Abstract
Genetic polymorphisms in ERCC1 are thought to contribute to altered sensitivity to platinum-based chemotherapy. Although ERCC1 N118N (500 C>T, rs11615) is the most studied polymorphism, the impact of this polymorphism on platinum-based chemotherapy remains unclear. This is the first study in which the functional impact of ERCC1 N118N on gene expression and platinum sensitivity was explored. The aim of this study is to investigate if the reduced codon usage frequency of AAT, which contains the variant allele of the silent mutation, has functional impact on ERCC1 in a well-controlled biological system. Specifically, the ERCC1 cDNA clone with either the C or T allele was introduced into an ERCC1 deficient cell line, UV20, and assayed for the effect of the two alleles on ERCC1 transcription, translation and platinum sensitivity. Both ERCC1 mRNA and protein expression levels increased upon cisplatin treatment, peaking at 4 hours post-treatment, however there were no differences between the two alleles (p>0.05). Cells complemented with ERCC1 showed significantly higher survival proportion than the parental cell line following platinum exposure (P<0.0001), although no differences were observed between the cells transfected with the wild type or the polymorphic allele. These data suggest that N118N itself is not related to the phenotypic differences in ERCC1 expression or function, but rather this polymorphism may be linked to other causative variants or haplotypes.
Keywords: ERCC1, silent mutation, codon bias, DNA repair, platinum drugs
INTRODUCTION
Platinum-based chemotherapies play an important role in the treatment of several solid malignancies but the application of these treatment methods has been limited by clinical resistance. DNA excision repair plays a significant part in platinum-based chemotherapy by removing DNA lesions caused by platinum-containing drugs. The nucleotide excision repair (NER) pathway is the mammalian DNA repair mechanism that removes UV-induced DNA lesions, as well as bulky DNA adducts induced by DNA damaging chemotherapeutic agents. The excision repair cross-complementing protein 1 (ERCC1) is the rate-limiting protein in the NER pathway; it forms a heterodimeric protein complex with Xeroderma pigmentosum group F-complementing protein (XPF) to carry out the 5’ incision in the presence of a DNA lesion. Reduction of ERCC1 function may predispose people to cancer due to the inefficiency of DNA damage removal. However, improved response to DNA-damaging chemotherapy in those individuals is anticipated.
The protein expression level of ERCC1 has been correlated to the outcome of platinum-based chemotherapies but the genetic contribution to the variation in ERCC1 expression has yet to be clarified. To date, more than 190 single nucleotide polymorphisms (SNPs) have been reported for ERCC1 (http://preview.ncbi.nlm.nih.gov/snp/?term=ercc1). Of these, a silent polymorphism in exon 4 (500 C>T in mRNA, N118N, rs11615) has been extensively studied and associated with an altered outcome in platinum-based chemotherapy in multiple malignancies, as summarized in Table 1. However, the results vary in different types of malignancies. For example, in non-small lung cancer, several studies [1–4] suggest that the C allele signifies a better response to platinum-based chemotherapy, while other studies [5,6] report no association. Alternatively, in metastatic colorectal cancer, both the C and T alleles were found to be associated with either an improved or impaired outcome.
Table 1.
Previous pharmacogenetics studies on ERCC1 N118N polymorphism in mutiple malignancies.
| Disease | Therapy | # of pts | Association | HR (95% CI) | P value | Ref. |
|---|---|---|---|---|---|---|
| mCRC | FOLFOX | 118 | T allele: worse progression-free survival | 2.62 (1.14–6.0) | 0.02 | [27] |
| FOLFOX | 168 | T allele: increased ERCC1 expression, lower response, and shorter OS and PFS | N/A | ≤ 0.01 | [28] | |
| Fluorouracil/oxaliplatin | 49 | C allele correlated with a shorter PFS | 1.96 (0.99–3.92) | 0.050 | [29] | |
| Platinum-based chemotherapy | 106 | CC genotype had longer median survival | N/A | N/A | [30] | |
| Oxaliplatin/5-fluorouracil | 91 | TT genotype had higher response rate | N/A | 0.018 | [31] | |
| Oxaliplatin/5-fluorouracil | 106 | CC genotype showed the most favorable survival | 2.29 (1.19, 4.41) for CT and 1.86 (0.91, 3.83) for TT | 0.021 | [32] | |
| NSCLC | Paclitaxel plus carboplatin | 153 | No association | 1.2 (0.74–1.96) | 0.45 | [5] |
| Platinum-based chemotherapy | 119 | C allele associated with better response | 0.10 (0.013–0.828) | 0.033 | [1] | |
| Cisplatin combination | 245 | CC had longer OS in patients having over 50 packs per year | N/A | 0.03 | [2] | |
| Docetaxel-cisplatin | 62 | CC genotype had longer OS | 0.01 | [3] | ||
| Platinum-based chemotherapy | 128 | No association | N/A | 0.41 | [6] | |
| Cisplatin combination chemotherapy | 109 | CC had better survival | 0.0058 | [4] | ||
| Platinum-based chemotherapy | 115 | T allele associated with elevated response | 0.008 | [33] | ||
| Pancreatic cancer | Cisplatin-based chemotherapy | 67 | T allele: longer PFS and OS | 0.006 for PFS and 0.03 for OS | [34] | |
| EOC | Platinum-based chemotherapy | 159 | TT signalized a better response | 0.026 | [35] | |
| Gastric cancer | Oxaliplatin-based adjuvant chemotherapy | 102 | No association | N/A | > 0.05 | [36] |
| Esophageal cancer | Cisplatin | 262 with or 108 without cisplatin | No association | N/A | 0.1 for cisplatin-treated and 0.49 for nocisplatin | [37] |
| NPC | Gemcitabine and oxaliplatin | 29 | No association | N/A | 0.76 | [38] |
mCRC: metastatic colorectal cancer, NPC: Nasopharyngeal Carcinoma, EOC: Epithelial Ovarian Cancer, NSCLC: Non-Small Cell Lung Cancer, PFS: Progression free Survival, OS: Overall Survival, HR: Hazard Ratio.
Silent mutations do not alter amino acid sequence, and therefore are not expected to change the function of the protein. However, it has been proposed that silent mutations may have functional consequences by changing translation kinetics and protein folding [7]. For example, a study showed that a naturally occurring rare silent mutation in the MDR1 gene affected the timing of co-translational folding and insertion of P-gp into the membrane, thereby altering the structural and functional properties of the gene product [8]. In addition, synonymous codons may have different codon usage, and therefore result in altered levels of gene expression [9].
Due to the lack of knowledge of the functional consequences of the ERCC1 N118N polymorphism, it is difficult to interpret the findings from genetic association studies. Therefore, to clarify the role that this silent mutation in ERCC1 plays in terms of gene expression and protein function, we introduced a ERCC1 cDNA clone with either the C or T at the specific position into an ERCC1 deficient cell line UV20. We then assayed for ERCC1 mRNA and protein expression levels upon cisplatin treatment, and cellular sensitivity to platinum-containing drugs.
MATERIALS AND METHODS
Cell culture
The ERCC1 deficient cell line UV20 was obtained from the American Type Culture Collection (Manassas, VA). UV20 is a UV sensitive mutant of CHO [10,11]. The cells were maintained in a humidified incubator at 37°C equilibrated with 5% CO2 and 95% air in Alpha minimum essential medium containing 10% fetal bovine serum and 1% Penicillin-Streptomycin. To determine the minimum concentration of Geneticin®, 5 ×104 cells were plated in 6-well dishes, and a range of Geneticin concentrations were tested. 750 ug/ml of Geneticin was used to select the stably transfected cell line. The plasmids containing the reference or variant allele of ERCC1 N118N, as well as the vector control, were transfected into UV-20 cells using the Lipofectamine™ LTX Reagent according to the manufacurer’s instructions (Invitrogen, Carlsbad, CA). The stably transfected cell lines were designated as UV20ERCC1_C and UV20ERCC1_T, respectively.
Plasmid construct
The ERCC1 Ultimate™ ORF clone (IOH5754) was purchased from Invitrogen and was introduced into the destination vector pcDNA™/V5-DEST by LR recombination reaction. The recombination reaction was performed using the Gateway® LR Clonase™ II Enzyme Mix according to the manufacture’s instructions (Invitrogen, Carlsbad, CA). The mutant was generated with the QuikChange® Lightning Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) using the ERCC1 ORF clone in pcDNA™/V5-DEST as the template and the following primers: Primer1: 5'-ACT GAA GTT CGT GCG CAA TGT GCC CTG GG-3' and Primer2: 5'-CCC AGG GCA CAT TGC GCA CGA ACT TCA GT-3'. The introduced allele is in italics. The proper construction of both the original ORF clone and the synthesized mutant strand were verified by direct sequencing using the Big Dye Terminator Cycle Sequencing Ready Reaction kit V3.1 (Applied Biosystems, Foster City, CA) and an ABI Prism 3130 Genetic Analyzer using the manufacturers instructions.
Real-time RT PCR
Expression of ERCC1 mRNA was measured by quantitative reverse transcription-PCR (RT-PCR) using the Stratagene Mx3005P™ Real-Time PCR System (La Jolla, CA). Briefly, cell lysates were collected at the appointed times following treatment of cisplatin (3.75×10−2 mg/ml) using the AllPrep RNA/Protein Kit (Qiagen, Valencia, CA). The total RNA was reverse-transcribed using the RT2 First Strand Kits (SABiosciences, Frederick, MD). Each sample was analyzed in duplicate and the results are an average of four analyses. Analysis of mRNA expression was conducted using the RT2 qPCR Primer Assay (PPH01539A, SABiosciences, Frederick, MD) for ERCC1 and normalized to the expression of CHO ACTB [12].
Western blotting
Expression of ERCC1 protein was assessed by Western blot analysis. Briefly, cells were washed in cold PBS and lysed in 100 µl of RIPA buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) containing 10 µl/ml of Halt™ Protease Inhibitor Cocktail (Pierce, Rockford, IL) at 4°C. Samples were then sonicated on ice and collected by centrifuging at 14,000 × g for 15 minutes. Protein concentration was determined using the BCA Protein Assay Kit (Pierce, Rockford, IL) and 20 µg of protein was subjected to electrophoresis on a NuPage® 4–12% Bis-Tris Gel (Invitrogen, Carlsbad, CA). Proteins were transferred electrophoretically to a PVDF membrane using the iBlot® Dry Blotting System (Invitrogen, Carlsbad, CA). The membrane was blocked in Odyssey blocking buffer (LI-COR, Lincoln, NE) followed by an overnight incubation with a 1: 200 dilution of ERCC1 primary antibody (FL-297, Santa Cruz Biotechnology, Santa Cruz, CA). For detection with the Odyssey imaging system, a 1:5,000 dilution of the infrared fluorophore conjugated antibody, IRDye™ 800-conjugated goat anti-rabbit IgG was used. Proteins were visualized using the Odyssey Infrared Imaging System and Odyssey software (LI-COR, Lincoln, NE). Quantification of western blots was performed using ImageJ (http://rsbweb.nih.gov/ij/) according to the developer’s instructions.
Cell cytotoxicity assays
The night before treatment, cells were plated in 96-well dishes at a density of 5000 cells/well. A range of cisplatin (2.56×10−6 to 0.2 µg/µl), carboplatin and oxaliplatin concentrations (3.2×10−4 to 1 µg/µl) was tested in triplicate. To mimic the clinical infusion time, cells were exposed to these concentrations of toxicants for 1 hour and then incubated with fresh medium without toxicants for another 72 hours. Cell viability was tested using CCK-8 (Dojindo, Rockville, MD) and CellTiter-Blue® (Promega Corporation, Madison, WI) assays according to the technical manuals.
Statistical analysis
Data are presented as the mean ± SD. To assess statistical significance of differences, Student's t test or one-way ANOVA was conducted. P values <0.05 were considered statistically significant. All statistical analyses were conducted using GraphPad Software (GraphPad Software, Inc., La Jolla, CA).
RESULTS
ERCC1 expression in UV20 cell lines
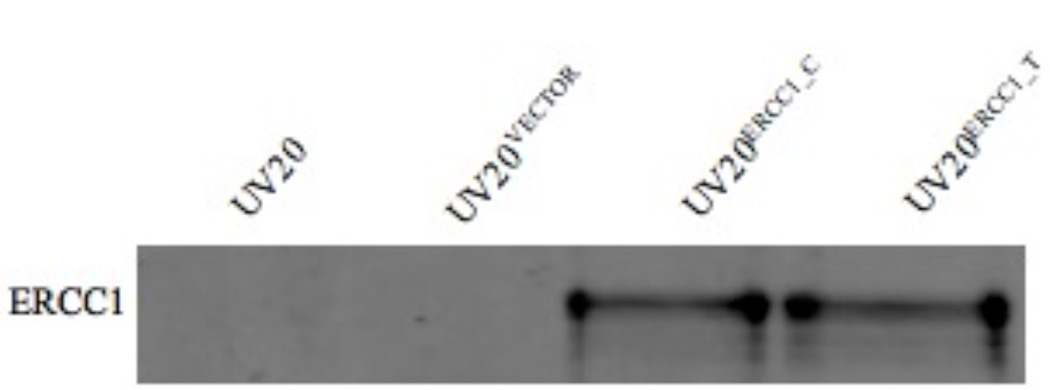
In order to compare the effects of the genetic polymorphism of ERCC1 N118N (500 C>T) on ERCC1 expression and platinum sensitivity in the same genetic background, the ERCC1 deficient cell line UV20, a derivative from CHO cell line, was employed. The UV20 cell lines stably expressing ERCC1 with C or T allele, denoted by UV20ERCC1_C and UV20ERCC1_T, were established. As shown in Figure 1, the parental UV20 cell line and the empty vector pcDNA™/V5-DEST transfected cells did not show detectable ERCC1 protein expression, while cells transfected with ERCC1 cDNA containing either genotype of 500 C>T showed comparable ERCC1 protein expression levels.
Figure 1.
ERCC1 over-expression in the UV20 cell line. The parental UV20 cell line and the empty vector pcDNA™/V5-DEST transfected cells (UV20VECTOR) did not show detectable ERCC1 protein expression, while cells transfected with ERCC1 cDNA containing either genotype of 500 C>T, designated as UV20ERCC1_C and UV20ERCC1_T, showed comparable ERCC1 protein expression levels.
Real-time quantitative PCR analysis reveals no differences in transcriptional levels in the UV20ERCC1_C and UV20ERCC1_T cell lines
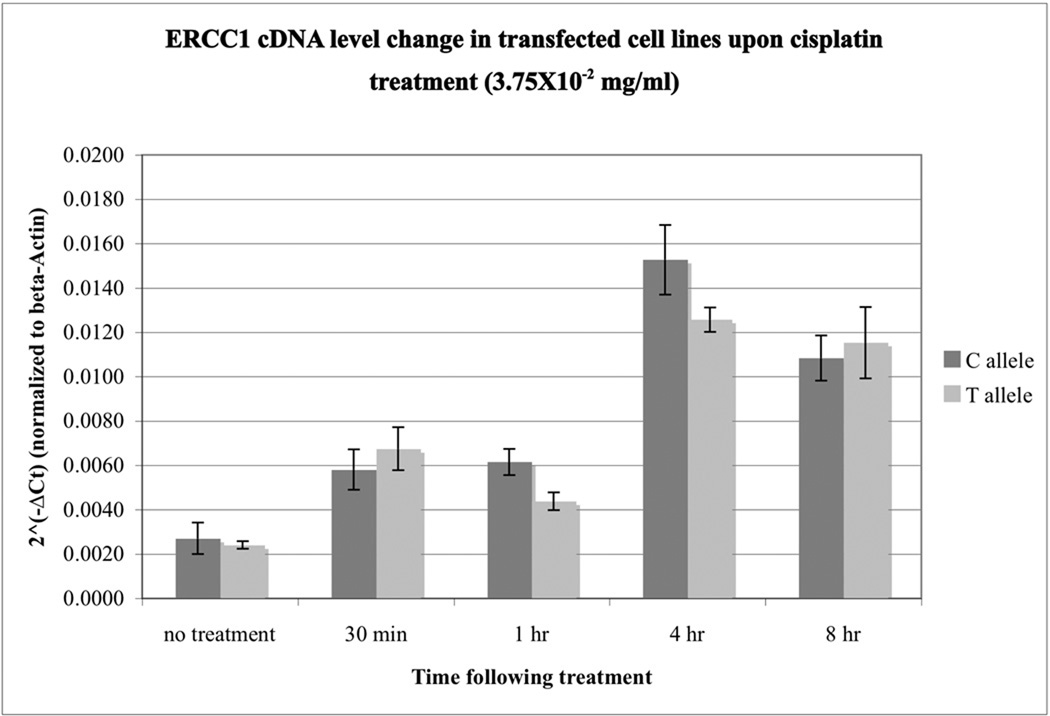
To study how exogenous stress induces ERCC1 transcription in the transfected UV20 cell lines, we assayed for changes in ERCC1 expression levels upon cisplatin treatment in UV20 cells transfected with the C or T allele. The transcription levels of ERCC1 in UV20ERCC1_C and UV20ERCC1_T cell lines were similar prior to cisplatin treatment, as normalized to beta-actin expression in each cell line (Figure 2). Upon cisplatin treatment, the expressional levels of ERCC1 transcripts increased in both cell lines immediately. This increase peaked at 4 hours following cisplatin treatment. However, there were no differences in ERCC1 transcription levels between the UV20ERCC1_C and UV20ERCC1_T cell lines (p=0.1737). At 8 hours after cisplatin treatment, the ERCC1 transcription level started to decrease. The two cell lines showed comparable levels of ERCC1 transcripts (p=0.6376).
Figure 2.
The transcription levels of ERCC1 in UV20ERCC1_C and UV20ERCC1_T cell lines remained the same upon cisplatin treatment. There is no difference in ERCC1 transcripts in the cells transfected with either C or T allele before treatment (p=0.495). ERCC1 transcription increased in both cell lines 30 minutes after cisplatin treatment. This increase in transcription peaked at 4 hours following cisplatin induction. However, there was no significant difference in ERCC1 gene expression levels between the two transfected cell lines, UV20ERCC1_C and UV20ERCC1_T (p=0.1737). Results are obtained from 4 independent experiments with duplicates in each experiment.
ERCC1 protein expression levels in UV-20 cells with different ERCC1 N118N (500 C>T) genotypes remained the same
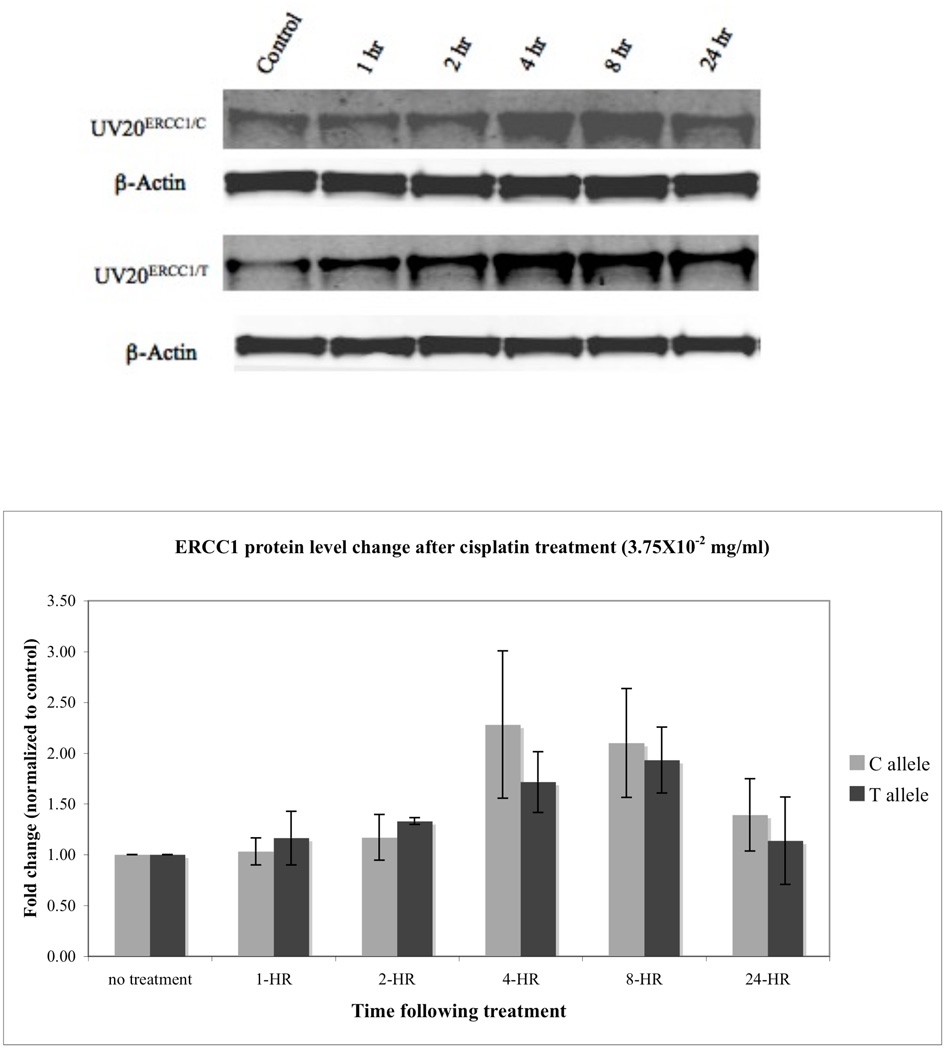
To investigate how the 500 C>T allele affects ERCC1 translation upon cisplatin treatment, we used Western blot analysis to examine ERCC1 protein levels in the UV20ERCC1_C and UV20ERCC1_T cell lines. Figure 3 shows that in both cell lines, ERCC1 protein expression levels increased upon cisplatin treatment, and peaked at 4 hours post-treatment, reaching a 2.28 fold increase in the UV20ERCC1_C cell line and a 1.71 fold increase in the UV20ERCC1_T cell line (p=0.5061). This increase in protein production dropped after 24 hours. However, there was no significant difference in ERCC1 expression upon the cisplatin challenge, although the UV20ERCC1_C cell line did show slightly more ERCC1 expression compared to that of the UV20ERCC1_T cell line.
Figure 3.
ERCC1 expression levels in UV20ERCC1_C and UV20ERCC1_T cell lines following cisplatin treatment did not show difference. Upper panel: Western blots for ERCC1. Lower panel: Quantification of ERCC1 expression change following cisplatin treatment. Data are mean ± SD obtained from three independent experiments. The expression levels of ERCC1 in the control cells pre-treatment were arbitrarily assigned 1. The change in ERCC1 level is expressed as the fold change compared to untreated cells. Data were normalized for equal loading.
Cellular sensitivity to platinum compounds did not exhibit difference in the UV20ERCC1_C and UV20ERCC1_T cell lines
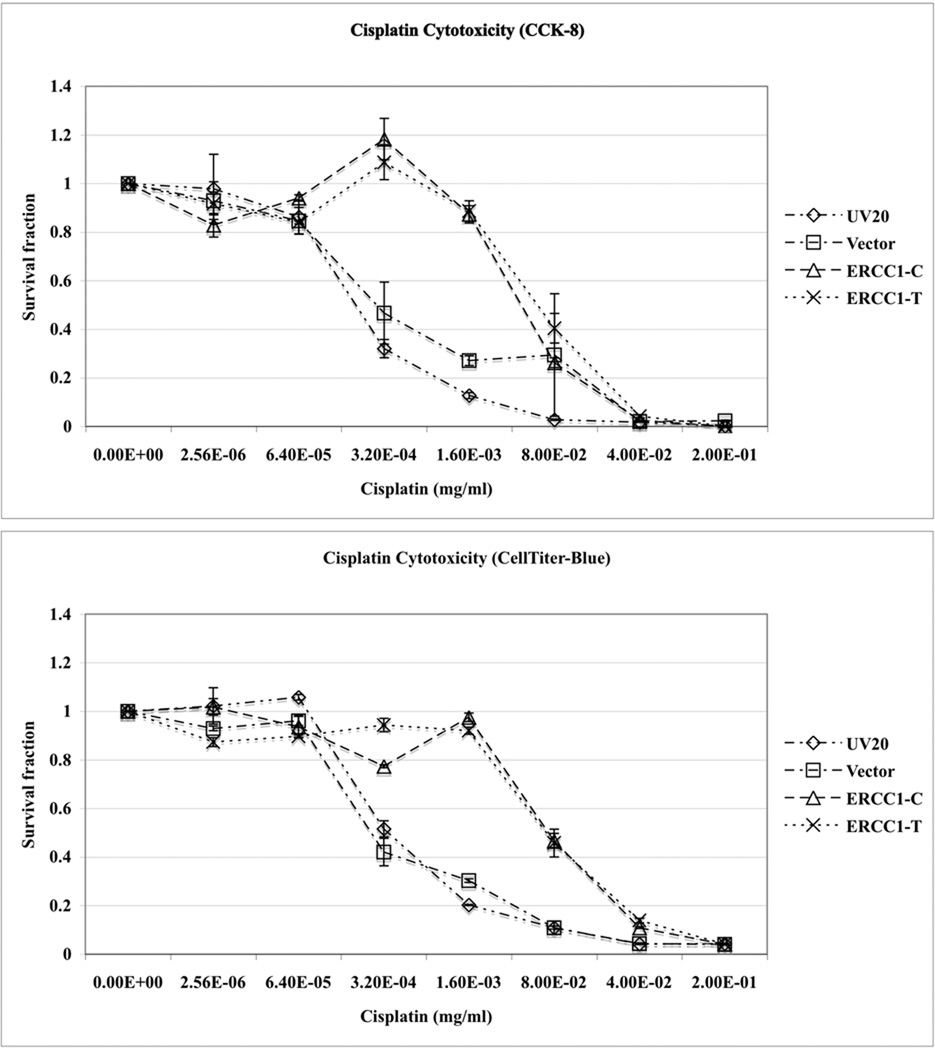
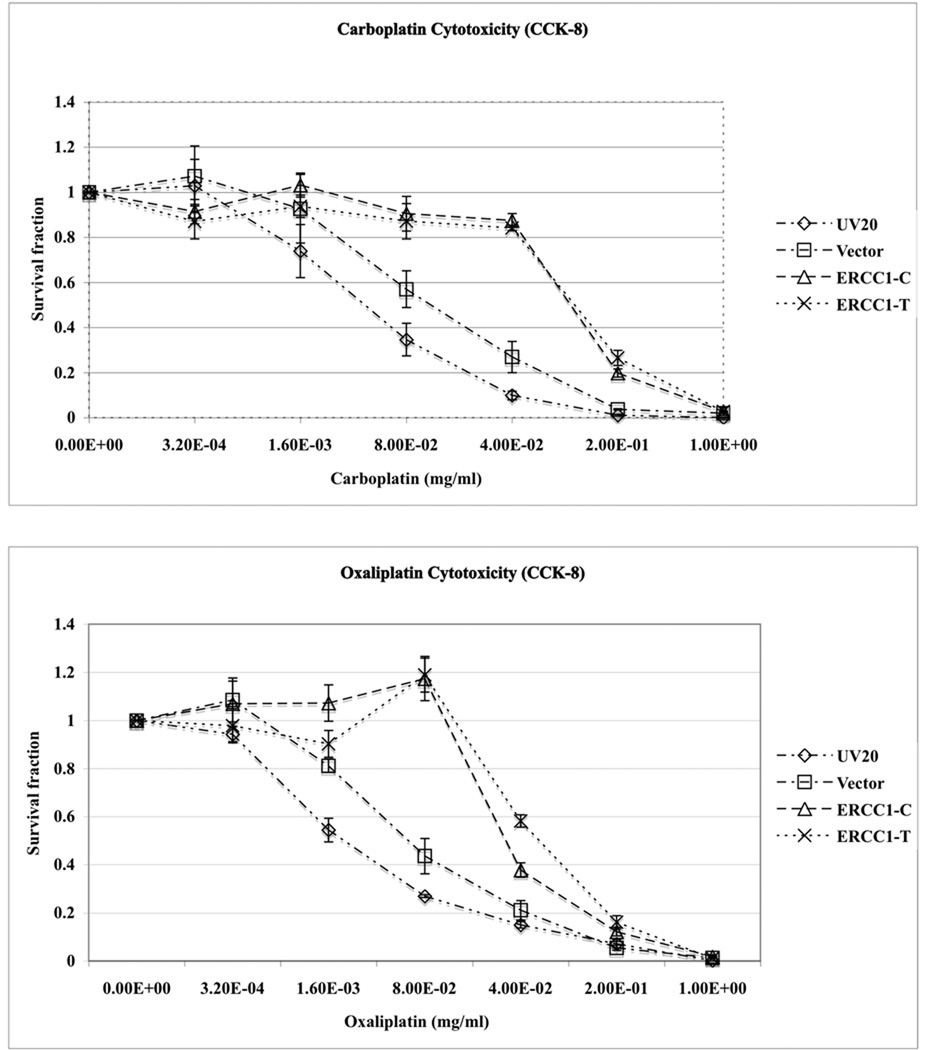
Lastly, cell viability assays were performed to determine if the cells transfected with different alleles of ERCC1 N118N would respond to platinum drugs differently. Both the UV20ERCC1_C and UV20ERCC1_T cells showed significantly increased viability upon cisplatin treatment compared to the parental cell line and vector transfected control cell line, as confirmed by CCK-8 and CellTiter-Blue assays independently (Figure 4, upper panel). The survival proportion of the UV20ERCC1_C and UV20ERCC1_T cells were not affected by cisplatin at a concentration of 3.2×10−4 mg/ml. However, only 32% of the parental cell line and 47% of cells transfected with the empty vector survived after treated by 3.2×10−4 mg/ml of cisplatin (P<0.0001). When the cisplatin concentration was increased to 0.0016 mg/ml, 13% and 27% cells survived for the parental and control cell lines while 87% of UV20ERCC1_C cells and 88% of UV20ERCC1_T cells survived, respectively (P<0.0001). Nonetheless, the UV20ERCC1_C and UV20ERCC1_T cell lines did not show differences in terms of cisplatin sensitivity at various cisplatin concentrations. Similar results were obtained for carboplatin and oxaliplatin (Figure 4, lower panel).
Figure 4.
Cell viability did not change in the UV20ERCC1_C and UV20ERCC1_T cell lines. The parental cell line UV20, empty vector transfected control cell line, and ERCC1 cDNA with either allele of 500 C>T transfected cell line UV20ERCC1_C and UV20ERCC1_T were treated with cisplatin (0, 2.56×10−6, 6.40×10−5, 3.2×10−4, 1.6×10−3, 8×10−2, 4×10−2, 0.2 µg/µl), carboplatin (0, 3.2×10−4, 1.6×10−3, 8×10−2, 4×10−2, 0.2 and 1.00 µg/µl) and oxaliplatin (0, 3.2×10−4, 1.6×10−3, 8×10−2, 4×10−2, 0.2 and 1.00 µg/µl). The cell cytotoxicity was tested by CCK-8 and CellTiter-Blue assayes. Only the results from CCK-8 assay for carboplatin and oxaliplatin are shown. Data are mean ± SD obtained from three independent experiments with triplicates in each experiment.
The results of this study confirm the pivotal role that ERCC1 plays in platinum sensitivity. However, the synonymous mutation ERCC1 N118N (500 C>T, rs11615) alone does not cause differential expression of the corresponding gene, nor does it change the cellular sensitivity to platinum drugs.
DISCUSSION
Personalized medicine is the use of both a patient’s genotypic and phenotypic data to choose a treatment or therapy that will best help the patient by maximizing benefit and minimizing harm [13]. Therefore, it is important to understand how inter-individual variations in the DNA sequence of specific genes affect drug responses. By introducing the cDNA clone of ERCC1 containing either the C or T allele of N118N into an ERCC1 deficient cell line UV20, we were able to examine the functional consequences of these genetic polymorphisms on ERCC1 expression and function. We found that this polymorphism did not contribute to altered ERCC1 expression upon cisplatin treatment or cellular sensitivity to platinum-containing drugs in vitro.
The ERCC1 N118N silent mutation was first described by genotyping a series of human cell lines and ovarian cancer tumor tissue specimens, and it was suggested that the conversion of the common codon AAC to an infrequently used codon AAT could affect protein translation rate and response to cisplatin [14]. A subsequent study suggested that this polymorphism could affect the DNA repair capacity in human ovarian cancer cell lines through a reduction in peak ERCC1 mRNA production and a consequent reduction in the translation of ERCC1 mRNA into protein. However, this conclusion was drawn from two independent studies using different cell lines, CP70 [15] and MCAS [16], which are equally resistant to cisplatin but differ at the synonymous mutation. The two studies used different assays to investigate ERCC1 transcripts, and the heterogeneous background of the two cell lines may also compromise the conclusion. However, a growing body of literature does show that ERCC1 expression levels correlate with responses to platinum containing reagents [17–21] while the results of genetic association studies are not always agreeable with one another (Table 1). The codon usage frequency for the SNP at position 500 with AAC changed to AAT (both encode Asn) changes moderately from 21.30 (The human codon usage and codon preference table, http://genome.crg.es/courses/genefinding/T4/main/) to 16.43 per thousand. This codon usage preference is conserved among species.
To test whether the synonymous codon of AAC and AAT in exon 4 of ERCC1 affects gene expression and function in response to platinum drugs, we established UV20 cell lines constitutively expressing ERCC1 with a C or T allele at the 500 position in the mRNA. Our results showed that the two cell lines had close basal ERCC1 transcription levels and performed similarly in ERCC1 transcription upon cisplatin induction. The UV20ERCC1_C cells showed slightly faster production of ERCC1 protein than the UV20ERCC1_T cells, corresponding with the moderate codon usage bias toward the AAC codon. However, it did not reach statistical significance. Nonetheless, the UV20ERCC1_C and UV20ERCC1_T cell lines exhibited the same level of sensitivity to platinum-containing drugs, including cisplatin, carboplatin and oxaliplatin.
The amino acid asparagine at this position is conserved among species, however, the wild type allele at the third position of this codon is not conserved, according to analysis performed using the UCSC Genome Browser (http://genome.ucsc.edu). Some species use AAC, such as rhesus, dog and opossum, etc., while others use AAT, such as human, mouse, elephant and zebrafish, etc. In addition, this SNP ERCC1 N118N (rs11615) is not under selective pressure in European, African or Chinese decent as indicated by low positive Tajima’s D values [22,23]. These results confirm that this SNP has little to no phenotypic effects although there is a clear preference for asparagine at the 118th amino acid. Furthermore, the linkage disequilibrium (LD) plot shows that this SNP is linked in a haplotype block of 18 kb within ERCC1 and the adjacent genomic region in European (TSI and CEU) population. But this is not true for African (YRI) or Asian (CHB and JPT) populations [24]. Although another NER gene, ERCC2 (also called XPD), is located in the same chromosome, the haplotype block does not extend to ERCC2 in any of the three populations, suggesting linkage disequilibrium with causative SNPs within ERCC1 might account for previous clinical associations with ERCC1 N118N. Mutations in ERCC1 that cause protein sequence change are not common and the only reported case is an infant possessing two point mutations that had severe developmental abnormality and did not thrive [25]. Therefore, instead of focusing on the ERCC1 N118N polymorphism, future genetic association studies should be expanded to include other polymorphisms linked to ERCC1 N118N, such as those in the regulatory regions of this gene.
In addition, the limitations and advantages of cell-based model should be taken into account. The cell line used in the study, UV20, is a UV sensitive derivative of a CHO cell line. Hamster cells are more sensitive to DNA damaging agents than human cell lines in general. The results, such as the IC50 of platinum compounds in the cells, should not be used to reflect the in vivo situations. Another limitation is that most cell lines do not express cytochrome P450 thus the contribution of metabolism and pharmacokinetics is not taken into account [26]. But the advantages are that the gene of interest is expressed in a homogenous genetic background and the cells are maintained under identical conditions. This allows the investigators to determine the genetic influence of human mutations in drug sensitivity.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCE
- 1.Kalikaki A, Kanaki M, Vassalou H, Souglakos J, Voutsina A, Georgoulias V, Mavroudis D. DNA repair gene polymorphisms predict favorable clinical outcome in advanced non-small-cell lung cancer. Clin Lung Cancer. 2009;10:118–123. doi: 10.3816/CLC.2009.n.015. [DOI] [PubMed] [Google Scholar]
- 2.Park SY, Hong YC, Kim JH, Kwak SM, Cho JH, Lee HL, Ryu JS. Effect of ERCC1 polymorphisms and the modification by smoking on the survival of non-small cell lung cancer patients. Med Oncol. 2006;23:489–498. doi: 10.1385/MO:23:4:489. [DOI] [PubMed] [Google Scholar]
- 3.Isla D, Sarries C, Rosell R, Alonso G, Domine M, Taron M, Lopez-Vivanco G, Camps C, Botia M, Nunez L, Sanchez-Ronco M, Sanchez JJ, Lopez-Brea M, Barneto I, Paredes A, Medina B, Artal A, Lianes P. Single nucleotide polymorphisms and outcome in docetaxel-cisplatin-treated advanced non-small-cell lung cancer. Ann Oncol. 2004;15:1194–1203. doi: 10.1093/annonc/mdh319. [DOI] [PubMed] [Google Scholar]
- 4.Ryu JS, Hong YC, Han HS, Lee JE, Kim S, Park YM, Kim YC, Hwang TS. Association between polymorphisms of ERCC1 and XPD and survival in non-small-cell lung cancer patients treated with cisplatin combination chemotherapy. Lung Cancer. 2004;44:311–316. doi: 10.1016/j.lungcan.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 5.Gandara DR, Kawaguchi T, Crowley J, Moon J, Furuse K, Kawahara M, Teramukai S, Ohe Y, Kubota K, Williamson SK, Gautschi O, Lenz HJ, McLeod HL, Lara PN, Jr, Coltman CA, Jr, Fukuoka M, Saijo N, Fukushima M, Mack PC. Japanese-US common-arm analysis of paclitaxel plus carboplatin in advanced non-small-cell lung cancer: a model for assessing population-related pharmacogenomics. J Clin Oncol. 2009;27:3540–3546. doi: 10.1200/JCO.2008.20.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou W, Gurubhagavatula S, Liu G, Park S, Neuberg DS, Wain JC, Lynch TJ, Su L, Christiani DC. Excision repair cross-complementation group 1 polymorphism predicts overall survival in advanced non-small cell lung cancer patients treated with platinum-based chemotherapy. Clin Cancer Res. 2004;10:4939–4943. doi: 10.1158/1078-0432.CCR-04-0247. [DOI] [PubMed] [Google Scholar]
- 7.Komar AA. Genetics. SNPs, silent but not invisible. Science. 2007;315:466–467. doi: 10.1126/science.1138239. [DOI] [PubMed] [Google Scholar]
- 8.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A "silent" polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 9.Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Busch DB, Cleaver JE, Glaser DA. Large-scale isolation of UV-sensitive clones of CHO cells. Somatic Cell Genet. 1980;6:407–418. doi: 10.1007/BF01542792. [DOI] [PubMed] [Google Scholar]
- 11.Thompson LH, Rubin JS, Cleaver JE, Whitmore GF, Brookman K. A screening method for isolating DNA repair-deficient mutants of CHO cells. Somatic Cell Genet. 1980;6:391–405. doi: 10.1007/BF01542791. [DOI] [PubMed] [Google Scholar]
- 12.Bahr SM, Borgschulte T, Kayser KJ, Lin N. Using microarray technology to select housekeeping genes in Chinese hamster ovary cells. Biotechnol Bioeng. 2009;104:1041–1046. doi: 10.1002/bit.22452. [DOI] [PubMed] [Google Scholar]
- 13.Zhou SF, Di YM, Chan E, Du YM, Chow VD, Xue CC, Lai X, Wang JC, Li CG, Tian M, Duan W. Clinical pharmacogenetics and potential application in personalized medicine. Curr Drug Metab. 2008;9:738–784. doi: 10.2174/138920008786049302. [DOI] [PubMed] [Google Scholar]
- 14.Yu JJ, Mu C, Lee KB, Okamoto A, Reed EL, Bostick-Bruton F, Mitchell KC, Reed E. A nucleotide polymorphism in ERCC1 in human ovarian cancer cell lines and tumor tissues. Mutat Res. 1997;382:13–20. doi: 10.1016/s1383-5726(97)00004-6. [DOI] [PubMed] [Google Scholar]
- 15.Li Q, Gardner K, Zhang L, Tsang B, Bostick-Bruton F, Reed E. Cisplatin induction of ERCC-1 mRNA expression in A2780/CP70 human ovarian cancer cells. J Biol Chem. 1998;273:23419–23425. doi: 10.1074/jbc.273.36.23419. [DOI] [PubMed] [Google Scholar]
- 16.Yu JJ, Lee KB, Mu C, Li Q, Abernathy TV, Bostick-Bruton F, Reed E. Comparison of two human ovarian carcinoma cell lines (A2780/CP70 and MCAS) that are equally resistant to platinum, but differ at codon 118 of the ERCC1 gene. Int J Oncol. 2000;16:555–560. doi: 10.3892/ijo.16.3.555. [DOI] [PubMed] [Google Scholar]
- 17.Qian XP, Liu BR, Shi MQ, Liu XZ, Hu WJ, Zou ZY, Wei J. Prognostic significance of ERCC1 mRNA expression in patients with non-small cell lung cancer receiving platinum-based chemotherapy. Zhonghua Zhong Liu Za Zhi. 2009;31:33–37. [PubMed] [Google Scholar]
- 18.Olaussen KA, Dunant A, Fouret P, Brambilla E, Andre F, Haddad V, Taranchon E, Filipits M, Pirker R, Popper HH, Stahel R, Sabatier L, Pignon JP, Tursz T, Le Chevalier T, Soria JC. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355:983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 19.Ren S, Zhou S, Zhang L, Xu J, Lv M, Zhang J, Zhou C. High-Level mRNA of Excision Repair Cross-Complementation Group 1 Gene Is Associated With Poor Outcome of Platinum-Based Doublet Chemotherapy of Advanced Nonsmall Cell Lung Cancer Patients. Cancer Invest. 2010 doi: 10.3109/07357901003735659. [DOI] [PubMed] [Google Scholar]
- 20.Ozkan M, Akbudak IH, Deniz K, Dikilitas M, Dogu GG, Berk V, Karaca H, Er O, Altinbas M. Prognostic value of excision repair cross-complementing gene 1 expression for cisplatin-based chemotherapy in advanced gastric cancer. Asian Pac J Cancer Prev. 2010;11:181–185. [PubMed] [Google Scholar]
- 21.Chen S, Zhang J, Wang R, Luo X, Chen H. The platinum-based treatments for advanced non-small cell lung cancer, is low/negative ERCC1 expression better than high/positive ERCC1 expression? A meta-analysis. Lung Cancer. 2010 doi: 10.1016/j.lungcan.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 22.Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 23.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 25.Jaspers NG, Raams A, Silengo MC, Wijgers N, Niedernhofer LJ, Robinson AR, Giglia-Mari G, Hoogstraten D, Kleijer WJ, Hoeijmakers JH, Vermeulen W. First reported patient with human ERCC1 deficiency has cerebro-oculofacio-skeletal syndrome with a mild defect in nucleotide excision repair and severe developmental failure. Am J Hum Genet. 2007;80:457–466. doi: 10.1086/512486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Innocenti F. Genomics and Pharmacogenomics in Anticancer Drug Development and Clinical Response Cancer Drug Discovery and Development. 2009:19–31. [Google Scholar]
- 27.Chua W, Goldstein D, Lee CK, Dhillon H, Michael M, Mitchell P, Clarke SJ, Iacopetta B. Molecular markers of response and toxicity to FOLFOX chemotherapy in metastatic colorectal cancer. Br J Cancer. 2009;101:998–1004. doi: 10.1038/sj.bjc.6605239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang PM, Tzeng CH, Chen PM, Lin JK, Lin TC, Chen WS, Jiang JK, Wang HS, Wang WS. ERCC1 codon 118 C-->T polymorphism associated with ERCC1 expression and outcome of FOLFOX-4 treatment in Asian patients with metastatic colorectal carcinoma. Cancer Sci. 2008 doi: 10.1111/j.1349-7006.2008.01031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Balibrea E, Abad A, Aranda E, Sastre J, Manzano JL, Diaz-Rubio E, Gomez-Espana A, Aparicio J, Garcia T, Maestu I, Martinez-Cardus A, Gines A, Guino E. Pharmacogenetic approach for capecitabine or 5-fluorouracil selection to be combined with oxaliplatin as first-line chemotherapy in advanced colorectal cancer. Eur J Cancer. 2008;44:1229–1237. doi: 10.1016/j.ejca.2008.03.025. [DOI] [PubMed] [Google Scholar]
- 30.Park DJ, Zhang W, Stoehlmacher J, Tsao-Wei D, Groshen S, Gil J, Yun J, Sones E, Mallik N, Lenz HJ. ERCC1 gene polymorphism as a predictor for clinical outcome in advanced colorectal cancer patients treated with platinum-based chemotherapy. Clin Adv Hematol Oncol. 2003;1:162–166. [PubMed] [Google Scholar]
- 31.Viguier J, Boige V, Miquel C, Pocard M, Giraudeau B, Sabourin JC, Ducreux M, Sarasin A, Praz F. ERCC1 codon 118 polymorphism is a predictive factor for the tumor response to oxaliplatin/5-fluorouracil combination chemotherapy in patients with advanced colorectal cancer. Clin Cancer Res. 2005;11:6212–6217. doi: 10.1158/1078-0432.CCR-04-2216. [DOI] [PubMed] [Google Scholar]
- 32.Stoehlmacher J, Park DJ, Zhang W, Yang D, Groshen S, Zahedy S, Lenz HJ. A multivariate analysis of genomic polymorphisms: prediction of clinical outcome to 5-FU/oxaliplatin combination chemotherapy in refractory colorectal cancer. Br J Cancer. 2004;91:344–354. doi: 10.1038/sj.bjc.6601975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li F, Sun X, Sun N, Qin S, Cheng H, Feng J, Chen B, Cheng L, Lu Z, Ji J, Zhou Y. Association between polymorphisms of ERCC1 and XPD and clinical response to platinum-based chemotherapy in advanced non-small cell lung cancer. Am J Clin Oncol. 2010;33:489–494. doi: 10.1097/COC.0b013e3181b9cedc. [DOI] [PubMed] [Google Scholar]
- 34.Kamikozuru H, Kuramochi H, Hayashi K, Nakajima G, Yamamoto M. ERCC1 codon 118 polymorphism is a useful prognostic marker in patients with pancreatic cancer treated with platinum-based chemotherapy. Int J Oncol. 2008;32:1091–1096. [PubMed] [Google Scholar]
- 35.Steffensen KD, Waldstrom M, Jeppesen U, Brandslund I, Jakobsen A. Prediction of response to chemotherapy by ERCC1 immunohistochemistry and ERCC1 polymorphism in ovarian cancer. Int J Gynecol Cancer. 2008;18:702–710. doi: 10.1111/j.1525-1438.2007.01068.x. [DOI] [PubMed] [Google Scholar]
- 36.Huang ZH, Hua D, Du X. Polymorphisms in p53, GSTP1 and XRCC1 predict relapse and survival of gastric cancer patients treated with oxaliplatin-based adjuvant chemotherapy. Cancer Chemother Pharmacol. 2009;64:1001–1007. doi: 10.1007/s00280-009-0956-2. [DOI] [PubMed] [Google Scholar]
- 37.Bradbury PA, Kulke MH, Heist RS, Zhou W, Ma C, Xu W, Marshall AL, Zhai R, Hooshmand SM, Asomaning K, Su L, Shepherd FA, Lynch TJ, Wain JC, Christiani DC, Liu G. Cisplatin pharmacogenetics, DNA repair polymorphisms, and esophageal cancer outcomes. Pharmacogenet Genomics. 2009;19:613–625. doi: 10.1097/FPC.0b013e32832f3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma BB, Hui EP, Wong SC, Tung SY, Yuen KK, King A, Chan SL, Leung SF, Kam MK, Yu BK, Zee B, Chan AT. Multicenter phase II study of gemcitabine and oxaliplatin in advanced nasopharyngeal carcinoma--correlation with excision repair cross-complementing-1 polymorphisms. Ann Oncol. 2009 doi: 10.1093/annonc/mdp065. [DOI] [PubMed] [Google Scholar]