer (50 mm MES, pH 6.5) containing 1 m NaCl. For solubilization of the membranes, 2% (w/v) n-dodecyl β-d-maltoside (DDM) was added, and the suspension was incubated for 30 min at 4 °C. The solubilized membranes were isolated by high speed centrifugation (100,000 × g for 30 min at 4 °C), and the supernatant was applied to a nickel affinity column (HisTrap, GE Healthcare) at 4 °C in the dark. Thereafter, the column was washed extensively with buffer (50 mm MES, pH 6.5) containing 1 m NaCl, 20 mm imidazole, and 0.05% w/v DDM to remove unspecifically bound proteins. The histidine-tagged proteins were then eluted using a linear gradient of up to 100% elution buffer (0.05% DDM, 1 m NaCl, 50 mm Tris-Cl, pH 7.0, and 500 mm imidazole). The eluted proteins were then further applied to a HiTrapQ ion-exchange column (GE Healthcare) at 4 °C in buffer (50 mm Tris-Cl, pH 7.5) containing 30 mm NaCl and 0.05% DDM. Thereafter, the column was washed extensively with the buffer to remove unspecifically bound proteins. The proteins were then eluted using a linear gradient of up to 100% elution buffer (0.05% DDM, 1 m NaCl, 50 mm Tris-Cl, pH 7.5). For the FTIR measurements, the eluted proteins were then further purified by a Sephacryl S-400 HR gel filtration column (Amersham Biosciences) in buffer containing 0.05% DDM, 1 m NaCl, and 50 mm Tris-Cl, pH 7.0. The purified samples were then reconstituted into phosphatidylcholine liposomes (protein/phosphatidylcholine = 1:30 molar ratio) by removing the detergent with SM2 Bio-Beads (Bio-Rad).
UV-Visible Spectroscopy, HPLC Analysis, and Time-resolved Flash Spectroscopy
The sample was concentrated and exchanged using an Amicon ultra concentrators (Millipore, Bedford, MA) against the media with compositions given below. UV-visible spectra were obtained using a UV2450 spectrophotometer with an ISR2200 integrating sphere (Shimadzu, Kyoto, Japan). High performance liquid chromatography (HPLC) analysis was performed following the published procedure (36). Briefly, the purified sample was analyzed in buffer (0.1% DDM, 1 m NaCl, 50 mm Tris-Cl, pH 7.0). The HPLC system consisted of a PU-2080 pump and an UV-2075 UV/visible detector (JASCO, Tokyo, Japan). The chromatograph was equipped with a silica column (6.0 × 150 mm; YMC-Pack SIL), and the solvent was 12% (v/v) ethyl acetate and 0.12% (v/v) ethanol in hexane, and the flow rate was 1.0 ml/min. Extraction of retinal oxime from the sample was carried out with hexane after denaturation in methanol and 500 mm hydroxylamine at 4 °C. The molar ratios of retinal isomers were calculated from the areas of the peaks monitored at 360 nm. The assignment of each peak was performed by comparing it with the HPLC pattern from retinal oximes of authentic all-trans- and 13-cis-retinals (36). For the assignment of the 11-cis-retinal peak, a mixture of retinal isomers was prepared by irradiating all-trans-retinal dissolved in acetonitrile overnight with a fluorescent lamp according to the standard method (37). For the time-resolved flash spectroscopy, the apparatus and the procedure for the analysis were described previously (38).
FTIR Spectroscopy
FTIR spectroscopy was performed following the established protocol (39). After hydration with H2O or D2O, the sample film was placed in a cell mounted in an Oxford Optistat-DN cryostat placed in a FTS-40 spectrometer (Bio-Rad). Spectra were constructed from 128 interferograms with a spectral resolution of 2 cm−1. To investigate the structural changes upon formation of the K-and M-intermediates, difference spectra were calculated from spectra before and after illumination. For HwBR, illumination with 520 nm light (KL52 filter) at 77 K for 2 min converted HwBR to HwBRK, and a subsequent illumination with >610 nm light (R63 filter) for 1 min reverted the HwBRK back to HwBR, as evidenced by the same (but inverted) spectral shape. The cycles of alternating illuminations with 520 and >610 nm light were repeated a number of times. Thirty two difference spectra obtained in this way were averaged for each HwBRK minus the HwBR spectrum. For MR, illumination with 500 nm light (KL50 filter) at 77 K for 2 min converted the MR to MRK. Because MRK was completely reconverted to MR upon illumination with >540 nm light (O56) for 1 min, as evidenced by the spectral shape, which is a mirror image of that for the MR-MRK transition, cycles of alternating illumination with 500 and >540 nm light were repeated a number of times. Twenty four difference spectra obtained in this way were averaged for each MRK minus MR spectrum.
Light-induced pH Changes
The proton transport activity of each protein was measured by monitoring pH changes using a glass electrode (40). Briefly, spheroplasts containing HwBR or MR protein were illuminated at >500 nm through a glass filter (AGC Techno Glass Y-52, Japan), and the pH value changes were monitored (F-55 pH meter, Horiba, Japan). The amount of protein in the spheroplast suspension was estimated by measuring the absorption spectra after solubilizing the protein with 2% DDM. The concentrations of the solutions with HwBR and MR were 0.06 and 0.04 μm, respectively.
Phototaxis Measurements
H. salinarum strain Pho81Wr−, which lacks the four native rhodopsins (BR, HR, SRI, and SRII) and the two transducer proteins (HtrI and HtrII), and is carotenoid pigment-deficient and restriction-deficient, was used for transformation by following the previously described protocol (22). The expression plasmid was modified from plasmid pYS001 that encodes the WT SRII-HtrII fusion gene (22). For preparation of the MR-HtrII expression plasmid, the 5′ and 3′ ends of the bopI gene were mutated by PCR to introduce NcoI and SpeI restriction sites, respectively. The resulting NcoI-SpeI fragments were ligated with the large fragment of the NcoI/SpeI-treated pYS001 vector. The stop codon was deleted during amplification, which generated a linker region between SRII and HtrII that contains 11 residues (Thr-Ser-Ala-Ser-Ala-Ser-Asn-Gly-Ala-Ser-Ala; 5′-CTAGTGCGTCGGCGTCGAACGGCGCGTCGGCG-3′) (22). The underlined part indicates the added restriction site for SpeI. The A201T/M211Y mutant gene of MR was constructed using the QuikChange site-directed mutagenesis method. All constructed plasmids were analyzed using an automated sequencer to confirm the expected nucleotide sequences. Phototaxis responses were measured as transient swimming reversal frequency changes of cell populations in response to pulse photostimuli. Swimming reversals were measured by stimulus-induced effects as the ratio of rate of change of direction to speed (22).
RESULTS AND DISCUSSION
Two Microbial Rhodopsin Proteins in the Genome of H. walsbyi
The genome sequence of H. walsbyi contains two genes that encode microbial rhodopsins (bopI and bopII) (Fig. 1a and supplemental Fig. S1) and also one gene that encodes a halorhodopsin-like protein (33). The amino acid sequence of bopII (HwBR) is distantly related to BR from H. salinarum (∼52% identity) and contains all amino acid residues (Asp-85, Asp-96, Asp-201, Glu-194, and Glu-204 in BR) identified as necessary for its function (Fig. 1a) (41). In contrast, bopI (MR) is distantly related to both BR and SRII (∼41% identity with BR and ∼35% identity for SRII) and contains the amino acid residues important for the proton pumping function (Asp-85, Asp-96, Asp-212, and Glu-204 in BR) except for the equivalent of Glu-194 in BR, and also those important for signal transduction (Tyr-174 and Thr-204 in SRII (28)). The identity of the amino acid sequences between HwBR and MR is 41%. The phylogenetic analysis showed that HwBR and MR can be classified as BR and an intermediate type between BR and SRII, respectively (Fig. 1a). The gene sequence of bopII is similar to that of a BR gene from Haloarcula vallismortis, and although bopI is located near bopII in the genome (33), the gene sequence of bopI is quite different from bopII, suggesting that bopI is gained by the lateral gene transfer. The supplemental Fig. S2 shows the deduced secondary structure of MR from H. walsbyi. Similar to other microbial rhodopsins, it has seven transmembrane segments. A lysine on the helix G and an aspartate residue (Asp-84) on the helix C, which are predicted to be necessary for the stabilization of the Schiff base and for the binding between protein and retinal, are completely conserved both in HwBR and in MR (supplemental Figs. S1 and S2). Sensory rhodopsin genes previously identified are followed by the genes of cognate transducers, in an operon that is under control of the same promoter (42). The transducer proteins HtrI for SRI and HtrII for SRII belong to a family of two transmembrane helical proteins similar to chemoreceptors Tar and Tsr in E. coli (31), and they are termed methyl-accepting chemotaxis proteins (31). It should be noted that no putative transducer protein genes were found, not only in the sequences downstream of HwBR or MR but in the whole genome of H. walsbyi (33).
Protein Expression, Purification, and Absorption Spectrum
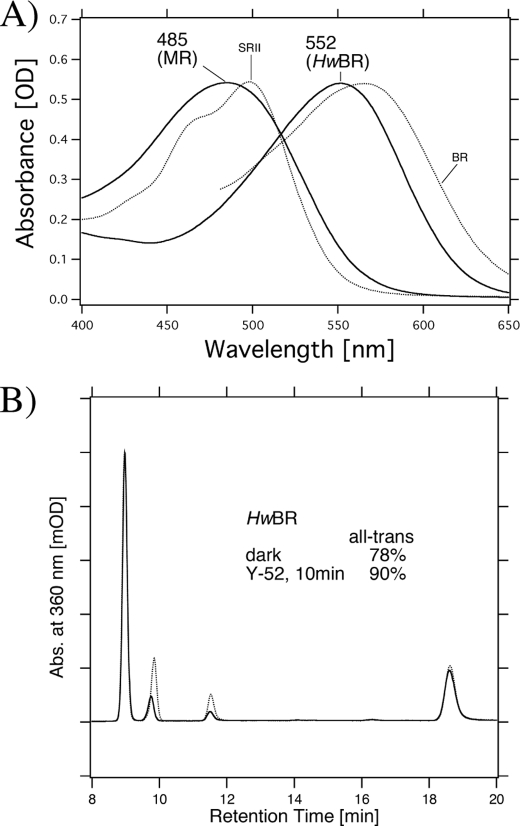
To determine whether bopI and bopII encode functional proteins, we cloned and expressed them in E. coli BL21(DE3) cells as recombinant proteins. Using PCR, we obtained a fragment containing the full length of the gene coding regions. Upon the addition of retinal, color changes of the transformed cells were obvious, and the color was distinct from cells transformed only with the plasmid vector, implying that the proteins were successfully expressed in E. coli. The absorption spectra of purified MR and HwBR in the dark were obtained (Fig. 2A) and were compared with those of BR and SRII (22). The absorption maximum of HwBR is located at 552 nm, which is similar to that of BR (566 nm), whereas the absorption maximum of MR is located at 485 nm, which is close to that of SRII (498 nm) and is significantly different from BR. Thus, MR is one of the most blue-shifted microbial rhodopsins identified to date.
FIGURE 2.
A, absorption spectra of MR and HwBR in buffer containing 1 m NaCl, 0. 05% DDM, and 50 mm Tris-HCl, pH 7.0. The temperature was kept at 20 °C. The data for BR and SRII were reproduced from Ref. 22 for comparison. B, conformation of retinal extracted from HwBR in 1 m NaCl, 0.05% DDM, and 50 mm Tris-HCl, pH 7.0, in the dark (dotted line) and after illumination with >500 nm light for 10 min (solid line). The detection beam was set at 360 nm. The molar composition of retinal isomers was calculated from the areas of the peaks in the HPLC patterns. One division of the y axis corresponds to 2000 absorbance units.
Retinal Configuration of MR and HwBR
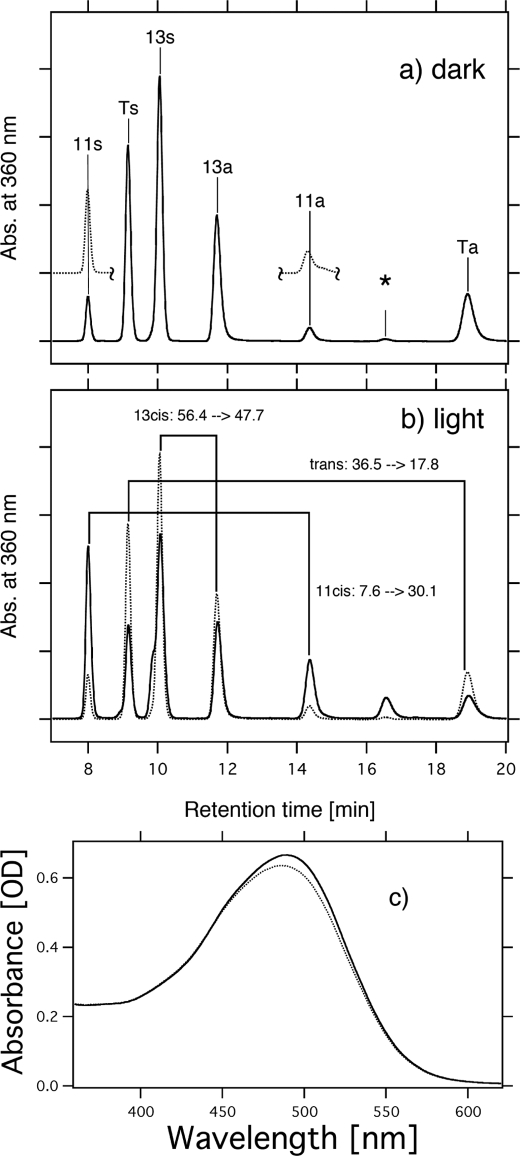
To investigate the retinal configuration of MR and HwBR, HPLC analysis was carried out. In the dark, light-driven ion pumps (BR and HR) have all-trans- and 13-cis-retinal, whereas the photosensors, HsSRI, HsSRII, and NpSRII, have only all-trans-retinal (43). Figs. 2B and 3 show the retinal isomeric composition of HwBR and MR, respectively. Assignment of each peak was performed by comparing it with the HPLC pattern from retinal oximes of authentic all-trans- and 13-cis-retinals (44), whereas 11-cis-retinal was obtained by irradiating all-trans-retinal dissolved in acetonitrile (37). As shown in Fig. 3a (dotted lines), the peaks of the extracted 11-cis-retinal coincide well with those of 11-cis-retinal obtained by irradiating all-trans-retinal, and therefore, they were assigned to 11-cis-retinal oxime. The isomeric composition of the retinal chromophore of HwBR is all-trans (78%) and 13-cis (22%), and it becomes predominantly all-trans (90%) with a small portion of 13-cis (10%) upon illumination with >500 nm light (Y52 filter) for 10 min (Fig. 2B). Although the ratio of all-trans and 13-cis forms is different from that in BR, the pattern is similar (41), indicating that the retinal composition of HwBR is markedly BR-like. On the other hand, MR has three retinal isomers, all-trans (36.5%), 11-cis (7.6%), and 13-cis (56.4%) in the dark (Fig. 3), and the retinal chromophore of MR becomes predominantly 11-cis (30.1%) and 13-cis (47.7%) upon illumination with >460 nm light (Y48 filter) for 10 min, suggesting the conversion from all-trans to 11-cis form. Significant changes were not observed following further illumination (data not shown), indicating the equilibrium between the dark- and the light-adapted forms is established within 10 min. Fig. 3c shows the absorption spectrum of dark-adapted MR (dotted line) and light-adapted MR (solid line). The absorption maximum of light-adapted MR is slightly shifted to a longer wavelength with an increase in intensity compared with the dark-adapted form, indicating that MR with 11-cis-retinal absorbs longer wavelengths of light with a larger molar extinction coefficient than all-trans-retinal. These light-induced changes recovered within 30 min to the initial states (11-cis, 30.1 → 7.5%; 13-cis, 47.7 → 51.3%; all-trans, 17.8 → 36.1%). Thus, the relatively broad absorption spectrum of MR (Fig. 2A) presumably originates from the coexistence of the three retinal isomers in the dark. Interestingly, MR is the first type 1 rhodopsin to be discovered that has 11-cis-retinal as a chromophore.
FIGURE 3.
Conformation of retinal extracted from MR in 1 m NaCl, 0.05% DDM, and 50 mm Tris-HCl, pH 7.0, in the dark (a) and upon illumination with >460 nm light for 10 min (b). The detection beam was set at 360 nm. Ts, Ta, 11s, 11a, 13s, and 13a stand for all-trans-15-syn-retinal oxime, all-trans-15-anti-retinal oxime, 11-cis-15-syn-retinal oxime, 11-cis-15-anti-retinal oxime, 13-cis-15-syn-retinal oxime, and 13-cis-15-anti-retinal oxime, respectively. The HPLC pattern of 11-cis-retinal obtained by irradiating all-trans-retinal is shown as dotted lines (a). One division of the y axis of a and b corresponds to 5000 absorbance units. An asterisk means an unknown peak, presumably originating from 9-cis-retinal oxime. The molar composition of retinal isomers was calculated from the areas of the peaks in the HPLC patterns. c, absorption spectra measured in the dark (dotted line) and upon illumination with >460 nm light for 10 min (solid line).
Photocycles of MR and HwBR as Revealed by FTIR and Time-resolved Flash Photolysis
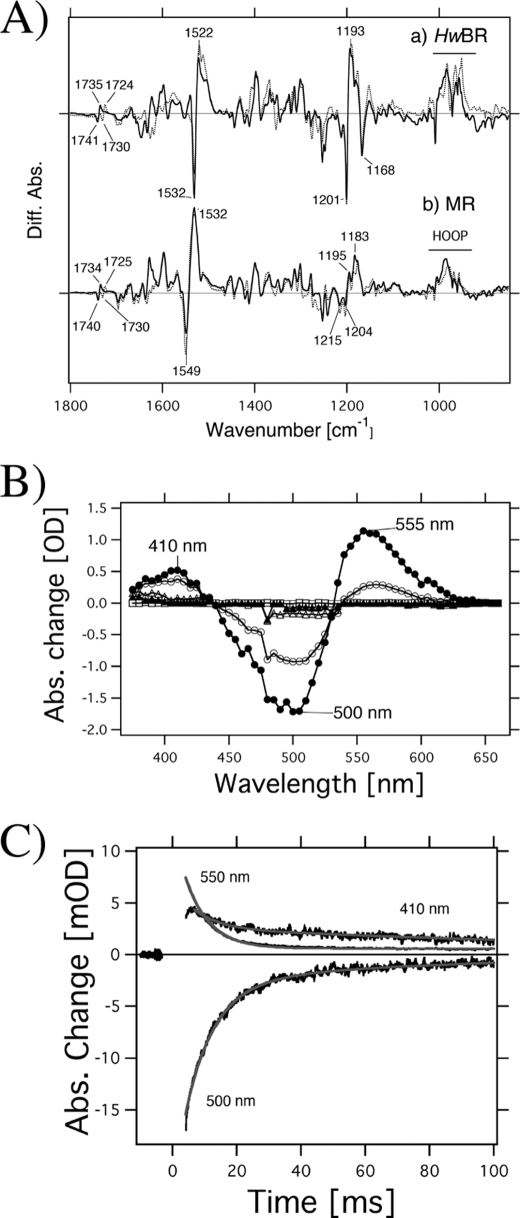
To investigate the structural changes and photocycle kinetics of MR and HwBR, we used low temperature FTIR and time-resolved flash photolysis (Figs. 4 and 5). Fig. 4A shows the photoproduct minus the initial state difference FTIR spectra of HwBR (a) and MR (b) hydrated with H2O (solid lines) and D2O (dotted lines) at 77 K in the 1800–900 cm−1 region. The horizontal bar in Fig. 4A indicates spectral changes in the 1100–900 cm−1 region, where hydrogen out-of-plane vibrations of the retinal molecule are mainly observed (45). The bands at 1532 (−)/1522 (+) cm−1 for HwBR and 1549 (−)/1532 (+) cm−1 for MR correspond to the ethylenic C=C stretching vibrations of the retinal chromophore. The lower frequency shift corresponds to the visible spectrum red shift, indicating formation of the K-intermediate both in HwBR and in MR. The frequency difference between MR (1549 cm−1) and HwBR (1532 cm−1) corresponds to the blue-shifted absorption spectrum of MR compared with HwBR (Fig. 2). The bands at 1201 (−), 1193 (+), and 1168 (−) cm−1 for HwBR, and at 1215 (−), 1204 (−), 1195 (+), and 1183 (+) cm−1 for MR correspond to the C–C stretching vibrations of the retinal chromophore. The frequency shift of HwBR represents retinal isomerization from the all-trans to 13-cis, and the spectral features are quite similar to those of BR (29), which indicates that the structure and structural changes of HwBR are similar to those of BR. Similarly, the bands at 1204 (−) and 1195 (+) cm−1 in the difference spectra of MR are typical for the isomerization from all-trans to 13-cis. However, it is likely that the bands at 1215 (−) and 1183 (+) cm−1 originate from C–C stretching vibrations of 11-cis- and/or 13-cis retinal, because the band originating from photoisomerization of 13-cis-retinal of BR appears at 1179 (+) cm−1 (46). In BR, the 1742(−)/1733(+) cm−1 bands in H2O are shifted to 1727(−)/1721(+) cm−1 in D2O, which was previously assigned to the C=O stretch of Asp-115 (47). The corresponding amino acid (an Asp residue) is conserved both in HwBR and in MR (supplemental Fig. S1), and similar spectral changes are observed in the 1750–1710 cm−1 region (Fig. 4A). Therefore, we tentatively assign the bands at 1741(−)/1735(+) cm−1 for HwBR and at 1740(−)/1734(+) cm−1 for MR as C=O stretching vibrations of the corresponding Asp residue.
FIGURE 4.
A, HwBRK minus HwBR (a) and MRK minus MR (b) difference infrared spectra measured at 77 K at pH 7.0 in the 1800–850 cm−1 region. The MRK minus MR spectrum (b) is multiplied by 3.8 for comparison. The sample was hydrated with H2O (solid lines) or D2O (dashed lines). One division of the y axis corresponds to 0.0055 absorbance units. B, flash-induced difference spectra of MR in a spectral range from 375 to 660 nm. The purified MR was resuspended in 50 mm Tris-HCl, pH 7.0, 0.05% DDM, and 1 m NaCl. Closed and open circles are spectra taken at 1.5 and 10.5 ms after the illumination, respectively. The temperature was kept at 25 °C. C, flash-induced kinetics of absorbance changes of MR at 410 nm representing the M-decay and at 500 nm representing the recovery of the original MR. The absorbance change at 550 nm presumably originates from a photointermediate of MR having 13-cis- and/or 11-cis-retinal chromophore. Gray lines show single exponential fits.
FIGURE 5.
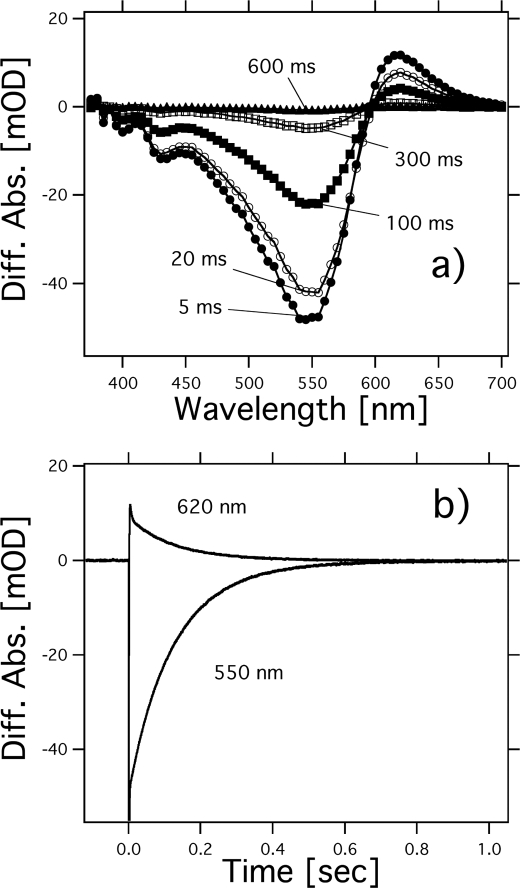
Photochemical reactions of HwBR. a, flash-induced difference spectra of HwBR in a spectral range from 380 to 700 nm. The purified HwBR was resuspended in 50 mm Tris-HCl, pH 7.0, 0.05% DDM, and 1 m NaCl. Curves represent the spectra taken at 5, 20, 100, 300, and 600 ms after the illumination. The temperature was kept at 25 °C. b, flash-induced kinetics of absorbance changes of HwBR at 620 nm, presumably representing the O-decay and at 550 nm representing the recovery of the original HwBR.
Fig. 4B shows the flash-induced difference spectra of MR in the spectral range from 375 to 660 nm. Before the measurements, the samples were irradiated with >460 nm light for 10 min for the light adaptation. Flash-induced bleaching and recovery of the original pigment was observed at around 500 nm, a value similar to the absorption maximum of MR as shown in Fig. 2. At 410 nm, an increase and a decrease of absorbance were observed, implying the formation and decay of a blue-shifted M-intermediate of MR (MRM) with an absorbance maximum located at 410 nm. In general, the microbial rhodopsins having all-trans-retinal as a chromophore produce M-intermediates, and therefore this photoreaction likely originates from MR with all-trans-retinal. Fig. 4C shows the time courses of the absorbance change of MR at selected wavelengths (410 nm for the M state, 500 nm for the unphotolyzed state, and 550 nm for the unknown state). The half-life of the M-intermediate is estimated as 0.014 s using a single exponential equation, which is close to that of BR (0.005 s). The fast M-decay was also observed in PC-reconstituted MR (data not shown). The fast photocycle is particularly important because a key difference between transport and sensory rhodopsins is the much slower kinetics of the photochemical reaction cycle of the sensors (43). The ion-pumping rhodopsins have been optimized by nature for fast photocycling rates to make them efficient pumps. In contrast, sensory rhodopsins SRI and SRII have slow photocycles (20, 31, 48), which allows the transient accumulation of long lived signaling states of the receptors to catalyze a sustained phosphorylation cascade controlling flagellar motor rotation (49). The fast photocycle of MR is therefore similar to those of ion pumping rhodopsins from other sources. It is likely that the formation and decay of an unknown state, with an absorption maximum located at 555 nm (Fig. 4B), corresponds to the photoreaction of MR having 13-cis- and/or 11-cis-retinal because its half-life is estimated as 0.005 s by a single exponential fit, which is different from that of the M-decay. The rate constant is similar to that of the photoreaction of BR having 13-cis-retinal (50). Similar experiments were performed with HwBR, as shown in Fig. 5. The photocycle of HwBR is slower than that of the proton-pumping rhodopsins, presumably caused by the slow O-intermediate decay, with the absorption maximum located at 620 nm. In addition, the M-like intermediate was not observed. Presumably, the M-decay is faster than the time resolution of the system (<5 ms), because we expect the M-intermediate to be present as it is essential for the proton pumping.
Proton Pumping Activities of HwBR and MR and Phototaxis Responses by the MR-HtrII Artificial Complex
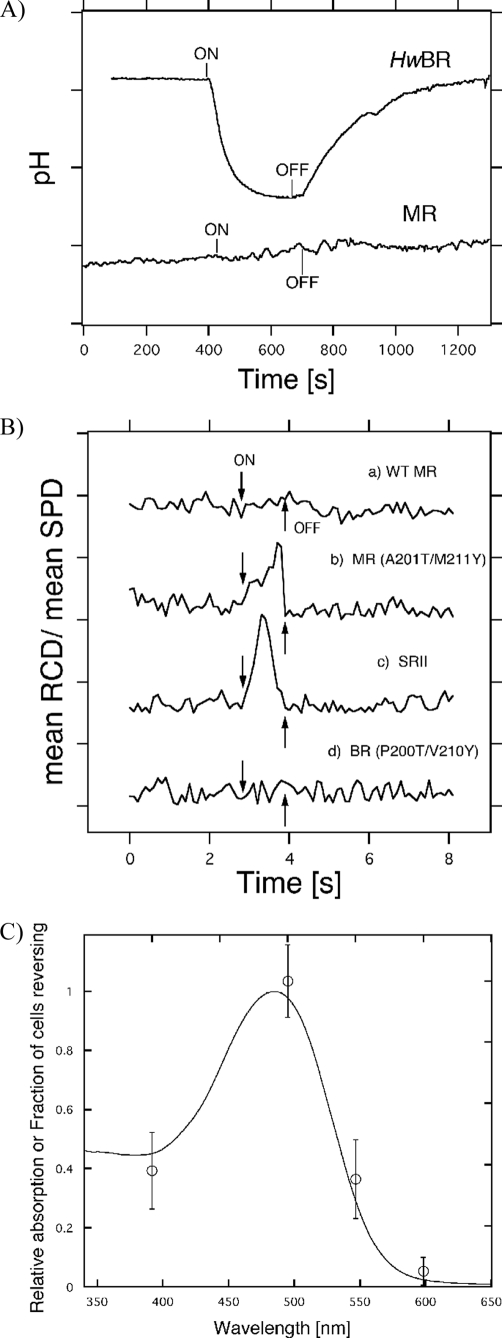
To investigate the functions of HwBR and MR, we measured the proton pumping activities of HwBR and MR. For proton transport measurements, we prepared spheroplast vesicles by removing the cell wall with lysozyme (40). Illumination caused a net outward transport of protons for HwBR, resulting in acidification of the medium (Fig. 6A). This light-induced acidification was absent in the presence of 10 μm carbonyl cyanide p-chlorophenylhydrazone (a proton ionophore which collapses the protonmotive force) (data not shown). These results demonstrate that HwBR can function as a light-driven outward proton transporter, similar to BR. On the other hand, no photo-induced pH change was observed in spheroplast vesicles containing MR, indicating the lack of proton transport activity. This is unexpected, considering that the residues important for proton transport in BR were conserved in MR, except for Glu-194 in the proton release complex of BR (51). To confirm whether MR is able to function as a photosensor, phototaxis responses of H. salinarum Pho81Wr− cells expressing the MR-HtrII artificial complex were measured (Fig. 6B). The wild-type MR/HtrII does not mediate phototaxis responses. Two strategically placed surface residues were then introduced into MR, enabling hydrogen bonding to HtrII to align MR with the transducer in the same manner as in SRII-HtrII complex (52). The x-ray crystallographic structure of the SRII-HtrII complex shows two critical intermolecular hydrogen-bonding sites at Tyr-199-SRII/Asn-74-HtrII and at Thr-189-SRII/Glu-43-HtrII/Ser-62-HtrII (53). Ala-201 and Met-211 in MR correspond to Thr-189 and Tyr-199 in SRII, respectively. The MR(A201T/M211Y)/HsHtrII construct produced a sufficiently robust response to >460 nm light (Fig. 6B), similar to SRII-NpHtrII (Fig. 6C). On the other hand, the homologous mutant of BR did not show such photoresponses (Fig. 6B). Presumably, the slower phototactic response of MR(A201T/M211Y) relative to that of SRII is caused by faster M-decay of MR(A201T/M211Y), as less of the active M-intermediate is accumulated under stationary illumination. In addition, the action spectrum of the motility of the cells containing MR(A201T/M211Y)/HsHtrII matches the visible absorption spectrum of MR (Fig. 6C). These results clearly show that MR is potentially capable of functioning as a sensory SRII-like protein. It should be noted that BR, HR, and SRI mutants having the corresponding two surface residues do not mediate detectable phototaxis responses (22)3 although they are able to form complexes with HtrII with sufficient binding affinity (52, 54). These results strongly suggest the close relationship between SRII and MR.
FIGURE 6.
A, light-driven pH changes in spheroplast vesicles containing HwBR or MR (50 mm MgSO4, 150 mm NaCl, initial pH ∼6.5). On and Off indicate the onset and offset of illumination (with yellow light, >500 nm for HwBR and >460 nm for MR), and the negative signal corresponds to a decrease in pH (outward proton transport). One division of the y axis corresponds to 0.1 pH unit. B, phototaxis responses of the MR-HtrII artificial complexes. On and Off indicate the onset and offset of illumination with >460 nm stimulus (1 s). Swimming reversal frequency responses of cell populations measured by stimulus effects on the ratio of rate of change of direction (RCD) to speed (SPD) by computer-assisted motion analysis. One division of the y axis corresponds to 50. C, action spectrum for the phototactic response in the transformant containing MR(P201T/M211Y)-HtrII. We measured phototaxis responses as swimming frequency changes to 1-s photo stimuli. Light intensities were normalized by using an actinometer, and we used four filters (400 ± 10, 500 ± 10, 550 ± 10, and 600 ± 10 nm).
Molecular Evolution from BR to SRII Revealed by Characterization of MR
The data obtained in this study are summarized in Table 1 and show the values determined previously for BR and SRII for comparison (22, 55). Two newly identified microbial rhodopsins, HwBR and MR, are characterized here. HwBR shows properties similar to those of BR from H. salinarum, except for the slow O-decay (Table 1); therefore, HwBR is classified into the BR-like protein subfamily, as expected. On the other hand, MR shows an SRII-like blue-shifted absorption maximum, a BR-like fast photocycle, and a unique retinal composition (Table 1). The vesicles containing MR do not show any light-induced ion transport activity (Fig. 6A), although residues important for proton pumping are completely conserved in MR. As it is known that at least one member of the SRII protein family, SRII from H. salinarum (HsSRII) (supplemental Fig. S1), does not show the light-induced proton pumping activity (56), this property of MR would be similar to that of HsSRII. Interestingly, the MR mutant having two critical hydrogen-bonding residues functions as a negative phototaxis sensor (Fig. 6B). Thus, MR shows properties intermediate between those of BR and SRII.
TABLE 1.
Characteristics of HwBR, MR, BR, and SRII
ND indicates not determined.
| Genotype | λmax | Retinal composition | Half-life of M-intermediate | Proton pumping | |
|---|---|---|---|---|---|
| HwBR | bop | 552 | All-trans, 13-cis | ND (fast?) | Yes |
| MR | bop/sop | 485 | 13-cis, all-trans, 11-cis | 0.014 s | No |
| BR | bop | 566 | All-trans, 13-cis | 0.005 s | Yes |
| SRII | sop | 498 | All-trans | 1.23 s | Weak |
On the basis of our results, combined with other findings, a possible mechanism for the molecular evolution from BR to SRII can be proposed. At the beginning of the molecular evolution from BR to SRII, color tuning and insertion of a critical Thr residue may have occurred. In previous studies, we demonstrated that the Thr residue is important not only for signal transduction (28) but also for color tuning (57). However, it is difficult to explain the large blue shift of the MR spectrum only by insertion of the Thr residue, because the T204A mutant of SRII and the A215T mutant of BR show only an ∼17 nm spectral shift (22, 57). The retinal composition of MR is a very unique feature. To induce such change of the isomeric composition, a large structural rearrangement around the retinal chromophore should have occurred in BR during the evolution. H. walsbyi does not have any known putative transducer proteins such as HtrI, HtrII, Anabaena sensory rhodopsin transducer, Tar, or Tsr. In addition, genes encoding Che proteins and flagellar motor proteins are missing in the genomic sequence of H. walsbyi. The H. walsbyi genome encodes a proton-pumping rhodopsin HwBR and a putative chloride-pumping rhodopsin HR, and MR absorbs shorter wavelengths of light than these ion-pumping rhodopsins. Thus, a novel signaling pathway was required for MR to function as a photosensor in the native cells. Consequently, SRII-HtrII had to be generated to become a functional unit for the negative phototaxis sensor with downstream proteins, an adaptor protein CheW and a kinase CheA. Interestingly, the retinal composition of MR is altered by illumination, shifting from all-trans- to 11-cis-retinal, suggesting the functional importance of the 11-cis- and/or all-trans-retinal chromophores. This indicates that MR may be a missing link in the evolution from type 1 rhodopsins (microorganisms) to type 2 rhodopsins (animals), because MR is the first microbial rhodopsin molecule discovered that has 11-cis-retinal as a chromophore similar to type 2 rhodopsins.
Supplementary Material
Acknowledgments
We thank Drs. Akira Kawanabe and Tatsuya Iwata for technical assistance in the measurements of proton pumping activity and FTIR measurements. We are grateful to Prof. John L. Spudich for useful comments on the manuscript.
This work was supported in part by grants-in-aid for scientific research (KAKENHI) on a priority area (area 477) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by grants from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (to Y. S., H. K., and M. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Y. Sudo and J. L. Spudich, unpublished data.
- BR
- bacteriorhodopsin
- MR
- middle rhodopsin
- HwBR
- bacteriorhodopsin-like protein from H. walsbyi
- SRII
- sensory rhodopsin II
- HtrII
- halobacterial transducer protein for SRII
- DDM
- n-dodecyl β-d-maltoside
- HR
- halorhodopsin.
REFERENCES
- 1. Spudich J. L. (2006) Trends Microbiol. 14, 480–487 [DOI] [PubMed] [Google Scholar]
- 2. Zhang F., Aravanis A. M., Adamantidis A., de Lecea L., Deisseroth K. (2007) Nat. Rev. Neurosci. 8, 577–581 [DOI] [PubMed] [Google Scholar]
- 3. Costanzi S., Siegel J., Tikhonova I. G., Jacobson K. A. (2009) Curr. Pharm. Des. 15, 3994–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ruiz-González M. X., Marín I. (2004) J. Mol. Evol. 58, 348–358 [DOI] [PubMed] [Google Scholar]
- 5. Sharma A. K., Spudich J. L., Doolittle W. F. (2006) Trends Microbiol. 14, 463–469 [DOI] [PubMed] [Google Scholar]
- 6. Sharma A. K., Zhaxybayeva O., Papke R. T., Doolittle W. F. (2008) Environ. Microbiol. 10, 1039–1056 [DOI] [PubMed] [Google Scholar]
- 7. Fuhrman J. A., Schwalbach M. S., Stingl U. (2008) Nat. Rev. Microbiol. 6, 488–494 [DOI] [PubMed] [Google Scholar]
- 8. Hofmann K. P., Scheerer P., Hildebrand P. W., Choe H. W., Park J. H., Heck M., Ernst O. P. (2009) Trends Biochem. Sci. 34, 540–552 [DOI] [PubMed] [Google Scholar]
- 9. Béjà O., Aravind L., Koonin E. V., Suzuki M. T., Hadd A., Nguyen L. P., Jovanovich S. B., Gates C. M., Feldman R. A., Spudich J. L., Spudich E. N., DeLong E. F. (2000) Science 289, 1902–1906 [DOI] [PubMed] [Google Scholar]
- 10. Brown L. S. (2004) Photochem. Photobiol. Sci. 3, 555–565 [DOI] [PubMed] [Google Scholar]
- 11. Waschuk S. A., Bezerra A. G., Jr., Shi L., Brown L. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 6879–6883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Balashov S. P., Imasheva E. S., Boichenko V. A., Antón J., Wang J. M., Lanyi J. K. (2005) Science 309, 2061–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jung K. H. (2007) Photochem. Photobiol. 83, 63–69 [DOI] [PubMed] [Google Scholar]
- 14. Walter J. M., Greenfield D., Liphardt J. (2010) Curr. Opin. Biotechnol. 21, 265–270 [DOI] [PubMed] [Google Scholar]
- 15. Sasaki J., Spudich J. L. (2008) Photochem. Photobiol. 84, 863–868 [DOI] [PubMed] [Google Scholar]
- 16. Spudich J. L., Bogomolni R. A. (1984) Nature 312, 509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spudich J. L., Bogomolni R. A. (1988) Annu. Rev. Biophys. Biophys. Chem. 17, 193–215 [DOI] [PubMed] [Google Scholar]
- 18. Spudich J. L., Yang C. S., Jung K. H., Spudich E. N. (2000) Annu. Rev. Cell Dev. Biol. 16, 365–392 [DOI] [PubMed] [Google Scholar]
- 19. Bogomolni R. A., Stoeckenius W., Szundi I., Perozo E., Olson K. D., Spudich J. L. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 10188–10192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sudo Y., Iwamoto M., Shimono K., Sumi M., Kamo N. (2001) Biophys. J. 80, 916–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schmies G., Engelhard M., Wood P. G., Nagel G., Bamberg E. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1555–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sudo Y., Spudich J. L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16129–16134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luecke H., Schobert B., Richter H. T., Cartailler J. P., Lanyi J. K. (1999) J. Mol. Biol. 291, 899–911 [DOI] [PubMed] [Google Scholar]
- 24. Luecke H., Schobert B., Lanyi J. K., Spudich E. N., Spudich J. L. (2001) Science 293, 1499–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Royant A., Nollert P., Edman K., Neutze R., Landau E. M., Pebay-Peyroula E., Navarro J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10131–10136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sudo Y., Furutani Y., Shimono K., Kamo N., Kandori H. (2003) Biochemistry 42, 14166–14172 [DOI] [PubMed] [Google Scholar]
- 27. Sudo Y., Furutani Y., Wada A., Ito M., Kamo N., Kandori H. (2005) J. Am. Chem. Soc. 127, 16036–16037 [DOI] [PubMed] [Google Scholar]
- 28. Sudo Y., Furutani Y., Kandori H., Spudich J. L. (2006) J. Biol. Chem. 281, 34239–34245 [DOI] [PubMed] [Google Scholar]
- 29. Sudo Y., Furutani Y., Spudich J. L., Kandori H. (2007) J. Biol. Chem. 282, 15550–15558 [DOI] [PubMed] [Google Scholar]
- 30. Ito M., Sudo Y., Furutani Y., Okitsu T., Wada A., Homma M., Spudich J. L., Kandori H. (2008) Biochemistry 47, 6208–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Suzuki D., Irieda H., Homma M., Kawagishi I., Sudo Y. (2010) Sensors 10, 4010–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Spudich J. L., Zacks D. N., Bogomolni R. A. (1995) Israel J. Chem. 35, 495–513 [Google Scholar]
- 33. Bolhuis H., Palm P., Wende A., Falb M., Rampp M., Rodriguez-Valera F., Pfeiffer F., Oesterhelt D. (2006) BMC Genomics 7, 169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marmur J. (1961) J. Mol. Biol. 3, 208–218 [Google Scholar]
- 35. Sudo Y., Okada A., Suzuki D., Inoue K., Irieda H., Sakai M., Fujii M., Furutani Y., Kandori H., Homma M. (2009) Biochemistry 48, 10136–10145 [DOI] [PubMed] [Google Scholar]
- 36. Suzuki D., Furutani Y., Inoue K., Kikukawa T., Sakai M., Fujii M., Kandori H., Homma M., Sudo Y. (2009) J. Mol. Biol. 392, 48–62 [DOI] [PubMed] [Google Scholar]
- 37. Maeda A., Schichida Y., Yoshizawa T. (1978) J. Biochem. 83, 661–663 [DOI] [PubMed] [Google Scholar]
- 38. Yagasaki J., Suzuki D., Ihara K., Inoue K., Kikukawa T., Sakai M., Fujii M., Homma M., Kandori H., Sudo Y. (2010) Biochemistry 49, 1183–1190 [DOI] [PubMed] [Google Scholar]
- 39. Sudo Y., Furutani Y., Iwamoto M., Kamo N., Kandori H. (2008) Biochemistry 47, 2866–2874 [DOI] [PubMed] [Google Scholar]
- 40. Kawanabe A., Furutani Y., Jung K. H., Kandori H. (2009) J. Am. Chem. Soc. 131, 16439–16444 [DOI] [PubMed] [Google Scholar]
- 41. Lanyi J. K. (2004) Annu. Rev. Physiol. 66, 665–688 [DOI] [PubMed] [Google Scholar]
- 42. Seidel R., Scharf B., Gautel M., Kleine K., Oesterhelt D., Engelhard M. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 3036–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Spudich J. L., Lanyi J. K. (1996) Curr. Opin. Cell Biol. 8, 452–457 [DOI] [PubMed] [Google Scholar]
- 44. Kitajima-Ihara T., Furutani Y., Suzuki D., Ihara K., Kandori H., Homma M., Sudo Y. (2008) J. Biol. Chem. 283, 23533–23541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Furutani Y., Sudo Y., Wada A., Ito M., Shimono K., Kamo N., Kandori H. (2006) Biochemistry 45, 11836–11843 [DOI] [PubMed] [Google Scholar]
- 46. Mizuide N., Shibata M., Friedman N., Sheves M., Belenky M., Herzfeld J., Kandori H. (2006) Biochemistry 45, 10674–10681 [DOI] [PubMed] [Google Scholar]
- 47. Braiman M. S., Mogi T., Marti T., Stern L. J., Khorana H. G., Rothschild K. J. (1988) Biochemistry 27, 8516–8520 [DOI] [PubMed] [Google Scholar]
- 48. Hoff W. D., Jung K. H., Spudich J. L. (1997) Annu. Rev. Biophys. Biomol. Struct. 26, 223–258 [DOI] [PubMed] [Google Scholar]
- 49. Yan B., Takahashi T., Johnson R., Spudich J. L. (1991) Biochemistry 30, 10686–10692 [DOI] [PubMed] [Google Scholar]
- 50. Hofrichter J., Henry E. R., Lozier R. H. (1989) Biophys. J. 56, 693–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mathias G., Marx D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6980–6985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sudo Y., Yamabi M., Kato S., Hasegawa C., Iwamoto M., Shimono K., Kamo N. (2006) J. Mol. Biol. 357, 1274–1282 [DOI] [PubMed] [Google Scholar]
- 53. Gordeliy V. I., Labahn J., Moukhametzianov R., Efremov R., Granzin J., Schlesinger R., Büldt G., Savopol T., Scheidig A. J., Klare J. P., Engelhard M. (2002) Nature 419, 484–487 [DOI] [PubMed] [Google Scholar]
- 54. Furutani Y., Ito M., Sudo Y., Kamo N., Kandori H. (2008) Photochem. Photobiol. 84, 874–879 [DOI] [PubMed] [Google Scholar]
- 55. Shimono K., Ikeura Y., Sudo Y., Iwamoto M., Kamo N. (2001) Biochim. Biophys. Acta 1515, 92–100 [DOI] [PubMed] [Google Scholar]
- 56. Sasaki J., Spudich J. L. (1999) Biophys. J. 77, 2145–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shimono K., Hayashi T., Ikeura Y., Sudo Y., Iwamoto M., Kamo N. (2003) J. Biol. Chem. 278, 23882–23889 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.