Abstract
The serine proteinase α-thrombin plays a pivotal role in the regulation of blood fluidity, and therefore constitutes a primary target in the treatment of various haemostatic disorders. Haemadin is a slow tight- binding thrombin inhibitor from the land-living leech Haemadipsa sylvestris. Here we present the 3.1 Å crystal structure of the human α-thrombin– haemadin complex. The N-terminal segment of haemadin binds to the active site of thrombin, forming a parallel β-strand with residues Ser214–Gly216 of the proteinase. This mode of binding is similar to that observed in another leech-derived inhibitor, hirudin. In contrast to hirudin, however, the markedly acidic C-terminal peptide of haemadin does not bind the fibrinogen-recognition exosite, but interacts with the heparin-binding exosite of thrombin. Thus, haemadin binds to thrombin according to a novel mechanism, despite an overall structural similarity with hirudin. Haemadin inhibits both free and thrombomodulin-bound α-thrombin, but not intermediate activation forms such as meizothrombin. This specific anticoagulant ability of haemadin makes it an ideal candidate for an antithrombotic agent, as well as a starting point for the design of novel antithrombotics.
Keywords: coagulation/crystal structure/heparin-binding exosite/hirudin/thrombin inhibitor
Introduction
Thrombin is a trypsin-like serine proteinase that plays a crucial role in the processes of thrombosis and haemostasis. Free α-thrombin performs several major functions leading to clot formation. It converts soluble fibrinogen into fibrin and activates the transglutaminase factor XIII, which thereafter cross-links adjacent fibrin monomers (Davie et al., 1991; Stubbs and Bode, 1993). Furthermore, thrombin induces platelet aggregation via proteolytic activation of G protein-coupled protease-activated receptors (PARs) (Vu et al., 1991; Coughlin, 1999). In addition, thrombin promotes its own production by activating the non-enzymatic cofactors V and VIII (Kane and Davie, 1988) as well as factor XI (Gailani and Broze, 1991). When bound to the endothelial membrane receptor thrombomodulin, thrombin becomes both a potent activator of protein C (Esmon, 1995) and of the procarboxypeptidase thrombin-activatable fibrinolysis inhibitor (Nesheim et al., 1997), thereby simultaneously deactivating the coagulation cascade and inhibiting fibrinolysis.
Myocardial infarction and stroke are rooted in arterial and venous thrombotic disorders. To treat these diseases, potent and specific antithrombotic agents are required, and their development continues to be an area of intensive research (Fenton et al., 1991; Bode et al., 1994; Stone and Tapparelli, 1995; Gulba et al., 1998). The search for selective antithrombotics has benefited from the isolation of inhibitors from haematophagous animals and the determination of their three-dimensional structures (for a recent review see Lombardi et al., 1999). These thrombin–inhibitor complexes have shown the adaptation of natural inhibitors to the unique molecular architecture of thrombin. Thrombin’s specific features include not only a rather inaccessible active site due to two striking loop insertions, but also two patches of positive surface potential that mediate intermolecular interactions: the fibrinogen-recognition exosite (also termed anion-binding exosite I) and the heparin-binding exosite (or anion-binding exosite II) (Bode et al., 1989, 1992).
Occupancy of exosite I alone effectively blocks thrombin’s cleavage of physiological substrates, as shown in the case of triabin, a lipocalin-like protein from the bug Triatoma pallidipennis (Noeske-Jungblut et al., 1995; Fuentes-Prior et al., 1997). Two related inhibitors present in snake venoms, bothrojaracin (Arocas et al., 1996) and bothroalternin (Castro et al., 1998), seem to interact with both thrombin exosites without blocking the active site. More prevalent, however, are two-domain thrombin inhibitors in which the N-terminal domain occupies or blocks the active-site cleft of the proteinase, while a markedly acidic C-terminal domain binds the basic fibrinogen-recognition exosite. This now paradigmatic mode of inhibition was first identified in hirudin (Rydel et al., 1990), a small (Mr ∼8 kDa) polypeptide isolated from the leech Hirudo medicinalis (Walsmann and Markwardt, 1985; Wallis, 1988). Rhodniin, a Kazal-type inhibitor isolated from the bug Rhodnius prolixus (van de Locht et al., 1995), and the Kunitz-type inhibitor ornithodorin purified from the soft tick Ornithodorus moubata (van de Locht et al., 1996) are double-headed inhibitors that contact both the active site and exosite I.
In spite of the diverse sources and inhibition mechanisms, in all crystallographically studied thrombin–inhibitor complexes one domain of the inhibitor contacts the fibrinogen-recognition exosite. In this regard, proteinaceous inhibitors mimic the binding mechanism of physiological substrates (e.g. fibrinogen, PARs) or the natural regulator of haemostasis, thrombomodulin. We have identified a slow–tight binding thrombin inhibitor (Ki ∼2 × 10–13 M) in the saliva of the land-living leech Haemadipsa sylvestris, haemadin (Strube et al., 1993). Surprisingly, haemadin displays several sequence and functional features that distinguish it clearly from hirudin: it is significantly smaller than hirudin, lacks a C-terminal sulfated tyrosine and, in contrast to hirudin, complex formation with thrombin is independent of ionic strength. These features suggested differences in the mode of inhibition, and prompted us to determine the three-dimensional structure of the thrombin–haemadin complex. In this work we show that haemadin constitutes a novel thrombin inhibitor.
Results
Overall structure
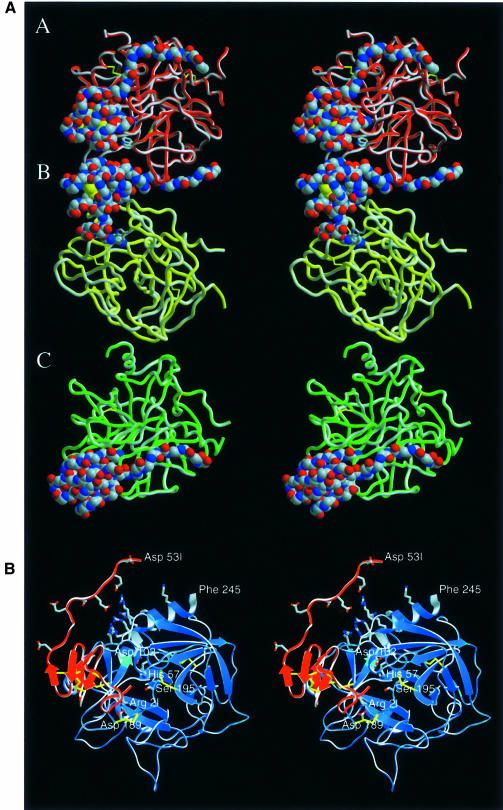
The human α-thrombin–haemadin complex crystallizes as a trimer in the monoclinic space group P2, with the crystallographically independent complexes (denoted A, B and C) aligned almost parallel to the a–c plane (Figure 1A). In all three complexes, the main body of the inhibitor binds to the target proteinase, adopting extremely similar conformations (described in detail below), while its C-terminal tail shows slightly different structures. This remarkably acidic peptide of haemadin ‘sandwiches’ between the heparin-binding exosite of the cognate thrombin molecule and the fibrinogen-recognition exosite of a neighbouring molecule, and is thus essential for formation of the crystallographic A–B–C trimer. As seen in Figure 1A, the C-terminal tail of haemadin in complex B fills the A–B interface by simultaneously binding to its cognate thrombin moiety (B) and to the neighbouring thrombin molecule (complex A). Similar contacts are made between the haemadin peptide of the boundary complex A and the symmetry-related molecule C′, while the tail of haemadin molecule C contacts both its ‘own’ thrombin (C) and the symmetry-related molecule A′ (not depicted in Figure 1A). Strong thrombin–thrombin contacts involving both charged and hydrophobic residues of the adjacent antiparallel stretches Ser11–Ile14K (light chains, chymotrypsinogen numbering) (Bode et al., 1992) and Ser20–Gly25 (heavy chains) in complexes B and C further stabilize the asymmetric unit trimer.
Fig. 1. Crystal structure of the human thrombin–haemadin complex. (A) Structure of the crystallographic trimer present in the asymmetric unit. Monomers are labelled A, B and C. Thrombin molecules are shown as red, yellow and green ribbons; the Cα traces of the three inhibitors are presented as colour-coded van der Waals spheres (red, oxygen; blue, nitrogen; grey, carbon). (B) Stereo diagram of complex molecule A. The protease is shown in its ‘standard orientation’ (Bode et al., 1992), i.e. with the active-site cleft facing the viewer and substrates running from left to right. Side chains of the catalytic triad residues are shown explicitly, as well as the side chains of the interacting residues Asp189 (thrombin) and Arg2I (haemadin) (colour coded as in Figure 1A). Also shown (unlabelled) are the side chains of the basic residues of the heparin-binding site (thrombin), as well as the side chains of the acidic residues of haemadin’s C-terminal tail. This figure and Figures 3, 5A, 7A and 8 were prepared with SETOR (Evans, 1993).
However, the remarkable polymeric arrangement of the thrombin–haemadin complexes does not seem to be relevant in diluted protein solutions and at more physiological conditions. Evidence discussed below entitles us to assume that the solution structure of the thrombin–haemadin complex corresponds to the monomeric 1:1 complex shown in more detail in Figure 1B.
Structure of thrombin
The structure of haemadin-bound thrombin is similar to that of thrombin in the human α-thrombin–hirudin structure (Grütter et al., 1990; Rydel et al., 1990); this also applies to the active-site catalytic triad, which is almost identical in both structures. Large deviations in the main chain are essentially limited to the so-called ‘autolysis loop’ or 149-loop (residues Thr147–Lys149E). This loop possesses a well reported conformational flexibility (Bode et al., 1992) and has missing or very weak electron density for up to four residues in the three thrombin–haemadin complexes. Electron density is also weak or missing in the light chain termini (residue Thr1H of complex C as well as residues Gly14M and Arg15 of complex A) and in the C-terminus of the heavy chain (residues Glu247 of all complexes, Gly246 of complexes A and B, and Phe245 of complex B). Weak density accounting for the first N-acetyl-glucosamine moiety attached to the amide group of Asn60G is present in all three complexes.
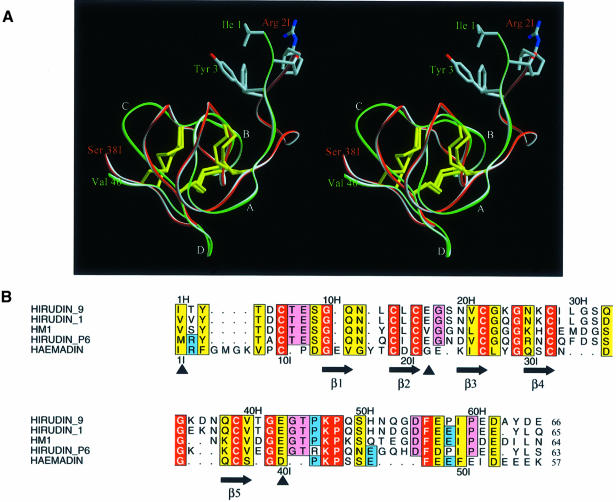
Structure of haemadin
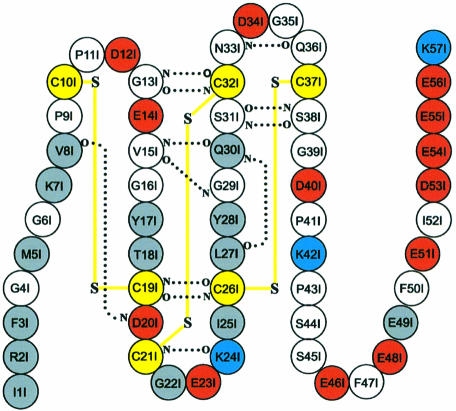
Haemadin residues Pro9I–Ser38I (the suffix ‘I’ identifies haemadin residues) are folded into a compact ellipsoidal core of approximate dimensions 15 × 16 × 18 Å. This core contains five short β-strands (β1–β5), which are arranged in two antiparallel distorted sheets formed by strands β1–β4–β5 and β2–β3 facing each other (for a schematic drawing of haemadin’s primary and secondary structures see Figure 2). This β-sandwich is stabilized by six enclosed cysteines arranged in a [1–2, 3–5, 4–6] disulfide pairing (Figure 3), resulting in a disulfide-rich hydrophobic core that is largely inaccessible to bulk solvent. The close proximity of disulfide bonds [3–5] and [4–6] organizes haemadin into four distinct loops. The largest of these loops (loop A, residues Pro11I–Thr18I) contains β-strand β1 (Gly13I–Val15I), which hydrogen-bonds residues Gln30I–Cys32I (β4). The shorter loops B (residues Gly22I–Ile25I), C (Leu27I–Ser31I) and D (Asn33I–Gln36I) are essentially β-hairpin turns connecting consecutive β-strands β2–β3, β3–β4 and β4–β5, respectively.
Fig. 2. Primary and secondary structure of haemadin. Acidic, basic and cysteine residues are represented by red, blue and yellow circles, respectively. Main chain–main chain hydrogen bonds formed in at least two of the three independent molecules forming the asymmetric unit are indicated with dotted lines. Disulfide bridges are shown with yellow lines. Residues involved in interactions with thrombin are represented by grey circles.
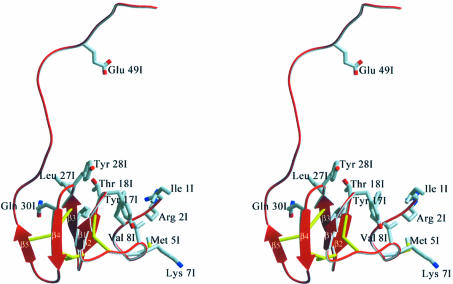
Fig. 3. Ribbon diagram of haemadin’s structure, highlighting elements of secondary structure and the disulfide-rich core. Side chains of residues involved in major interactions with thrombin are shown explicitly (colour coded as in Figure 1).
The conformations adopted by both the N- (Ile1I–Val8I) and especially the C-terminal segments (Gly39I–Lys57I) seem to be largely determined by contacts to the cognate thrombin and/or to neighbouring molecules in the crystal (see below). In particular, the acidic C-terminal tail protrudes away from the main body of the inhibitor (Figures 1 and 3). This extended stretch lacks secondary structure and adopts slightly different conformations in each of the three crystallographically independent complexes (Figure 1A). Moreover, the last four C-terminal residues Glu54I–Lys57I are not defined in the electron density, suggesting enhanced conformational flexibility.
Binding of the inhibitor
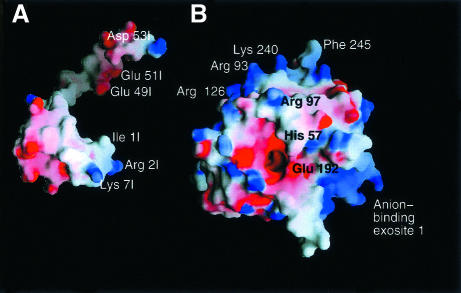
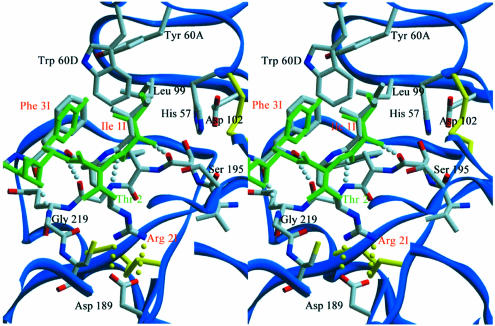
Analysis of the interacting surfaces of haemadin and thrombin reveals a striking complementarity of their electrostatic surface potentials (Figure 4), which is most certainly a major driving force for complex formation. Solvent-accessible molecular surfaces of 780 Å2 and 200–230 Å2 are buried at the active-site and exosite II regions, respectively. Beyond the association of thrombin’s highly basic heparin-binding exosite and the acidic C-terminal tail of haemadin, the negatively charged active-site region is compensated by the N-terminal alkaline side chains of Arg2I and Lys7I. In the complex, the first three N-terminal residues of haemadin are inserted into the active-site cleft of thrombin, forming a parallel β-sheet with thrombin residues Ser214–Gly216. Asides from several intermolecular main chain-to-main chain hydrogen bonds that stabilize this β-sheet, the side chains of these three residues make numerous contacts with thrombin’s active site (Figure 5).
Fig. 4. Space-filling models of human α-thrombin and haemadin, showing the surface potential of the two molecules. Positive charges are displayed in blue, negative charges in red, with darkest blue and red colours corresponding to electrostatic potentials beyond –10 and +10 kT/e, respectively. The plot was prepared with GRASP (Nicholls et al., 1993). The thrombin component (B) is shown in the standard orientation, while haemadin (A) is rotated along the y-axis to present the thrombin binding surface to the viewer. Some of the residues of the interacting interfaces are labelled.
Fig. 5. Close-up stereoview of the active-site cleft of haemadin-bound human α-thrombin. Side chains of selected residues are depicted colour coded. Intermolecular hydrogen bonds are shown as white dotted lines for the parallel interaction of haemadin’s N-terminal loop with thrombin residues Ser214–Gly216 or yellow dotted lines for contacts made inside the S1 pocket. Some residues of thrombin and the inhibitors along with all water molecules are omitted for the sake of simplicity. The three N-terminal amino acid residues of hirudin (green) are shown for comparison.
Close to the active site of thrombin is Ile1I, with its isobutyl group occupying thrombin’s S2 pocket, while its amino group hydrogen-bonds the carbonyl of Ser214. The protruding side chain of Arg2I occupies the S1 specificity pocket, where it forms a hydrogen-bonded ion pair with the carboxylate of Asp189, but enters this pocket from a position that in canonically binding inhibitors is occupied by P3 residues. In addition, Arg2I Nη1 donates a hydrogen bond to the carbonyl of Gly219. The phenyl moiety of Phe3I protrudes up into the S4 pocket of thrombin; this aromatic side chain and the aliphatic Ile1I also participate in intramolecular van der Waals contacts with Val8I as well as with some residues belonging to the compact core of haemadin (e.g. Thr18I, Ile25I). These interactions stabilize the N-terminal peptide in its characteristic loop shape whilst bound to thrombin (Figures 1B and 3).
Emerging from the active site, the carbonyl of Gly4I accepts a hydrogen bond from the side chain of thrombin’s Arg221A, while the side chain of Met5I is interspersed between the thrombin carboxylates of Glu146 and Glu192. The prominent Glu192 residue is in turn ‘sandwiched’ between the side chains of Met5I and Lys7I, experiencing the positive electrostatic potential of the latter (Figure 4). However, the distance of ∼4.1 Å between Lys7I Nζ and Glu192 Oε1 and their unfavourable relative orientation preclude formation of a direct hydrogen-bonded salt bridge. Val8I makes hydrophobic interactions with the indole moiety of Trp60D; this insertion (60-)loop acts as a centre of hydrophobicity and places residues Phe3I and Tyr17I within the realms of van der Waals contacts.
Solvent-exposed side chains of haemadin’s core, in particular those of loops A and C, participate in several major interactions with surface loops of thrombin that surround the ‘west’ and ‘north’ sides of the active site (standard orientation, Figure 1B). At the end of loop A, Tyr17I edges thrombin’s 60- and 96-loops, making close van der Waals contacts with residue Pro60C and the indole moiety of Trp96, while the Thr18I amide hydrogen-bonds Arg97 O. The guanidinium group of the latter donates a hydrogen bond to Gly16I’s carbonyl, thus mimicking formation of a short parallel β-sheet. The exposed side chain of Ile25I (loop B) makes strong van der Waals contacts with thrombin’s Glu217, as well as a weaker interaction with Ile174. Emerging from loop C, Tyr28I is tilted against the aliphatic parts of Arg97 and Glu97A. Furthermore, the side chain of Leu27I is in close van der Waals distance to both the backbone and side chains of Arg173–Ile174. The guanidinium group of Arg173 donates hydrogen bonds to both the carbonyl of Leu25I and Gln30I Oε1.
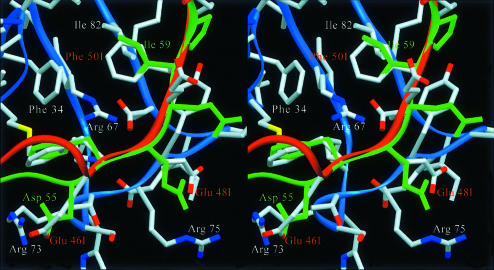
The extended C-terminal peptide of haemadin covers the heparin-binding exosite of thrombin. This surface patch of the proteinase is dominated by unbalanced positive charges (e.g. Arg93, Arg97, Arg101, Arg233, Lys240; see Figure 4), whereas the C-terminal domain of haemadin harbours a large number of negatively charged residues (8 out of 19 residues are aspartates or glutamates). In spite of this overall electrostatic complementarity, the only strong intermolecular salt bridges observed in the crystal are those linking the carboxylate of Glu49I to the guanidinium groups of Arg93 and Arg101 (Figure 1B).
Binding studies in solution
Band shift studies were performed to analyse the behaviour of the human α-thrombin–haemadin complex in solution. Addition of the exosite I-binding inhibitor triabin to a sample of human α-thrombin–haemadin complex in a 1:1 molar ratio resulted in the appearance of a new band in non-denaturing polyacrylamide electrophoresis gels (Figure 6A, lane 5). This band does not correspond to the human thrombin–triabin complex (lane 9) or to any of the individual proteins. Addition of triabin in ratios >1:1 did not alter the appearance of this new band (lanes 6 and 7), and no additional interactions were detected between triabin and haemadin. These findings indicate that the new band corresponds to the ternary triabin–thrombin–haemadin complex, and that both inhibitors do not compete with each other for thrombin binding. Moreover, haemadin inhibited the amidolytic activity of the degraded thrombin form γ-thrombin (data not shown), which is cleaved by trypsin behind Arg75, Arg77A and Lys149E, resulting in a largely distorted exosite I and therefore in a greatly reduced affinity for hirudin (Chang, 1986). Also, haemadin could not be displaced from its thrombin complex by hirugen (hirudin residues Asp55–Glu65), since no thrombin chromogenic activity could be detected after adding up to 1000 M excess of hirugen (data not shown).
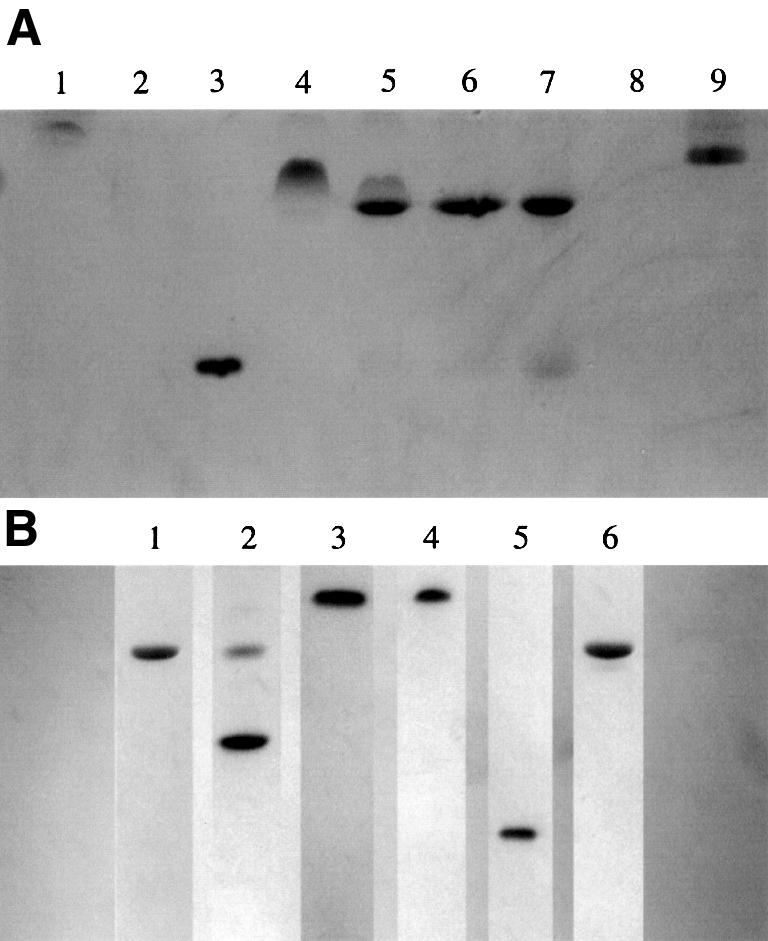
Fig. 6. Binding studies in solution. (A) Formation of the ternary complex haemadin–thrombin–triabin, as followed using non-denaturant PAGE. Lane 1, human α-thrombin (5 µg); lane 2, haemadin (10 µg); lane 3, triabin (20 µg); lane 4, thrombin–haemadin complex (10 µg); lanes 5–7, 10 µg of thrombin–haemadin complex incubated with increasing amounts of triabin (1:1, 1:1.5 and 1:2 equivalents); lane 9, human α-thrombin–triabin complex (10 µg). (B) Haemadin binds to thrombomodulin-bound thrombin, but not to meizothrombin. Lane 1, 10 µg of human thrombin–TME456; lane 2: 10 µg of the complex incubated with 1 µg haemadin; lane 3, 10 µg of meizothrombin; lane 4, 10 µg meizothrombin incubated with 1.5 µg haemadin; lane 5, 20 µg triabin; lane 6, human thrombin–triabin complex (10 µg).
In contrast, we could not detect haemadin binding to meizothrombin (Figure 6B, lanes 3 and 4), while hirudin forms a stable complex with this intermediate activation form of thrombin, which possesses a blocked exosite II (Fischer et al., 1998). Formation of the 1:1 thrombin– haemadin complex was not inhibited by the presence of up to 100 U/ml sulfated glycosaminoglycan heparin. However, under low-salt conditions the human α-thrombin–haemadin complex bound partially to heparin–Sepharose.
The smallest active fragment of thrombomodulin (termed TME456) binds only to thrombin’s exosite I, without altering the active-site region of the enzyme (Fuentes-Prior et al., 2000). Lack of interaction between haemadin and exosite I of thrombin suggested the ability of haemadin also to block the anticoagulant activity of thrombomodulin-bound thrombin. This previously uncharacterized inhibitory activity of haemadin was investigated using slow-binding inhibition kinetics, and an intrinsic dissociation constant (Ki) of 296.3 ± 94.2 × 10–15 M was calculated for the inhibition of the thrombin–TME456 complex.
We examined next the contribution of several basic thrombin residues to the binding of the inhibitor. Kinetic studies employed glutamate mutants of thrombin that have been shown previously not to affect the chromogenic activity of the proteinase (Wu et al., 1991; Sheehan and Sadler, 1994). The results of this study are summarized in Table I. The highest decrease in inhibitory potency (one order of magnitude) is observed with thrombin mutant Arg93Glu; substitution of two other exosite II residues by glutamate (Lys233, Lys240) results in 3- and 5-fold reductions in activity, respectively. In contrast, no significant decrease in activity is associated with mutations Arg73→Glu or Lys149E→Glu (exosite I).
Table I. Haemadin binding to mutant thrombins.
| Mutant | Ki × 1013 (M) | Fold increase compared to native |
|---|---|---|
| Native | 2.435 ± 0.138 | 1.00 |
| Arg73Glu | 3.365 ± 0.523 | 1.38 |
| Arg93Glu | 24.081 ± 0.119 | 9.89 |
| Lys149EGlu | 2.337 ± 0.347 | 0.96 |
| Arg233Glu | 7.333 ± 0.448 | 3.01 |
| Lys236Glu | 4.221 ± 0.302 | 1.73 |
| Lys240Glu | 12.905 ± 0.872 | 5.30 |
Comparison of haemadin with hirudin
The disulfide-rich cores of haemadin (residues Pro9I–Ser38I) and H.medicinalis hirudin (Thr4H– Val40H; the suffix ‘H’ denotes hirudin residues) can be overlaid with a root-mean-square deviation of 1.15 Å for 22 pairs of equivalent Cα atoms. As shown in Figure 7A, all three disulfide bonds are spatially similar, but the four loops described earlier for haemadin are somewhat offset in the two structures. Some of the differences can be accounted for by loop size discrepancies, but in the case of loop C, which is of identical size, the displacement is due to Gly23H following the disulfide bond [4–6] (Cys22H–Cys39H) in hirudin. A structure-based sequence alignment of haemadin with four hirudin variants is presented in Figure 7B; it highlights the fact that the overall conservation of the three-dimensional structure is only marginally matched at the sequence level.
Fig. 7. (A) Stereoview of the main chain of haemadin (red, residues Ile1I–Ser38I) and hirudin (green, residues Ile1H–Val40H) after optimal least-squares fit; only the side chains of the first three residues of both molecules are shown explicitly. Note the different location of the N-terminal segments, indicating divergent arrangements of the compact domains relative to thrombin (compare Figure 5). (B) Structure-based alignment of the amino acid sequences of haemadin and of four representative hirudin variants. Nomenclature follows the work of Steiner et al. (1992). Residues with particularly close homologies are boxed in yellow, identities in red. Residues conserved in hirudin but not haemadin are shadowed pink; those common to haemadin and some hirudin variants are shadowed blue. Numbers refer to the sequences of hirudin (above) and haemadin (below the alignment). The secondary structure of haemadin is also given. The intron–exon boundaries (full arrows) are those determined for Hirudinaria manillensis (Scacheri et al., 1993). The aligned sequences were formatted using the program ALSCRIPT (Barton, 1993).
The considerable similarities of the C-terminal tails manifest themselves in the binding of the C-terminal peptides of haemadin to the fibrinogen-recognition exosites of neighbouring thrombin molecules in the current crystal structure (Figures 1A and 8). The main chains of residues Glu46I–Glu51I and Asp55H–Pro60H can be superimposed, with Cα atoms deviating <1.3 Å. This similarity extends to the conformation of several side chains and thus to the contacts made with thrombin (Figure 8).
Fig. 8. Close-up stereoview comparing the interactions of the C-terminal tails of haemadin (red) and hirudin (green) with the fibrinogen-recognition exosite of a neighbouring thrombin molecule (blue) (see text for details). Side chains of interacting thrombin/inhibitor residues are labelled explicitly. Notice the close agreement between the phenyl moieties of Phe47I and Phe56H; also the side chain pairs Phe50I–Ile59H and Glu48I–Glu57H occupy similar positions.
Discussion
Serine proteinase substrates bind to the active-site cleft of their cognate proteinase by building an antiparallel β-strand with residues Ser214–Gly216 (chymotrypsinogen numbering) (Bode and Huber, 1992). Although this ‘canonical’ mode of binding has been encountered in a natural thrombin inhibitor, rhodniin (van de Locht et al., 1995), evolutionary pressure appears to have favoured a different type of thrombin–inhibitor association, in which the inhibitor forms instead a parallel β-strand with enzyme residues Ser214–Gly216. This parallel retro-binding alignment was first observed in the leech-derived inhibitor hirudin (Grütter et al., 1990; Rydel et al., 1990), and subsequently also in the double-headed Kunitz-type inhibitor ornithodorin (van de Locht et al., 1996) and in the small molecule nazumamide A (Nienaber and Amparo, 1996). The parallel mode of binding has been exploited by developing potent thrombin inhibitors, termed hirunorms (De Simone et al., 1998). In spite of a definite sequence variability within hirudin variants isolated from different leeches (Steiner et al., 1992; Scacheri et al., 1993) (Figure 7B), the hirudin structure and thus its mode of inhibition appeared to be essentially conserved (Folkers et al., 1989; Haruyama and Wüthrich, 1989; Grütter et al., 1990; Rydel et al., 1990; Vitali et al., 1992; Priestle et al., 1993).
Preliminary biochemical studies conducted with another leech-derived inhibitor, haemadin, pointed to some differences in the detailed thrombin–inhibitor contacts, within an overall conserved inhibition mechanism. The current crystal structure shows that the N-terminal loop of haemadin indeed forms a non-canonical, hirudin-like, parallel β-sheet with thrombin residues Ser214–Gly216. Interestingly, the N-terminal amino acid residues of haemadin seem to be better adapted to the active-site features of thrombin than those of H.medicinalis hirudin (Figure 5). In particular, Arg2I is strongly preferred over a valine due to its favourable interaction with Asp189 at the bottom of the S1 specificity pocket. Experimental data confirm the preference for a basic arginine side chain, as the recombinant hirudin variant Val2H→Arg possesses a 9-fold higher affinity to thrombin compared with the wild-type form (Betz et al., 1992). The following Phe3I seems to be more appropriate than the conserved Tyr3H of hirudin to occupy the hydrophobic S4 pocket. Once again, mutational analyses are consistent with this proposal, as the Tyr3Phe hirudin mutant possesses a 6-fold lower Ki than wild-type hirudin (Lazar et al., 1991). Amino acid residues at positions two and three are therefore optimized in haemadin for non-canonical binding to the active site of thrombin.
Additional interactions made by haemadin’s unique four-residue insertion Gly4I–Met–Gly–Lys7I, in particular by residues Met5I and Lys7I, contribute to strengthen the binding of the inhibitor to thrombin. Unexpectedly, the presence of this insertion and the divergence of the main chains en route to the different exosites exclude any further similarities between both thrombin–inhibitor complexes. Most strikingly, the shorter C-terminal tail of haemadin would not be able to reach exosite I of its cognate thrombin molecule. This essential binding site for thrombin substrates and inhibitors is occupied by hirudin’s C-terminal tail (Grütter et al., 1990; Rydel et al., 1990); the same is also true for the isolated C-terminal peptide, hirugen (Skrzypczak-Jankun et al., 1991). The interaction of the corresponding haemadin tail with exosite II clearly differentiates haemadin from other members of the hirudin family. This constitutes one of the most dramatic structural and functional rearrangements observed in inhibitors from organisms so closely related phylogenetically.
It is noteworthy that the disulfide-rich core of haemadin is more than just a rigid connector between the extended N- and C-terminal peptides, since it is involved in major contacts with thrombin’s 60-, 96-, 174- and 222-loops. The presence of a unique triplet of glutamate residues Glu54I–Glu56I further points to the adaptation of haemadin to the more basic exosite II of thrombin. However, in spite of the overall complementarity of electrostatic potentials, only Glu49I is involved in direct ion pairs with thrombin residues Arg93 and Arg101 (Figure 1B). Both residues form, together with Arg97, a positive surface patch that mediates physiologically important interactions with negatively charged glycosaminoglycans such as heparin (Sheehan and Sadler, 1994; Ye et al., 1994). It is tempting to speculate that optimal adaptation to thrombin’s active-site features along with several insertions–deletions within a rather conserved disulfide-stabilized core (Figure 7A) have transformed hirudin into an inhibitor exhibiting a markedly different mechanism, haemadin.
We have noted previously that ionic interactions between thrombin and haemadin are not a rate-limiting factor for complex formation (Strube et al., 1993). This contrasts with the large effect of ionic strength on the thrombin–hirudin interaction (Stone et al., 1989), as well as with the large effect of salt concentration on the thrombin interaction with platelet glycoprotein Ibα (GP Ibα), which is mediated by thrombin’s exosite II (De Cristofaro et al., 2000). It is conceivable that the electrostatic complementarity of the N- and C-terminal peptides of haemadin with the active site and exosite II regions of the cognate thrombin, respectively (compare Figure 4), largely mitigate solvent effects on complex formation. Thus, haemadin binding to thrombin appears to occur via nearly simultaneous docking of both pairs of complementary regions, in contrast to the two-step mechanism found for the thrombin–hirudin interaction. Several additional factors might contribute to the discrepancy in salt concentration dependency. First, since exosite II is a more electropositive region than the fibrinogen recognition exosite (Bode et al., 1992), coupled with the fact that the C-terminal tail of haemadin is more acidic than the corresponding hirudin peptide, putative solvent shielding effects might only become relevant at higher salt concentrations than those used in our previous study (0.3 M NaCl; Strube et al., 1993). Secondly, almost all hirudin variants (in particular the most thoroughly studied variant form from H.medicinalis) possess a sulfated tyrosine residue in the third position from the C-terminus; loss of this sulfate group results in a 2-fold reduction in potency. Curiously, the ‘hirudin-like’ acidic peptide of GP Ibα (residues Asp272–Glu282) also contains three sulfated tyrosine residues, which are essential for thrombin binding. In contrast, haemadin lacks any sulfated tyrosine residues. Finally, we cannot exclude the possibility that minor effects of the salt concentration had been obscured by allosteric effects mediated by sodium binding to thrombin (Di Cera, 1998).
Even though the shorter C-terminal tail of haemadin would not allow simultaneous binding to the active site and anion-binding exosite I, the involvement of this acidic peptide in ‘hirudin-like’ contacts with a neighbouring thrombin molecule (Figures 1A and 8) prompted us to analyse the character of the thrombin–haemadin interactions in solution. We have recently shown that the thrombin inhibitor triabin binds solely to the fibrinogen-recognition exosite of the proteinase, leaving its active site unoccupied (Fuentes-Prior et al., 1997). Therefore, haemadin and triabin would not compete with each other for thrombin binding. Band shift studies showed that this is indeed the case, and that a ternary haemadin–thrombin–triabin complex could be isolated.
A similar line of reasoning applies to the active thrombomodulin fragment comprising the three C-terminal epidermal growth factor (EGF)-like repeats (TME456). Exosite I is blocked by domains TME5 and TME6, but no contacts are made with either the active-site region or with exosite II (Fuentes-Prior et al., 2000). Again, observation of a ternary haemadin–thrombin– TME456 complex is in accord with the occupation of exosite II by haemadin. More relevant for future therapeutic applications, the Ki determined for inhibition of the thrombin–TME456 complex by recombinant haemadin is of the same order of magnitude as that determined for free human α-thrombin. This observation is in line with the absence of major allosteric rearrangements in the active site of thrombomodulin-bound thrombin (Fuentes-Prior et al., 2000). The slight decrease in inhibitory potency might result from minor clashes between the C-terminal residues of the inhibitor and thrombomodulin residues located in close proximity to the ‘upper’ part of exosite I.
The contribution of positively charged residues of thrombin exosite II to haemadin binding seems to correspond closely to the interactions observed in the thrombin–haemadin crystal structure. In particular, the replacement of three basic amino acids in exosite II (Arg93, Lys240 and Lys233) with glutamic acid led to 10-, 5- and 3-fold reductions in the inhibition constants, whereas exosite I mutants show virtually no change in inhibition constants.
Our preliminary results also indicate that haemadin does not bind to the important intermediate activation form, meizothrombin. This observation is explained by the occupation of exosite II by the second Kringle domain of meizothrombin (Martin et al., 1997), which covers a surface on the catalytic domain ∼6-fold larger than the interface between haemadin’s tail and thrombin. Superposition of the meizothrombin(desF1) structure onto that of α-thrombin–haemadin reveals that residues of the inhibitor C-terminal to Pro43I would not be able to contact exosite II, besides severe clashes involving exposed side chains of the inhibitor (e.g. Tyr28I) and Kringle 2 of meizothrombin. In contrast, exosite I remains free in meizothrombin, and hirudin binds to this intermediate form as strongly as it does to α-thrombin (Fischer et al., 1998).
The search for heparin substitutes, in light of the drawbacks of heparinotherapy, is an area of intensive research. A recent study reports the synthesis of novel heparin mimetics (Petitou et al., 1999). Peptides and peptide mimetics that target protease regions distant from the active site (exosites) may also play an important role as antithrombotic drugs. This opens promising avenues, which may play an important role in antithrombotic drug development, as has been shown for coagulation factor VIIa recently (Dennis et al., 2000).
The combined crystallographic and biochemical evidence establishes conclusively that haemadin binds to the heparin-binding exosite but does not interact with the fibrinogen-recognition exosite. This feature allows haemadin to inhibit not only free α-thrombin, but also the thrombomodulin-bound proteinase. From the viewpoint of the development of novel antithrombotics, haemadin offers therefore a unique advantage over current concepts based on hirudin and hirudin-like molecules. Membrane-bound active intermediates formed during the conversion of prothrombin to α-thrombin seem to play major physiological roles (Doyle and Mann, 1990; Côté et al., 1997). More relevantly, meizothrombin possesses a 6-fold higher protein C cofactor activity than α-thrombin, pointing to a probable anticoagulant role in vivo (Côté et al., 1997). Haemadin binding only to forms possessing a freely accessible exosite II would target circulating α-thrombin selectively, without interfering with the anticoagulant and possibly also antifibrinolytic activities of meizothrombin. Finally, the ability of the C-terminal peptide of haemadin to bind a second thrombin molecule could become relevant at high thrombin concentrations, as might be found in the clot.
Materials and methods
Protein purification
Human α-thrombin was prepared from frozen serum following standard protocols. Recombinant haemadin was expressed as a periplasmic fusion protein with maltose-binding protein and was cleaved with factor Xa (Roche Diagnostics) and purified as previously described (Strube et al., 1993). Lyophilized haemadin was added to human α-thrombin (4.1 mg/ml, 10 mM MES pH 6.0, 500 mM NaCl) in slight excess of a 1:1 molar ratio. After dialysis against 20 mM Tris–HCl pH 8.0, the complex was purified on a DEAE-5PW column (TosoHaas, Germany) using a linear gradient of 0–250 mM NaCl, in 20 mM Tris–HCl pH 8.0. Fractions corresponding to the equimolar complex were exhaustively dialysed against 10 mM Tris–HCl pH 8.0, 20 mM NaCl, 0.02% (w/v) NaN3, and concentrated in Centricon 30 tubes (Amicon) to ∼5 mg/ml.
Crystallization and data collection
Crystals of approximate dimensions 0.3 × 0.15 × 0.15 mm3 were grown by vapour diffusion from 3 µl droplets consisting of 2 µl of protein solution and 1 µl of precipitant [100 mM sodium citrate pH 5.6, 14% (w/v) PEG 4000, 12.5% (v/v) isopropanol], equilibrated against 500 µl of precipitant solution at 22°C. Crystals were directly mounted in thin-walled glass capillaries, and diffraction data were collected at 16°C on a 300 mm MAR image plate system (MAR Research, Hamburg) using monochromatized CuKα radiation produced by a Rigaku rotating anode generator. Crystals diffracted initially to below 2.8 Å, but showed rapid decay due to radiation damage, which could not be attenuated by cryo-cooling, as cryo-cooling irreversibly damaged the crystals under all conditions tested so far. Diffraction data from two crystals were evaluated using the MOSFLM package (Leslie, 1991), and merged and scaled using programs of the CCP4 suite (CCP4, 1994). The crystals belong to the monoclinic space group P2, and contain three complexes per asymmetric unit. Self-rotational searches with GLRF (Tong and Rossmann, 1997) failed to reveal rotational solutions, but a Patterson search indicated two pseudo-origins, suggesting a linear arrangement of three monomers within the asymmetric unit.
Structure determination
The structure was solved using Patterson search techniques. Rotational and translational searches to determine the orientation and position of the thrombin components were performed with the program AMoRe (Navaza, 1994) using diffraction data from 10.0 to 3.5 Å and the coordinates of human α-thrombin from the thrombin–hirudin complex (Rydel et al., 1990), from which residues Thr1H–Asp1A and Asp14L–Arg15 of the A chain and most of the residues forming the autolysis loop (Glu146–Glu149E) were excluded. As expected from the Matthews parameter for a trimer, three rotation solutions were found. After translational search and rigid body refinement, these solutions resulted in a correlation value of 0.67 and an R-factor of 0.34 (next highest peak: c = 0.46, R = 0.44). A 3.1 Å (2Fobs – Fcalc) Fourier map calculated after appropriate positioning of the thrombin molecules in the crystal cell allowed us to build some of the omitted parts of thrombin and the haemadin segments in contact with thrombin, using the program MAIN (Turk, 1992). The complexes were crystallographically refined with X-PLOR (Brünger, 1991) using the target parameters of Engh and Huber (1991). This process was repeated cyclically a few times until most of the inhibitor sequence could be fitted into the density. Seventy-one water molecules were added at stereochemically reasonable positions where the (2Fobs – Fcalc) density and (Fobs – Fcalc) difference density maps were above 1σ and 2σ levels, respectively. The R-factor of the final model is 0.208 (with a free R-factor of 0.255). The data statistics are given in Table II. The atomic coordinates of the complex have been deposited in the Protein Data Bank with the entry code 1E0F.
Table II. Summary of crystallographic data.
| Space group | P2 |
| Cell constants (Å) | |
| a = 121.67 | |
| b = 50.57 | |
| c = 129.74 | |
| β (°) | 114.76 |
| Limiting resolution (Å) | 3.1 |
| Rmergea (%) | 10.9 |
| Completeness (∞–3.1 Å) (%) | 81.2 |
| Non-hydrogen protein atoms | 8381 |
| Solvent molecules | 68 |
| Reflections collected | 126 754 |
| Unique reflections | 23 938 |
| Reflections used for refinement | 22 278 |
| Resolution range (Å) | 10.0–3.1 |
| R valueb (%) | 20.8 |
| Rfreec (%) | 25.5 |
| Standard deviations | |
| bond lengths (Å) | 0.006 |
| main chain bond angles (°) | 1.52 |
| side chain bond angles (°) | 23.55 |
aRmerge = [ΣhΣi|I(h,i) – <I(h)>|/ΣhΣiI(h,i)] × 100, where I(h,i) is the intensity value of the ith measurement of h and <I(h)> is the corresponding mean value of h for all i measurements of h. The summation is over all measurements.
bR value = {Σ(|Fo| – |Fc|)/Σ|Fo|} × 100, where |Fo| is the observed and |Fc| the calculated structure factor amplitude of reflection hkl.
cRfree was calculated randomly omitting 2% of the observed reflections from refinement.
Binding studies in solution
Triabin was a gift of Dr Noeske-Jungblut (Research Laboratories of Schering AG, Germany). Thrombomodulin fragment TME456 (residues Val345–Cys462 of human TM, with residues Arg456 and His457 mutated to Gly and Gln and the N-terminus extended by a Phe-Pro dipeptide) was kindly provided by Dr David Light (Berlex Biosciences, Richmond, CA). Recombinant meizothrombin was a kind gift of Dr Alireza Rezaie (University of St Louis, MO). Complex formation in solution was analysed using non-denaturing PAGE. After pre-incubation for 2 min in 20 mM Tris–HCl pH 6.9, the protein solutions were subjected to electrophoresis in 10% polyacrylamide gels containing 365 mM Tris–HCl pH 8.8. The migration buffer consisted of 100 mM glycine and 25 mM Tris–HCl pH 8.8.
Chromogenic amidolytic assays of thrombin activity were performed using 150 µM Tos-Gly-Pro-Arg–pNA as a substrate and a standard thrombin concentration of 6 nM. Reactions were performed at 25°C in 100 mM Tris–HCl buffer pH 7.8, containing 100 mM NaCl and 0.1% PEG 6000. Reaction progress was monitored at 405 nm on a Uvikon 943 double beam spectrophotometer. Slow and tight binding assays were performed at 25°C as described previously (Strube et al., 1993). Thrombin mutants were prepared as described previously (Sheehan and Sadler, 1994). Slow binding assays were conducted using 25 µM Tos-Gly-Pro-Arg-AMC as a substrate in 50 mM Tris–HCl pH 8.3, 50 mM NaCl and 0.1% PEG 6000 with varying inhibitor concentrations (20–70 × [E0]); the reaction was started by the addition of 10 pM human α-thrombin–TME456 complex (final concentration). Tight binding assays were performed with a final concentration of 500 pM human α-thrombin (or mutant), being pre-incubated for 10 min with inhibitor (0.4–2.4 × [E0]), in 50 mM Tris–HCl pH 8.3, 50 mM NaCl and 0.1% PEG 6000, with the steady-state velocity being measured at varying substrate concentrations (100–500 μM S-2238). The kinetic constants koff, kon and Ki were determined according to the theory of tight-binding and slow-binding inhibition, by non-linear regression analysis of the data as described previously (Strube et al., 1993).
References
- Arocas V., Zingali,R.B., Guillin,M.-C., Bon,C. and Jandrot-Perrus,M. (1996) Bothrojaracin: a potent two-site-directed thrombin inhibitor. Biochemistry, 35, 9083–9089. [DOI] [PubMed] [Google Scholar]
- Barton G.J. (1993) ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng., 6, 37–40. [DOI] [PubMed] [Google Scholar]
- Betz A., Hofsteenge,J. and Stone,S.R. (1992) Interaction of the N-terminal region of hirudin with the active-site cleft of thrombin. Biochemistry, 31, 4557–4562. [DOI] [PubMed] [Google Scholar]
- Bode C., Nordt,T.K. and Runge,M.S. (1994) Thrombolytic therapy in acute myocardial infarction—selected recent developments. Ann. Hematol., 69, S35–40. [DOI] [PubMed] [Google Scholar]
- Bode W. and Huber,R. (1992) Natural protein proteinase inhibitors and their interaction with proteinases. Eur. J. Biochem., 204, 433–451. [DOI] [PubMed] [Google Scholar]
- Bode W., Mayr,I., Baumann,U., Huber,R., Stone,S.R. and Hofsteenge,J. (1989) The refined 1.9 Å crystal structure of human α-thrombin: interaction with d-Phe-Pro-Arg chloromethylketone and significance of the Tyr-Pro-Pro-Trp insertion segment. EMBO J., 8, 3467–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode W., Turk,D. and Karshikov,A. (1992) The refined 1.9 Å X-ray crystal structure of d-Phe-Pro-Arg chloromethylketone-inhibited human α-thrombin: structure analysis, overall structure, electrostatic properties, detailed active-site geometry, and structure–function relationships. Protein Sci., 1, 426–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. (1991) Crystallographic phasing and refinement of macromolecules. Curr. Opin. Struct. Biol., 1, 1016–1022. [Google Scholar]
- Castro H.C., Dutra,D.L., Oliveira-Carvalho,A.L. and Zingali,R.B. (1998) Bothroalternin, a thrombin inhibitor from the venom of Bothrops alternatus. Toxicon, 36, 1903–1912. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chang J.Y. (1986) The structures and proteolytic specificities of autolysed human thrombin. Biochem. J., 240, 797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté H.C., Bajzar,L., Stevens,W.K., Samis,J.A., Morser,J., MacGillivray,R.T. and Nesheim,M.E. (1997) Functional characterization of recombinant human meizothrombin and meizothrombin(desF1). Thrombomodulin-dependent activation of protein C and thrombin-activatable fibrinolysis inhibitor (TAFI), platelet aggregation, antithrombin-III inhibition. J. Biol. Chem., 272, 6194–6200. [DOI] [PubMed] [Google Scholar]
- Coughlin S.R. (1999) How the protease thrombin talks to cells. Proc. Natl Acad. Sci. USA, 96, 11023–11027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie E.W., Fujikawa,K. and Kisiel,W. (1991) The coagulation cascade: initiation, maintenance and regulation. Biochemistry, 30, 10363–10370. [DOI] [PubMed] [Google Scholar]
- De Cristofaro R., De Candia,E., Rutella,S. and Weitz,J.I. (2000) The Asp272–Glu282 region of platelet glycoprotein Ibα interacts with the heparin-binding site of α-thrombin and protects the enzyme from the heparin-catalyzed inhibition by antithrombin III. J. Biol. Chem., 275, 3887–3895. [DOI] [PubMed] [Google Scholar]
- Dennis M.S., Eigenbrot,C., Skelton,N.J., Ultsch,M.H., Santell,L., Dwyer,M.A., O’Connell,M.P. and Lazarus,R.A. (2000) Peptide exosite inhibitors of factor VIIa as anticoagulants. Nature, 404, 465–470. [DOI] [PubMed] [Google Scholar]
- De Simone G., Lombardi,A., Galdiero,S., Nastri,F., Della Morte,R., Staiano,N., Pedone,C., Bolognesi,M. and Pavone,V. (1998) Hirunorms are true hirudin mimetics. The crystal structure of human α-thrombin–hirunorm V complex. Protein Sci., 7, 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cera E. (1998) Anticoagulant thrombins. Trends Cardiovasc. Med., 8, 340–350. [DOI] [PubMed] [Google Scholar]
- Doyle M.F. and Mann,K.G. (1990) Multiple active forms of thrombin. IV. Relative activities of meizothrombins. J. Biol. Chem., 265, 10693–10701. [PubMed] [Google Scholar]
- Engh R. and Huber,R. (1991) Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. A, 47, 392–400. [Google Scholar]
- Esmon C.T. (1995) Thrombomodulin as a model of molecular mechanisms that modulate protease specificity and function at the vessel surface. FASEB J., 9, 946–955. [DOI] [PubMed] [Google Scholar]
- Evans S.V. (1993) SETOR: hardware lighted three-dimensional solid model representations of macromolecules. J. Mol. Graph., 11, 134–138. [DOI] [PubMed] [Google Scholar]
- Fenton J.W., Ofosu,F.A., Moon,D.G. and Maraganore,J.M. (1991) Thrombin structure and function: why thrombin is the primary target for antithrombotics. Blood Coagul. Fibrinolysis, 2, 69–75. [PubMed] [Google Scholar]
- Fischer B.E., Schlokat,U., Himmelspach,M. and Dorner,F. (1998) Binding of hirudin to meizothrombin. Protein Eng., 11, 715–721. [DOI] [PubMed] [Google Scholar]
- Folkers P.J., Clore,G.M., Driscoll,P.C., Dodt,J., Kohler,S. and Gronenborn,A.M. (1989) Solution structure of recombinant hirudin and the Lys-47→Glu mutant: a nuclear magnetic resonance and hybrid distance geometry–dynamical simulated annealing study. Biochemistry, 28, 2601–2617. [DOI] [PubMed] [Google Scholar]
- Fuentes-Prior P., Noeske-Jungblut,C., Donner,P., Schleuning,W.D., Huber,R. and Bode,W. (1997) Structure of the thrombin complex with triabin, a lipocalin-like exosite-binding inhibitor derived from a triatomine bug. Proc. Natl Acad. Sci. USA, 94, 11845–11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Prior P., Iwanaga,Y., Huber,R., Pagila,R., Rumennik,G., Seto,M., Morser,J., Light,D.R. and Bode,W. (2000) Structural basis for the anticoagulant activity of the thrombin–thrombomodulin complex. Nature, 404, 518–525. [DOI] [PubMed] [Google Scholar]
- Gailani D. and Broze,G.J.,Jr (1991) Factor XI activation in a revised model of blood coagulation. Science, 253, 909–912. [DOI] [PubMed] [Google Scholar]
- Grütter M.G., Priestle,J.P., Rahuel,J., Grossenbacher,H., Bode,W., Hofsteenge,J. and Stone,S.R. (1990) Crystal structure of the thrombin–hirudin complex: a novel mode of serine protease inhibition. EMBO J., 9, 2361–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulba D.C., Bode,C., Runge,M.S. and Huber,K. (1998) Thrombolytic agents: an updated overview. Fibrin. Proteol., 12, 39–58. [Google Scholar]
- Haruyama H. and Wüthrich,K. (1989) Conformation of recombinant desulfatohirudin in aqueous solution determined by nuclear magnetic resonance. Biochemistry, 28, 4301–4312. [DOI] [PubMed] [Google Scholar]
- Kane W.H. and Davie,E.W. (1988) Blood coagulation factors V and VIII: structural and functional similarities and their relationship to hemorrhagic and thrombotic disorders. Blood, 71, 539–555. [PubMed] [Google Scholar]
- Lazar J.B., Winant,R.C. and Johnson,P.H. (1991) Hirudin: amino terminal residues play a major role in the interaction with thrombin. J. Biol. Chem., 266, 685–688. [PubMed] [Google Scholar]
- Leslie A. (1991) Macromolecular data processing. In Moras,D., Podjarny,A.D. and Thierry,J.C. (eds), Crystal. Computing V. Oxford University Press, Oxford, UK, pp. 27–38. [Google Scholar]
- Lombardi A., De Simone,G., Galdiero,S., Staiano,N., Nastri,F. and Pavone,V. (1999) From natural to synthetic multisite thrombin inhibitors. Biopolymers, 51, 19–39. [DOI] [PubMed] [Google Scholar]
- Martin P.D., Malkowski,M.G., Box,J., Esmon,C.T. and Edwards,B.F. (1997) New insights into the regulation of the blood clotting cascade derived from the X-ray crystal structure of bovine meizothrombin des F1 in complex with PPACK. Structure, 5, 1681–1693. [DOI] [PubMed] [Google Scholar]
- Navaza J. (1994) An automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Nesheim M., Wang,W., Boffa,M., Nagashima,M., Morser,J. and Bajzar,L. (1997) Thrombin, thrombomodulin and TAFI in the molecular link between coagulation and fibrinolysis. Thromb. Haemost., 78, 386–391. [PubMed] [Google Scholar]
- Nicholls A., Bharadwaj,R. and Honig,B. (1993) GRASP—graphical representation and analysis of surface properties. Biophys. J., 64, A166. [Google Scholar]
- Nienaber V.L. and Amparo,E.C. (1996) A noncleavable retro-binding peptide that spans the substrate binding cleft of serine proteases. Atomic structure of nazumamide A: human thrombin. J. Am. Chem. Soc., 118, 6807–6810. [Google Scholar]
- Noeske-Jungblut C., Haendler,B., Donner,P., Alagon,A., Possani,L. and Schleuning,W.D. (1995) Triabin, a highly potent exosite inhibitor of thrombin. J. Biol. Chem., 270, 28629–28634. [DOI] [PubMed] [Google Scholar]
- Petitou M., Hérault,J.-P., Bernat,A., Driguez,P.-A., Duchaussoy,P., Lormeau,J.-C. and Herbert,J.-M. (1999) Synthesis of thrombin-inhibiting heparin mimetics without side effects. Nature, 398, 417–422. [DOI] [PubMed] [Google Scholar]
- Priestle J.P., Rahuel,J., Rink,H., Tones,M. and Grütter,M.G. (1993) Changes in interactions in complexes of hirudin derivatives and human α-thrombin due to different crystal forms. Protein Sci., 2, 1630–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydel T.J., Ravichandran,K.G., Tulinsky,A., Bode,W., Huber,R., Roitsch,C. and Fenton,J.W. (1990) The structure of a complex of recombinant hirudin and human α-thrombin. Science, 249, 277–280. [DOI] [PubMed] [Google Scholar]
- Scacheri E., Nitti,G., Valsasina,B., Orsini,G., Visco,C., Ferrera,M., Sawyer,R.T. and Sarmientos,P. (1993) Novel hirudin variants from the leech Hirudinaria manillensis. Amino acid sequence, cDNA cloning and genomic organization. Eur. J. Biochem., 214, 295–304. [DOI] [PubMed] [Google Scholar]
- Sheehan J.P. and Sadler,J.E. (1994) Molecular mapping of the heparin-binding exosite of thrombin. Proc. Natl Acad. Sci. USA, 91, 5518–5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzypczak-Jankun E., Carperos,V.E., Ravichandran,K.G., Tulinsky,A., Westbrook,M. and Maraganore,J.M. (1991) Structure of the hirugen and hirulog 1 complexes of α-thrombin. J. Mol. Biol., 221, 1379–1393. [PubMed] [Google Scholar]
- Steiner V., Knecht,R., Bornsen,K.O., Gassmann,E., Stone,S.R., Raschdorf,F., Schlaeppi,J.M. and Maschler,R. (1992) Primary structure and function of novel O-glycosylated hirudins from the leech Hirudinaria manillensis. Biochemistry, 31, 2294–2298. [DOI] [PubMed] [Google Scholar]
- Stone S.R. and Tapparelli,C. (1995) Thrombin inhibitors as antithrombotic agents: the importance of rapid inhibition. J. Enzyme Inhib., 9, 3–15. [DOI] [PubMed] [Google Scholar]
- Stone S.R., Dennis,S. and Hofsteenge,J. (1989) Quantitative evaluation of the contribution of ionic interactions to the formation of the thrombin–hirudin complex. Biochemistry, 28, 6857–6863. [DOI] [PubMed] [Google Scholar]
- Strube K.H., Kroger,B., Bialojan,S., Otte,M. and Dodt,J. (1993) Isolation, sequence analysis, and cloning of haemadin. An anticoagulant peptide from the Indian leech. J. Biol. Chem., 268, 8590–8595. [PubMed] [Google Scholar]
- Stubbs M.T. and Bode,W. (1993) A player of many parts: the spotlight falls on thrombin’s structure. Thromb. Res., 69, 1–58. [DOI] [PubMed] [Google Scholar]
- Tong L. and Rossmann,M.G. (1997) Rotation function calculations with GLRF program. Methods Enzymol., 276, 594–611. [PubMed] [Google Scholar]
- Turk D. (1992) Weiterentwicklung eines Programms für Molekülgraphik und Elektronendichte-Manipulation und seine Anwendung auf verschiedene Protein-Strukturaufklärungen. PhD thesis, Technische Universität, München, Germany.
- van de Locht A., Lamba,D., Bauer,M., Huber,R., Friedrich,T., Kröger,B., Höffken,W. and Bode,W. (1995) Two heads are better than one: crystal structure of the insect derived double domain Kazal inhibitor rhodniin in complex with thrombin. EMBO J., 14, 5149–5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Locht A., Stubbs,M.T., Bode,W., Friedrich,T., Bollschweiler,C., Höffken,W. and Huber,R. (1996) The ornithodorin–thrombin crystal structure, a key to the TAP enigma? EMBO J., 15, 6011–6017. [PMC free article] [PubMed] [Google Scholar]
- Vitali J., Martin,P.D., Malkowski,M.G., Robertson,W.D., Lazar,J.B., Winant,R.C., Johnson,P.H. and Edwards,B.F. (1992) The structure of a complex of bovine α-thrombin and recombinant hirudin at 2.8-Å resolution. J. Biol. Chem., 267, 17670–17678. [PubMed] [Google Scholar]
- Vu T.K., Hung,D.T., Wheaton,V.I. and Coughlin,S.R. (1991) Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell, 64, 1057–1068. [DOI] [PubMed] [Google Scholar]
- Wallis R.B. (1988) Hirudins and the role of thrombin: lessons from leeches. Trends Pharmacol. Sci., 9, 425–427. [DOI] [PubMed] [Google Scholar]
- Walsmann P. and Markwardt,F. (1985) On the isolation of the thrombin inhibitor hirudin. Thromb. Res., 40, 563–569. [DOI] [PubMed] [Google Scholar]
- Wu Q., Sheehan,J.P., Tsiang,M., Lentz,S.R., Birktoft,J.J. and Sadler,J.E. (1991) Single amino acid substitutions dissociate fibrinogen-clotting and thrombomodulin-binding activities of human thrombin. Proc. Natl Acad. Sci. USA, 88, 6775–6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Rezaie,A.R. and Esmon,C.T. (1994) Glycosaminoglycan contributions to both protein C activation and thrombin inhibition involve a common arginine-rich site in thrombin that includes residues arginine 93, 97, and 101. J. Biol. Chem., 269, 17965–17970. [PubMed] [Google Scholar]