Abstract
Microbial cytochromes c′ contain a 5-coordinate His-ligated heme that forms stable adducts with nitric oxide (NO) and carbon monoxide (CO), but not with dioxygen. We report the 1.95 and 1.35 Å resolution crystal structures of the CO- and NO-bound forms of the reduced protein from Alcaligenes xylosoxidans. NO disrupts the His–Fe bond and binds in a novel mode to the proximal face of the heme, giving a 5-coordinate species. In contrast, CO binds 6-coordinate on the distal side. A second CO molecule, not bound to the heme, is located in the proximal pocket. Since the unusual spectroscopic properties of cytochromes c′ are shared by soluble guanylate cyclase (sGC), our findings have potential implications for the activation of sGC induced by the binding of NO or CO to the heme domain.
Keywords: cytochrome c′/guanylate cyclase/heme/nitric oxide/X-ray crystallography
Introduction
Gas-sensing heme proteins play a central role in the regulation of important biological processes in mammals, plants and bacteria. Nitric oxide (NO)-, O2- and carbon monoxide (CO)-sensing systems have been identified (Rodgers, 1999; Hou et al., 2000). The regulation of gene expression by O2 in plant-associated nitrogen-fixing bacteria is well characterized and involves the heme protein FixL (Gong et al., 1998). CO regulates the transcription of enzymes that allow the photosynthetic microorganism Rhodospirillum rubrum to grow on CO as the sole energy source by binding to the heme protein CooA (He et al., 1996). NO plays a role in regulating a diverse range of physiological processes in higher organisms, such as the immune response to tumor cells, vasodilation and neuronal synaptic transmission (Denninger and Marletta, 1999). Many of these responses to NO are triggered by an increase in the level of the cellular second messenger cGMP. The latter is produced by soluble guanylate cyclase (sGC), which is activated by binding of NO to the heme domain. Particular interest surrounds the mechanism of activation and selective ligand recognition by these various sensor proteins.
Cytochromes (cyt) c′ are found in the periplasmic space of a number of photosynthetic, nitrogen-fixing and denitrifying bacteria. In denitrifiers, they have been proposed to have a role in mediating NO transfer and protecting the organism from the potentially toxic levels of NO that may otherwise accumulate (Moir, 1999). The strong similarities in the distinctive reactivity and spectroscopic properties of the heme center in cyt c′ and that of sGC have been noted previously (Stone and Marletta, 1994). The ligand binding properties of both these proteins are anomalous when compared with other high spin hemoproteins since both sGC and cyt c′ form stable complexes with both NO and CO but not O2. In addition, it has been shown that NO forms 5-coordinate heme adducts with both sGC (Vogel et al., 1999) and cyt c′ (Iwasaki et al., 1991). In the absence of a crystal structure for any 5-coordinate NO–heme complex, the structural interpretation of spectroscopic data for NO binding to sGC is based solely on comparisons with the structures of 6-coordinate hemoprotein–ligand adducts and model complexes. Implicit in these comparisons is the assumption that NO binding is restricted to the distal side of the heme, a view that so dominates the interpretation of studies of ligand binding to hemoproteins that it allows the term ‘distal dogma’ to be used.
The structural data for Alcaligenes xylosoxidans cytochrome c′ (Axcyt c′) we report here show that on binding NO, cleavage of the proximal Fe–His bond occurs and a 5-coordinate heme–NO complex is formed. However, contrary to expectations, we find NO to be bound on the proximal side of the heme. We also show that CO binding does not result in cleavage of the His–Fe heme bond and is ligated 6-coordinate on the distal side of the heme. These structures are the first for any gaseous ligands bound to cyt c′ and also the first for any 5-coordinate NO-ligated hemoprotein. Our findings that Axcyt c′ binds NO and CO to opposite sides of the heme center have clear implications for the mode of binding of NO and CO to sGC.
Results and discussion
Structure determination
Axcyt c′ was isolated from the denitrifying bacterium A.xylosoxidans NCIMB 11015 essentially as described (Ambler, 1973). All the X-ray data presented here were collected under cryogenic conditions, where the unit cell typically shrinks by ∼5% relative to its volume at ambient temperature. Thus, for each structure, molecular replacement was used to place correctly the ambient temperature structure of the oxidized protein determined at 1.8 Å resolution (Dobbs et al., 1996) into the cryogenic coordinate frame prior to refinement and model building. In order to define potential structural changes to the protein and heme environment associated with the binding of ligands, we have determined the structure of the reduced and the CO- and NO-bound forms of Axcyt c′. However, since all X-ray data were collected at cryogenic temperatures, it was considered appropriate to re-determine the oxidized structure under these conditions as well.
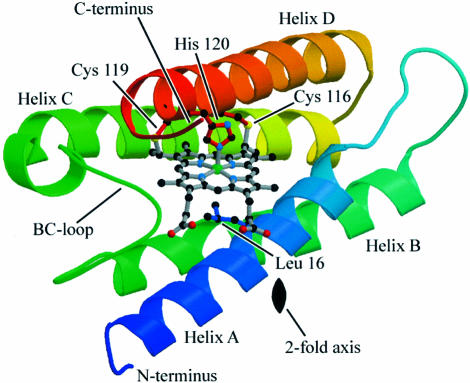
Axcyt c′ is a homodimer of 27.2 kDa with each subunit being comprised of an antiparallel four α-helix bundle containing a 5-coordinate c-type heme that is partially exposed to solvent (Figure 1). The subunits are arranged in a head-to-tail fashion, with both hemes on the same face of the dimer. Intersubunit contacts are mediated through residues in the A and B helices. The observations made by Shibata et al. (1998) place cyt c′ into two families according to the nature of the surfaces of these helices. Axcyt c′ belongs to Type 1, which have hydrophobic surfaces to their A and B helices and usually form globular, X-shaped dimers. It is notable, however, that the Chromatium vinosum protein will monomerize when CO is bound (Doyle et al., 1986), as will several others for which there are no structures (Ren et al., 1993). In contrast, the Type 2 proteins have A and B helices that are hydrophilic and they are either monomeric or form flattened dimers.
Fig. 1. Ribbon representation of a single subunit of the reduced Axcyt c′ structure showing the position of the heme. Also depicted are the side chains of the proximal His, Leu16 and the two Cys residues that form thioether bridges to the heme. The Leu blocks access to the vacant sixth coordination site in the distal pocket. The location of the crystallographic 2-fold axis is indicated, which is perpendicular to the plane of the paper. A 180° rotation of this subunit about the 2-fold axis generates the second subunit of the functional dimer. The structure is colored with respect to sequence number, starting with blue at the N-terminus and finishing with red at the C-terminus. This figure was produced using MOLSCRIPT (Kraulis, 1991) and Raster3D (Merritt and Bacon, 1997).
The heme lies towards one end of the subunit and is essentially sandwiched between helices A and D, but also has interactions with helix C and the BC loop. Consistent with a body of spectroscopic data, it is 5-coordinate and covalently bound to the protein via two Cys–thioether bonds that are provided by a conserved Cys-X-X-Cys-His motif, where the His (residue 120) is the fifth ligand to the heme iron. This His lies in the proximal heme pocket, which is solvent exposed. In contrast, the distal pocket is buried and a hydrophobic residue, Leu16, blocks access to the vacant sixth coordination site of the heme. These are characteristics of Group 2 cyt c′, according to the distinctions made by Tahirov et al. (1996b), while Group 1 proteins have an aromatic residue in this position (Phe or Tyr) and the distal pocket is exposed to solvent (due to a channel between helices B and C).
A more extensive description of the overall Axcyt c′ structure is not warranted here as this has been presented elsewhere (Dobbs et al., 1996). In this paper the focus will be on the heme environment in both the oxidized and reduced states, and the NO- and CO-bound forms.
Oxidized and reduced structures
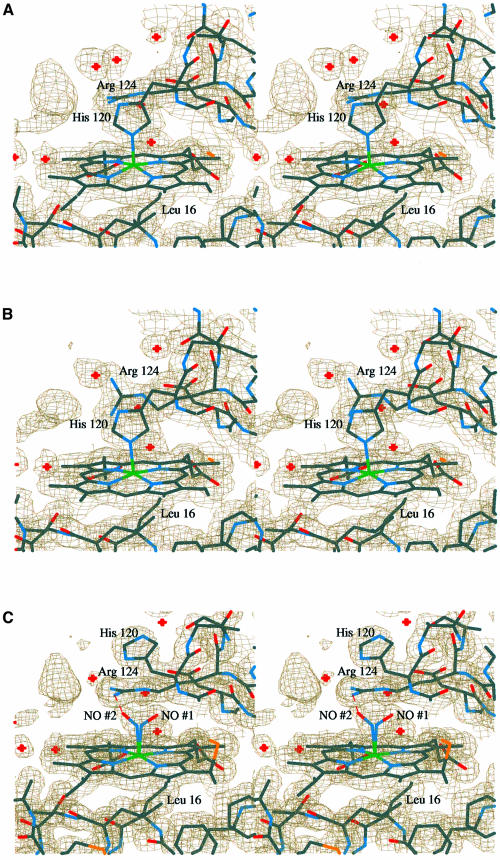
In order to monitor potential redox-dependent changes in the heme environment, data were collected on both oxidized and reduced crystals of Axcyt c′ to resolutions of 2.05 and 1.90 Å, respectively. Overall, there was very little difference between the resultant models, having a root mean square (r.m.s.) deviation of 0.13 Å over all main chain atoms after least squares superposition. In both oxidation states, the proximal ligand, His120, is additionally hydrogen bonded through Nδ1 to an ordered water molecule that is accessible to bulk solvent. However, there was a clear difference in the position of the Arg124 side chain, which lies adjacent to the heme, being well defined in both structures. In the oxidized structure, the planar guanidinium moiety is perpendicular to the imidazole ring of His120 and almost parallel to the A pyrrole ring of the heme. In this position, the positive charge of the Arg presumably overlaps with the negative charge of the heme π system. In contrast, in the reduced structure this side chain is parallel to the His (Figure 2). Clearly, this residue senses the oxidation state of the iron, although the mechanism is not apparent. The possibility that the oxidized Axcyt c′ crystal may have been photo-reduced during X-ray data collection has been considered, even though the experiment was performed at 100 K. However, this cannot have occurred to any significant extent because of the unambiguous differences described above. At the resolutions of these two structures, the observed difference of 0.1 Å in the His Nε2–Fe bond lengths, which are 2.0 and 2.1 Å, respectively, in the oxidized and reduced models, may not be significant. In both structures the heme group is distinctly puckered and the Fe is displaced out of the heme plane by ∼0.3 Å towards the proximal His. This is consistent with the Fe remaining high spin in both structures, as would be expected given that the crystals were grown under alkaline conditions. Moreover, the sixth coordination site remains vacant in both models.
Fig. 2. Stereo diagrams of the final 2mFobs – dFcalc electron density maps contoured at 1σ for the (A) oxidized, (B) reduced, (C) NO-bound and (D) CO-bound Axcyt c′ structures. The atoms are colored as follows: Fe, green; S, yellow; O, red; N, blue; C, gray. The red crosses indicate water molecules. In this view, both the left-hand and rear edges of the heme are accessible to solvent, as is evident from the presence of water molecules in these regions. In the interests of clarity, the thioether bridges to the heme group, which would be in the foreground, have been omitted. In the oxidized and NO-bound structures in particular, there is a large unassigned region of electron density within hydrogen bonding distance of the side chain of Arg124. Attempts to model buffer and cryoprotectant components here were not convincing. However, a reasonable fit was obtained with a sulfate anion (present in the precipitant), but this gave high temperature factors after refinement. (E) A least squares superposition of all four structures based on all main chain atoms. They are colored as follows: oxidized, yellow; reduced, red; NO-bound, green; CO-bound, blue. In the interests of clarity, all bonds to the heme Fe have been omitted. All images were produced using the programs O and OPLOT (Jones et al., 1991).
NO-bound structure
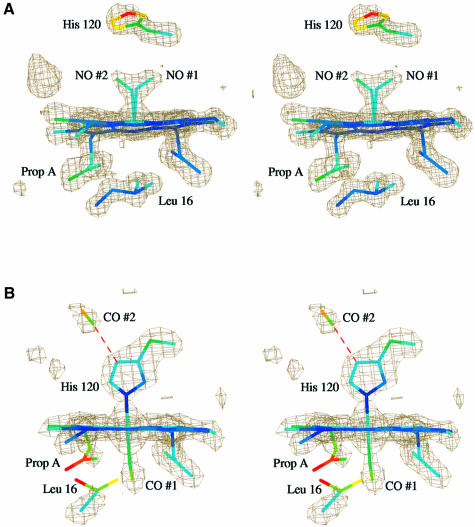
We originally determined the NO-bound structure using X-ray data to 1.95 Å resolution (not shown). The resultant electron density maps clearly indicated the binding of NO as a 5-coordinate adduct. This is consistent with the interpretation of earlier spectroscopic studies, which indicated that at physiological pH values the His–Fe bond is readily cleaved on NO binding (Yoshimura et al., 1986). However, unexpectedly, binding occurs on the proximal face of the heme with the concomitant displacement of His120, mainly through a 111° rotation about the Cα–Cβ bond. Unfortunately, at this resolution the absolute conformation of the ligand could not be modeled confidently. In order to resolve this issue, data were collected to near atomic resolution. In electron density maps subsequently calculated at 1.35 Å resolution, it was possible to model NO binding in two alternative bent conformations, each with half occupancy, with Fe–N–O angles of 124 and 132° for the two conformers and an average Fe–NO bond length of 2.0 Å (Figure 2). In one binding mode, the NO has no specific interactions with the protein (NO 1), whilst in the other a hydrogen bond forms with Nη1 of Arg124 (NO 2). This interaction is possible because the Arg side chain has adopted a conformation similar to that observed in the oxidized structure, where it stacks against the heme plane. It is notable that the side chain of His120 in this structure is less well defined in the electron density, presumably because it has lost the covalent bond to the iron, instead having only a single hydrogen bond to a solvent molecule through atom Nε2. After refinement, the two alternative conformations for NO had essentially equivalent thermal parameters (Table I; Figure 3), suggesting that the assignment of half occupancy to each was valid. This is surprising since conformation 2 would appear to be the more favorable because it has a hydrogen bonding interaction with the protein, whilst conformation 1 does not.
Table I. Summary of X-ray data and model parameters for A.xylosoxidans cytochrome c′.
| Oxidized | Reduced | NO-bound | CO-bound | |
|---|---|---|---|---|
| Data collectiona | ||||
| site | PX7.2 (SRS) | ID14-2 (ESRF) | ID14-2 (ESRF) | BM30 (ESRF) |
| detector | Mar300 image plate | ADSC Quantum-4 CCD | ADSC Quantum-4 CCD | Mar345 image plate |
| wavelength (Å) | 1.488 | 0.933 | 0.933 | 0.979 |
| cell parameters: a = b, c (Å) | 52.9, 182.3 | 52.8, 182.6 | 53.0, 180.9 | 53.6, 180.9 |
| resolution range (Å) | 40.0–2.05 | 40.0–1.90 | 40.0–1.35 | 40.0–1.95 |
| unique reflections | 10 205 | 12 387 | 33 925 | 12 158 |
| completeness (%) | 99.0 (98.9) | 97.1 (84.5) | 98.8 (93.1) | 98.7 (86.8) |
| redundancy | 8.1 | 4.9 | 6.8 | 9.9 |
| Rmergeb | 0.109 (0.349) | 0.041 (0.122) | 0.039 (0.227) | 0.066 (0.310) |
| <I>/<σI> | 18.8 (5.2) | 31.6 (8.3) | 39.8 (3.9) | 33.2 (5.8) |
| Refinement | ||||
| Rcrystc (based on 95% of data) (%) | 19.2 | 21.7 | 19.4 | 20.3 |
| Rfreec (based on 5% of data) (%) | 25.2 | 27.3 | 22.0 | 25.8 |
| DPId (based on Rfree) (Å) | 0.18 | 0.16 | 0.06 | 0.16 |
| residues with most favored φ/ψe (%) | 93.6 | 94.5 | 94.4 | 95.4 |
| r.m.s. deviation bond distances (Å) | 0.017 | 0.016 | 0.011 | 0.016 |
| r.m.s. deviation angle distances (Å) | 0.037 | 0.033 | 0.024 | 0.034 |
| Average temperature factors (Å2) | ||||
| main chain atoms | 20 | 19 | 14 | 20 |
| side chain atoms | 23 | 21 | 17 | 23 |
| heme | 17 | 15 | 13 | 18 |
| gaseous ligandsf (1/2) | – | – | 16/17 | 27/34 |
| overall | 23 | 22 | 18 | 23 |
| R.m.s. deviation versus reduced structureg (Å) | 0.13 | – | 0.15 | 0.60 |
aThe figures in parentheses indicate the values for the outer resolution shell.
bRmerge = ∑(|Ij – <Ij> |)/∑<Ij>, where Ij is the intensity of an observation of reflection j and <Ij> is the average intensity for reflection j.
cThe R factors Rcryst and Rfree were calculated as follows: R = ∑(|Fobs – Fcalc|)/∑|Fobs| × 100, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively.
dDiffraction component precision index (Cruickshank, 1999), an estimate of the overall coordinate errors calculated in REFMAC.
eAs calculated using PROCHECK (Laskowski et al., 1993).
fSee Figure 3. Note, the two NO molecules have half occupancy whilst the two CO molecules have unit occupancy.
gAfter least squares superposition based on all main chain atoms.
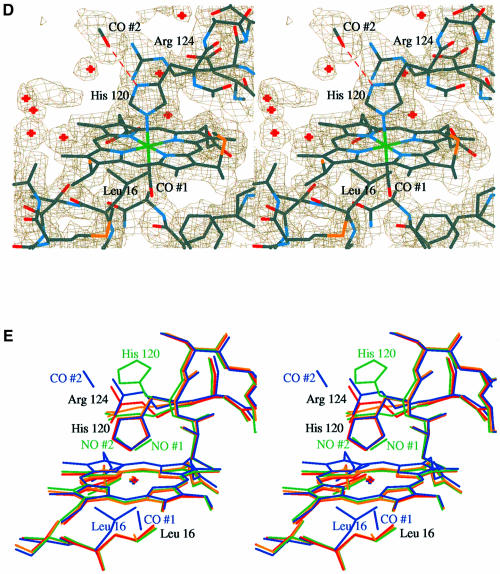
Fig. 3. Omit difference maps for the (A) NO- and (B) CO-bound Axcyt c′ structures contoured at 3.5σ. All the displayed atoms were omitted from the refinements (see Materials and methods). However, Arg124 was retained in the refinement, in order to remove distracting density, in particular from the NO ligand. The view is chosen to emphasize the flattening of the heme plane upon binding of CO. The atoms are colored according to their temperature factors, which all lie roughly in the range 10–40 Å2, where dark blue indicates a low value increasing through light blue, dark green, light green, yellow and orange, to red, which indicates a high value. Note that the side chain of His120 in the NO-bound structure and the side chain of Leu16 and the propionate group A in the CO-bound structure are less well defined in the electron density and have relatively high thermal parameters. See main text for a further explanation.
CO-bound structure
In the 1.95 Å resolution structure of the CO-bound adduct, the heme is 6-coordinate with the CO bound to the distal face of the reduced heme. The CO binds in an almost linear configuration with an Fe–C–O angle of 167° and Fe–CO bond length of 2.1 Å. This binding induces a cascade of conformational changes. The side chain of Leu16, which mediates access to the distal heme face, is displaced to one side, largely by a simple 134° rotation about the Cα–Cβ bond. In doing so, it pushes pyrrole ring A of the heme group upward into the proximal pocket (maximum displacement ∼1 Å). This movement effectively flattens the heme group. The positions of the Fe and the proximal His also move in the same direction, but to a lesser extent, with the net effect of restoring the Fe into the heme plane. The conformation adopted by Arg124 is similar to that found in the reduced structure, although it is slightly higher due to the upward movement of the heme group (Figure 2). The side chain of Leu16, the propionate group A of the heme and the CO molecule all have relatively high thermal parameters compared with the overall value for the heme (Figure 3). Indeed, in maps calculated from a previous data set (not shown) there was electron density for Leu16 in both the conformation seen here and in the conformation observed in the other three structures overlapping with density for CO. Thus, the elevated thermal parameters of the CO may indicate that it is not fully occupied here as well.
During the process of model building, an elongated peak of electron density was observed adjacent to and within hydrogen bonding distance of Nδ1 of the proximal His residue. The single water molecule observed in both the reduced and oxidized structures (having temperature factors of 27 and 31 Å2, respectively) was insufficient to account for all the electron density. However, the inclusion of a second CO molecule at this position gave a good fit. After refinement at full occupancy, the CO had tem perature factors of 37 and 31 Å2 for the C and O atoms, respectively, as compared with the value of 18 Å2 for the Nδ1 atom of His120, with a hydrogen bond length of 2.9 Å. This interaction with CO can be likened to that seen in many hemoproteins where the proximal His forms a hydrogen bond with either a main chain carbonyl or a side chain carboxyl group (Poulos and Kraut, 1980; Harutyunyan et al., 1996). The presence of this additional CO shows that, at least in this structure, the proximal His must be protonated. The possibility that this CO has arisen from X-ray-induced photo-dissociation of heme-bound CO during data collection cannot be excluded and may provide an explanation for the reduced occupancy of the CO bound to heme. Recent experiments with carbonmonoxymyoglobin crystals have shown that laser irradiation induces photolysis of the Fe–CO bond, resulting in CO becoming trapped in alternative positions in both the proximal and distal pockets (Brunori et al., 2000; Chu et al., 2000; Ostermann et al., 2000). In these cases, the CO migrates to specific sites but does not hydrogen bond to the protein.
Of the two ligand-bound Axcyt c′ structures presented here, the CO-bound form differs the most with respect to the reduced structure after least squares superposition (Table I). The overall r.m.s. deviations were 0.15 and 0.60 Å for the NO- and CO-bound models, respectively. CO binding in other cyt c′ has been shown to induce dimer dissociation (Ren et al., 1993). This has not been reported for Axcyt c′, although these differences could indicate that such a dissociation might occur in this protein outside the constraints of the crystal lattice. However, the differences did not exceed 1.2 Å for main chain atoms and were largely restricted to the N-terminus and the AB, BC and CD loops, whilst the dimer interface remained virtually unchanged.
The ligand-bound structures presented here are not the first for a cyt c′. The structure of reduced Rhodobacter capsulatus cyt c′ (Rccyt c′) with n-butylisocyanide (BIC) bound to the heme has been determined at 2.4 Å resolution (Tahirov et al., 1996a). This structure has much in common with the CO-bound Axcyt c′ structure. First, the heme is 6-coordinate, with the ligand binding in the distal pocket with an average Fe–C–N angle of 165° (there are two subunits per asymmetric unit). This binding displaces Phe14 (equivalent to Leu16 in Axcyt c′), which mediates access to the sixth coordination site. Furthermore, this movement pushes pyrrole ring A of the heme upward into the proximal pocket and the Fe moves back into the porphyrin plane. In this BIC-bound structure, Arg126 (equivalent to Arg124 in Axcyt c′) is parallel to the plane of the imidazole ring of the proximal His and perpendicular to the heme plane. This contrasts with its position in the ligand-free oxidized Rccyt c′ structure, where it stacks against the heme plane (Tahirov et al., 1996b), as it does in Axcyt c′. The authors inferred that this side chain flipping is attributable to the effects of BIC binding (Tahirov et al., 1996a), whereas our Axcyt c′ structures indicate that this is simply the result of the transition from an oxidized to a reduced state. The side chain is merely shifted upward by <1 Å upon CO binding to the ferrous heme of Axcyt c′ (Figure 2). The effects of BIC binding in Rccyt c′ are more widespread than those of CO binding in Axcyt c′, possibly because a larger ligand is involved. In particular, at least three other residues in the distal pocket become re-oriented and propionate group A flips around such that it resides on the proximal side of the heme (Tahirov et al., 1996a). This results in the loss of a water molecule that was hydrogen bonded between the two propionate groups in the oxidized Rccyt c′ structure. This water is structurally conserved in several other cyt c′ structures and has been suggested to play an important functional role (Tahirov et al., 1996a). In contrast, this water is retained in all four Axcyt c′ structures reported here.
Steric constraints within the distal heme pocket most likely favor linearly bound adducts such as CO and discriminate against those ligands that prefer a bent configuration, like O2 and NO. O2 binding needs the proximal histidine as a strong electron donor and therefore can only bind to the distal side in 6-coordinate heme complexes. However, the steric hindrence, possibly involving Leu16, may prevent this in Axcyt c′. These constraints could also help to explain the transient nature of the 6-coordinate NO species of Axcyt c′ discussed below.
Structural comparison of the mode of NO binding with other hemoproteins
5-coordinated nitrosyl heme was first discovered in a spectroscopic study of the α-hemes of human deoxyhemoglobin (Szabo and Perutz, 1976). However, previous crystallographic studies of the NO complexes of hemoproteins are restricted to 6-coordinate adducts with the proximal histidine still ligated to the Fe atom. In addition to this mode of binding, in hemoglobin NO has been reported to bind to a Cys residue (β93) outside the heme pocket, to form an S-nitroso adduct (Chan et al., 1998), and is therefore not directly relevant to this discussion. This modification cannot occur in Axcyt c′ as there are no free Cys residues. Significantly, none of these structures has revealed the binding of NO to the proximal face of the heme that we find for Axcyt c′.
The first reported nitrosyl hemoprotein structure was that of ferrous horse hemoglobin (Deatheridge and Moffat, 1979) to a resolution of 2.8 Å. In this structure the ligand was modeled into a difference electron density map with an Fe–NO bond length of 1.7 Å and Fe–N–O angle of 145°, but was not refined. More recently, a ferrous nitrosyl leghemoglobin structure refined at 1.7 Å resolution gave similar values of 1.7 Å and 147°, respectively, for the Fe–NO bond length and the Fe–N–O angle (Harutyunyan et al., 1996), whilst average values of 1.8 Å and 127° were obtained for the Fe–NO bond length and the Fe–N–O angle, respectively, from a 1.8 Å resolution ferrous nitrosyl hemoglobin structure (Chan et al., 1998). In contrast, a 1.7 Å resolution structure of ferrous nitrosyl myoglobin (Brucker et al., 1998) showed a very acute Fe–N–O angle of 112° with an Fe–NO distance of 1.9 Å. In the latter study, the authors postulated that this acute angle may have been influenced by the strength of the proximal bond, hydrogen bonding interactions between the ligand and the distal histidine and, possibly, crystal packing forces.
Nitrophorins are a class of NO transport proteins found in the saliva of blood-feeding insects and function as vasodilators and anticoagulants (Weichsel et al., 2000). The Fe is 5-coordinate with a His residue as the proximal ligand and spectroscopic studies indicate that both the ferric and ferrous oxidation states of nitrophorins bind NO. However, the ferric form of the heme in these proteins is stabilized in some way to prevent autoreduction by NO, allowing the freely reversible binding of NO from this 6-coordinate species. The crystal structure of the NO- complexed form of nitrophorin 1 has been determined at 2.3 Å resolution (Ding et al., 1999). The structure contains two molecules per asymmetric unit and shows the Fe–NO moiety to be bent by an average of 130° with an Fe–NO bond length of 2.0 Å. Although formally ferric, since the X-ray data were collected at ambient temperature, it is possible that the heme Fe may have been photo-reduced during the experiment, thereby accounting for the acuteness of the Fe–N–O angle. Very recently, a 1.4 Å resolution crystal structure of the 6-coordinate NO adduct of nitrophorin 4 was presented in which the NO was discretely disordered (Weichsel et al., 2000). In one conformation, the Fe–N–O bond angle was 177° with an Fe–NO bond length of 1.5 Å, whilst in the other the Fe–N–O bond angle was 110° with an Fe–NO bond length of 2.6 Å. Rather than a mixture of ferric and ferrous states of the heme Fe, the authors interpreted this as a mixture of bound and unbound or loosely ligated NO molecules.
The cd1 heme-containing nitrite reductase of Thiosphaera pantoteopha forms NO as a product of nitrite reduction and has been studied by time-resolved crystallography. The catalytic site is a 6-coordinate d1 heme with axial ligands His and Tyr in the resting state. During catalysis, the Tyr residue is displaced on reduction of the heme by internal electron transfer from the c heme. Nitrite is N-bonded to the reduced d1 heme and is reduced to NO, which has an Fe–N–O bond angle of 131° and an Fe–NO bond length of 2.0 Å in a structure refined at 1.8 Å resolution (Williams et al., 1997). Spectroscopic measurements of the nitrite reductase crystals indicated that the heme–NO-bound product is in the ferrous oxidation state. In structural studies on the same enzyme from Pseudomonas aeruginosa, the d1 heme in the resting enzyme was not directly ligated by a Tyr, but is linked to one via a hydroxide ion (Nurizzo et al., 1998). In the structure of the NO-bound form of this enzyme determined at 2.65 Å resolution, both the hydroxide ion and the Tyr are displaced, and the NO binds with an Fe–N–O angle of 135° and an Fe–NO distance of 1.8 Å.
In a comparative study of diatomic ligand binding to cytochrome c peroxidase, the NO-bound structure of the enzyme was determined at 1.85 Å resolution (Edwards and Poulos, 1990) and represents another example of a nitrosyl hemoprotein structure containing a discretely disordered ligand. In this case, the Fe–N–O angles of the alternative conformers differ by only 10°, being 125 and 135°, respectively, and are separated from the heme Fe by a distance of 1.8 Å. Although the heme Fe was formally ferric, it may have been photo-reduced during the X-ray data collection because the experiment was performed at ambient temperature. It seems unlikely that the bent configuration results entirely from steric clashes with the protein environment, since in the CO adduct of the enzyme the ligand is linear (Edwards and Poulos, 1990).
The crystal structure of the NO-bound form of the oxygen sensor FixL was recently determined at 2.5 Å resolution (Gong et al., 2000). In this model, a value of 146° for the Fe–N–O angle was reported, but the Fe–NO distance was not given. Although the crystal was not chemically reduced prior to addition of NO, the authors suggested that the heme Fe subsequently became photo-reduced in the X-ray beam.
Clearly, there is significant variation in the ligand binding geometries of nitrosyl hemoprotein complexes. In fact, the ranges spanned by the two alternative NO conformations observed in a single structure, that of nitrophorin 4, completely encompass all other reported values (Weichsel et al., 2000). However, as one of these conformations may represent only a loosely bound state, these values will not be considered further. The Fe–NO bond length of 2.0 Å seen in Axcyt c′ falls at the upper end of the range of values reported above (1.7–2.0 Å). With the exception of the myoglobin (Brucker et al., 1998) structure, in the remaining ferrous heme complexes the Fe–N–O angles fall within the range 125–147°, as compared with the average value of 128° we observe for Axcyt c′. This is to be expected because when the Fe is ferrous, NO is an electron acceptor and, consequently, electron back-donation from the Fe to the NO through a π orbital is favorable, resulting in a highly bent NO coordination. Conversely, when the Fe is ferric, the bound NO donates an electron to the Fe through a σ orbital, favoring a more linear coordination, as seen in the nitric oxide reductase ferric NO complex, which has an Fe–N–O angle of 161° (Shimizu et al., 2000). It should not be forgotten, however, that the protein environment also affects the ligand binding geometry. It is likely that the difference in energy between the bent and linear binding modes is small enough that other effects, such as steric hindrance, electrostatic interactions and crystal packing forces, exert a significant influence on the orientation of the ligand. For a discussion of the comparison of Fe–NO distances and angles derived from multiple scattering analysis of XAFS data with those derived from crystallographic studies of several hemoproteins and model compounds, see Rich (1998).
Biological implications of proximal NO binding to heme
The two main findings of our structural studies on Axcyt c′ are: (i) NO binds to the proximal face of the heme, following disruption of the His–Fe bond; (ii) CO binds to the distal side of the heme with retention of the trans histidine ligand. Mechanistic analyses of the heme-based gas sensors sGC and CooA have been hindered to date by the absence of crystal structures. The environment of the heme and the potential conformational changes that have been proposed to occur on heme–ligand binding have only been investigated by spectroscopic techniques. These proteins share many of the spectroscopic and ligand-binding properties of cyt c′, which are unusual compared with other ferrohemoproteins. All three show similar spectral changes on binding CO or NO to the reduced heme. The strong similarities of the unusual spectroscopic properties of cyt c′ and sGC have been noted previously (Stone and Marletta, 1994). On binding of NO to these proteins, the Soret band is blue shifted and decreases in intensity, and the α and β bands shift in position and become equal in intensity (Table II). Despite the dogma that exogenous ligands coordinate to the heme centers only from the distal pocket, the possibility that NO and CO could bind to opposite faces of the heme to form stable adducts has recently been raised for CooA (Reynolds et al., 2000), but has yet to be proven. Moreover, in an earlier study of the reductive nitrosylation of ferric NO– hemoglobin, a transient dinitrosyl ferric heme intermediate was postulated (Addison and Stephanos, 1986). The structures we describe have implications for NO binding to sGC and the discrimination of CO and NO by both sGC and CooA.
Table II. Comparison of electronic absorption spectral parameters of Axcyt c′ with CooA and sGC.
| Soret (nm) | β (nm) | α (nm) | Reference | |
|---|---|---|---|---|
| Fe(II)Axcyt c′ | 426, 434a | ∼550 | ∼570 | Cusanovich et al. (1970) |
| Fe(II)CooA | 425 | ∼540 | ∼570 | Reynolds et al. (2000) |
| Fe(II)sGC | 431 | 555b | 555b | Stone and Marletta (1994) |
| Fe(II)Axcyt c′–NO | 397, 415a | 541 | 565a | Yoshimura et al. (1986) |
| Fe(II)CooA–NO | 399 | 544 | 572 | Reynolds et al. (2000) |
| Fe(II)sGC–NO | 398 | 537 | 572 | Stone and Marletta (1994) |
aShoulder.
bPeaks not resolved.
CooA
The photosynthetic bacterium R.rubrum can grow using the energy associated with the oxidation of CO to CO2 as the sole energy source (He et al., 1996). This ability is controlled by CooA, which is a hemoprotein member of the cAMP receptor family of single-component regulatory proteins (Shelver et al., 1997). It has a role in the CO-mediated control of transcription of the coo operon, which enables CO-dependent growth of R.rubrum. In addition to CO, CooA binds NO, but does not bind a number of other potential heme probes. The binding of CO, but not NO, allows CooA to bind to its target DNA (Reynolds et al., 2000). The interaction of CooA with CO and NO has been studied to provide insight as to how the heme coordination environment modulates the conformational change that confers this specificity of interaction with DNA. Spectroscopic studies have shown that NO binds to reduced CooA to form a 5-coordinate adduct, in contrast to CO, which forms a 6-coordinate species. Since in CooA as isolated the heme is 6-coordinate, CO must displace one of the ligands, and in this respect differs from cyt c′.
Guanylate cyclase
The sGC of bovine lung is a heterodimer that binds one protoporphyrin IX and catalyzes the formation of cGMP from GTP (Hoenicka et al., 1999). The NO-sensing domain is localized in the N-terminal region of the β subunit (Zhao et al., 1998), which regulates activity of the catalytic site located at the interface of the α and β subunits. UV-vis and resonance Raman spectroscopy of the resting enzyme indicate that the enzyme can be isolated in two forms: a 6-coordinate species with bis-His ligation and a 5-coordinate species with mono-His ligation (Vogel et al., 1999). The 5-coordinate species is a high spin histidyl system resembling those of myoglobin and hemoglobin. However, significant differences are observed in the reactivity of O2 with sGC compared with hemoglobin, since O2 is reported not to bind to sGC even under 1 atmosphere pressure (Stone and Marletta, 1994). The lack of a stable oxy adduct allows excess NO to bind to the ferrous deoxy form of sGC under aerobic conditions, unlike ferrous oxyhemoglobin, where NO binding results in the formation of nitrate and methemoglobin, although the physiological relevance of this reaction has recently been questioned (Gow et al., 1999).
The enzymatic activity of sGC is stimulated some 200-fold when NO is bound to the heme, and activation has been proposed to arise from a conformational change in the catalytic domain. The binding of CO results in a 5-fold stimulation of activity, although in the presence of YC-1, a xenobiotic benzylimidazole derivative, CO can be as effective an activator as NO (Sharma and Magde, 1999). Extensive spectroscopic studies have shown that when NO binds, the His–Fe bond is cleaved and a 5-coordinate species is formed. In contrast, when CO binds to sGC, the His–Fe bond remains intact and a 6-coordinate species is formed (Vogel et al., 1999).
It has been proposed that the environment of the heme favors the binding of NO and CO to the sterically hindered distal face of the heme, trans to the proximal histidine residue (Zhao et al., 1998). The chemical basis for the different effects of NO and CO on sGC activation has been attributed to the opposite nature of their trans effects. Thus, studies on model porphyrins have shown that the affinity of heme for CO increases with a basic ligand at the trans position, whereas the affinity for NO is greater when the fifth position is vacant (Traylor and Sharma, 1992). This so-called repulsive trans effect of NO, together with the intrinsic weakness of the Fe–His bond in sGC, is believed to account for cleavage of the heme proximal linkage.
The current models for NO and CO binding to the heme of sGC are dominated by the assumption that these ligands always bind on the distal face of the heme (Zhao et al., 1998), the so-called ‘distal dogma’. As a consequence of this, the binding of exogenous imidazole to a 5-coordinate NO–sGC complex has been proposed to occur on the proximal side and involve a conformational change in the protein (Zhao et al., 1998). Given the results presented here, the binding of imidazole to the distal face of the heme in the NO–sGC complex is a distinct possibility.
Kinetic studies on the binding of NO to ferrous sGC have shown that formation of the 5-coordinate product involves a transient 6-coordinate species (Makino et al., 1999; Zhao et al., 1999). In the 6-coordinate species, the trans effect of NO is assumed to facilitate displacement of histidine to form the 5-coordinate product. An unexpected and intriguing finding of the kinetic experiments was the NO concentration dependence of the 6- to 5-coordination conversion required for enzyme activation. It has been suggested as one possibility that attack by a second NO occurs on the proximal face of the 6-coordinate NO complex, but it was still assumed that NO must finally reside on the distal side in the 5-coordinate adduct (Zhao et al., 1999). We have observed similar interconversion of 6- to 5-coordinated species and an NO concentration dependence for these reactions of Axcyt c′ with NO (S.J.George and C.R.Andrew, unpublished observations). Our demonstration that NO can bind on the proximal side of the heme provides a ready explanation for this dependence, if a second NO molecule were to attack on the proximal side, to displace both the histidine residue and the distally bound NO. The mode of binding we observe also has implications for the differences in activation of sGC by NO and CO. The predominant model to explain the differences in activation by NO and CO relies on the fact that the proximal Fe–His bond is severed in the reaction with NO (but not CO). Our discovery that NO can bind on the opposite side of the heme to that of CO provides an additional structural mechanism for the differences in activation and also accounts for the inability of CO to inhibit activation of sGC by NO (Vogel et al., 1999).
Materials and methods
Sample preparation
Axcyt c′ was purified and crystallized using procedures similar to those described previously (Ambler, 1973; Dobbs et al., 1996). The protein was concentrated to ∼8 mg/ml in 20 mM Tris–HCl pH 7.2, and filtered through a 0.1 µM Ultrafree centrifugal filter (Millipore) prior to use. All crystallizations were performed under aerobic conditions at 4°C by hanging drop vapor diffusion from a mother liquor consisting of 55–65% saturated (NH4)2SO4 in 100 mM HEPES pH 7.5. The crystals belonged to space group P6522, with approximate cell parameters a = b = 53 Å, c = 182 Å (when measured at 100 K), and were cryoprotected in mother liquor to which 25–30% (v/v) glycerol had been added in place of buffer before flash cooling to 100 K. For the oxidized data set, other than the cryoprotection, the crystals were not pre-treated at all. For all subsequent experiments, the crystals were handled under anaerobic conditions after being reduced using mother liquor containing 20 mM sodium dithionite. In all cases, the final step was a short soak in argon-sparged cryoprotectant prior to flash cooling. No further manipulations were performed for the ‘reduced’ crystal. For the NO-bound data set, a reduced crystal was incubated for 6 days in mother liquor saturated with NO (∼2 mM), whilst for the CO-bound data set a reduced crystal was incubated for 6 days in mother liquor saturated with CO (∼1 mM).
X-ray data collection and molecular replacement
X-ray data were recorded at either the Synchrotron Radiation Source (SRS; Daresbury, UK) or the European Synchrotron Radiation Facility (ESRF; Grenoble, France). The data were processed using DENZO and SCALEPACK (Otwinowski and Minor, 1997) and all subsequent downstream processing and statistical analysis were effected using programs from the CCP4 suite (CCP4, 1994) unless stated otherwise. Data collection and processing statistics are summarized in Table I. All structures were solved by molecular replacement using the 1.8 Å resolution ambient temperature structure of Axcyt c′ (Dobbs et al., 1996) as the search model (PDB accession code 1CGO). In all cases, molecular replacement gave a single clear peak consistent with one subunit per asymmetric unit.
Refinement and model building
In all data sets, an equivalent subset of the data comprising 5% of the reflections was set aside for the calculation of ‘free’ (Rfree) crystallographic R-factors (Brünger, 1993) during model refinement. Throughout refinement, neither low resolution nor amplitude cut-offs were applied. Simulated annealing refinement of positional parameters of the molecular replacement solutions was performed with the program X-PLOR (Brünger, 1992) from a starting temperature of 4000 K. Both positional and thermal parameters of the models were subsequently refined using REFMAC. The Fe atoms of the heme groups were refined with anisotropic thermal parameters, and geometrical restraints to the bonds between the Fe and its various ligands were explicitly switched off. Final model parameters are summarized in Table I. Model building was performed by interactive computer graphics using the program O (Jones et al., 1991) by inspection of 2mFobs – dFcalc and mFobs – dFcalc Fourier electron density maps. Omit difference maps were produced as follows. To reduce the effects of model bias, small random shifts were applied to all coordinates of the final models and all the temperature factors were reset to the overall average for each structure. Then the atoms of interest were omitted from their respective models before subjecting them to five cycles of restrained refinement in REFMAC. Maps with coefficients mFobs – dFcalc were then calculated and inspected for residual positive electron density.
Coordinates
The X-ray coordinates for the oxidized, reduced, NO-bound and CO-bound Axcyt c′ structures have been deposited in the Protein Data Bank with accession codes 1E83–1E86.
Acknowledgments
Acknowledgements
We are grateful for support and access to the SRS in Daresbury and the ESRF in Grenoble and would particularly like to thank beamline scientists P.Rizkallah and R.Kehoe (SRS) and W.Burmeister, S.McSweeney, E.Mitchell and J.-L.Ferrer (ESRF). L.Delarbre, S.Mayer and L.Mitchenall are acknowledged for assistance during X-ray data collection. We are indebted to B.Smith for critically reading this manuscript. This work was funded by the BBSRC as part of the competitive strategic grant to the John Innes Centre.
References
- Addison A.W. and Stephanos,J.J. (1986) Nitrosyliron(III) hemoglobin: autoreduction and spectroscopy. Biochemistry, 25, 4104–4113. [DOI] [PubMed] [Google Scholar]
- Ambler R.P. (1973) The amino acid sequence of cytochrome c′ from Alcaligenes sp. NCIMB 11015. Biochem. J., 135, 751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brucker E.A., Olson,J.S., Ikeda-Saito,M. and Phillips,G.N.,Jr (1998) Nitric oxide myoglobin: crystal structure and analysis of ligand geometry. Proteins, 30, 352–356. [PubMed] [Google Scholar]
- Brünger A.T. (1992) X-PLOR: Version 3.1. A System for X-ray Crystallography and NMR. Yale University Press, New Haven, CT. [Google Scholar]
- Brünger A.T. (1993) Assessment of phase accuracy by cross validation—the free R-value—methods and applications. Acta Crystallogr. D, 49, 24–36. [DOI] [PubMed] [Google Scholar]
- Brunori M., Vallone,B., Cutruzzola,F., Travaglini-Allocatelli,C., Berendzen,J., Chu,K., Sweet,R.M. and Schlichting,I. (2000) The role of cavities in protein dynamics: crystal structure of a photolytic intermediate of a mutant myoglobin. Proc. Natl Acad. Sci. USA, 97, 2058–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chan N.L., Rogers,P.H. and Arnone,A. (1998) Crystal structure of the S-nitroso form of liganded human hemoglobin. Biochemistry, 37, 16459–16464. [DOI] [PubMed] [Google Scholar]
- Chu K., Vojtchovsky,J., McMahon,B.H., Sweet,R.M., Berendzen,J. and Schlichting,I. (2000) Structure of a ligand-binding intermediate in wild-type carbonmonoxy myoglobin. Nature, 403, 921–923. [DOI] [PubMed] [Google Scholar]
- Cruickshank D.W.J. (1999) Remarks about protein structure precision. Acta Crystallogr. D, 55, 583–601. [DOI] [PubMed] [Google Scholar]
- Cusanovich M.A., Tedro,S.M. and Kamen,M.D. (1970) Pseudomonas denitrificans cytochrome cc′. Arch. Biochem. Biophys., 141, 557–570. [DOI] [PubMed] [Google Scholar]
- Deatheridge J.F. and Moffat,K. (1979) Stucture of nitric oxide hemoglobin. J. Mol. Biol., 134, 401–417. [DOI] [PubMed] [Google Scholar]
- Denninger J.W. and Marletta,M.A. (1999) Guanylate cyclase and the NO/cGMP signaling pathway. Biochim. Biophys. Acta, 1411, 334–350. [DOI] [PubMed] [Google Scholar]
- Ding X.D., Weichsel,A., Andersen,J.F., Shokhireva,T.K., Balfour,C., Pierik,A.J., Averill,B.A., Montfort,W.R. and Walker,F.A. (1999) Nitric oxide binding to the ferri- and ferroheme states of nitrophorin 1, a reversible NO-binding heme protein from the saliva of the blood-sucking insect, Rhodnius prolixus. J. Am. Chem. Soc., 121, 128–138. [Google Scholar]
- Dobbs A.J., Anderson,B.F., Faber,H.R. and Baker,E.N. (1996) Three-dimensional structure of cytochrome c′ from two Alcaligenes species and the implications for four-helix bundle structures. Acta Crystallogr. D, 52, 356–368. [DOI] [PubMed] [Google Scholar]
- Doyle M.L., Gill,S.J. and Cusanovich,M.A. (1986) Ligand controlled dissociation of Chromatium vinosum cytochrome c′. Biochemistry, 25, 2509–2516. [DOI] [PubMed] [Google Scholar]
- Edwards S.L. and Poulos,T.L. (1990) Ligand binding and structural perturbations in cytochrome c peroxidase. A crystallographic study. J. Biol. Chem., 265, 2588–2595. [PubMed] [Google Scholar]
- Gong W., Hao,B., Mansy,S., Gonzalez,G., Gilles-Gonzalez,M.A. and Chan,M.K. (1998) Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc. Natl Acad. Sci. USA, 95, 15177–15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W., Hao,B. and Chan,M.K. (2000) New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL. Biochemistry, 39, 3955–3962. [DOI] [PubMed] [Google Scholar]
- Gow A.J., Luchsinger,B.P., Pawloski,J.R., Singel,D.J. and Stamler,J.S. (1999) The oxyhemoglobin reaction of nitric oxide. Proc. Natl Acad. Sci. USA, 96, 9027–9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harutyunyan E.H., Safonova,T.N., Kuranova,I.P., Popov,A.N., Teplyakov,A.V., Obmolova,G.V., Valnshtein,B.K., Dodson,G.G. and Wilson,J.C. (1996) The binding of carbon monoxide and nitric oxide to leghaemoglobin in comparison with other haemoglobins. J. Mol. Biol., 264, 152–161. [DOI] [PubMed] [Google Scholar]
- He Y., Shelver,D., Kerby,R.L. and Roberts,G.P. (1996) Characterization of a CO-responsive transcriptional activator from Rhodospirillum rubrum. J. Biol. Chem., 271, 120–123. [DOI] [PubMed] [Google Scholar]
- Hoenicka M., Becker,E.M., Apeler,H., Sirichoke,T., Schroder,H., Gerzer,R. and Stasch,J.P. (1999) Purified soluble guanylyl cyclase expressed in a baculovirus/Sf9 system: stimulation by YC-1, nitric oxide and carbon monoxide. J. Mol. Med., 77, 14–23. [DOI] [PubMed] [Google Scholar]
- Hou S., Larsen,R.W., Boudko,D., Riley,C.W., Karatan,E., Zimmer,M., Ordal,G.W. and Alam,M. (2000) Myoglobin-like aerotaxis transducers in Archaea and Bacteria. Nature, 403, 540–544. [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Yoshimura,T., Suzuki,S. and Shidara,S. (1991) Spectral properties of Achromobacter xylosoxidans cytochromes c′ and their NO complexes. Biochim. Biophys. Acta, 1058, 79–82. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. D, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Laskowski R.A., Macarthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Makino R., Matsuda,H., Obayashi,E., Shiro,Y., Iizuka,T. and Hori,H. (1999) EPR characterization of axial bond in metal center of native and cobalt-substituted guanylate cyclase. J. Biol. Chem., 274, 7714–7723. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Moir J.W.B. (1999) Cytochrome c′ from Paracoccus denitrificans: spectroscopic studies consistent with a role for the protein in nitric oxide metabolism. Biochim. Biophys. Acta, 1430, 65–72. [DOI] [PubMed] [Google Scholar]
- Nurizzo D., Cutruzzola,F., Arese,M., Bourgeois,D., Brunori,M., Cambillau,C. and Tegoni,M. (1998) Conformational changes occurring upon reduction and NO binding in nitrite reductase from Pseudomonas aeruginosa. Biochemistry, 37, 13987–13996. [DOI] [PubMed] [Google Scholar]
- Ostermann A., Waschipky,R., Parak,F.G. and Nienhaus,G.U. (2000) Ligand binding and conformational motions in myoglobin. Nature, 404, 205–208. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Poulos T.L. and Kraut,J. (1980) The stereochemistry of peroxidase catalysis. J. Biol. Chem., 255, 8199–8205. [PubMed] [Google Scholar]
- Ren Z., Meyer,T. and McRee,D.E. (1993) Atomic structure of a cytochrome c′ with an unusual ligand-controlled dimer dissociation at 1.8 Å resolution. J. Mol. Biol., 234, 433–445. [DOI] [PubMed] [Google Scholar]
- Reynolds M.F., Parks,R.B., Burstyn,J.N., Shelver,D., Thorsteinsson,M.V., Kerby,R.L., Roberts,G.P., Vogel,K.M. and Spiro,T.G. (2000) Electronic absorption, EPR and resonance Raman spectroscopy of CooA, a CO-sensing transcription activator from R. rubrum, reveals a five-coordinate NO-heme. Biochemistry, 39, 388–396. [DOI] [PubMed] [Google Scholar]
- Rich A.M. (1998) Determination of the Fe–ligand bond lengths and Fe–N–O bond angles in horse heart and ferrous nitrosylmyoglobin using multiple-scattering XAFS analyses. J. Am. Chem. Soc., 120, 10827–10836. [Google Scholar]
- Rodgers K.R. (1999) Heme-based sensors in biological systems. Curr. Opin. Chem. Biol., 3, 158–167. [DOI] [PubMed] [Google Scholar]
- Sharma V.S. and Magde,D. (1999) Activation of soluble guanylate cyclase by carbon monoxide and nitric oxide: a mechanistic model. Methods, 19, 494–505. [DOI] [PubMed] [Google Scholar]
- Shelver D., Kerby,R.L., He,Y. and Roberts,G.P. (1997) CooA, a CO-sensing transcription factor from Rhodospirillum rubrum, is a CO-binding heme protein. Proc. Natl Acad. Sci. USA, 94, 11216–11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N., Iba,S., Misaki,S., Meyer,T.E., Bartsch,R.G., Cusanovich,M.A., Morimoto,Y., Higuchi,Y. and Yasuoka,N. (1998) Basis for monomer stabilization in Rhodopseudomonas palustris cytochrome c′ derived from the crystal structure. J. Mol. Biol., 284, 751–760. [DOI] [PubMed] [Google Scholar]
- Shimizu H., Obayashi,E., Gomi,Y., Arakawa,H., Park,S.Y., Nakamura,H., Adachi,S., Shoun,H. and Shiro,Y. (2000) Proton delivery in NO reduction by fungal nitric-oxide reductase. Cryogenic crystallography, spectroscopy and kinetics of ferric–NO complexes of wild-type and mutant enzymes. J. Biol. Chem., 275, 4816–4826. [DOI] [PubMed] [Google Scholar]
- Stone J.R. and Marletta,M.A. (1994) Soluble guanylate cyclase from bovine lung: activation with nitric oxide and carbon monoxide and spectral characterization of the ferrous and ferric states. Biochemistry, 33, 5636–5640. [DOI] [PubMed] [Google Scholar]
- Szabo A. and Perutz,M.F. (1976) Equilibrium between six- and five-coordinated hemes in nitrosylhemoglobin: interpretation of electron spin resonance spectra. Biochemistry, 15, 4427–4428. [DOI] [PubMed] [Google Scholar]
- Tahirov T.H., Misaki,S., Meyer,T.E., Cusanovich,M.A., Higuchi,Y. and Yasuoka,N. (1996a) Concerted movement of side chains in the haem vicinity observed on ligand binding in cytochrome c′ from Rhodobacter capsulatus. Nature Struct. Biol., 3, 459–464. [DOI] [PubMed] [Google Scholar]
- Tahirov T.H., Misaki,S., Meyer,T.E., Cusanovich,M.A., Higuchi,Y. and Yasuoka,N. (1996b) High resolution crystal structures of two polymorphs of cytochrome c′ from the purple phototrophic bacterium Rhodobacter capsulatus. J. Mol. Biol., 259, 467–479. [DOI] [PubMed] [Google Scholar]
- Traylor T.G. and Sharma,V.S. (1992) Why NO? Biochemistry, 31, 2847–2849. [DOI] [PubMed] [Google Scholar]
- Vogel K.M., Hu,S.Z., Spiro,T.G., Dierks,E.A., Yu,A.E. and Burstyn,J.N. (1999) Variable forms of soluble guanylyl cyclase: protein–ligand interactions and the issue of activation by carbon monoxide. J. Biol. Inorg. Chem., 4, 804–813. [DOI] [PubMed] [Google Scholar]
- Weichsel A., Andersen,J.F., Roberts,S.A. and Montfort,W.R. (2000) Nitric oxide binding to nitrophorin 4 induces complete distal pocket burial. Nature Struct. Biol., 7, 551–554. [DOI] [PubMed] [Google Scholar]
- Williams P.A., Fulop,V., Garman,E.F., Saunders,N.F.W., Ferguson,S.J. and Hajdu,J. (1997) Haem-ligand switching during catalysis in crystals of a nitrogen-cycle enzyme. Nature, 389, 406–412. [DOI] [PubMed] [Google Scholar]
- Yoshimura T., Suzuki,S., Nakahara,A., Iwasaki,H., Masuko,M. and Matsubara,T. (1986) Spectral properties of nitric oxide complexes of cytochrome c′ from Alcaligenes sp. NCIMB 11015. Biochemistry, 25, 2436–2442. [Google Scholar]
- Zhao Y.D., Hoganson,C., Babcock,G.T. and Marletta,M.A. (1998) Structural changes in the heme proximal pocket induced by nitric oxide binding to soluble guanylate cyclase. Biochemistry, 37, 12458–12464. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Brandish,P.E., Ballou,D.P. and Marletta,M.A. (1999) A molecular basis for nitric oxide sensing by soluble guanylate cyclase. Proc. Natl Acad. Sci. USA, 96, 14753–14758. [DOI] [PMC free article] [PubMed] [Google Scholar]