Abstract
Developing a new drug from original idea to the launch of a finished product is a complex process which can take 12–15 years and cost in excess of $1 billion. The idea for a target can come from a variety of sources including academic and clinical research and from the commercial sector. It may take many years to build up a body of supporting evidence before selecting a target for a costly drug discovery programme. Once a target has been chosen, the pharmaceutical industry and more recently some academic centres have streamlined a number of early processes to identify molecules which possess suitable characteristics to make acceptable drugs. This review will look at key preclinical stages of the drug discovery process, from initial target identification and validation, through assay development, high throughput screening, hit identification, lead optimization and finally the selection of a candidate molecule for clinical development.
Keywords: drug discovery, high throughput screening, target identification, target validation, hit series, assay development, screening cascade, lead optimization
Introduction
A drug discovery programme initiates because there is a disease or clinical condition without suitable medical products available and it is this unmet clinical need which is the underlying driving motivation for the project. The initial research, often occurring in academia, generates data to develop a hypothesis that the inhibition or activation of a protein or pathway will result in a therapeutic effect in a disease state. The outcome of this activity is the selection of a target which may require further validation prior to progression into the lead discovery phase in order to justify a drug discovery effort (Figure 1). During lead discovery, an intensive search ensues to find a drug-like small molecule or biological therapeutic, typically termed a development candidate, that will progress into preclinical, and if successful, into clinical development (Figure 2) and ultimately be a marketed medicine.
Figure 1.
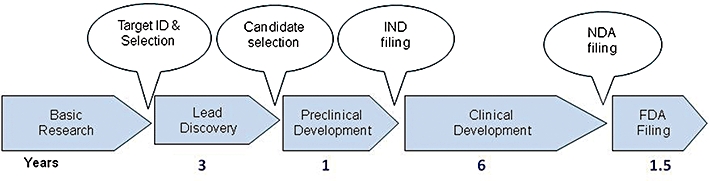
Drug discovery process from target ID and validation through to filing of a compound and the approximate timescale for these processes. FDA, Food and Drug Administration; IND, Investigational New Drug; NDA, New Drug Application.
Figure 2.
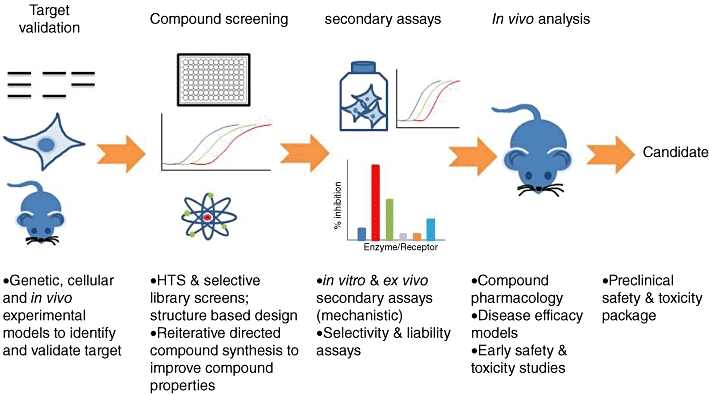
Overview of drug discovery screening assays.
Target identification
Drugs fail in the clinic for two main reasons; the first is that they do not work and the second is that they are not safe. As such, one of the most important steps in developing a new drug is target identification and validation. A target is a broad term which can be applied to a range of biological entities which may include for example proteins, genes and RNA. A good target needs to be efficacious, safe, meet clinical and commercial needs and, above all, be ‘druggable’. A ‘druggable’ target is accessible to the putative drug molecule, be that a small molecule or larger biologicals and upon binding, elicit a biological response which may be measured both in vitro and in vivo. It is now known that certain target classes are more amenable to small molecule drug discovery, for example, G-protein-coupled receptors (GPCRs), whereas antibodies are good at blocking protein/protein interactions. Good target identification and validation enables increased confidence in the relationship between target and disease and allows us to explore whether target modulation will lead to mechanism-based side effects.
Data mining of available biomedical data has led to a significant increase in target identification. In this context, data mining refers to the use of a bioinformatics approach to not only help in identifying but also selecting and prioritizing potential disease targets (Yang et al., 2009). The data which are available come from a variety of sources but include publications and patent information, gene expression data, proteomics data, transgenic phenotyping and compound profiling data. Identification approaches also include examining mRNA/protein levels to determine whether they are expressed in disease and if they are correlated with disease exacerbation or progression. Another powerful approach is to look for genetic associations, for example, is there a link between a genetic polymorphism and the risk of disease or disease progression or is the polymorphism functional. For example, familial Alzheimer's Disease (AD) patients commonly have mutations in the amyloid precursor protein or presenilin genes which lead to the production and deposition in the brain of increased amounts of the Abeta peptide, characteristic of AD (Bertram and Tanzi, 2008). There are also examples of phenotypes in humans where mutations can nullify or overactivate the receptor, for example, the voltage-gated sodium channel NaV1.7, both mutations incur a pain phenotype, insensitivity or oversensitivity respectively (Yang et al., 2004; Cox et al., 2006).
An alternative approach is to use phenotypic screening to identify disease relevant targets. In an elegant experiment, Kurosawa et al. (2008) used a phage-display antibody library to isolate human monoclonal antibodies (mAbs) that bind to the surface of tumour cells. Clones were individually screened by immunostaining and those that preferentially and strongly stained the malignant cells were chosen. The antigens recognized by those clones were isolated by immunoprecipitation and identified by mass spectroscopy. Of 2114 mAbs with unique sequences they identified 21 distinct antigens highly expressed on several carcinomas, some of which may be useful targets for the corresponding carcinoma therapy and several mAbs which may become therapeutic agents.
Target validation
Once identified, the target then needs to be fully prosecuted. Validation techniques range from in vitro tools through the use of whole animal models, to modulation of a desired target in disease patients. While each approach is valid in its own right, confidence in the observed outcome is significantly increased by a multi-validation approach (Figure 3).
Figure 3.
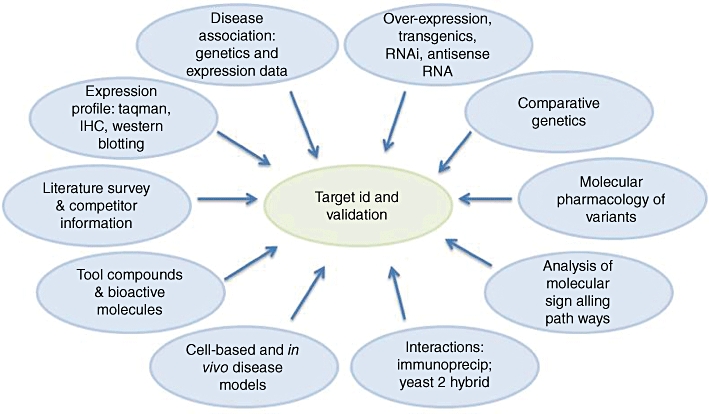
Target ID and validation is a multifunctional process. IHC, immunohistochemistry.
Antisense technology is a potentially powerful technique which utilizes RNA-like chemically modified oligonucleotides which are designed to be complimentary to a region of a target mRNA molecule (Henning and Beste, 2002). Binding of the antisense oligonucleotide to the target mRNA prevents binding of the translational machinery thereby blocking synthesis of the encoded protein. A prime example of the power of antisense technology was demonstrated by researchers at Abbott Laboratories who developed antisense probes to the rat P2X3 receptor (Honore et al., 2002). When given by intrathecal minipump, to avoid toxicities associated with bolus injection, the phosphorothioate antisense P2X3 oligonucleonucleotides had marked anti-hyperalgesic activity in the Complete Freund's Adjuvant model, demonstrating an unambiguous role for this receptor in chronic inflammatory states. Interestingly, after administration of the antisense oligonucleonucleotides was discontinued, receptor function and algesic responses returned. Therefore, in contrast to the gene knockout approach, antisense oligonucleotide effects are reversible and a continued presence of the antisense is required for target protein inhibition (Peet, 2003). However, the chemistry associated with creating oligonucleotides has resulted in molecules with limited bioavailability and pronounced toxicity, making their in vivo use problematic. This has been compounded by non-specific actions, problems with controls for these tools and a lack of diversity and variety in selecting appropriate nucleotide probes (Henning and Beste, 2002).
In contrast, transgenic animals are an attractive validation tool as they involve whole animals and allow observation of phenotypic endpoints to elucidate the functional consequence of gene manipulation. In the early days of gene targeting animals were generated that lacked a given gene's function from inception and throughout their lives. This work yielded great insights into the in vivo functions of a wide range of genes. One such example is through use of the P2X7 knockout mouse to confirm a role for this ion channel in the development and maintenance of neuropathic and inflammatory pain (Chessell et al., 2005). In mice lacking P2X7 receptors, inflammatory and neuropathic hypersensitivity is completely absent to both mechanical and thermal stimuli, while normal nociceptive processing is preserved. These transgenic animals were also used to confirm the mechanism of action for this ablation in vivo as the transgenic mice were unable to release the mature pro-inflammatory cytokine IL-1beta from cells although there was no deficit in IL-1beta mRNA expression. An alternative to gene knockouts are gene knock-ins, where a non-enzymatically functioning protein replaces the endogenous protein. These animals can have a different phenotype to a knockout, for example when the protein has structural as well as enzymatic functions (Abell et al., 2005) and these mice should ostensibly mimic more closely what happens during treatment with drugs, that is, the protein is there but functionally inhibited.
More recently, the desire to be able to make tissue-restricted and/or inducible knockouts has grown. Although these approaches are technically challenging, the most obvious reason for this is the need to overcome embryonic lethality of the homozygous null animals. Other reasons include avoidance of compensatory mechanisms due to chronic absence of a gene-encoded function and avoidance of developmental phenotypes. However, the use of transgenic animals is expensive and time-consuming. So in order to circumvent some of these issues, the use of small interfering RNA (siRNA) has become increasingly popular for target validation. Double-stranded RNA (dsRNA) specific to the gene to be silenced is introduced into a cell or organism, where it is recognized as exogenous genetic material and activates the RNAi pathway. The ribonuclease protein Dicer is activated which binds and cleaves dsRNAs to produce double-stranded fragments of 21–25 base pairs with a few unpaired overhang bases on each end. These short double-stranded fragments are called siRNAs. These siRNAs are then separated into single strands and integrated into an active RNA-induced silencing complex (RISC). After integration into the RISC, siRNAs base-pair to their target mRNA and induce cleavage of the mRNA, thereby preventing it from being used as a translation template (reviewed in Castanotto and Rossi, 2009). However, RNAi technology still has the major problem of delivery to the target cell, but many viral and non-viral delivery systems are currently under investigation (for review see Whitehead et al., 2009).
Monoclonal antibodies are an excellent target validation tool as they interact with a larger region of the target molecule surface, allowing for better discrimination between even closely related targets and often providing higher affinity. In contrast, small molecules are disadvantaged by the need to interact with the often more conserved active site of a target, while antibodies can be selected to bind to unique epitopes. This exquisite specificity is the basis for their lack of non-mechanistic (or ‘off-target’) toxicity – a major advantage over small-molecule drugs.
However, antibodies cannot cross cell membranes restricting the target class mainly to cell surface and secreted proteins. One impressive example of the efficacy of a mAb in vivo is that of the function neutralizing anti-TrkA antibody MNAC13, which has been shown to reduce both neuropathic pain and inflammatory hypersensitivity (Ugolini et al., 2007), thereby implicating NGF in the initiation and maintenance of chronic pain. Finally, the classic target validation tool is the small bioactive molecule that interacts with and functionally modulates effector proteins.
More recently, chemical genomics, a systemic application of tool molecules to target identification and validation has emerged. Chemical genomics can be defined as the study of genomic responses to chemical compounds. The goal is the rapid identification of novel drugs and drug targets embracing multiple early phase drug discovery technologies ranging from target identification and validation, over compound design and chemical synthesis to biological testing. Chemical genomics brings together diversity-oriented chemical libraries and high-information-content cellular assays, along with the informatics and mining tools necessary for storing and analysing the data generated (reviewed in Zanders et al., 2002). The ultimate goal of this approach is to provide chemical tools against every protein encoded by the genome. The aim is to use these tools to evaluate cellular function prior to full investment in the target and commitment to a screening campaign
The hit discovery process
Following the process of target validation, it is during the hit identification and lead discovery phase of the drug discovery process that compound screening assays are developed. A ‘hit’ molecule can vary in meaning to different researchers but in this in review we define a hit as being a compound which has the desired activity in a compound screen and whose activity is confirmed upon retesting. A variety of screening paradigms exist to identify hit molecules (see Table 1). High throughput screening (HTS) involves the screening of the entire compound library directly against the drug target or in a more complex assay system, such as a cell-based assay, whose activity is dependent upon the target but which would then also require secondary assays to confirm the site of action of compounds (Fox et al., 2006). This screening paradigm involves the use of complex laboratory automation but assumes no prior knowledge of the nature of the chemotype likely to have activity at the target protein. Focused or knowledge-based screening involves selecting from the chemical library smaller subsets of molecules that are likely to have activity at the target protein based on knowledge of the target protein and literature or patent precedents for the chemical classes likely to have activity at the drug target (Boppana et al., 2009). This type of knowledge has given rise, more recently, to early discovery paradigms using pharmacophores and molecular modelling to conduct virtual screens of compound databases (McInnes, 2007). Fragment screening involves the generation of very small molecular weight compound libraries which are screened at high concentrations and is typically accompanied by the generation of protein structures to enable compound progression (Law et al., 2009). Finally, a more specialized focused screening approach can also be taken, physiological screening. This is a tissue-based approach and looks for a response more aligned with the final desired in vivo effect as opposed to targeting one specific molecular component.
Table 1.
Screening strategies
| Screen | Description | Comments |
|---|---|---|
| High throughput | Large numbers of compounds analysed in a assay generally designed to run in plates of 384 wells and above | Large compound collections often run by big pharma but smaller compound banks can also be run in either pharma or academia which can help reduce costs. Companies also now trying to provide coverage across a wide chemical space using computer assisted analysis to reduce the numbers of compounds screened. |
| Focused screen | Compounds previously identified as hitting specific classes of targets (e.g. kinases) and compounds with similar structures | Can provide a cheaper avenue to finding a hit molecule but completely novel structures may not be discovered and there may be difficulties obtaining a patent position in a well-covered IP area |
| Fragment screen | Soak small compounds into crystals to obtain compounds with low mM activity which can then be used as building blocks for larger molecules | Can join selected fragments together to fit into the chemical space to increase potency. Requires a crystal structure to be available |
| Structural aided drug design | Use of crystal structures to help design molecules | Often used as an adjunct to other screening strategies within big pharma. In this case usually have docked a compound into the crystal and use this to help predict where modifications could be added to provide increased potency or selectivity |
| Virtual screen | Docking models: interogation of a virtual compound library with the X-ray structure of the protein or, if have a known ligand, as a base to develop further compounds on | Can provide the starting structures for a focused screen without the need to use expensive large library screens. Can also be used to look for novel patent space around existing compound structures |
| Physiological screen | A tissue-based approach fordetermination of the effects of a drug at the tissue rather than the cellular or subcellular level, for example, muscle contractility | Bespoke screens of lower throughput. Aim to more closely mimic the complexity of tissue rather than just looking at single readouts. May appeal to academic experts in disease area to screen smaller number of compounds to give a more disease relevant readout |
| NMR screen | Screen small compounds (fragments) by soaking into protein targets of known crystal or NMR structure to look for hits with low mM activity which can then be used as building blocks for larger molecules | Use of NMR as a structure determining tool |
NMR, nuclear magnetic resonance.
High throughput and other compound screens are developed and run to identify molecules that interact with the drug target, chemistry programmes are run to improve the potency, selectivity and physiochemical properties of the molecule, and data continue to be developed to support the hypothesis that intervention at the drug target will have efficacy in the disease state. It is this series of activities that are the subject of intense activity within the pharmaceutical industry and increasingly within academia to identify candidate molecules for clinical development. Pharmaceutical companies have built large organizations with the objective of identifying targets, assembling compound collections and the associated infrastructure to screen those compounds to identify initially hit molecules from HTS or other screening paradigms and to optimize those screening ‘hits’ into clinical candidates. In recent years the academic sector has become increasingly interested in the activities traditionally performed within the lead discovery phase in the pharmaceutical industry. Academic scientists are now formatting assays for drug discovery which are passed onto academic drug discovery centres for compound screening. These centres, as exemplified by the NIH Roadmap initiative in the USA (Frearson and Collie, 2009), have established compound libraries, screening infrastructure and the appropriate expertise traditionally found within the industrial sector to screen target proteins to identify so-called chemical probes for use in target validation and disease biology studies and increasingly to identify chemical start points for drug discovery programmes. The success of these efforts has been facilitated by the transfer of skills between the industrial and academic sectors.
A typical programme critical path within the lead discovery phase consists of a number of activities and begins with the development of biological assays to be used for the identification of molecules with activity at the drug target. Once developed, such assays are used to screen compound libraries to identify molecules of interest. The output of a compound screen is typically termed a hit molecule, which has been demonstrated to have specific activity at the target protein. Screening hits form the basis of a lead optimization chemistry programme to increase potency of the chemical series at the primary drug target protein. During the lead discovery, phase molecules are also screened in cell-based assays predictive of the disease state and in animal models of disease to characterize both the efficacy of the compound and its likely safety profile (Figure 2). The following paragraphs describe in more detail the requirements and application of compound screening assays within hit and lead discovery.
Assay development
In the recombinant era the majority of assays in use within the industry rely upon the creation of stable mammalian cell lines over-expressing the target of interest or upon the over-expression and purification of recombinant protein to establish so-called biochemical assays although in recent years there has been an increase in the number of reports describing the use of primary cell systems for compound screening (Dunne et al., 2009). Generally, cell-based assays have been applied to target classes such as membrane receptors, ion channels and nuclear receptors and typically generate a functional read-out as a consequence of compound activity (Michelini et al., 2010). In contrast, biochemical assays, which have been applied to both receptor and enzyme targets, often simply measure the affinity of the test compound for the target protein. The relative merits of biochemical and cell-based assays have been debated extensively and have been reviewed elsewhere (Moore and Rees, 2001). Both assay paradigms have been used successfully to identify hit and candidate molecules.
A plethora of assay formats have been enabled to support compound screening. The choice of assay format is dependent upon the biology of the drug target protein, the equipment infrastructure in the host laboratory, the experience of the scientists in that laboratory, whether an inhibitor or activator molecule is sought and the scale of the compound screen. For example compound screening assays at GPCRs have been configured to measure the binding affinity of a radio- or fluorescently labelled ligand to the receptor, to measure guanine nucleotide exchange at the level of the G-protein, to measure compound-mediated changes in one of a number of second messenger metabolites including calcium, cAMP or inositiol phosphates or to measure the activation of downstream reporter genes. Whatever the assay format that is selected, it is a requirement that the following factors are considered:
Pharmacological relevance of the assay. If available, studies should be performed using known ligands with activity at the target under study, to determine if the assay pharmacology is predictive of the disease state and to show that the assay is capable of identifying compounds with the desired potency and mechanism of action.
Reproducibility of the assay. Within a compound screening environment it is a requirement that the assay is reproducible across assay plates, across screen days and, within a programme that may run for several years, across the duration of the entire drug discovery programme.
Assay costs. Compound screening assays are typically performed in microtitre plates. Within academia or for relatively small numbers of compounds assays are typically formatted in 96-well or 384-well microtitre plates whereas in industry or in HTS applications assays are formatted in 384-well or 1536-well microtire plates in assay volumes as small as a few microlitires. In each case assay reagents and assay volumes are selected to minimize the costs of the assay.
Assay quality. Assay quality is typically determined according to the Z' factor (Zhang et al., 1999). This is a statistical parameter that in addition to considering the signal window in the assay also considers the variance around both the high and low signals in the assay. The Z factor has become the industry standard means of measuring assay quality on a plate bases. The Z factor has a range of 0 to 1; an assay with a Z factor of greater than 0.4 is considered appropriately robust for compound screening although many groups prefer to work with assays with a Z factor of greater than 0.6. In addition to the Z factor assay quality is also monitored through the inclusion of pharmacological controls within each assay. Assays are deemed acceptable if the pharmacology of the standard compound(s) falls within predefined limits. Assay quality is affected by many factors. Generally, high-quality assays are created through implementing simple assay protocols with few steps, minimizing wash steps or plate to plate reagent transfers within the assay, through the use of stable reagents and biologicals, and through ensuring that all the instrumentation used to perform the assay is performing optimally. This is typically achieved through developing quality control practices for all items of laboratory automation (see http://www.ncgc.nih.gov/guidance/section2.html#replicate-experiment-study-summary-acceptance).
Effects of compounds in the assay. Chemical libraries are typically stored in organic solvents such as ethanol or dimethyl sulphoxide (DMSO). Thus, assays need to be configured that are not sensitive to the concentrations of solvents used in the assay. Typically, cell-based assays are intolerant to solvent concentrations of greater than 1% DMSO whereas biochemical assays can be performed in solvent concentrations of up to 10% DMSO. Studies are also performed to establish the false negative and false positive hit rates in the assay. If these are unacceptably high then the assay will need to be reconfigured. Finally some consideration should be made to the screening concentration. Compound screening assays for hit discovery are typically run at 1–10 µM compound concentration. At these concentrations compounds with activities of up to 40 µM can be identified. The test concentration can be varied to identify compounds with higher or lower activity.
One example of an HTS technology implemented for the identification of hit molecules with activity at GPCRs is the aequorin assay (Stables et al., 2000). Aequorin is a calcium-sensitive bioluminescent protein cloned from the jellyfish Aequorea victorea. Stable mammalian cell lines have been created transfected to express the GPCR drug target and the aequorin biosensoer. For receptors capable of coupling to heterotrimeric G-proteins of the Gαq/11 family, ligand activation results in an increase in intracellular calcium concentration. When aequorin is expressed in the same cells, this increase in intracellular calcium concentration is detected as a consequence of calcium binding to the aequorin photoprotein, which in the presence of the cofactor coelenterazine, results in the generation of a flash of light that can be detected within a microtitre plate-based luminometer such as the Lumilux™ platform (PerkinElmer, Waltham, MA, USA). The aequorin assay has a very simple protocol and has been developed for HTS in 1536-well plate format in assay volumes of 6 µL and for compound profiling activities in 384-well plate format.
When developing any HTS assay, which can involve the screening of several million molecules over several weeks, it is best practice to screen training sets of compounds to verify that the assay is performing acceptably. Figure 4 shows the screening of a 12 000 compound training set against the histamine H1 receptor expressed in Chinese hamster ovary cells in a 1536-well format HTS assay. The training set is typically run on two or three occasions to identify the hit rate in the assay, the reproducibility of the assay and the false positive and false negative hit rates in the assay. Typically, statistical packages have been developed to identify these parameters. When screened to detect agonist ligands the hit rates in the aequorin assay are typically less than 0.5% of compounds screened with a statistical assay cut-off of 5% or less of the agonist signal seen with a standard agonist ligand. In this assay format false positive and false negative hit rates are very low. For antagonist screening the hit rate in the aequorin assay is typically of 2–3% of compounds screened with an activity cut-off of greater than 25% inhibition. This is a common phenomenon of all screening assays. Hit rates in antagonist or inhibitor format tend to be higher than hit rates in agonist assays as antagonist assays, which are defined by detection of a decrease in assay signal, will also detect compounds that interfere in signal generation. Following completion of robustness testing an assay moves into HTS. During HTS, up to 200 assay plates are screened each day, often using complex laboratory automation. During the screen, assay performance is measured according to the Z' on the assay plate and the variance in the pharmacology of a standard compound, with assay plates being failed and rescreened if these quality control measures fall outside predefined limits (Figure 5).
Figure 4.
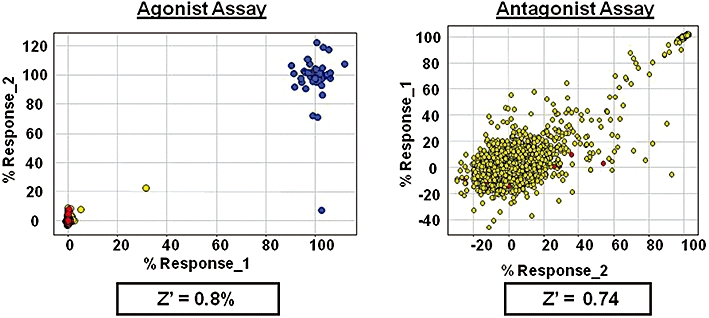
Aequorin high throughput screening: validation testing GPCR antagonist assay (1536-well). Assay validation of a GPCR drug screening assay for the identification of agonist and antagonist ligands. Cells expressing the histamine H1 receptor and the calcium-sensitive photoprotein aequorin were dispensed into 1536-well microtitre plates. A total of 12 000 compounds were screened in duplicate to detect agonist ligands (left panel) and antagonist ligands (right panel). In the agonist assay (left panel), no drug response is represented in red, the response to a maximal concentration of the ligand histamine in blue and compound data in yellow. As is typically seen in agonist assays, the hit rate is very low due to the absence of false positives. In the antagonist assay (right panel), the response to histamine in the absence of test compound is represented in red (basal response), the response to a maximal concentration of a histamine antagonist in blue (100% inhibition) and compound data in yellow. As is typically seen in a cell-based inhibitor assay, there is significant spread of the compound data due to a combination of assay interference and compound activity. True actives correlate in the range 40% to 100% inhibition. Both assays have excellent Z'. GPCR, G-protein-coupled receptor.
Figure 5.
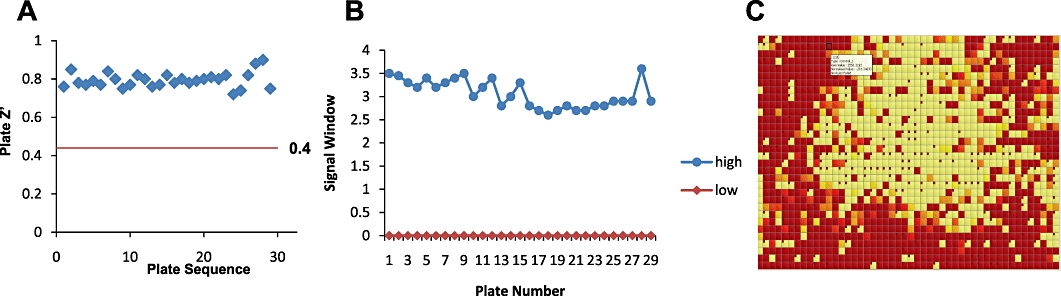
Quality control (QC) in high throughput screening. To ensure the control of screening data in compound screening campaigns each assay plate typically contains a number of pharmacological control compounds. (A) Each 384-well plate contains 16 wells containing a low control and a further 16 wells containing an EC100 concentration of a pharmacological standard which are used to calculate the Z' factor (reference Zhang et al., 1999). Plates that generate a Z' factor below 0.4 are rescreened. (B) Each plate also contains 16 wells of an EC50 concentration of a pharmacological standard to monitor the variance in the assay (diamonds). (C) A heat map is generated for all plates that pass the pharmacological standard QC to monitor the distribution of activity across the assay plate. One would expect to see a random distribution of activity across the screening plate. A plate such as the one presented would be failed and rescreened due to the active wells clustering in the centre of the plate.
Defining a hit series
Compound libraries have been assembled to contain small molecular weight molecules that obey chemical parameters such as the Lipinski Rule of Five (Lipinski et al., 2001), and more often have molecular weights of less than 400 and clogP (a measure of lipophilicity which affects absorption into the body) of less than 4. Molecules with these features have been termed ‘drug-like’, in recognition of the fact that the majority of clinically marketed drugs have a molecular weight of less than 350 and a cLogP of less than 3. It is critically important to initiate a drug discovery programme with a small simple molecule as lead optimization, to improve potency and selectivity, typically involves an increase in molecular weight which in turn can lead to safety and tolerability issues.
Once a number of hits have been obtained from virtual screening or HTS, the first role for the drug discovery team is to try to define which compounds are the best to work on. This triaging process is essential as, from a large library, a team will likely be left with many possible hits which they will need to reduce, confirm and cluster into series. There are several steps to achieving this. First, although this is less of a problem as the quality of libraries have improved, compounds that are known by the library curators to be to be frequent hitters in HTS campaigns need to be removed from further consideration. Second, a number of computational chemistry algorithms have been developed to group hits based on structural similarity to ensure that a broad spectrum of chemical classes are represented on the list of compounds taken forward. Analysis of the compound hit list using these algorithms allows the selection of hits for progression based on chemical cluster, potency and factors such as ligand efficiency which gives an idea of how well a compound binds for its size (log potency divided by number of ‘heavy atoms’ i.e. non-hydrogen atoms, in a molecule).
The next phase in the initial refinement process is to generate dose–response curves in the primary assay for each hit, preferably with a fresh sample of the compound. Showing normal competitive behaviour in hits is important. Compounds which give an all or nothing response are not acting in a reversible manner and indeed may not be binding to the target protein at all, with the activity at high concentrations arising from an interaction between the sample and another component of the assay system. Reversible compounds are favoured because their effects can be more easily ‘washed-out’ following drug withdrawal, an important consideration when using in patients. Obtaining a dose–response curve allows the generation of a half maximal inhibitory concentration which is used to compare of the potencies of candidate compounds. Sourcing and using fresh samples of compounds for this exercise is highly desirable. Nearly all HTS libraries are stored as frozen DMSO solutions with the result that, after some time, the compound can become degraded or modified. Virtually anyone who has worked with libraries of this type has got anecdotes about how potent activity has disappeared when the compound was resynthesized and used in re-testing, although occasionally identification of potent impurities has allowed progress to be made.
With reliable dose–response curves generated in the primary assay for the target, the stage is set to examine the surviving hits in a secondary assay, if one is available, for the target of choice. This need not be an assay in a high throughput format but will involve looking at the affect of the compounds in a functional response, for example in a second messenger assay or in a tissue-or cell-based bioassay. Activity in this setting will give reassurance that compounds are able to modulate more intact systems rather than simply interacting with the isolated and often engineered protein used in the primary assay. Throughout the confirmation process, medicinal chemists would be looking to cluster compounds into groups which could form the basis of lead series. As part of this process, consideration will be given to the properties of each cluster such as whether there is an identifiable structure–activity relationship (SAR) evolving over a number of compounds, that is, identification of a group of compounds which have some section or chemical motif in common and the addition of different chemical groups to this core structure results in different potencies. Issues of chemical synthesis would also be examined. Thus, ease of preparation, potential amenability to parallel synthesis and the ability to generate diversity from late-stage intermediates would be assessed.
With defined clusters in place an exercise can now take place on several groups of compounds in parallel. This phase will include the rapid generation of rudimentary SAR data and defining the essential elements in the structure associated with activity. At the same time, representative examples of each of these mini-series will be subjected to various in vitro assays designed to provide important information with regard to absorption, distribution, metabolism and excretion (ADME) properties as well as physicochemical and pharmacokinetic (PK) measurements (see Table 2). Selectivity profiling, especially against the types of targets, if any, for which the compounds were originally made, is also useful to carry out at this time. For example you may want to inhibit kinase X but avoid kinase Y to reduce unwanted in vivo side effects. This exercise will reveal the strengths and flaws of each series and allow a decision to be taken about the most promising series of compounds to be progressed. The numbers of series taken forward at this stage will depend on the resource available but ideally several should be taken into the hit-to-lead stage to allow for attrition in the coming phase.
Table 2.
Key in vitro assays in early drug discovery
| Assays | Target value | Comments |
|---|---|---|
| Aqueous solubility | >100 µM | Important for running in vitro assays and for in vivo delivery of drug |
| Log D7.4 | 0–3 (for BBB penetration ca 2) | A measure of lipophilicity hence movement across membranes |
| Microsomal stability Clint | <30 µL·min−1·mg−1 protein | Liver microsomes contain membrane bound drug metabolizing enzymes. This assay measures compound clearance and can give an idea of how fast it will be cleared out in vivo |
| CYP450 inhibition | >10 µM | Main enzymes in body which metabolize drugs and their inhibition can cause toxicity |
| Caco-2 permeability Papp | >1 × 10−6 cm−1 (asymmetry <2) | Caco-2 colon carcinoma cell line used to estimate permeability across intestinal epithelium, important for drug absorption from gut |
| MDR1-MDCK permeability Papp | >10 × 10−6 cm−1 (asymmetry <2) | MDCK cells transfected with the MDR1 gene, which encodes the efflux protein P glycoprotein (P-gp). An important efflux transporter in many tissues including intestine, kidney and brain, P-gp can be used to predict intestinal and brain permeability |
| Hep G2 hepatotoxicity | No effect at 50 × IC50 or EC50 | Human HepG2 cells can act as a surrogate for effects of toxicity on human liver, an important cause of drug failure in the clinic |
| Cytotoxicity in suitable cell line | No effect at 50 × IC50 or EC50 | Reduce the likelyhood of cellular toxicity in vivo |
IC50, half maximal inhibitory concentration.
Whatever the screening paradigm, the output of the hit discovery phase of a lead identification programme is a so-called ‘hit’ molecule, typically with a potency of 100 nM–5 µM at the drug target. A chemistry programme is initiated to improve the potency of this molecule.
Hit-to-lead phase
The aim of this stage of the work is to refine each hit series to try to produce more potent and selective compounds which possess PK properties adequate to examine their efficacy in any in vivo models that are available.
Typically, the work now consists of intensive SAR investigations around each core compound structure, with measurements being made to establish the magnitude of activity and selectivity of each compound. This needs to be carried out systematically and, where structural information about the target is known, structure-based drug design techniques using molecular modelling and methodologies such as X-ray crystallography and NMR can be applied to develop the SAR faster and in a more focused way. This type of activity will also often give rise to the discovery of new binding sites on the target proteins.
A screening cascade at this time would generally consist of a relatively high throughput assay establishing the activity of each molecule on the molecular target, together with assays in the same format for sites where selectivity might be known, or expected to be, an issue (Figure 6). A compound meeting basic criteria at this stage would be escalated into a further bank of assays. These should include higher order functional investigations against the molecular target and also whether the compounds were active in primary assays in different species. The HTS assay is generally carried out on protein encoded by human DNA sequences but as animal models are used to validate the activity of compounds in in vivo disease models, in pharmacodynamic (PD)/PK modelling and in preclinical toxicity studies, it is important to have data on activity in vitro on orthologues. This is also particularly important as it will assist in minimizing dosing levels in toxicology studies which are chosen on the basis multiples of the pharmacologically effective doses.
Figure 6.
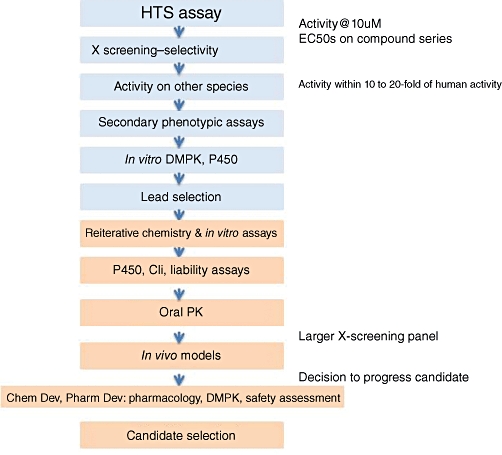
Hypothetical screening cascade. Examples of assays along the screening cascade from high throughput screening (HTS) to candidate selection are shown. DMPK, drug metabolism pharmacokinetics.
Attention in this phase has to also turn to more detailed profiling of physicochemical and in vitro ADME properties and this series of studies is carried out in parallel, with key compounds being selected for assessment. The sort of assays to be considered, with targets that have been found to be appropriate are shown in Table 2.
Solubility and permeability assessments are crucial in ruling in or out the potential of a compound to be a drug, that is, drug substance often needs access to a patient's circulation and therefore may be injected or more generally has to be adsorbed in the digestive system. Deficiency in one or other parameter in a molecule can sometimes be put right. For example formulation strategies can be used to design a tablet such that it dissolves in a particular region of the gut at a pH in which the compound is more soluble. A compound that lacks both these properties is very unlikely to become a drug no matter how potent it is in the primary screening assay. Microsomal stability is a useful measure of the ability of in vivo metabolizing enzymes to modify and then remove a compound. Hepatocytes are sometimes used in this sort of study instead and these will give more extensive results but are not used routinely as they need to be prepared freshly on a regular basis. CYP450 inhibition is examined as, among other things, it is an important predictor of whether a new compound might have an influence on the metabolism of an existing drug with which it may be co-administered.
If one or more of these properties is less than ideal, then it might be necessary to screen many more compounds specifically for those properties. Each programme will end up subtly different in this regard. For example in one recent project to identify novel GPCR antagonists, a number of sub-micromolar hit compounds were identified. The main issues associated with these molecules was that they showed some speciation with poorer receptor affinities in rodent receptors, a general lack of selectivity with >50% inhibition at 10 µM at 30 out of 63 GPCRs and transporters tested in a cross-screening panel as well as broad CYP450 inhibitory activity. It was felt that a number of these deficiencies were associated with the nature of the base common to all the initial structures. Modification of the basic residue resulted in a number of compounds which were as potent as the initial hits at the principal receptor but which were more selective in their actions. In common with many programmes, as potency at the principal target improved selectivity issues in this series were left behind.
Key compounds which are beginning to meet the target potency and selectivity, as well as most of the physicochemical and ADME targets, should be assessed for PK in rats. Here one would normally be aiming for a half-life of >60 min when the compound is administered intravenously and a fraction in excess of 20% absorbed following oral dosing although sometimes, different targets require very different PK profiles. In large pharma with inhouse drug metabolism pharmacokinetics (DMPK) departments numerous compounds might be profiled while in academic environments there may be funds for only a predefined number of these expensive investigations As the receptor antagonist programme, described above, advanced through the hit-to-lead phase, a number of compounds were prepared which had potency in the nanomolar range and a benign selectivity profile except for some potency at the hERG channel, a potassium voltage-gated ion channel important for cardiac function and inhibition at which can cause cardiac liability. Ideally for hERG we were aiming for an activity over 30 uM or at least a 1000-fold selectivity for the target. A number of these compounds were examined in PK studies and were found to have a reasonable half-life following intravenous dosing but poor plasma levels were noted when the compound was given orally to rats. It was felt that some of these compounds, representing the end of the hit-to-lead phase of the project were, although not likely themselves to be progressed, capable of answering questions in disease models. Thus, compounds were administered intra-peritoneally and results from the experiments gave substantial credence to the developing programme.
Lead optimization phase
The object of this final drug discovery phase is to maintain favourable properties in lead compounds while improving on deficiencies in the lead structure. Continuing with example above, the aim of the programme was now to modify the structure to minimize hERG liability and to improve the absorption of the compound. Thus, more regular checks of hERG affinity and CACO2 permeation were undertaken and compounds were soon available which maintained their potency and selectivity at the principal target but which had a much reduced hERG affinity and a better apparent permeation than initial lead compounds. When examined for PK properties in rat one of these compounds, with 8 nM affinity at the receptor of interest, had an oral bioavailability of over 40% in rats and about 80% in dogs.
Compounds at this stage may be deemed to have met the initial goals of the lead optimization phase and are ready for final characterization before being declared as preclinical candidates. Discovery work does not cease at this stage. The team has to continue to explore synthetically in order to produce potential back up molecules, in case the compound undergoing further preclinical or clinical characterization fails and, more strategically, to look for follow-up series.
The stage at which the various elements that constitute further characterization are carried out will vary from company to company and parts of this process may be incorporated into the lead optimization phase. However, in general molecules need to be examined in models of genotoxicity such as the Ames test and in in vivo models of general behaviour such as the Irwin's test. High-dose pharmacology, PK/PD studies, dose linearity and repeat dosing PK looking for drug-induced metabolism and metabolic profiling all need to be carried out by the end of this stage. Consideration also needs to be given to chemical stability issues and salt selection for the putative drug substance.
All the information gathered about the molecule at this stage will allow for the preparation of a target candidate profile which with together with toxicological and chemical manufacture and control considerations will form the basis of a regulatory submission to allow human administration to begin.
The process of hit generation to preclinical candidate selection often takes a long time and cannot in any way be considered a routine activity. There are rarely any short cuts and significant, intellectual input is required from scientists from a variety of disciplines and backgrounds. The quality of the hit-to-lead starting point and the expertise of the available team are the key determinants of a successful outcome of this phase of work. Typically, within industry for each project 200 000 to >106 compounds might be screened initially and during the following hit-to-lead and lead optimization programmes 100's of compounds are screened to hone down to one or two candidate molecules, usually from different chemical series. In academia screens are more likely to be of a focused nature due to the high cost of an extensive HTS or compounds are derived from a structure-based approach. Only 10% of small molecule projects within industry might make the transition to candidate, failing at multiple stages. These can include the (i) inability to configure a reliable assay; (ii) no developable hits obtained from the HTS; (iii) compounds do not behave as desired in secondary or native tissue assays; (iv) compounds are toxic in vitro or in vivo; (v) compounds have undesirable side effects which cannot be easily screened out or separated from the mode of action of the target; (vi) inability to obtain a good PK or PD profile in line with the dosing regeme required in man, for example, if require a once a day tablet then need the compound to have a half-life in vivo suitable to achieve this; and (vii) inability to cross the blood brain barrier for compounds whose target lies within the central nervous system. The attrition rate for protein therapeutics, once the target has been identified, is much lower due to less off target selectivity and prior experience of PK of some proteins, for example, antibodies.
Although relatively less costly than many processes carried out later on in the drug development and clinical phases, preclinical activity is sufficiently high risk and remote from financial return to often make funding it a problem. Ensuring transparency of the cost of each stage/assay within large pharma may help reduce some of their costs and there are some movements towards this as companies instigate a ‘biotech’ mentality and accountability for costs.
Once a candidate is selected, the attrition rate of compounds entering the clinical phase is also high, again only one in 10 candidates reaching the market but at this stage the financial consequences of failure are much higher. There has been considerable debate in industry as to how to improve the success rate, by ‘failing fast and cheap’. Once a candidate reaches the clinical stage, it can become increasingly difficult to kill the project, as at this stage the project has become public knowledge and thus termination can influence confidence in the company and shareholder value. Carrying out more studies prior to clinical development such as improved toxicology screens (using failed drugs to inform these assays), establishing predictive translational models based on a thorough disease understanding and identifying biomarkers may help in this endeavour. It is particularly in these later two areas where academic-industry partnerships could really add value preclinically and eventually help bring more effective drugs to patients.
Acknowledgments
Karen Philpott is supported by the Medical Research Council and Guys and St Thomas' Charity.
S. Barret Kalindjian is supported by a Seeding Drug Discovery Wellcome Trust grant.
Glossary
Abbreviations
- ADME
absorption, distribution, metabolism and excretion
- DMPK
drug metabolism pharmacokinetics
- DMSO
dimethyl sulphoxide
- GPCRs
G-protein-coupled receptors
- HTS
high throughput screening
- mAbs
human monoclonal antibodies
- PD
pharmacodynamic
- PK
pharmacokinetic
- SAR
structure–activity relationship
Conflict of interest
Jane Hughes is employed by MedImmune, Steve Rees is employed by GSK and Karen Philpott was previously employed by GSK.
Supplementary material
Supporting Information: Teaching Materials; Figs 1–6 as PowerPoint slide.
References
- Abell AN, Rivera-Perez JA, Cuevas BD, Uhlik MT, Sather S, Johnson NL, et al. Ablation of MEKK4 kinase activity causes neurulation and skeletal patterning defects in the mouse embryo. Mol Cell Biol. 2005;25:8948–8959. doi: 10.1128/MCB.25.20.8948-8959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Tanzi RE. Thirty years of Alzheimer's disease genetics: the implications of systematic meta-analyses. Nat Rev Neurosci. 2008;9:768–778. doi: 10.1038/nrn2494. [DOI] [PubMed] [Google Scholar]
- Boppana K, Dubey PK, Jagarlapudi SARP, Vadivelan S, Rambabu G. Knowledge based identification of MAO-B selective inhibitors using pharmacophore and structure based virtual screening models. Eur J Med Chem. 2009;44:3584–3590. doi: 10.1016/j.ejmech.2009.02.031. [DOI] [PubMed] [Google Scholar]
- Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chessell IP, Hatcher JP, Bountra C, Michel AD, Hughes JP, Green P, et al. Disruption of the P2X7 purinoceptor gene abolishes chronic inflammatory and neuropathic pain. Pain. 2005;114:386–396. doi: 10.1016/j.pain.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Cox JJ, Reimann F, Nicholas AK. An SCN9A channelopathy causes congenital inability to experience pain. Nature. 2006;444:894–898. doi: 10.1038/nature05413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne A, Jowett M, Rees S. Use of primary cells in high throughput screens. Meth Mol Biol. 2009;565:239–257. doi: 10.1007/978-1-60327-258-2_12. [DOI] [PubMed] [Google Scholar]
- Fox S, Farr-Jones S, Sopchak L, Boggs A, Nicely AW, Khoury R, et al. High-throughput screening; Update on practices and success. J Biol Screen. 2006;11:864–869. doi: 10.1177/1087057106292473. [DOI] [PubMed] [Google Scholar]
- Frearson JA, Collie IT. HTS and hit finding in academia – from chemical genomics to drug discovery. Drug Discov Today. 2009;14:1150–1158. doi: 10.1016/j.drudis.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning SW, Beste G. Loss-of-function strategies in drug target validation. Curr Drug Discov. 2002;May:17–21. [Google Scholar]
- Honore P, Kage K, Mikusa J, Watt AT, Johnston JF, Wyatt JR, et al. Analgesic profile of intrathecal P2X3 antisense oligonucleotide treatment in chronic inflammatory and neuropathic pain states. Pain. 2002;99:11–19. doi: 10.1016/s0304-3959(02)00032-5. [DOI] [PubMed] [Google Scholar]
- Kurosawa G, Akahori Y, Morita M, Sumitomo M, Sato N, Muramatsu C, et al. Comprehensive screening for antigens overexpressed on carcinomas via isolation of human mAbs that may be therapeutic. Proc Natl Acad Sci U S A. 2008;105:7287–7292. doi: 10.1073/pnas.0712202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law R, Barker O, Barker JJ, Hesterkamp T, Godemann R, Andersen O, et al. The multiple roles of computational chemistry in fragment-based drug design. J Comput Aided Mol Des. 2009;23:459–473. doi: 10.1007/s10822-009-9284-1. [DOI] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- McInnes C. Virtual screening strategies in drug discovery. Curr Opin Chem Biol. 2007;11:494–502. doi: 10.1016/j.cbpa.2007.08.033. [DOI] [PubMed] [Google Scholar]
- Michelini E, Cevenini L, Mezzanotte L, Coppa A, Roda A. Cell Based Assays: fuelling drug discovery. Anal Biochem. 2010;397:1–10. doi: 10.1007/s00216-010-3933-z. [DOI] [PubMed] [Google Scholar]
- Moore K, Rees S. Cell-based versus isolated target screening: how lucky do you feel? J Biomol Scr. 2001;6:69–74. doi: 10.1177/108705710100600202. [DOI] [PubMed] [Google Scholar]
- Peet NP. What constitutes target validation? Targets. 2003;2:125–127. [Google Scholar]
- Stables J, Mattheakis LC, Chang TR, Rees S. Recombinant aequorin as a reporter of changes in intracellular calcium concentration in mammalian n cells. Meth Enzymol. 2000;327:456–471. doi: 10.1016/s0076-6879(00)27296-0. [DOI] [PubMed] [Google Scholar]
- Ugolini G, Marinelli S, Covaceuszach S, Cattaneo A, Pavone F. The function neutralizing anti-TrkA antibody MNAC13 reduces inflammatory and neuropathic pain. Proc Natl Acad Sci U S A. 2007;104:2985–2990. doi: 10.1073/pnas.0611253104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nature Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang Y, Li S. Mutations in SCN9A, encoding a sodium channel alpha subunit, in patients with primary erythermalgia. J Med Genet. 2004;41:171–174. doi: 10.1136/jmg.2003.012153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Adelstein SJ, Kassis AI. Target discovery from data mining approaches. Drug Discov Today. 2009;14:147–154. doi: 10.1016/j.drudis.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Zanders ED, Bailey DS, Dean PM. Probes for chemical genomics by design. Drug Discov Today. 2002;7:711–718. doi: 10.1016/s1359-6446(02)02325-5. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung DY, Oldenberg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Scr. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


