Abstract
Retrovirus vectors are de novo methylated and transcriptionally silent in mammalian stem cells. Here, we identify epigenetic modifications that mark retrovirus-silenced transgenes. We show that murine stem cell virus (MSCV) and human immunodeficiency virus type 1 (HIV-1) vectors dominantly silence a linked locus control region (LCR) β-globin reporter gene in transgenic mice. MSCV silencing blocks LCR hypersensitive site formation, and silent transgene chromatin is marked differentially by a histone code composed of abundant linker histone H1, deacetylated H3 and acetylated H4. Retrovirus-transduced embryonic stem (ES) cells are silenced predominantly 3 days post-infection, with a small subset expressing enhanced green fluorescent protein to low levels, and silencing is not relieved in de novo methylase-null [dnmt3a–/–;dnmt3b–/–] ES cells. MSCV and HIV-1 sequences also repress reporter transgene expression in Drosophila, demonstrating establishment of silencing in the absence of de novo and maintenance methylases. These findings provide mechanistic insight into a conserved gene silencing mechanism that is de novo methylase independent and that epigenetically marks retrovirus chromatin with a repressive histone code.
Keywords: chromatin/gene silencing/methylase/stem cells
Introduction
Transcription from retrovirus long terminal repeats (LTRs) is silenced in hematopoietic and embryonic stem (ES) cells of mammals (Jahner et al., 1982; Challita and Kohn, 1994; Yoder et al., 1997). This stem cell-specific gene silencing event is accompanied by heavy de novo cytosine methylation at CpG dinucleotides in the retrovirus LTR (Jahner et al., 1982; Stewart et al., 1982; Bednarik et al., 1990; Challita and Kohn, 1994). It is widely, but not universally, accepted that methylation is the first step in a gene silencing mechanism that ultimately controls chromatin structure (Eden et al., 1998; Razin, 1998). The developmental timing of the gene silencing event can be accounted for by the presence of a postulated stem cell-specific de novo methyltransferase (Lei et al., 1996; Tucker et al., 1996) such as the recently described murine dnmt3 proteins (Okano et al., 1998). Indeed [dnmt3a–/–;dnmt3b–/–] ES cells fail to methylate newly integrated exogenous retrovirus, although partial methylation of endogenous retrovirus sequences is still observed (Okano et al., 1999).
Evidence that supports a functional role for methylases in retrovirus silencing includes the demonstration that the methylation inhibitor 5-azacytidine (5-azaC) can reactivate expression of silenced murine retroviruses in embryonic carcinoma (EC) cells (Stewart et al., 1982). In addition, dnmt1 null mice fail to silence the intracisternal A particle (IAP) class of endogenous retrovirus that has no extracellular infectious stage (Walsh and Bestor, 1999). These findings support a genome defense model in which de novo methylases have evolved in higher eukaryotes to silence foreign parasitic DNA or to direct allele-specific gene expression (Walsh and Bestor, 1999).
The action of methylases has direct and indirect effects on gene expression (Razin, 1998). Methyl groups on modified DNA directly affect the ability of some transcription factors to bind DNA, and methylases indirectly accomplish the same result by provoking condensation of chromatin structure. Such chromatin structure alterations are mediated by the binding of MeCP2 factor to methylated DNA. MeCP2 in turn interacts with a complex of several co-repressor molecules including mSin3A and the histone deacetylases HDAC1 and HDAC2 (Jones et al., 1998; Nan et al., 1998). Therefore, de novo methylases are responsible for the primary tag that marks a DNA sequence for MeCP2 binding, and recruitment of the mSin3A–HDAC complex results in histone deacetylation, which condenses chromatin and prevents transcription factor access.
Some retrovirus silencing results are not easily explained by de novo methylase-dependent silencing pathways. Specifically, the infectious C-type retrovirus Moloney murine leukemia virus (MoMLV) is silenced in murine EC cells 2 days post-infection, but methylation at the LTRs is not detectable until 8–16 days post-infection (Gautsch and Wilson, 1983; Niwa et al., 1983; Kempler et al., 1993). Unlike EC cells, more differentiated cell types such as murine erythroleukemia (MEL) cells express MoMLV-based vectors initially, but these vectors are silenced progressively by an HDAC-mediated mechanism that precedes methylation density-dependent silencing (Lorincz et al., 2000). In addition, methylation does not correlate with silencing of endogenous genes and is not targeted to retrovirus LTRs in the sea squirt (Simmen et al., 1999).
An example of a methylase-independent pathway has been described that acts to silence reporter genes linked to adeno-associated virus (AAV) sequences in HeLa or MEL cells. This pathway permits reactivation of the silenced gene by treatment with the histone deacetylase inhibitors trichostatin A (TSA) and sodium butyrate, but not by the methylation inhibitor 5-azaC (Chen et al., 1997). Therefore, it appears that, in some circumstances methylase action is not a prerequisite for virus silencing. Chromatin immunoprecipitation analyses of the AAV-silenced reporter gene demonstrated that histone H4 is deacetylated at K8 and the chromatin condensed (Chen and Townes, 2000). These results suggest that H4 deacetylation is a component of the combinatorial histone code that marks silent AAV sequences. The histone code is composed of multiple post-translational modifications on histone tails that protrude from nucleosomes and on the linker histone H1 (Strahl and Allis, 2000). Modifications to H3 and H4 include acetylation of an array of lysine residues, phosphorylation of serines, and methylation. Silent chromatin is usually considered to be bound by H1 and hypoacetylated H3 and H4. The broad histone code that marks silent retrovirus vectors in embryonic or hematopoietic stem cells has not been described.
In contrast to the complex situation in mammals, methylation is not detectable in Drosophila (Urieli-Shoval et al., 1982), and embryonic gene silencing in flies is dependent on altered chromatin structures or silencing factors. For example, the Polycomb group (Pc-G) genes are silencing factors that maintain patterns of repression established in the early embryo (Pirrotta, 1998). Reversal of such Pc-G silencing is accompanied by H4 hyperacetylation (Cavalli and Paro, 1999). Given the high degree of conservation of Pc-G and HDAC genes between eukaryotes (Pazin and Kadonaga, 1997), it is likely that gene silencing mechanisms themselves are also widely conserved. In fact, several homologs of methyl-binding proteins are conserved in Drosophila that do not bind methylcytosine but retain the ability to interact with HDACs, such as those in the Mi-2–NuRD complex (Tweedie et al., 1999). Therefore, Drosophila melanogaster presents a unique system for the study of methylase-independent silencing mechanisms that are highly conserved.
MoMLV contains several cis-acting sequences that function as silencers. The finding that MoMLV-based vectors containing all of these elements are silenced in hematopoietic stem cells has limited their potential effectiveness for gene therapy purposes (Challita and Kohn, 1994). In an effort to relieve silencing effects, the murine stem cell virus (MSCV) (Hawley et al., 1994) and HSC1 vectors (Osborne et al., 1999) were developed containing mutations in the LTR direct repeat and primer-binding site, or all known silencer elements, respectively. Our assay for gene silencing takes advantage of a powerful human β-globin reporter gene regulated by the locus control region (LCR) in transgenic mice (Ellis et al., 1997). The human β-globin LCR is composed of at least four erythroid-specific DNase I-hypersensitive sites (HS) (Grosveld et al., 1987). These HS are nucleosome-free regions of open chromatin that are highly accessible to trans-acting factors (Tuan and London, 1984; Forrester et al., 1986). LCR activity confers copy number-dependent expression in transgenic mice (Grosveld et al., 1987; Ryan et al., 1989), and transgene expression is accompanied by the establishment of HS at the LCR and the β-globin promoter (Ellis et al., 1996).
Here, we show that addition of MoMLV and human immunodeficiency virus type 1 (HIV-1) sequences completely silences the LCRβ-globin reporter transgene at all integration sites. MSCV sequences also silence the LCRβ-globin reporter transgene and prevent the formation of HS at the LCR. The reduced chromatin accessibility of the silent transgene is code marked by linker H1, deacetylated H3 and acetylated H4. Retrovirus vectors also silence reporter genes in [dnmt3a–/–;dnmt3b–/–] ES cells and transgenic Drosophila, showing that retrovirus silencing does not require de novo methylases. These independent and complementary data demonstrate that gene silencing of retrovirus sequences occurs in mice and Drosophila, and are consistent with the existence of a conserved retrovirus gene silencing mechanism that involves repressive histone code marks.
Results
Retrovirus vectors silence LCRβ-globin transgenes in mice
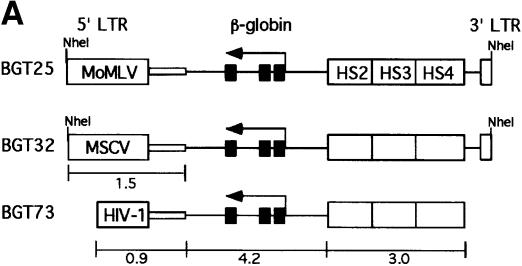
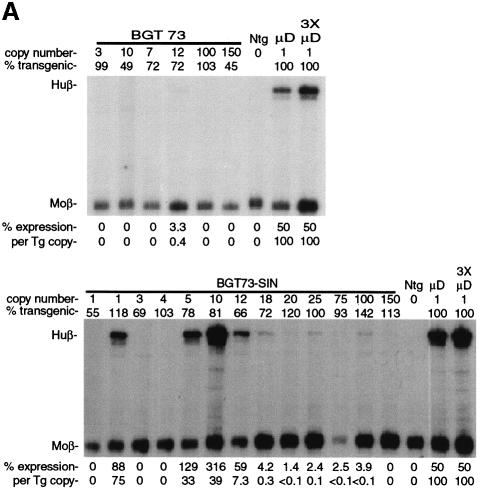
To determine whether retrovirus sequences can silence a powerful LCRβ-globin transgene in mice, we linked MoMLV and MSCV sequences to an antisense orientation of the BGT14 reporter cassette, which expresses at 16–71% per transgene copy and at all integration sites (Ellis et al., 1997). This orientation is preferred in β-globin retrovirus vectors in order to preserve the introns during retrovirus replication (Leboulch et al., 1994). The BGT14 reporter gene contains a 3.0 kb LCR composed of 5′HS2–4 that is linked to a 4.2 kb human β-globin gene regulated by its 815 bp promoter. Its ability to express β-globin reproducibly and consistently in transgenic mice makes it an ideal reporter for silencing activity. The microinjection fragments used in the transgenic mouse silencing assays are described in Figure 1. BGT25 contains 1.5 kb of MoMLV 5′ LTR and extended packaging sequences, while BGT32 contains the equivalent 1.5 kb sequence derived from MSCV (Figure 1A).
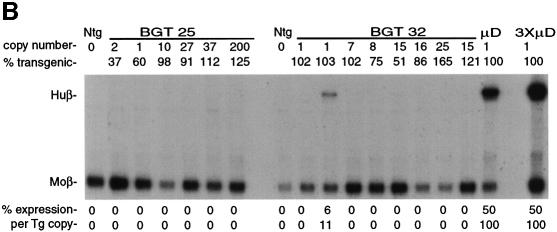
Fig. 1. Silencing by C-type retrovirus sequences in transgenic mice. (A) Structure of LCRβ-globin reporter transgenes. MSCV contains several mutations in viral silencers. HS2, HS3, HS4: human β-globin LCR elements. (B) S1 nuclease analysis of globin expression in RNA of 15.5 day fetal livers showing that BGT25 (MoMLV) and BGT32 (MSCV) transgenes are completely silenced, with one exception. Huβ, human β-globin protected probe fragment; Moβ, mouse β-major protected probe fragment; Ntg, non-transgenic; µD, one-copy µD14 microlocus transgenic line; 3XµD, probe excess control.
The BGT25 and BGT32 transgenes were introduced into mice, and founder fetuses were collected at embryonic day 15.5. Transgenic animals were identified by Southern blot analysis with the βivs2 probe on fetal head DNA. Transgene copy number and intactness were determined by DNA digestion with restriction enzymes that reveal end fragments (EcoRI and BamHI for copy number) or internal fragments (for intactness). S1 nuclease analysis of fetal liver RNA showed complete silencing of the human β-globin transgene in all BGT25 (6/6) transgenic mice, and complete silencing in seven of eight BGT32 transgenic animals (Figure 1B). High level expression of a human β-globin transgene is detectable in the positive control fetal liver RNA from the µD14 transgenic mouse line (Ellis et al., 1996). Therefore, silencing mediated by MoMLV (McCune and Townes, 1994) and MSCV sequences is dominant to the activating functions of the β-globin LCR.
Silenced transgenes are in inaccessible chromatin
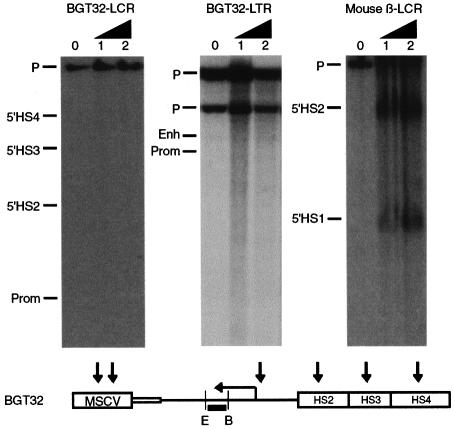
To examine the chromatin structure of the LCR and β-globin promoter in a silenced BGT32 transgene, we generated a silenced mouse line and performed HS mapping (Forrester et al., 1990) on F1 fetal liver nuclei (Figure 2). Digestion with EcoRI and hybridization with the βivs2 probe did not detect any HS in the transgene β-globin promoter or LCR. In contrast, HS in the endogenous mouse β-globin LCR (Ellis et al., 1996) in the same samples was detected. Hence, the chromatin of the silenced transgene is inaccessible to the factors that cause HS formation, and this inaccessible structure could itself prevent activation of the reporter transgene.
Fig. 2. DNase I-hypersensitive site mapping of 15.5 day fetal liver nuclei from a silenced four-copy BGT32 line showing that the transgene β-globin promoter, LCR and retrovirus LTR elements are in an inaccessible chromatin conformation. The endogenous mouse β-globin LCR elements in the same samples are hypersensitive. Expected transgene HS are shown below the vertical arrows. E, EcoRI; B, BamHI; the black rectangle indicates the βivs2 probe.
Silencing does not occur by transcriptional interference
Transgene silencing by retrovirus sequences could be due to transcriptional interference initiated from the viral 5′ LTR promoter (Cullen et al., 1984). Actively transcribed 5′ LTRs possess an HS at both the initiation site and the enhancer, but the 3′ LTR forms only the enhancer HS (Thompson and Fan, 1985; Rasmussen and Gilboa, 1987). We used the same DNase I-treated samples described above to search for HS formation on the viral LTR of the MSCV-silenced transgene. Digestion with BamHI and hybridization with the βivs2 probe demonstrated no HS elements on the β-globin 3′ enhancer, nor on the 5′ LTR (Figure 2). These data are inconsistent with transgene silencing mediated by transcriptional interference. Rather than being in an expressed conformation, the retrovirus sequences are themselves in an inaccessible chromatin structure.
Silenced LCRβ-globin transgenes have a repressive histone code
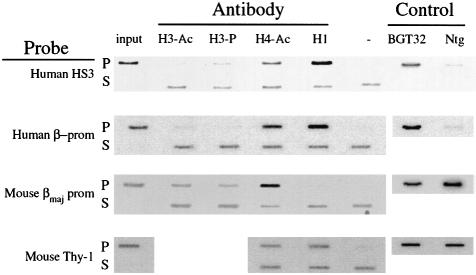
Inaccessible chromatin structures present on silent genes are believed to have a combinatorial histone code that includes bound H1 and hypoacetylated H3 and H4. Phosphorylated Ser10 on H3 is also present on condensed mitotic chromosomes (Wei et al., 1999), suggesting that it may also be found on silent chromatin. To discover the broad histone code of MSCV-silenced retrovirus transgenes, we performed chromatin immunoprecipitation (ChIP) assays on day 15.5 fetal liver cells explanted from the BGT32 transgenic mouse line. Cross-linked chromatin was immunoprecipitated with commercially available antibodies that specifically recognize biacetylated H3 (H3-Ac), phospho-Ser10 H3 (H3-P), hyperacetylated H4 (H4-Ac) or unmodified H1. Specific transgene DNA fragments in the input, pellet and supernatant DNA fractions were detected by Southern hybridization after equal loading onto slot-blots. A representative ChIP experiment for each antibody and probe is shown in Figure 3. Probes for both the human 5′HS3 LCR element and the silent human β-globin promoter did not detect signals in the H3-Ac pellet, but visible signals of increasing intensity were observed in the H4-Ac and H1 pellet fractions. A probe for the expressed endogenous mouse β-major promoter detected moderate H3-Ac and strong H4-Ac signals, but little or no H1 signal. These data suggest that the histone code of the silent BGT32 transgene includes abundant H1, deacetylated H3 and acetylated H4. In contrast, the histone code for the expressing mouse β-major promoter includes depleted H1, acetylated H3 and hyperacetylated H4. This expressing histone code also differs from the acetylated H4 and bound H1 found on the silent mouse thy-1 gene. ChIPs with the H3-P antibody showed more variability, with moderate signals detected with the human 5′HS3 probe and mouse β-major promoter probe, but not with the human β-globin promoter probe. These results suggest that H3-P does not in general correlate with silent chromatin states.
Fig. 3. ChIP assays detect a repressed histone code on the MSCV-silenced LCRβ-globin reporter in BGT32 transgenic mice. ChIP was performed with antibodies that detect H3-Ac, H3-P, H4-Ac and H1. Histone modifications at specific DNA fragments were detected by slot-blot hybridization with the probes listed on the left.
To identify the specific histone code marks that correlate best with the silenced transgene state, the ChIP results were quantified using a PhosphorImager. The intensity of the band in the no antibody (–) slot was subtracted from that of each pelleted band, and this normalized intensity was expressed as a percentage of the normalized input DNA. To calculate relative fold differences in histone code marks, the mean average percentage of input values obtained from at least two independent immunoprecipitations was calculated, and fold differences were determined with respect to the expressed mouse β-major gene (Table I). These calculations show that the silent human β-globin promoter is marked by a 9.1-fold enrichment of H1, an 8.8-fold depletion of H3-Ac, but only a 2.3-fold depletion of H4-Ac. Similar code marks tag the human 5′HS3 element, although depletion of H4-Ac is increased to 6.1-fold. We conclude that the most dramatically altered histone code marks at both 5′HS3 and the silent transgene promoter are abundant H1 and deacetylated H3. These data are the first description of the histone code of retrovirus-silenced transgenes in mice, and are consistent with the formation of inaccessible chromatin structures. The histone code described above suggests that retrovirus silencing in transgenic mice involves either the action of an HDAC that has a greater impact on H3 acetylation ratios than it does on H4, and/or reflects reduced accessibility by H3-specific HATs to the transgene chromatin.
Table I. Quantitation of ChIP data on individual histone code marks in silenced BGT32 transgenic mice compared with the expressed mouse β-major globin gene.
| H3-Ac | H3-P | H4-Ac | H1 | |
|---|---|---|---|---|
| Hβ-HS3 | –9.5× (5.0) | –2.5× (11.2) | –6.1× (69.5) | 7.4× (136.1) |
| Hβ-Prom | –8.8× (5.4) | –284× (0.1) | –2.3× (181.3) | 9.1× (167.7) |
| Mβmaj-Prom | 1.0× (47.4) | 1.0× (28.4) | 1.0× (422.7) | 1.0× (18.5) |
| MThy1 | ND | ND | –7.0× (60.3) | 4.7× (86.1) |
Figures in parentheses denote the mean average percentage of input DNA values for each pellet fraction from at least two independent ChIP experiments.
ND, not done.
Retrovirus vectors silence GFP reporters in de novo methylase-null ES cells
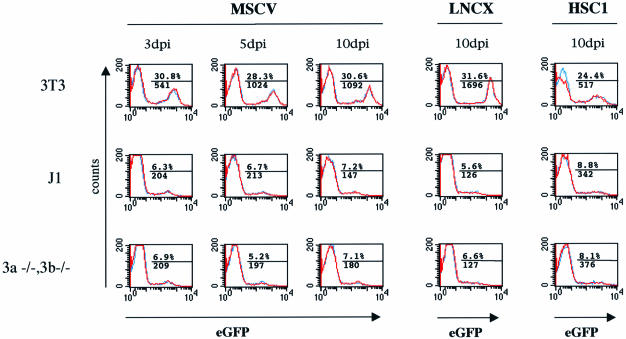
Could de novo methylases play a crucial role in targeted recognition of the retrovirus sequences and subsequent recruitment of the postulated HDAC as suggested in the genome defense model? Previous gene targeting procedures have generated ES cells that are compound homozygous null mutants for the de novo methylases dnmt3a and dnmt3b. In particular, the 7aabb [dnmt3a–/–; dnmt3b–/–] ES cell line has residual maintenance methylase activity, but is incapable of de novo methylation of newly infected exogenous MoMLV containing a SupF marker (Okano et al., 1999). To determine the importance of de novo methylases in retrovirus silencing, we compared expression of several retrovirus vectors containing an internal phosphoglycerate kinase (PGK)–enhanced green fluorescent protein (EGFP) reporter cassette in 7aabb and wild-type J1 ES cells over time. NIH 3T3, J1 and 7aabb cells were infected simultaneously in duplicate with each of the virus stocks, and GFP expression monitored over time by flow cytometry. Infections with MSCV–EGFP virus generated 30% GFP+ NIH 3T3 cells versus ∼7% GFP+ J1 and 7aabb cells at all time points (Figure 4). Mean fluorescence in expressing NIH 3T3 cells at day 10 was 1092 compared with 147–180 in the expressing ES cells. Therefore, the apparent titer in ES cells was 4.2-fold less than in NIH 3T3 cells, and mean fluorescence decreased by 7.4-fold. Assuming similar infection frequencies in NIH 3T3 and ES cells, these data suggest that retrovirus silencing is established by day 3 with equal efficiency in J1 and 7aabb ES cells. Infections with the LNCX–EGFP virus produced almost identical data (Figure 4, earlier time points not shown), indicating that a subset of infected ES cells permit low level expression of both wild-type and MSCV vectors.
Fig. 4. Flow cytometry demonstrates that retrovirus vectors are silenced in wild-type and de novo methylase-null ES cells. The PGK–EGFP cassette was delivered using MSCV, LNCX and HSC1 retrovirus vectors to NIH 3T3, wild-type ES cells (J1) and [dnmt 3a–/–;dnmt3b–/–] ES cells, and flow cytometry was performed 3, 5 and 10 days post-infection. Each infection was performed in duplicate (blue and red lines) and the data merged. The average percentage of GFP+ cells is shown above the line, and mean fluorescence below the line, indicating the gate used.
We then tested whether the SIN deletion in the HSC1–EGFP vector relieves silencing in ES cells. HSC1–EGFP infection generated 24% GFP+ NIH 3T3 cells compared with 8% GFP+ J1 and 7aabb cells at all time points, with mean fluorescence at day 10 of 517 in NIH 3T3 compared with 342–376 in the ES cells (Figure 4, earlier time points not shown). The apparent titer in ES cells was 3-fold less than in NIH 3T3 cells, and mean fluorescence decreased by only 1.5-fold, remaining higher than either MSCV- or LNCX virus-transduced ES cells. As the percentage of expressing cells and their mean fluorescence levels were indistinguishable in J1 and 7aabb cells for LNCX, MSCV and HSC1 retrovirus vectors, we conclude that retrovirus silencing is established with equal efficiency in the presence or absence of the de novo methylases dnmt3a and dnmt3b.
Drosophila reporter transgenes detect conserved silencing mechanisms
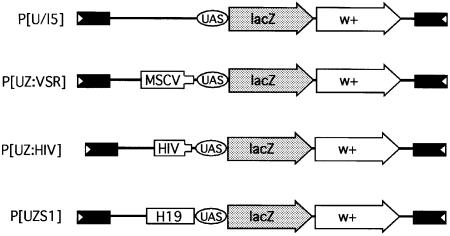
If chromatin structure alterations are a primary cause of retrovirus silencing, then the factors involved may be conserved in other organisms that lack methylases. To this end, we employed a transgenic Drosophila reporter system developed for the characterization of Polycomb-responsive elements (PREs). The P[U/l5] reporter transgene contains GAL4-UAS sites upstream of reporter lacZ and mini-white genes (Figure 5). In the presence of GAL4, the lacZ reporter gene is highly expressed. In contrast, the P[UZS1] transgene contains the mouse H19 imprinting center that functions as a silencer in Drosophila and prevents lacZ reporter expression (Lyko et al., 1997). Silencing in this system has been attributed to chromatin condensation effects that block GAL4 binding (Zink and Paro, 1995). Hence, this reporter system is ideal for investigating conserved silencing mechanisms.
Fig. 5. Structure of reporter transgenes used in Drosophila to assay for conserved silencing mechanisms. The construct P[U/l5] contains GAL4-UAS sites regulating the lacZ gene and the mini-white transformation marker. In the presence of GAL4, the lacZ gene is expressed to high levels. P[UZVSR] contains MSCV sequences, P[UZHIV] contains HIV-1 sequences and P[UZS1] contains the mouse H19 imprinting center, which functions as a silencer in Drosophila and blocks lacZ activation by GAL4.
Retrovirus vectors silence transgenes in Drosophila
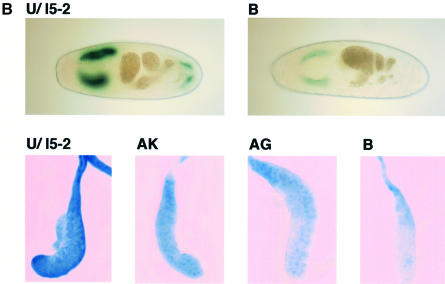
To test whether C-type retrovirus sequences are silenced in the absence of methylases but the presence of conserved silencing factors, we linked an MSCV target sequence to the GAL4-UAS-lacZ and mini-white reporter genes (Figure 5) and introduced the resulting P[UZVSR] construct into Drosophila. Seven independent P[UZVSR] transgenic lines containing MSCV sequences were generated by P-element-mediated germline transformation. The eye color of these lines ranged from orange to yellow, consistent with reduced to very low mini-white expression (Figure 6A). None of the flies demonstrated variegated eye color, and flies with white eyes were assumed to be non-transgenic. We crossed the P[UZVSR] lines to the 1032.hx line that expresses GAL4 in salivary glands and stained late stage embryos of the line VSR:B for β-galactosidase expression (Figure 6B). VSR:B expresses β-galactosidase to very low but uniform levels in the salivary glands, and the non-silenced P[U/l5-2] control expresses to high levels as expected (Figure 6B, upper panels). Dissected salivary glands from third instar larvae also show uniformly reduced β-galactosidase expression that is restricted primarily to the nucleus in lines VSR:AK and AG, and very low levels in VSR:B (Figure 6B, lower panels).
Fig. 6. MSCV sequences silence linked mini-white and lacZ reporter genes in Drosophila. (A) Heads of age-matched adult flies showing that the white gene is expressed at high levels in non-silenced P[U/l5] control flies, at intermediate levels in lines VSR:AK and AG, and at low levels in lines VSR:AS, BA and B. (B) Stage 16 embryos stained with X-Gal show high level β-galactosidase expression in salivary glands of P[U/l5] but low levels in VSR:B (upper panels). Salivary glands dissected from wandering third instar larvae stained with X-Gal show high level β-galactosidase expression in P[U/l5], intermediate levels in VSR:AK and AG, and low levels in VSR:B (lower panels).
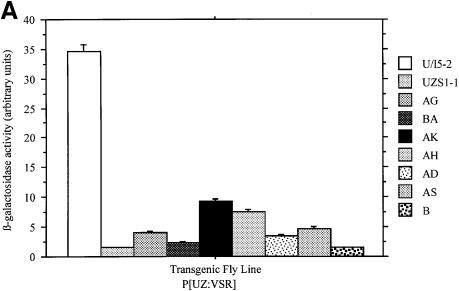
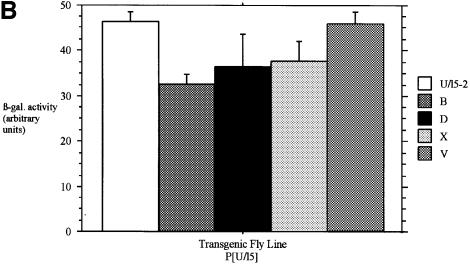
Levels of β-galactosidase were quantified in each line from homogenates of third instar larvae. In the presence of GAL4, β-galactosidase enzymatic activity was significantly reduced in all seven lines (Figure 7A), and lines VSR:B and VSR:BA were equivalent to the control silenced P[UZS1] line. We also created four additional non-silenced P[U/l5] control lines, all of which had red eyes (data not shown) and expressed significant β-galactosidase activity in the presence of GAL4 ranging from 70 to 99% of the levels of the published U/l5-2 line (Figure 7B). These results demonstrate that the reporter transgene is minimally subject to position effects, and argue that lacZ and mini-white silencing are specific consequences of MSCV sequences.
Fig. 7. MSCV sequences significantly reduce β-galactosidase enzymatic activity in transgenic Drosophila. (A) The P[UZVSR] lines express a range of 4–26% of the non-silenced P[U/l5-2] line. (B) The P[U/l5] lines exhibit only minor position effects and express from 70 to 99% of the levels of the control P[U/l5-2] line. Error bars indicate the standard error from the mean values shown. (C) RT–PCR with primer sets (above) show no MSCV transcription (below) in line VSR:BA, or transcription initiated upstream of MSCV in line VSR:B.
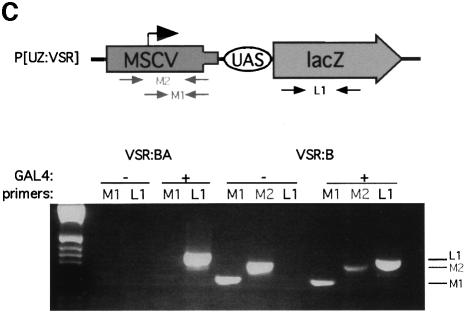
To examine the potential involvement of transcriptional interference from the MSCV promoter, we performed RT–PCR (Figure 7C) to detect transcripts that initiate at the MSCV promoter (M1 primer pair) or upstream of the MSCV promoter (M2 primer pair). We observed no expression from the MSCV promoter with the M1 primers in line VSR:BA, and detected GAL4-independent upstream transcripts with both M2 and M1 primers in line VSR:B. As expected, RT–PCR with the L1 primer pair detected GAL4-dependent lacZ mRNA in both these lines. These results are inconsistent with transcriptional interference from the MSCV promoter. We therefore conclude that MSCV sequences silence both closely linked lacZ and more distant white genes via a methylase-independent pathway in transgenic Drosophila.
Lentivirus vectors silence transgenes in mice and Drosophila
To determine whether transcriptional silencing occurs in other C-type retroviruses that have diverged from the murine retroviruses tested above, we inserted the 5′ LTR and minimal packaging site of HIV-1 as a 0.9 kb fragment into the LCRβ-globin reporter transgene to create the BGT73 construct (Figure 1A). Mice bearing the BGT73 construct were generated and characterized as described above. S1 nuclease analysis of 15.5 day fetal liver RNA (Figure 8A, upper panel) shows that the β-globin transgene is completely silenced in all the BGT73 mice obtained (6/6). We also purified an HIV-SIN injection fragment that deletes the U3 region to –18 bp (Zufferey et al., 1998) directly from the BGT73 plasmid and tested this construct in transgenic mice. S1 nuclease analysis of 15.5 day fetal liver RNA (Figure 8A, lower panel) shows that the β-globin transgene is completely silenced in 7/13 HIV-SIN mice. We conclude that the HIV-SIN construct ameliorates transgene silencing but retains some novel silencer elements.
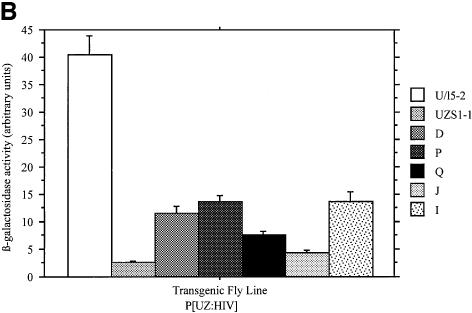
Fig. 8. HIV-1 sequences silence reporter transgene expression in mice and Drosophila. (A) S1 nuclease analysis of globin expression in RNA of 15.5 day fetal livers showing that BGT73 (HIV-1) transgenes are completely silenced in mice, while silencing is ameliorated but not eliminated in the HIV-SIN construct. (B) HIV-1 sequences significantly reduce β-galactosidase enzymatic activity in transgenic Drosophila. The P[UZHIV] lines express a range of 12–31% of the non-silenced P[U/l5-2] line.
We obtained five transgenic Drosophila lines containing the P[UZHIV] construct (Figure 5A) to examine whether HIV-1 sequences are silenced by a methylase-independent pathway. Eye color of these lines was not reduced observably, similar to the P[UZS1] H19 control but in contrast to the P[UZVSR] lines that contain MSCV. In the presence of GAL4, β-galactosidase activity was reduced in all five P[UZHIV] lines (Figure 8B) to levels comparable with that by MSCV. These results show that although HIV-1 silencing has been correlated with high methylation levels in mammals (Bednarik et al., 1990), it can also occur by a conserved pathway in the absence of methylases.
Discussion
One of the first epigenetic models of mammalian gene silencing was proposed to account for the behavior of C-type retroviruses in murine stem cells and embryos (Jahner et al., 1982). The genome defense model specifically proposes that de novo methylases are responsible for recognizing newly integrated viral DNA, thereby creating binding sites for methyl-binding proteins that recruit chromatin remodeling complexes such as HDACs. Some features of this model have not been tested in vivo. Here, we demonstrate that chromatin on silent retrovirus vectors in vivo is inaccessible and is marked differentially by a repressive histone code in transgenic mice. We also show that the efficiency of retrovirus silencing in vivo is not altered in de novo methylase-null ES cells, and that a methylase-independent retrovirus silencing pathway is conserved in Drosophila. These data argue that chromatin modifications play a role in C-type retrovirus silencing as predicted, and that de novo methylases are not required for retrovirus silencing by stem cells.
Role of chromatin structure in retrovirus silencing
We sought to obtain direct in vivo evidence that inaccessible chromatin is established by retroviruses in embryonic cells. To this end, we tested C-type retrovirus silencing in transgenic mice. An LCRβ-globin cassette was chosen as the reporter gene because it opens chromatin and expresses at all ectopic integration sites tested. MoMLV, MSCV and HIV-1 vectors dominantly silence transcription by this reporter, indicating that viral sequences establish inaccessible chromatin that spreads to prevent trans-acting factor access to the regulatory elements on the reporter transgene. Direct evidence of such chromatin structure was obtained by demonstrating that HS do not form on the retrovirus LTR in BGT32 transgenic mice. Moreover, this inaccessible structure spreads over 4 kb to prevent HS formation on the β-globin reporter LCR, promoter and enhancer elements.
Inaccessible chromatin structure may be formed by deacetylation of the nucleosomal core histones. Chromatin immunoprecipitation experiments on the silent transgene in BGT32 fetal liver cells confirm that retrovirus silencing is marked differentially by a histone code of deacetylated H3, acetylated H4 and abundant H1. In contrast to the silenced LCRβ-globin transgene, the endogenous mouse β-major promoter has a histone code composed of acetylated H3 and hyperacetylated H4. Our finding that an 8.8-fold depletion in H3 acetylation levels best distinguishes silent from expressing globin genes agrees with a similar report on the importance of H3 acetylation in the human β-globin locus (Schubeler et al., 2000). Overall, our data provide direct evidence for the establishment and spread of inaccessible chromatin structures from silent retroviruses, and the ChIP results describe a repressive histone code that localizes to silent retroviruses in mouse embryos.
Role of de novo methylases in retrovirus silencing
The transgenic mouse results correlate chromatin structure and histone modifications with retrovirus silencing but do not address the importance of de novo methylases in this process. To this end, we examined silencing of an internal PGK–EGFP reporter transgene by MoMLV, MSCV and HSC1 retrovirus vectors. Infection of wild-type ES cells and de novo methylase-null [dnmt3a–/–;dnmt3b–/–] ES cells with these vectors failed to reveal any differences in GFP silencing assayed by flow cytometry. Silencing was established by day 3 prior to completion of de novo methylation in wild-type cells, and was maintained essentially unaltered until day 10 when methylation is complete in wild-type cells (Okano et al., 1999). Some proviruses in ES cells do escape complete silencing and express low levels of GFP. These data suggest occasional position-dependent proviral expression, and perhaps reflect integration into highly transcribed regions of the genome. Nevertheless, GFP fluorescence from these sites is reduced 7.4-fold relative to the levels in NIH 3T3 cells. We conclude that the de novo methylases dnmt3a and dnmt3b are not required for retrovirus silencing in undifferentiated stem cells.
Evidence for a conserved silencing mechanism
Transgenic Drosophila lack both de novo and maintenance methylases, but share other conserved gene silencing pathways with mammalian cells. Therefore, we created transgenic Drosophila containing MSCV and HIV-1 vector sequences to examine their ability to silence a Gal4-UAS-lacZ reporter gene by methylation-independent conserved pathways. In the absence of any silencer element, five P[Ul/5] transgenic fly lines express β-galactosidase to high and reproducible levels with only minimal position effects. Addition of MSCV sequences to the transgene decreased β-galactosidase expression to 4–26% of the levels with uniform expression of lacZ in salivary glands and also reduced expression of the mini-white transformation marker in eyes. Silencing is not mediated by transcriptional interference from the MSCV retrovirus LTR in mice or flies. It is notable that MSCV does not completely silence Drosophila transgenes. The reasons for this are unclear, but may be related to the exclusion of completely silenced transgenes by selection for fly lines that express mini-white. The reduced levels of lacZ expression in flies mimic expression levels observed in the GFP+ MSCV-transduced ES cells. Although we cannot formally exclude the possibility that silencing occurs by two different mechanisms in mice and flies, it is unlikely given the evidence for de novo methylase independence in both organisms.
Taken together, these data implicate a conserved mechanism that silences MSCV in the absence of de novo methylases in ES cells and in the methylase-free environment of Drosophila. We infer that this putative pathway is not restricted to murine retroviruses because HIV-1 sequences also reduce β-galactosidase expression to 12–31% of the levels in transgenic Drosophila, completely silence the LCRβ-globin reporter transgene in mice, and a self-inactivating HIV-1 vector ameliorates transgene silencing in mice to the same degree as the HSC1 murine retrovirus. The involvement of highly conserved factors would suggest that silencing is not directed specifically at retroviruses, but rather reflects a novel use of a developmentally important gene silencing mechanism.
As de novo methylases are not essential for silencing in undifferentiated ES cells and Drosophila, we postulate that methylation is either a consequence of silencing, or is a redundant parallel pathway that is not essential with regard to C-type retroviruses but functions to tag IAP retroviruses or other foreign CpG-rich DNA elements (Walsh and Bestor, 1999). We propose that the conserved pathway is responsible for causing C-type retrovirus silencing, and that this pathway ultimately dictates the repressive histone code described here by invoking HDACs or HATs that preferentially deplete H3 acetylation levels and by loading H1.
Whether these repressive histone code marks cause silencing or are a consequence of the conserved mechanism has not been determined. Lorincz et al. (2000) have shown that extinction of retrovirus expression in MEL cells can be overcome initially by TSA alone but requires 5-azaC at later time points. Our preliminary ChIP studies show that TSA treatment of explanted BGT32 transgenic fetal liver cells partially restores H3 acetylation levels but fails to displace H1, and β-globin transcription is not reactivated (S.Yao, C.S.Osborne and J.Ellis, unpublished data). In addition, GFP expression in transduced ES and F9 EC cells is not enhanced measurably by TSA and/or 5-azaC treatment (T.Sukonnik and J.Ellis, unpublished data). These results are consistent with the original 5-azaC treatments that show limited reactivation of retrovirus expression in murine EC cells to <1% of the level of non-silenced control cells (Stewart et al., 1982). We therefore suggest that reactivation in embryonic cells will only occur once H1 is displaced, and that the above drugs are not capable of this function in our hands.
Implications for gene therapy
Gene therapy trials indicate that retrovirus vector silencing occurs in hematopoietic stem cells derived from human bone marrow or cord blood (Weinberg and Kohn, 1998). Although it is not clearly established that the same stem cell-specific pathway is involved, dissection of retrovirus silencing mechanisms in transgenic mice, ES cells and Drosophila may suggest strategies to overcome this block to sustained expression of therapeutic genes in transduced hematopoietic stem cells.
Materials and methods
Plasmid construction
BGT25 contains the 7.1 kb LCRβ-globin NotI–EcoRV fragment from BGT14 (Ellis et al., 1997) in the 4.7 kb EcoRI–NcoI p141 vector (Leboulch et al., 1994) that includes the MoMLV backbone. BGT32 contains the same 7.1 kb LCRβ-globin fragment in the 5.2 kb ClaI–HpaI MSCVneoEB (Hawley et al., 1994) plasmid backbone. BGT73 contains the 0.9 kb DraI–NruI HIV-1 5′ LTR and packaging signal fragment from pHR′ (Naldini et al., 1996) cloned into the NdeI site 3′ of the β-globin gene in BGT14. MSCV-PGK–EGFP plasmid DNA was provided by P.Leboulch. The PGK–EGFP cassette was also subcloned into LNCX (Clontech) and HSC1 vectors. P[UZVSR] contains the 5′ LTR and gag sequences of MSCVneoEB cloned as a 1.5 kb NheI–HpaI fragment in p[UZ] (Lyko et al., 1997). P[UZHIV] contains the 0.9 kb HIV-1 fragment cloned in p[UZ].
Transgenic mice
Transgenic mice were produced in FVB fertilized eggs as previously described (Ellis et al., 1997). BGT25, BGT32 and BGT73 were gel purified as 8.5 kb NheI, 8.6 kb NheI and 8.0 kb XmnI–NheI fragments, respectively. The 7.4 kb BGT73-SIN injection fragment was purified after digestion of BGT73 plasmid DNA with PvuII and EcoRV. Day 15.5 post-injection, fetal mice were dissected and DNA extracted from head tissue, while the fetal liver was saved frozen in two halves for future analyses. Transgenic fetuses were identified by Southern blot analysis of fetal head DNA hybridized with the βivs2 probe, and transgene copy number determined using a Molecular Dynamics PhosphorImager. The presence of the transgene in each fetal liver was determined by Southern blot analysis of fetal liver DNA digested with AccI, which cuts twice within the human β-globin gene (Ellis et al., 1997).
RNA analysis
Day 15.5 fetal liver RNA was extracted using Trizol reagent (Gibco-BRL); 1 µg was hybridized to [γ-32P]ATP-labeled double-stranded probes, and digested with 75 U of S1 nuclease (Boehringer Mannheim), and protected bands on a 6% sequencing gel were quantitated on a Molecular Dynamics PhosphorImager as described (Ellis et al., 1996).
RT–PCR was performed on third instar Drosophila larvae RNA extracted with Trizol using the Superscript One Step RT–PCR system (Gibco-BRL) and the listed primers for the following products: LI (5′-GAGCCTGCTAAAGCAAAAAAGAAGTCACC-3′ and 5′-CGTAACCGTGCATCTGCCAGTTTGAGG-3′), M1 (5′-CGGGTACCCGTATTCCCAAT-3′ and 5′-CGCAAACATTAGATGCCGGC-3′) and M2 (5′TGTTCGCGCGCTTCTGCTCCCCG3′ and 5′CGCAAACATTAGATGCCGGC3′).
Chromatin immunoprecipitation
The ChIP assay was performed on day 15.5 fetal liver cell suspensions using kits (Upstate Biotechnology) as described. Chromatin was cross-linked in vivo with 1% formaldehyde at 37°C for 10 min, cells washed once with phosphate-buffered saline (PBS) plus EDTA-free complete proteinase inhibitor (Boehringer Mannheim), resuspended in 200 µl of SDS lysis buffer with proteinase inhibitor on ice for 10 min, and sonicated four times (10 s pulses) at 35% output. After centrifugation, the soluble chromatin in the supernatant was either frozen in liquid nitrogen or used immediately by diluting 200 µl of soluble chromatin into 5.5 ml of ChIP dilution buffer with proteinase inhibitors for pre-cleaning with 200 µl of salmon sperm DNA/protein A–agarose slurry. An input control of 0.5 ml of supernatant was set aside. The remaining 5 ml were split into five Eppendorf tubes. Antibodies (Upstate Biotechnology) including anti-acetylated H3, anti-phospho-H3, anti-acetyl-H4 and anti-H1 were added to four tubes, with one tube for the no antibody control. Immunoprecipitations were performed overnight and washed as described; 0.5 ml of the supernatant of each sample was kept for unbound DNA isolation, and the bound DNA was eluted from the beads, the cross-links reversed and the DNA extracted. A 5 µg aliquot of DNA from each fraction was transferred to a slot-blot for hybridization with random-primed radiolabeled probes recognizing the human β-globin 5′ HS3 region (850 bp SacI–PvuII fragment), human β-globin promoter plus exon 1/exon 2 cDNA (520 bp AccI fragment), endogenous mouse β-globin promoter plus exon 1 and 2 (495 bp NcoI fragment) and endogenous mouse Thy-1 gene (1.1 kb AccI fragment). The slot-blot result was imaged by PhosphorImager, and quantitated with IQMac v1.2 software.
Retrovirus infection and flow cytometry
Virus was harvested from transfected PA317 producer cell populations, and 2 ml of undiluted virus plus polybrene used to infect 1 × 105 cells. GFP expression was detected by flow cytometry using a FacScan (Becton Dickinson) after gating to exclude dead cells.
Transgenic flies
Transgenic flies were raised at 25°C on standard medium unless specified otherwise. Transgenic flies were obtained using white1118 as host and pHSπ as an exogenous source of transposase. Transformants were screened on the basis of their ability to rescue white eye color.
β-galactosidase expression
Quantitation of β-galactosidase activity was performed in quadruplicate for each transgenic line using a chlorophenol red β-D-galactopyranoside (CPRG) assay as described (Lyko et al., 1997). One male and one female wandering third instar larvae were homogenized, aliquoted into a cuvette containing 1 mM CPRG, and ΔOD574/min/mg whole protein determined over the course of an hour. β-galactosidase in situ staining was performed on embryos that were fixed, washed and stained in X-Gal for 5 h at 37°C in the dark. Salivary glands from wandering third instar larvae were dissected and stained for 3 min with X-Gal as described (Zink and Paro, 1995). All lines shown were processed and photographed simultaneously.
Acknowledgments
Acknowledgements
We thank R.Paro and F.Lyko for providing advice, flies and plasmids; D.Allis for ChIP advice; and P.Leboulch, R.Hawley and A.Bukovsky for retrovirus and lentivirus vectors. T.Davies and A.Bashirullah provided advice on β-galactosidase staining. S.Soodeen-Karamath, G.Knowles and L.Wei provided assistance with ES cell culture, flow cytometry and transgenic mice, respectively. This work was supported by grants from the Medical Research Council (MRC) of Canada to J.E. and to H.D.L., the Bayer/CRCS Research and Development Fund to J.E., an Ontario Thalassemia Foundation Fellowship to C.S.O., and Hospital for Sick Children Foundation, OGS and MRC Doctoral Research Awards to D.P.
References
- Bednarik D.P., Cook,J.A. and Pitha,P.M. (1990) Inactivation of the HIV LTR by DNA CpG methylation: evidence for a role in latency. EMBO J., 9, 1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G. and Paro,R. (1999) Epigenetic inheritance of active chromatin after removal of the main transactivator. Science, 286, 955–958. [DOI] [PubMed] [Google Scholar]
- Challita P.M. and Kohn,D.B. (1994) Lack of expression from a retroviral vector after transduction of murine hematopoietic stem cells is associated with methylation in vivo. Proc. Natl Acad. Sci. USA, 91, 2567–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.Y. and Townes,T.M. (2000) Molecular mechanism for silencing virally transduced genes involves histone deacetylation and chromatin condensation. Proc. Natl Acad. Sci. USA, 97, 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.Y., Bailey,E.C., McCune,S.L., Dong,J.Y. and Townes,T.M. (1997) Reactivation of silenced, virally transduced genes by inhibitors of histone deacetylase. Proc. Natl Acad. Sci. USA, 94, 5798–5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B.R., Lomedico,P.T. and Ju,G. (1984) Transcriptional interference in avian retroviruses—implications for the promoter insertion model of leukaemogenesis. Nature, 307, 241–245. [DOI] [PubMed] [Google Scholar]
- Eden S., Hashimshony,T., Keshet,I. and Cedar,H. (1998) DNA methylation models histone deacetylation. Nature, 394, 842. [DOI] [PubMed] [Google Scholar]
- Ellis J., Tan-Un,K.C., Harper,A., Michalovich,D., Yannoutsos,N., Philipsen,S. and Grosveld,F. (1996) A dominant chromatin-opening activity in 5′ hypersensitive site 3 of the human β-globin locus control region. EMBO J., 15, 562–568. [PMC free article] [PubMed] [Google Scholar]
- Ellis J., Pasceri,P., Tan-Un,K.C., Wu,X., Harper,A., Fraser,P. and Grosveld,F. (1997) Evaluation of β-globin gene therapy constructs in single copy transgenic mice. Nucleic Acids Res., 25, 1296–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester W.C., Thompson,C., Elder,J.T. and Groudine,M. (1986) A developmentally stable chromatin structure in the human β-globin gene cluster. Proc. Natl Acad. Sci. USA, 83, 1359–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester W.C., Epner,E., Driscoll,M.C., Enver,T., Brice,M., Papayannopoulou,T. and Groudine,M. (1990) A deletion of the human β-globin locus activation region causes a major alteration in chromatin structure and replication across the entire β-globin locus. Genes Dev., 4, 1637–1649. [DOI] [PubMed] [Google Scholar]
- Gautsch J.W. and Wilson,M.C. (1983) Delayed de novo methylation in teratocarcinoma suggests additional tissue-specific mechanisms for controlling gene expression. Nature, 301, 32–37. [DOI] [PubMed] [Google Scholar]
- Grosveld F., van Assendelft,G.B., Greaves,D.R. and Kollias,G. (1987) Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell, 51, 975–985. [DOI] [PubMed] [Google Scholar]
- Hawley R.G., Lieu,F.H., Fong,A.Z. and Hawley,T.S. (1994) Versatile retroviral vectors for potential use in gene therapy. Gene Ther., 1, 136–138. [PubMed] [Google Scholar]
- Jahner D., Stuhlmann,H., Stewart,C.L., Harbers,K., Lohler,J., Simon,I. and Jaenisch,R. (1982) De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature, 298, 623–628. [DOI] [PubMed] [Google Scholar]
- Jones P.L., Veenstra,G.J., Wade,P.A., Vermaak,D., Kass,S.U., Landsberger,N., Strouboulis,J. and Wolffe,A.P. (1998) Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature Genet., 19, 187–191. [DOI] [PubMed] [Google Scholar]
- Kempler G., Freitag,B., Berwin,B., Nanassy,O. and Barklis,E. (1993) Characterization of the Moloney murine leukemia virus stem cell-specific repressor binding site. Virology, 193, 690–699. [DOI] [PubMed] [Google Scholar]
- Leboulch P., Huang,G.M., Humphries,R.K., Oh,Y.H., Eaves,C.J., Tuan,D.Y. and London,I.M. (1994) Mutagenesis of retroviral vectors transducing human β-globin gene and β-globin locus control region derivatives results in stable transmission of an active transcriptional structure. EMBO J., 13, 3065–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H., Oh,S.P., Okano,M., Juttermann,R., Goss,K.A., Jaenisch,R. and Li,E. (1996) De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development, 122, 3195–3205. [DOI] [PubMed] [Google Scholar]
- Lorincz M.C., Schubeler,D., Goeke,S.C., Walters,M., Groudine,M. and Martin,D.I. (2000) Dynamic analysis of proviral induction and de novo methylation: implications for a histone deacetylase-independent, methylation density-dependent mechanism of transcriptional repression. Mol. Cell. Biol., 20, 842–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F., Brenton,J.D., Surani,M.A. and Paro,R. (1997) An imprinting element from the mouse H19 locus functions as a silencer in Drosophila. Nature Genet., 16, 171–173. [DOI] [PubMed] [Google Scholar]
- McCune S.L. and Townes,T.M. (1994) Retroviral vector sequences inhibit human β-globin gene expression in transgenic mice. Nucleic Acids Res., 22, 4477–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L., Blomer,U., Gallay,P., Ory,D., Mulligan,R., Gage,F.H., Verma,I.M. and Trono,D. (1996) In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science, 272, 263–267. [DOI] [PubMed] [Google Scholar]
- Nan X., Ng,H.H., Johnson,C.A., Laherty,C.D., Turner,B.M., Eisenman,R.N. and Bird,A. (1998) Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature, 393, 386–389. [DOI] [PubMed] [Google Scholar]
- Niwa O., Yokota,Y., Ishida,H. and Sugahara,T. (1983) Independent mechanisms involved in suppression of the Moloney leukemia virus genome during differentiation of murine teratocarcinoma cells. Cell, 32, 1105–1113. [DOI] [PubMed] [Google Scholar]
- Okano M., Xie,S. and Li,E. (1998) Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nature Genet., 19, 219–220. [DOI] [PubMed] [Google Scholar]
- Okano M., Bell,D.W., Haber,D.A. and Li,E. (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell, 99, 247–257. [DOI] [PubMed] [Google Scholar]
- Osborne C.S., Pasceri,P., Singal,R., Sukonnik,T., Ginder,G.D. and Ellis,J. (1999) Amelioration of retroviral vector silencing in locus control region β-globin-transgenic mice and transduced F9 embryonic cells. J. Virol., 73, 5490–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazin M.J. and Kadonaga,J.T. (1997) What’s up and down with histone deacetylation and transcription? Cell, 89, 325–328. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. (1998) Polycombing the genome: PcG, trxG and chromatin silencing. Cell, 93, 333–336. [DOI] [PubMed] [Google Scholar]
- Rasmussen J.A. and Gilboa,E. (1987) Significance of DNase I-hypersensitive sites in the long terminal repeats of a Moloney murine leukemia virus vector. J. Virol., 61, 1368–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A. (1998) CpG methylation, chromatin structure and gene silencing—a three-way connection. EMBO J., 17, 4905–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan T.M., Behringer,R.R., Martin,N.C., Townes,T.M., Palmiter,R.D. and Brinster,R.L. (1989) A single erythroid-specific DNase I super-hypersensitive site activates high levels of human β-globin gene expression in transgenic mice. Genes Dev., 3, 314–323. [DOI] [PubMed] [Google Scholar]
- Schubeler D., Francastel,C., Cimbora,D.M., Reik,A., Martin,D.I. and Groudine,M. (2000) Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human β-globin locus. Genes Dev., 14, 940–950. [PMC free article] [PubMed] [Google Scholar]
- Simmen M.W., Leitgeb,S., Charlton,J., Jones,S.J., Harris,B.R., Clark,V.H. and Bird,A. (1999) Nonmethylated transposable elements and methylated genes in a chordate genome. Science, 283, 1164–1167. [DOI] [PubMed] [Google Scholar]
- Stewart C.L., Stuhlmann,H., Jahner,D. and Jaenisch,R. (1982) De novo methylation, expression and infectivity of retroviral genomes introduced into embryonal carcinoma cells. Proc. Natl Acad. Sci. USA, 79, 4098–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Thompson T. and Fan,H. (1985) Mapping of DNase I-hypersensitive sites in the 5′ and 3′ long terminal repeats of integrated Moloney murine leukemia virus proviral DNA. Mol. Cell. Biol., 5, 601–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan D. and London,I.M. (1984) Mapping of DNase I-hypersensitive sites in the upstream DNA of human embryonic ε-globin gene in K562 leukemia cells. Proc. Natl Acad. Sci. USA, 81, 2718–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker K.L., Beard,C., Dausmann,J., Jackson-Grusby,L., Laird,P.W., Lei,H., Li,E. and Jaenisch,R. (1996) Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev., 10, 1008–1020. [DOI] [PubMed] [Google Scholar]
- Tweedie S., Ng,H.H., Barlow,A.L., Turner,B.M., Hendrich,B. and Bird,A. (1999) Vestiges of a DNA methylation system in Drosophila melanogaster? Nature Genet., 23, 389–390. [DOI] [PubMed] [Google Scholar]
- Urieli-Shoval S., Gruenbaum,Y., Sedat,J. and Razin,A. (1982) The absence of detectable methylated bases in Drosophila melanogaster DNA. FEBS Lett., 146, 148–152. [DOI] [PubMed] [Google Scholar]
- Walsh C.P. and Bestor,T.H. (1999) Cytosine methylation and mammalian development. Genes Dev., 13, 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Yu,L., Bowen,J., Gorovsky,M.A. and Allis,C.D. (1999) Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell, 97, 99–109. [DOI] [PubMed] [Google Scholar]
- Weinberg K.I. and Kohn,D.B. (1998) Gene therapy for congenital lymphoid immunodeficiency diseases. Semin. Hematol., 35, 354–366. [PubMed] [Google Scholar]
- Yoder J.A., Walsh,C.P. and Bestor,T.H. (1997) Cytosine methylation and the ecology of intragenomic parasites. Trends Genet., 13, 335–340. [DOI] [PubMed] [Google Scholar]
- Zink D. and Paro,R. (1995) Drosophila Polycomb-group regulated chromatin inhibits the accessibility of a trans-activator to its target DNA. EMBO J., 14, 5660–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R., Dull,T., Mandel,R.J., Bukovsky,A., Quiroz,D., Naldini,L. and Trono,D. (1998) Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol., 72, 9873–9880. [DOI] [PMC free article] [PubMed] [Google Scholar]