Abstract
Chlamydiae contain a rough type lipopolysaccharide (LPS) of 3-deoxy-α-d-manno-oct-2-ulopyranosonic acid residues (Kdo). Two Kdo trisaccharides, 2.8/2.4- and 2.4/2.4-linked, and a branched 2.4[2.8]2.4-linked Kdo tetrasaccharide occur in Chlamydiaceae. While the 2.8/2.4-linked trisaccharide contains a family-specific epitope, the branched Kdo oligosaccharide occurs only in Chlamydophila psittaci and antibodies against it will be useful in human and veterinarian diagnostics. To overcome the generation of cross-reactive antibodies that bind with high affinity to a dominant epitope formed by 2.4/2.4-linked Kdo, we immunized mice with a synthetic 2.4[2.8]-linked branched Kdo trisaccharide and used phage display of scFv to isolate recombinant antibody fragments (NH2240-31 and SAG506-01) that recognize the branched Kdo oligosaccharide with a KD of less than 10 nM. Importantly, although these antibodies used germline genes coding for an inherited Kdo recognition site they were able clearly to distinguish between 2.4[2.8]2.4- and 2.4/2.4-linked Kdo. Sequence determination, binding data, and X-ray structural analysis revealed the basis for the improved discrimination between similar Kdo ligands and indicated that the alteration of a stacking interaction from a phenylalanine residue in the center of the combining site to a tyrosine residue facing away from the center favours recognition of branched 2.4[2.8]2.4-linked Kdo residues. Immunofluorescence tests of infected cell monolayers using this antibody show specific staining of C. psittaci elementary bodies that allow it to be distinguished from other pathogenic chlamydiae.
Introduction
A recent evaluation of over 2000 carbohydrate-protein interactions revealed that more than half of the investigated anti-carbohydrate antibodies cross-reacted with other glycans (Manimala et al. 2007); however, despite its biological and medical importance, there is only limited structural information describing cross-reactivity and specificity in carbohydrate recognition by antibodies. Low affinity and molecular flexibility associated with these interactions typically hamper structural analysis, and we have begun a systematic investigation on the structural level of cross-reactivity and specificity using antibodies that display high affinities for different closely related oligosaccharides of 3-deoxy-α-d-manno-oct-2-ulopyranosonic acid (Kdo) (Müller-Loennies et al. 2000; Nguyen et al. 2003; Brooks et al. 2008b; Brooks et al. 2009a). Similar studies have contributed to the generation of an antibody library against negatively charged carbohydrates and to the successful application of phage display for the clinically relevant isolation of antibodies against heparan sulfate and sulfated sialyl-Lewis X (Schoonbroodt et al. 2008).
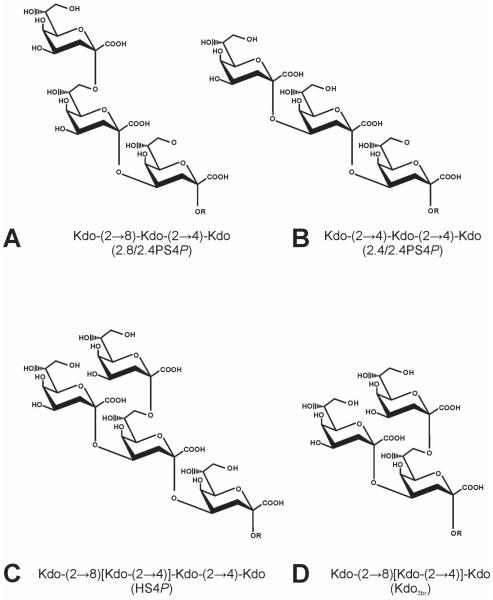
The genera Chlamydophila and Chlamydia belong to the family of Chlamydiaceae that contains important human pathogens such as Chlamydophila pneumoniae and Chlamydia trachomatis (Corsaro et al. 2003). Chlamydophila psittaci is primarily a pathogen of psittacine birds but can also cause zoonotic infections with symptoms ranging from mild pneumonia to severe systemic disease in humans. Like all Chlamydiaceae, C. psittaci is an obligate intracellular Gram-negative pathogen with a unique development cycle during which an infectious elementary body is formed (Moulder 1991). This elementary body contains a lipopolysaccharide (LPS) composed of a lipid A and a short chain of Kdo residues containing a family specific epitope found in all Chlamydiaceae, Kdo(2→8)Kdo(2→4)Kdo trisaccharide [2.8/2.4Kdo3, Fig. 1A, reviewed in (Brade 1999)]. In addition to this structure, a Kdo(2→4)Kdo(2→4)Kdo trisaccharide (2.4/2.4Kdo3, Fig. 1B) and, in large amounts, a branched Kdo tetrasaccharide Kdo(2→8)[Kdo(2→4)]Kdo(2→4)Kdo (Kdo4, Fig. 1C) are made by C. psittaci (Rund et al. 2000).
Figure 1. Kdo oligosaccharides from LPS of Chlamydiae (A-C) and the synthetic branched Kdo oligosaccharide used for immunization (D).
Oligosaccharides obtained by alkaline deacylation of LPS (A to C) contain the acylated lipid A backbone →6)-β-GlcN4P-(1→6)-α-GlcN1P (R) and have been abbreviated as 2.8/2.4PSBP (A), 2.4/2.4PSBP (B), and HSBP (C). For the generation of neoglyconjugates these oligosaccharides were dephosphorylated at the anomeric position and after introduction of an isothiocyanate spacer conjugated to BSA by reductive amination (Müller-Loennies et al. 2003). These structures have been abbreviated in the text as 2.8/2.4PS4P (A), 2.4/2.4PS4P (B), and HS4P (C). The corresponding Kdo epitopes in these oligosaccharides have been abbreviated as 2.8/2.4Kdo3 (A), 2.4/2.4Kdo3 (B), and Kdo4 (C). The Kdo oligosaccharide containing only the branched Kdo trisaccharide (D, Kdo3br) has been chemically synthesized as the allyl derivative which was conjugated to BSA as described (Kosma et al. 2009).
The recent report on the isolation of C. pneumoniae and C. psittaci from 30% of trachoma patients with ocular infections (Dean et al. 2008) indicates the need for the development of additional reliable diagnostic tools, and an antibody for the diagnosis of C. psittaci would be very valuable. Recently, we have obtained monoclonal antibody (mAb) S69-4 after immunization of mice with a synthetic neoglycoconjugate containing the branched Kdo4 and have shown that this antibody can be used for the specific staining of C. psittaci elementary bodies in infected cell monolayers (Müller-Loennies et al. 2006). This antibody had a relatively low affinity towards its natural antigen (KD = 10 μM) in comparison to other Kdo binding antibodies (Müller-Loennies et al. 2000) and considerable cross-reactivity at high concentration in immunofluorescence tests. This raised the general question of whether it would be possible to obtain high affinity antibodies specific for Kdo4 or whether an increase in specificity would always be accompanied by a loss of affinity.
The high degree of sequence similarity between the previously crystallized mAb S45-18, (Nguyen et al. 2003) and mAb S69-4 suggests that the observed cross-reactivity of S69-4 may have been due to an epitope formed by the 2.4/2.4Kdo3 (Fig. 1B) moiety of the branched Kdo4 (Fig. 1C). Based on this assumption we have now i) investigated the role of different parts of VH CDR3 in the recognition of 2.4/2.4Kdo3 and attempted to improve the affinity of S69-4 while retaining its higher specificity for Kdo4 by transferring parts of VH CDR3 of S45-18 into S69-4, ii) immunized mice with a novel conjugate containing only the terminal branched Kdo(2→8)[Kdo(2→4)]Kdo trisaccharide [Kdo3br, Fig. 1D, (Kosma et al. 2009)] in an attempt to avoid the induction of cross-reactive antibodies and iii) employed phage display for the isolation of antibodies specific for C. psittaci. We now report the isolation of scFvs from a library displayed on phage, their biochemical characterization in binding assays of the native scFvs and mutant derivatives thereof and their structural analysis in complex with Kdo. In order to explore their potential use as diagnostic tools, we have further determined their reactivity with elementary bodies by ELISA and immunofluorescence detection of infected cell monolayers.
To extend the current knowledge about carbohydrate recognition by antibodies and to contribute to the elucidation of structural mechanisms underlying the development of antibody specificity vs cross-reactivity we have determined the primary structures of several antibodies against LPS from Chlamydia, their affinities to related ligands by surface plasmon resonance (SPR), and compared X-ray crystal structures in complex with Kdo to those obtained earlier of an affinity-matured monoclonal antibody (mAb S45-18) and an antibody close to the germline sequence (mAb S25-2) (Nguyen et al. 2003; Brooks et al. 2008b). This study contributes to the general understanding of mechanisms for affinity maturation of anti-carbohydrate antibodies at the structural level by revealing how specific changes of VH CDR3 confer cross-reactivity or specificity to antibodies against Kdo containing oligosaccharides that have shown promiscuous recognition of structurally diverse ligands.
Results
Isolation of anti-carbohydrate scFv by Phage display
In order to amplify antibodies that are able to discriminate the antigens Kdo4 and 2.4/2.4Kdo3 we generated a scFv library displayed on filamentous phage of a mouse immunized with the BSA-neoglycoconjugate of Kdo3br. The initial size of the scFv phage library obtained after immunization consisted of 2.7 × 107 transformants, and colony PCR and sequencing revealed that 19 out of 20 clones contained an insert of the expected size.
It has been generally noted that the isolation of anti-carbohydrate antibodies by phage display is difficult due to their low affinity with only few successful examples published (Schoonbroodt et al. 2008). Optimization of panning conditions is one of the keys to successful phage display and temperature and length of incubation have been suggested to influence the kinetic properties of antibodies selected by phage display. Taking into account the lower affinity of carbohydrate-protein interactions in comparison to protein-protein-interactions, we have compared two different conditions for panning. Panning performed throughout at 4 °C allowed the rapid isolation of consensus sequences after three rounds; however, none of the isolated clones bound to immobilized C. psittaci in phage ELISA. In contrast, panning at 37 °C during the initial three rounds resulted in enrichment of consensus sequences as judged by BstOI digest and sequencing, accompanied by an increase of phage titer in ELISA against C. psittaci. Two further rounds of panning at 4 °C resulted in further enrichment of phage binding to C. psittaci.
Since we were interested in obtaining high affinity scFvs that specifically bind C. psittaci and do not cross-react with other Chlamydiae, 45 monoclonal scFvs bound to phage were selected and their reactivity with immobilized C. psittaci 6BC tested in single wells. Eighteen clones resulted in an OD405 above 0.1 and were thus considered as potential binders. These were investigated in single well ELISA against immobilized C. psittaci, C. pneumoniae, C. trachomatis and the neoglycoconjugate used for panning. Of the individual amplified phages several clones reacted specifically with C. psittaci. A second group of clones showed cross-reactivity between C. psittaci and C. pneumoniae. Two representative clones specific for C. psittaci, designated scFv NH2240-31 and SAG506-01, and a third cross-reactive clone, scFv NH2240-39, were expressed as soluble scFvs and investigated further.
Comparison of Antibody Combining Sites
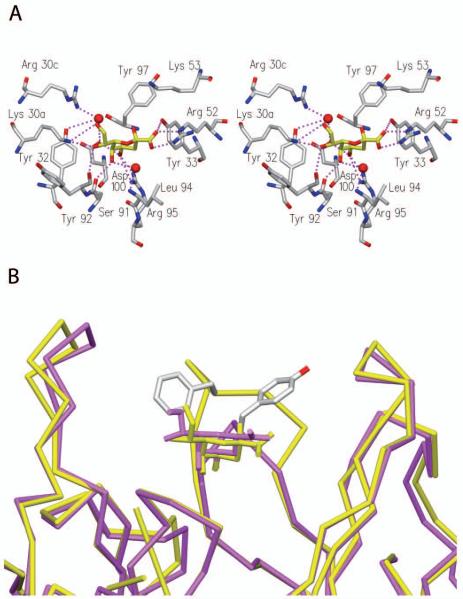
The determination of primary structures (Fig. 2) revealed that scFv antibodies isolated by phage display were homologous to the germline mAb S25-2 (Fu et al. 1992; Nguyen et al. 2003; Brooks et al. 2008b) and to mAbs S73-2 (Brooks et al. 2009b), S69-4 (Müller-Loennies et al. 2006), and S45-18 (Brade et al. 2000; Nguyen et al. 2003). Crystal structures of mAbs S25-2, S45-18 and related mAbs in complex with various Kdo ligands revealed that these antibodies contain an inherited germline-encoded combining site specific for Kdo (Nguyen et al. 2003; Brooks et al. 2008b). Characteristic contacts between antibody and antigen include charged-residue interactions with ArgL30c, ArgH52, and LysH53 and hydrogen bonds to SerL91, TyrL92, ArgL95, and TyrH33 (Fig. 3A). The antibody S45-18 contains PheH99 in the centre of the VH CDR3 loop that has been proposed to be the major determinant for 2.4/2.4Kdo3 specificity. This residue functions by restricting the conformational space available for the ligand and so excludes 2.8-linked Kdo residues and provides hydrophobic stacking interactions with all three Kdo-residues to give a combining site with high specificity and affinity (Nguyen et al. 2003). In the same complex AspH100c has been shown to form a bridging hydrogen bond to the ligand through a structured water molecule. A hydrogen bond between the MetH100e carbonyl and the TrpH103 side chain leads to the formation of a kink in the backbone structure of VH CDR3 loops (Shirai et al. 1996; Morea et al. 1998; Shirai et al. 1999) that is further stabilized by salt bridge between ArgH94 and AspH100f. This salt bridge is missing in the germline mAb S25-2, leading to higher flexibility and adaptation of the loop that allows this antibody to bind oligosaccharides that contain 2.8-linked Kdo residues, such as 2.8/2.4Kdo3 (Nguyen et al. 2003). Both, ArgH94 and the motif Ala-Met-Asp (H100d-f) at the C-terminal side of the loop are present in all these Kdo antibodies apart from S25-2.
Figure 2. Amino acid sequence alignment of VL and VH domains of monoclonal antibodies.
The numbering of residues and assignment of CDR (shaded) was done according to Chothia with structurally corrected framework indels as described by Andrew C. Martin (University College London, http://www.bioinf.org.uk/) using the software AbNum (Abhinandan and Martin 2008).
Figure 3. Crystal structure of SAG506-01 in complex with Kdo.
Stereo-view of the SAG506-01 antibody combining site in complex with Kdo (A) and SAG506-01 (PDB 3DV6, yellow) overlaid with S45-18 (PDB 1Q9W, magenta) (B). Shown are the side chain positions of TyrH97 in SAG506-01and of PheH99 S45-18.
The VL and VH of all investigated antibodies originated from the same germline and joining genes (IGKV8-21*01, IGKJ1*01, IGHV7-3*02 and IGHJ4*01). Differences were confined to length and composition of the VH CDR3 due to different diversity gene usage. Antibodies S69-4 and S45-18 used IGHD2-4*01 and IGHD1-1*01, respectively, which both have residue H99 mutated to phenylalanine. The combining site surface of the cross-reactive clone NH2240-39 was very similar to those of mAbs S69-4 and S45-18. Despite significant sequence dissimilarity in this loop the length of VH CDR3 was the same and contained PheH99 (Fig 2).
Clones NH2240-31 and SAG506-01, which were obtained after immunization of mice with Kdo3br and showed high specificity towards C. psittaci, used IGHD2-3*01 to give a much shortened VH CDR3 that did not contain PheH99 (Fig. 2). Instead, an isoleucine (NH2240-31) or valine (SAG506-01) occupied this position, which may have indicated that a weakened hydrophobic interaction was the basis for the higher specificity for C. psittaci. SAG506-01 was nearly identical in sequence to NH2240-31 but contained additional mutations at two positions in the framework region of VL and four positions in the VH domain (Brooks et al. 2008a). All amino acids known to be important for binding were conserved apart from seven mutations of the germline gene in VL CDR1 according to IMGT/V-Quest analysis (Brochet et al. 2008; Lefranc et al. 2009) leading to a sequence change from LNSRNR to FKSRNQ and a replacement of AsnL30a by Lys (Fig. 2). In the cocrystal structure of S45-18, AsnL30a is involved in an electrostatic interaction with the carboxyl group of the Kdo residue proximal to the lipid A. The crystal structure of SAG506-01 in complex with Kdo (Fig. 3) revealed that the sequence change in VL CDR1 changes this interaction. This residue now extends the inherited Kdo binding site by forming a hydrogen bond to the hydroxyl at position 7. The shortened CDR3 loop positions residue H99 outside the combining site close to the interface between VL and VH. Instead this position is occupied by TyrH97 whose aromatic ring is displaced in comparison to PheH99 of S45-18, which faces away from the center of the combining site (Fig. 3B).
ELISA binding assays
The question of the feasibility of using soluble monovalent fragments like scFv and Fab in ELISA has been addressed using neoglycoconjugates comprised of BSA and purified Kdo oligosaccharides (Fig. 4). In order to avoid differences due to valence effects, monomer scFv was purified by gel filtration just prior to the analyses. Binding to the ligands 2.8/2.4PS4P, 2.4/2.4PS4P, and HS4P (Fig. 1A to C) was tested after coupling to BSA by reductive amination of the 1-dephosphorylated compounds.
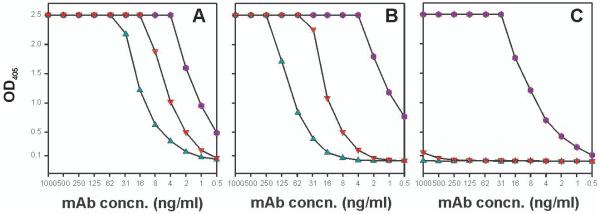
Figure 4. ELISA binding of mAb S45-18 (●) and Fab S45-18 (▼), and mAb S69-4 (▲) binding to HS4P (A), 2.4/2.4PS4P (B) and 2.8/2.4PS4P (C).
Neoglycoconjugates were immobilized (20 pmol oligosaccharide ligand/well) and incubated with indicated concentrations of protein. Binding was detected using a HRP labelled goat anti-mouse IgG(H+L).
Binding of mAb S45-18, its Fab and mAb S69-4 (Fig. 4A-C) was first compared at a high surface coating of 20 pmol ligand/well. While the mAb S45-18 bound to all antigens under such conditions, mAb S69-4 and the Fab only bound to HS4P and 2.4/2.4PS4P. The affinity of S45-18 towards 2.4/2.4PS4P and HS4P was similarly high, but significantly reduced against the 2.8/2.4PS4P. As expected, a reduced affinity was observed for the Fab fragment for all ligands (4-fold towards HS4P and 16-fold towards 2.4/2.4PS4P). MAb S69-4 bound HS4P 4- to 8-fold better than 2.4/2.4PS4P but showed considerably lower affinity than S45-18 (16-fold against HS4P and 128-fold against 2.4/2.4PS4P).
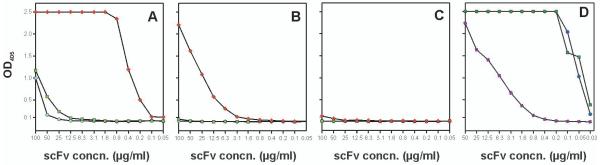
S69-4 and S45-18 both contain PheH99 but differ in stretches of four amino acids on either side of this residue [Fig. 2, (Nguyen et al. 2003; Müller-Loennies et al. 2006)]. In order to reveal the structural role of VH CDR3 for high affinity binding S69-4 was expressed as soluble scFv and parts of the CDR3 sequence were transferred from S45-18 into S69-4. While the scFv of S69-4 showed no binding in ELISA, a mutant containing amino acids following PheH99 (but not preceding) bound with very high affinity to HS4P and with lower affinity to 2.4/2.4PS4P (Fig. 5A-C). The affinity of this mutant towards HS4P was comparable to that of the S45-18 Fab (Fig 4A). Against the same ligands the scFv NH2240-31 showed a very high affinity towards the HS4P (Fig. 5D), only weak binding to 2.4/2.4PS4P and no binding towards the other ligands (data not shown). While an Ala/ProH52c substitution in CDR2 of this scFv had no effect, a Lys/AsnL30a replacement in CDR1 almost abrogated binding (Fig. 5D).
Figure 5. ELISA binding data of S69-4 wildtype scFv (■) and VH CDR3 mutants S69-4 H3ante (●) and H3post (◆) to neoglycoconjugates of HS4P (A), 2.4/2.4PS4P (B), 2.8/2.4PS4P (C), and binding of NH2240-31 (●) and Lys/AsnL30a (◆) and Ala/ProH52 (■) mutants to HS4P (D) neoglycoconjugate.
Immobilized neoglycoconjugates (20 pmol/well) were incubated with indicated concentrations of myc-tagged scFv followed by incubation with anti-c-myc antibody and HRP labelled goat anti-mouse IgG(H+L). S69-4 H3ante and H3post are mutant scFv in which amino acids in VH CDR3 have been exchanged with those of S45-18. The subscript indicates the position relative to PheH99 (for alignment see Fig. 2).
SPR Analyses
To obtain an insight into the kinetics of binding and to compare with the results of the ELISA analysis SPR analyses were performed (Fig. 6 and 7, Table I) using the same oligosaccharides (Fig. 1). First, attempts were made to determine the binding constants with natural and artificial oligosaccharides as solutes, but this resulted in a large percentage of the antibodies being inactivated upon immobilization. Instead, soluble scFv or Fab analytes were isolated as monovalent fragments by gel filtration in SPR-buffer just prior to the analysis. Binding constants were determined for the same neoglycoconjugates after immobilization on the sensor chips as those used for the ELISA.
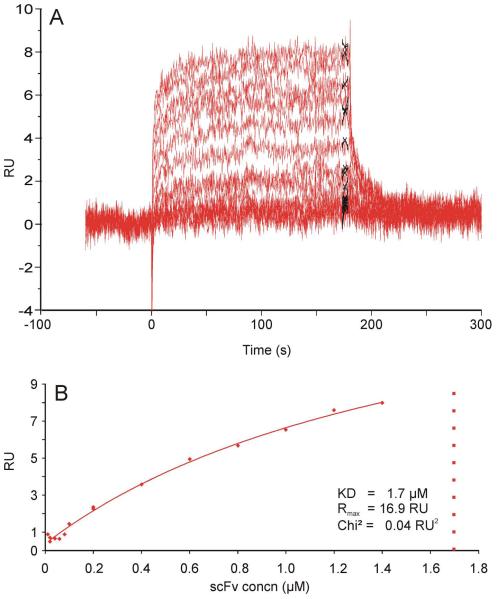
Figure 6. SPR sensorgrams of scFv 69-4 against immobilized 2.4/2.4PS4P neoglycoconjugate.
Sensorgrams were recorded for protein concentrations ranging from 0.01 μM to 1.4 μM (A). Results of a standard steady state analysis using the Biacore T100 Evaluation Software v. 1.1 at report points 4 s before stop of analyte injection and with a window of 5 s (B). The Rmax and KD values predicted from the fitting routine are given. The scFv was purified by gel filtration immediately prior to the analysis.
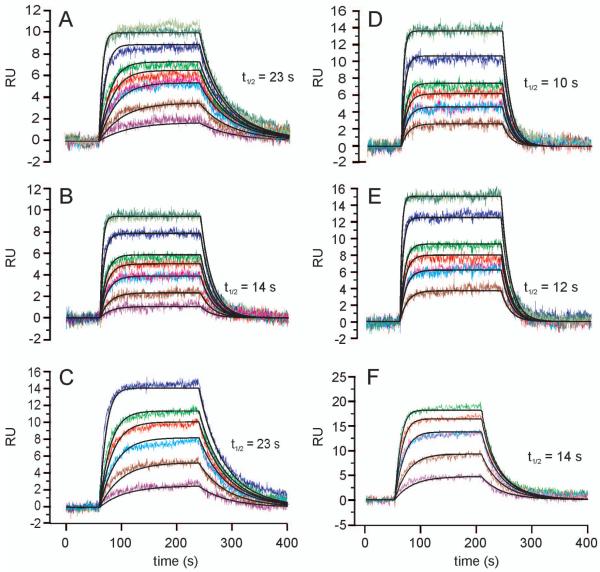
Figure 7. SPR sensorgrams of scFv against immobilized neoglycoconjugates.
NH2240-31 (A and D), scFv S69-4 H3post (B and E) and Fab S45-18 (C and F) against HS4P (A, B, C) and 2.4/2.4PS4P (D, E, and F). Sensorgrams recorded for protein concentrations of 1 nM, 2.5 nM, 5 nM, 7.5 nM, 10 nM, 20 nM, and 40 nM are shown in A and B. Identical concentrations are displayed with same colors. Proteins were purified by gel filtration immediately prior to the analysis.
Table I.
SPR binding data of scFv against immobilized neoglycoconjugates
| sample | ligand | kon (1/Ms ×106) |
SE kon (×104) |
koff (1/s ×10−3) |
SE koff (×10−3) |
KD (M ×10−9) |
Rmax (RU) | SE(Rmax) | t1/2 (s) |
|---|---|---|---|---|---|---|---|---|---|
| NH2240-31 | HS4P | 5.4 | 16 | 30 | 0.9 | 6 | 11 | 0.03 | 23 |
| 2.4/2.4PS4P | 4.6 | 7.1 | 70 | 1.1 | 15 | 19 | 0.02 | 10 | |
| scFv S69-4 H3post | HS4P | 4.7 | 2.4 | 50 | 0.2 | 10 | 11 | 0.01 | 14 |
| 2.4/2.4PS4P | 6.2 | 4.9 | 60 | 0.5 | 10 | 19 | 0.03 | 12 | |
| Fab S45-18 | HS4P | 4.1 | 15 | 30 | 0.9 | 7 | 19 | 0.09 | 23 |
| 2.4/2.4PS4P | 12.0 | 110 | 50 | 5 | 4 | 27 | 0.20 | 14 |
The binding of Fab S45-18, scFv of S69-4 and its VH CDR3 mutants to HS4P were compared (Fig. 7, Table I). The Fab showed a very high affinity with a KD of 7 nM and fast kinetic rate constants (kon = 4.1 × 106 M−1s−1 and koff = 0.03 s−1), the off rate corresponding to a half life (t1/2) of only 23 s (Fig. 7C). The scFv S69-4 had a much weaker affinity with very fast kinetics precluding determination of rate constants by SPR confirming previous analyses (Müller-Loennies et al. 2006). A low μM KD was determined by steady state analysis (Fig. 6A and B). Exchanging four amino acids in CDR3 preceding PheH99 (S69-4 H3ante) to those of S45-18 did not alter the affinity, however, changing four amino acids following this residue (S69-4 H3post), improved the affinity considerably to a KD of 10 nM (kon = 4.7 × 106 M−1s−1 and koff = 0.05 s−1, t1/2 = 14 s; Fig. 7B).
The binding of the Fab S45-18 to 2.4/2.4PS4P was almost identical to HS4P with a low KD of 4 nM (Fig. 7C and F). Binding constants for both scFv S69-4 and the mutant (S69-4 H3ante) could not be determined for this ligand due to a too weak interaction. The second mutant (S69-4 H3post) had a KD of 10 nM (kon = 6.2 × 106 M−1s−1 and koff = 0.06 s−1, t1/2 = 12 s), as good as for binding to HS4P (Fig. 7B and E) and thus confirming the results of the ELISA analysis.
The scFv NH2240-31 bound HS4P and 2.4/2.4PS4P ligands with similar KD values of 6 nM and 15 nM (Fig. 7A and D), respectively and binding to 2.8/2.4PS4P was too weak to determine KD values.
Immunofluorescence test
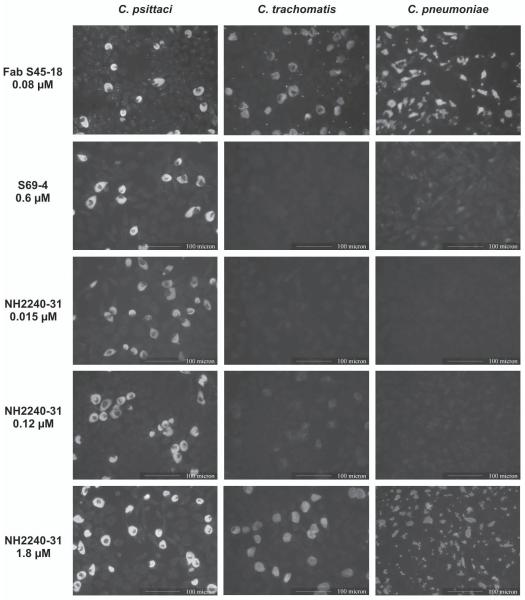
Whether scFv NH2240-31 could be used for the specific detection of C. psittaci in infected cell monolayers was examined by immunofluorescence tests. Cell cultures grown in 96 well ELISA plates were infected with C. pneumoniae, C. psittaci or C. trachomatis, and incubated with serially diluted soluble scFv, followed by incubation with mouse anti-c-myc antibody and FITC-labeled goat anti-mouse IgG for fluorescence microscopy. As shown in Fig. 8, the Fab fragment of S45-18 was strongly cross-reactive at a concentration 80 nM to all three chlamydiae. In contrast, scFv NH 2240-31 was highly specific for C. psittaci with clear staining at a concentration of 15 nM (0.4 μg/ml) and no cross-reaction even at 120 nM. The reaction with other chlamydiae observed at 1.8 μM (50 μg/ml, 100-fold) was still clearly distinguishable from the reaction with C. psittaci.
Figure 8. Detection of chlamydial inclusions in tissue culture with monoclonal antibodies S5-10, S69-4, Fab of S45-18 and NH2240-31.
Tissue culture monolayers were infected with C. psittaci, C. pneumoniae or C. trachomatis, and after fixation incubated with the indicated concentrations of antibody or fragments thereof. Reactions with scFv were developed with a c-myc specific second antibody (clone 9E10) and a third IgG H+L binding antibody conjugated to FITC. For analyses using Fab or mAb the second antibody was omitted.
Discussion
C. psittaci is the only chlamydial species containing a LPS with a branched Kdo tetrasaccharide (Fig. 1C, Kdo4), and is therefore an attractive target for generating species-specific antibodies useful for the diagnosis of C. psittaci infections. Because of their high affinity, antibodies against Kdo oligosaccharides are also a good model system to study carbohydrate recognition by antibodies in general and the recognition of negatively charged carbohydrates in particular. Such knowledge has contributed to the generation of an antibody library directed against negatively charged carbohydrates (Schoonbroodt et al. 2008). We have obtained a number of mAbs with different epitope specificities that recognize Kdo oligosaccharides and display varying degrees of cross-reactivity. One such antibody, mAb S25-2, has been shown to be close to the germline sequence and has not experienced significant mutational affinity maturation. This antibody shows broader cross-reactions than other Kdo antibodies but preferentially binds 2.8Kdo2 in native LPS (Fu et al. 1992; Nguyen et al. 2003; Brooks et al. 2008b).
We have investigated whether specific antibodies can be generated against the branched Kdo oligosaccharide present in LPS from C. psittaci, i.e. antibodies that do not cross-react with 2.4/2.4Kdo3, while simultaneously maintaining high affinity. In a previous study, mAb S69-4 obtained from a mouse immunized with the BSA-neoglycoconjugate of Kdo4 was able to distinguish between C. psittaci and other Chlamydiae in immunofluorescence detection but displayed a considerably lower affinity than other Kdo binding antibodies (Müller-Loennies et al. 2000; Müller-Loennies et al. 2006). In contrast, the homologous mAb S45-18 was shown to bind 2.4/2.4Kdo3 (Brade et al. 2000; Nguyen et al. 2003) with a very high affinity KD = 10 nM for the monovalent interaction as determined by SPR of its Fab fragment (this study) and does not cross-react with 2.8Kdo2 containing oligosaccharides, but is unable to distinguish between 2.4/2.4Kdo3 and Kdo4. The exceptionally high affinity of S45-18 and the low affinities of S69-4 [(Müller-Loennies et al. 2006), this study] and NH2240-39 (data not shown) were surprising considering their high sequence homology (Fig. 2). In order to explore the affinity differences between S69-4 and S45-18 we have created CDR3 hybrids. Transferring only three amino acids from the S45-18 D-gene into S69-4 improved the affinity in ELISA 250-fold towards HS4P, whereas the exchange of the N-terminal part of the CDR3 had no influence. Thus, although affinity maturation introduced a Phe at position H99 in both antibodies high affinity was seen only the context of the IGHD1-1*01 sequence (i.e., in S45-18), emphasizing the importance of the D-gene for generating specificity in the presence of the highly promiscuous inherited Kdo binding pocket. Using this hybrid antibody and 2.4/2.4PS4P as antigen, a 60-fold higher concentration was needed to obtain an OD405 of 1.0 in ELISA than for the branched ligand. There was only a 15-fold difference for the S45-18 Fab in the recognition of these two antigens and the bivalent mAb S45-18 did not discriminate between these structures, indicating that the discriminating properties of S69-4 have been partly retained in the mutant. Thus, other sequence differences contribute to the increased specificity of S69-4 for the branched HS4P. One way of achieving specificity for the latter structure could have been due to a reduction of affinity, rendering this antibody more sensitive towards minor structural conformational differences between these two closely related oligosaccharide structures. Confirmatory evidence came from the transfer of three amino acids from the VH CDR3 of S45-18 into S69-4, which led to a remarkable enhancement of affinity in binding assays and the similar affinities for both ligands determined by SPR. However, ELISA data did not agree as it showed that the mutant S69-4 retained a higher specificity for branched Kdo residues.
The difficulty in isolating a mAb specific for the branched hexasaccharide that does not cross-react with 2.4/2.4Kdo3 is illustrated by the crystal structure of mAb S45-18 in complex with 2.4/2.4PSBP (Nguyen et al. 2003), where the latter structure is recognized in a way that may allow the presence of an additional 2.8-linked Kdo residue that need not contact the antibody and therefore supports cross-reaction with the branched tetrasaccharide. To obtain high affinity antibodies against HS4P mice were immunized with a synthetic neoglycoconjugate of BSA and Kdo3br (Kosma et al. 2009) and antibodies isolated using hybridoma technology as well as phage display. The former yielded mAb S73-2 that bound HS4P with high affinity but was unable to distinguish between PS4P and HS4P (Brooks et al. 2009b). This antibody showed an unusual mode of binding by recognizing the inner Kdo of 2.8/2.4PS4P in a binding pocket occupied by the terminal Kdo residue when binding 2.4/2.4PS4P.
An attempt was made to use phage display technology to isolate HS4P specific antibodies useful for specific staining of C. psittaci. Successful panning of a scFv phage library against a neoglycoconjugate containing the branched Kdo oligosaccharide led to the isolation of the two closely related scFv NH2240-31 and SAG506-1, which were specific for C. psittaci in ELISA with whole bacteria. In addition, we have obtained scFv NH2240-39 that cross-reacted weakly between C. psittaci and C. pneumoniae in immunofluorescence experiments.
Genetic analysis (Brochet et al. 2008; Lefranc et al. 2009) revealed that the heavy chains of scFv NH2240-31, -39, and SAG506-01 originated from the same germline (IGHV7-3*02) and J-gene (IGHJ4*01) as mAbs S45-18, S25-2, S69-4, S73-2, but employed different D-genes. Both phage clones (which specifically bound to Kdo3br) possessed a much shortened CDR3 while the CDR3 of the cross-reactive clone NH2240-39 was of the same length as in S45-18 and S69-4. The longer CDR3 allows the positioning of residue PheH99 in the center of the combining site leading to binding of 2.4/2.4-linked Kdo residues and thus cross-reaction between linear 2.4/2.4PS4P and the branched HS4P. The much shortened CDR3 sequence in antibody SAG506-01 brings the aromatic ring of TyrH97 into a different position as compared to PheH99 of S45-18 (Fig. 3), and may be the source of the high affinity interaction with a branching Kdo. Unfortunately, cocrystallization trials with larger Kdo oligosaccharides have been unsuccessful, and the crystal packing of the current structures did not permit successful soaking experiments. An important additional structural alteration induced by affinity maturation was observed for the VL CDR1 sequence of these clones, where AsnL30a was replaced by a Lys residue. The importance of this interaction became clear after mutating this residue back to Asn, as the corresponding ELISA showed considerably reduced affinity towards HS4P.
Hydrophobic stacking interactions are known to play an important role in the recognition of carbohydrates by proteins. Our data show that this is also the case for highly negatively charged ligands such as Kdo4 (Fig. 1C). The positioning of a single aromatic residue within the VH CDR3 dictates specificity and cross-reactive properties towards Kdo containing ligands. However, a reduction of affinity by a weakening of hydrophobic stacking interactions appeared to be one possible factor by which an increased specificity can be obtained.
Immunization of mice with a synthetic neoglycoconjugate containing only the branching Kdo residues and phage display resulted in the isolation of a scFv which specifically recognizes C. psittaci in infected cell monolayers and which will be useful in the identification of this bacterium - particularly in veterinarian medicine, taxonomy and epidemiology.
Materials and methods
Bacteria
Partially purified elementary bodies of C. pneumoniae strain TWAR, C. trachomatis serotype L2 and C. psittaci strain 6BC (Rund et al. 1999; Rund et al. 2000) were prepared as described in the respective references. Bacteria used in ELISA or panning of the scFv library (see below) were inactivated with 0.02% formaldehyde, washed twice in PBS and finally suspended in phosphate buffered saline, pH 7.2.
Oligosaccharide antigens
For binding assays, we have used neoglycoconjugates containing oligosaccharides of Kdo which have been isolated from LPS of a recombinant E. coli strain (Re-mutant F515) with plasmid pUM140 (waaA of C. psittaci) or pFEN207 (Mamat et al. 1993; Holst et al. 1993) after deacylation and separation by high performance anion exchange chromatography as described (Holst et al. 1993). For ELISA, such oligosaccharides (Fig. 1) were conjugated to BSA after dephosphorylation at the anomeric position of the lipid A and reaction with allylamine and isothiocyanate as published (Müller-Loennies et al. 2002). The amount of ligand present in the conjugates was determined by measuring the amount of protein (Bradford assay, Bio-Rad) and Kdo (thiobarbiturate assay). The chemical structures are depicted in Fig. 1 and the abbreviations used in the text are indicated in Table 1. Sodium (3-deoxy-α-d-manno-oct-2-ulopyranosyl)onate-(2→8)-[sodium (3-deoxy-α-d-manno-oct-2-ulopyranosyl)onate-(2→4)]-sodium (allyl 3-deoxy-α-d-manno-oct-2-ulopyranosid)onate was chemically synthesized and conjugated to BSA as published (Kosma et al. 2009).
Immunization and monoclonal antibodies
Four Balb/c were mice immunized as described for the generation of mAb S73-2 (Brooks et al. 2009b). The isolation and characterization of mAbs S73-2, S45-18 (Brade et al. 2000) and S69-4 (Müller-Loennies et al. 2006) has been described in the respective references. MAb S45-18 preferentially binds 2.4/2.4Kdo (Brade et al. 2000; Nguyen et al. 2003), this study] with high affinity. MAb S69-4 also binds the latter antigen albeit with lower affinity despite a high homology to S45-18. It recognizes its epitope with slightly higher affinity in the branched Kdo4.
Phage display
For assembly of an scFv library displayed on phage, total RNA was isolated from 3.3 × 108 spleen cells of a mouse immunized with a synthetic neoglycoconjugate using the peqGold TriFast RNA isolation kit (PeqLab Biotechnologie GmbH, Erlangen, Germany). An aliquot (20 μg) of RNA was used for first strand cDNA synthesis by reverse transcription using SuperScript II reverse transcriptase (Invitrogen) and the supplied oligo (dT)12-18 primer. ScFv were assembled on M13 phage following the protocol of Barbas et al. using the pComb3XSS phagemid vector and the long linker mouse primer sets (Barbas III et al. 2001).
The panning procedure was done using a BSA neoglycoconjugate of the hexasaccharide bisphosphate (HSBP) obtained by reductive amination with glutardialdehyde (HSBP-GA-BSA, 4 μMol of Kdo per 1 mg of BSA). The concentration of the stock solution was 1.1 mg/ml based on the BSA content. All subsequent procedures were performed as described (Barbas III et al. 2001). Briefly, for panning of scFv-displaying phage 0.5 μl of neoglycoconjugate in PBS was immobilized on Maxisorp Lockwell U8 wells (Nunc, Germany) after addition of TBS (pH 7.5, 25 μl) for 16 h at 4 °C. After blocking (3% BSA in TBS), scFv-phage were added (2 times 6 × 1011 first round) and after washing with 0.5 % Tween 20 in TBS (5 times 150 μl each) bound phage were eluted using glycine/HCl, pH 2.2. We have compared two different conditions for panning during the initial three rounds, i.e. incubation at 4 °C for 16 h or for 2 h at 37 °C. Eluted phages after three rounds at 37 °C were then further panned for two rounds at 4 °C. Titers of the amplified phages used for the second to fifth round of panning were 3.7 × 1011, 2.6 × 1011, 2.4 × 1012, and 2.2 × 1012, respectively. Single colonies were used for colony PCR using Taq-Polymerase and primers ompseq and gback (Barbas III et al. 2001) at an annealing temperature of 58 °C. Amplified products were analyzed by BstOI restriction digest and isolated plasmids of 10 clones were sequenced each round of panning.
Sequencing
Sequencing was performed using the Blue Dye RR Terminator Cycle Sequencing kit (Applied Biosystems) according to the suppliers instructions. The sequencing reactions were purified with Centri-Sep spin columns (Applied Biosystems) prior to sequencing on a ABI PRISM 377 DNA Sequencer (Perkin Elmer).
Cloning and isolation of soluble scFv
For expression as soluble scFv, NH 2240-31 was cloned into the vector pSJF8 (MacKenzie and To 1998). After PCR amplification using the primers 5′-GGGGGGAATTCATGAAAAAGACAGCTATCGCGATTGC-3′ (reverse) and 5′-GGGGGATCCCTTCAAATCTTCCTCACTGATTAGCTTCTGTTCAGATCTTGAGGAGACGGTGACTGAGGT-3′ (forward) both scFv were cloned by EcoRI and BamHI restriction sites (underlined) introducing a myc-tag at 3′, replacing the HA-tag present in pComb3XSS. After transformation of E. coli TG-1 scFv were expressed as described (Anand et al. 1991). Soluble scFv was extracted from the periplasm of cultures grown over a period of five days at 24 °C using polymyxin B and purified by affinity chromatography on Ni2+-charged HiTrap columns (Amersham Biosciences, Uppsala, Sweden) as described (Müller-Loennies et al. 2006). Cloning of the scFv fragment of S69-4 has been described previously (Müller-Loennies et al. 2006).
Site-directed mutagenesis
The antibody combining sites of scFv S69-4 and NH2240-31 were mutated by site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) following the suppliers instructions. Double stranded plasmid DNA of pSJF8 (70 ng) which contained the insert of interest was used for each PCR reaction. In scFv 69-4, primers SAG175-01 5′-GTGCAAGAGATCTGATCTACTTTGATTACGACGATGCTATGGACTACTGGGGTCAAGGAACC-3′ and SAG175-02 5′-GGTTCCTTGTCCCCAGTAGTCCATCCCATCCCTGCTTCCAAAGTAGATCAGATCTCTTGCAC-3′ (annealing temperature 65 °C) changed the four amino acid stretch behind PheH99 from DYDD in VH CDR3 to GSRD (S69-4 H3post). Similarly, to change the three amino acid stretch before PheH99 from LIY to IYS (S69-4 H3ante) the primers SAG61-03 5′-GGATAGCGCCACTTATTACTGTGTGAGAGATATCTATTCCTTTGATTACGACGA-3′ and SAG61-13 5′-TCGTCGTAATCAAAGGAATAGATATCTCTCACACAGTAATAAGTGGCGCTATCC-3′ were used at an annealing temperature of 50 °C. PCR was performed with PfuTurbo DNA polymerase (Stratagene) and the following conditions: 1 min at 95 °C, 25 × (30 s at 95 °C/1 min at 65 °C or 50 °C/3.5 min at 72 °C), and final extension for 10 min at 72 °C.
An Ala/ProH52c mutation (CDR2) of scFv NH2240-31 was achieved as above using primers 5′-TGAGTGGTTGGGGTTCATTAGAAACAAGCCGAAAGGTTATACA-3′ and 5′-TGTATAACCTTTCGGCTTGTTTCTAATGAACCCCAACCACTCA-3′ and primers 5′-ATGAGCTGCAAATCCAGTCAGAGTCTGTTTAATAGTAG-3′ and 5′-CTACTATTAAACAGACTCTGACTGGATTTGCAGCTCAT-3′ for the mutation Lys/AsnL30a.
ELISA
For ELISA with scFv bound to phage elementary bodies from chlamydiae were immobilized over 48 h at 4 °C in 50 mM sodium carbonate buffer, pH 9.2 and blocked with 5% skim milk in PBS pH 7.2. Phage solutions were first diluted in PBS containing 5 % skim milk (1:3 v/v), 100 μl added to the first well and serially diluted. After washing bound phage was detected with horseradish peroxidase (HRP) conjugated mouse anti-M13 IgG (Amersham Biosciences, 1:5000 in PBS containing 5 % skim milk) and substrate and measured photometrically at 405 nm wavelength. ELISA using mAb and neoglycoconjugates was performed as described (Müller-Loennies et al. 2006). Binding of soluble scFv (conc. 50 μg/ml in PBS at start) was determined by ELISA against immobilized bacteria and immobilized neoglycoconjugates (20 pmol/cup) using unlabelled mouse anti-myc (clone 9E10) as second antibody. Antibody binding was then detected with horseradish peroxidase (HRP) conjugated anti-mouse IgG (H+L, Dianova) and ABTS substrate and measured photometrically at 405 nm. Experiments were done in quadruplicate and mean values were calculated. Confidence values did not exceed 10 %.
Fluorescence microscopy
HL cells infected with C. pneumoniae, and L929 fibroblast cells infected with C. psittaci 6BC or C. trachomatis L2; cells were grown in 96-well microtiter plate (Nunc) in Iskove’s medium containing 10 % fetal calf serum and 1 μg/ml cycloheximide. The plates were centrifuged for 1 h at 2000 rpm in a Hettich Rotanta AP centrifuge (450 x g). ScFv NH2240-31 or NH2240-39 (40 μg/ml in PBS) was added to the first row and serially diluted into the remaining wells. After incubation for 90 min at 37 °C, anti-myc IgG (clone 9E10) ascites (diluted 1:250) was added and after additional 90 min at 37 °C, detected by carbocyanin labeled goat anti-mouse IgG (H+L, Cy2 TM, Jackson ImmunoResearch Lab. Inc., Canada, used 1:80, 1 h, 37 °C). MAb S5-10 from unpurified supernatant diluted 1:2 and binding 2.8/2.4Kdo3 with high affinity (Brade et al. 1990), served as a positive control. Between incubations the plates were carefully washed 5 times with PBS, pH 7.2.
Surface plasmon resonance (SPR)
Measurements were performed by double referencing against a neoglycoconjugate of 1,4′-bisphosphorylated β1.6-linked GlcN disaccharide (lipid A backbone of LPS, BBP) on a Biacore T100 instrument (GE Healthcare Biosciences AB, Uppsala, Sweden) at analyte concentrations ranging from 1 nM to 100 nM. In each run duplicates for two concentrations were included and controls without analyte were performed before and after each series of concentrations. Neoglycoconjugates of oligosaccharides and BSA were immobilized using the amine coupling kit as described by the manufacturer on research grade CM5-sensor chips. The immobilized surface densities were 44 RU, 32 RU and 70 RU for HS4P, 2.4/2.4PS4P and 2.8/2.4PS4P, respectively, and BBP conjugated to BSA immobilized at similar surface densities of 32, 11, and 40 RU served as a reference. Analyses were carried out at 25 °C in 10 mM HEPES, pH 7.4, containing 150 mM NaCl, 0.05% (v/v) surfactant P-20 and 3 mM EDTA. Prior to the analysis the surface of the chip was flushed with a 6 s injection at 20 μl/min of 10 mM NaOH leading to an improved baseline stability. Analyses were performed at a flow rate of 30 μl/min and association and dissociation data were recorded for 3 min (90 μl) and 6 min (180 μl), respectively. Surface regeneration was generally necessary and was achieved by a 2 μl pulse injection of 10 mM NaOH at 20 μl/min. Data were evaluated by global fitting to a 1:1 interaction model using the BIAevaluation 3.0 software (Biacore, Inc.).
X-ray crystallography
The crystallization and structure determination of SAG506-01 has been previously reported (Brooks et al. 2008a)
Acknowledgments
We thank N. Harmel, Ute Agge, V. Susott, S. Cohrs and T. Hirama for technical assistance and the Deutsche Forschungsgemeinschaft (grant SFB470, C1) and Fonds zur Förderung der wissenschaftlichen Forschung (Grant P 13843) for financial support.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft [Grant SFB 470 C1 to SML and HB], the Austrian Science Fund FWF [Grant 13843 and 17407 to PK], and the Natural Sciences and Engineering Research Council of Canada [to SVE]. SVE is a Michael Smith Foundation for Health Research Senior Scholar.
Footnotes
Conflict of interest statement
None declared.
Abbreviations
- BSA
- bovine serum albumin
- CDR
- complementarity determining region
- D-gene
- diversity gene
- ELISA
- enzyme-linked immunosorbent assay
- FITC
- fluoresceine-isothiocyanate
- GlcN
- 2-amino-2-deoxy-glucopyranose
- HBS
- HEPES buffered saline
- HRP
- horseradish peroxidase
- Ig
- immunoglobulin
- J-gene
- joining gene
- Kdo
- 3-deoxy-α-D-manno-oct-2-ulopyranosonic acid
- LPS
- lipopolysaccharide
- mAb
- monoclonal antibody
- scFv
- single chain variable fragment
- SPR
- surface plasmon resonance
- VL
- variable domain of the light chain
- VH
- variable domain of the heavy chain
References
- Abhinandan KR, Martin AC. Analysis and improvements to Kabat and structurally correct numbering of antibody variable domains. Mol Immunol. 2008;45:3832–3839. doi: 10.1016/j.molimm.2008.05.022. [DOI] [PubMed] [Google Scholar]
- Anand NN, Mandal S, MacKenzie CR, Sadowska J, Sigurskjold B, Young NM, Bundle DR, Narang SA. Bacterial expression and secretion of various single-chain Fv genes encoding proteins specific for a Salmonella serotype B O-antigen. J Biol Chem. 1991;266:21874–21879. [PubMed] [Google Scholar]
- Barbas CF, III, Burton DR, Scott JK, Silverman GJ. Phage Display: A Laboratory Manual. Cold Spring Harbor Laboratory Press; New York: 2001. [Google Scholar]
- Brade H. Chlamydial lipopolysaccharide. In: Brade H, Opal SM, Vogel SN, Morrison DC, editors. Endotoxin in Health and Disease. Marcel Dekker Inc.; New York/Basel: 1999. pp. 229–242. [Google Scholar]
- Brade L, Holst O, Kosma P, Zhang YX, Paulsen H, Krausse R, Brade H. Characterization of murine monoclonal and murine, rabbit, and human polyclonal antibodies against chlamydial lipopolysaccharide. Infect Immun. 1990;58:205–213. doi: 10.1128/iai.58.1.205-213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brade L, Rozalski A, Kosma P, Brade H. A monoclonal antibody recognizing the 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo) trisaccharide αKdo(2-->4)αKdo(2-->4)αKdo of Chlamydophila psittaci 6BC lipopolysaccharide. J Endotoxin Res. 2000;6:361–368. [PubMed] [Google Scholar]
- Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CL, Blackler RJ, Gerstenbruch S, Kosma P, Müller-Loennies S, Brade H, Evans SV. Pseudo-symmetry and twinning in crystals of homologous antibody Fv fragments. Acta Crystallogr D Biol Crystallogr. 2008a;64:1250–1258. doi: 10.1107/S0907444908033453. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Blackler RJ, Sixta G, Kosma P, Müller-Loennies S, Brade L, Hirama T, MacKenzie CR, Brade H, Evans SV. The role of CDR H3 in Antibody Recognition of a Synthetic Analogue of a Lipopolysaccharide Antigen. Glycobiology. 2009a doi: 10.1093/glycob/cwp150. in press. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Müller-Loennies S, Borisova SN, Brade L, Kosma P, Hirama T, MacKenzie CR, Brade H, Evans SV. Antibodies raised against chlamydial lipopolysaccharide antigens reveal convergence in germline gene usage and differential epitope recognition. Biochemistry. 2009b doi: 10.1021/bi9011308. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CL, Müller-Loennies S, Brade L, Kosma P, Hirama T, MacKenzie CR, Brade H, Evans SV. Exploration of specificity in germline monoclonal antibody recognition of a range of natural and synthetic epitopes. J Mol Biol. 2008b;377:450–468. doi: 10.1016/j.jmb.2008.01.018. [DOI] [PubMed] [Google Scholar]
- Corsaro D, Valassina M, Venditti D. Increasing Diversity within Chlamydiae. Critical Reviews in Microbiology. 2003;29:37–78. doi: 10.1080/713610404. [DOI] [PubMed] [Google Scholar]
- Dean D, Kandel RP, Adhikari HK, Hessel T. Multiple Chlamydiaceae species in trachoma: implications for disease pathogenesis and control. PLoS Med. 2008;5:e14. doi: 10.1371/journal.pmed.0050014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Baumann M, Kosma P, Brade L, Brade H. A synthetic glycoconjugate representing the genus-specific epitope of chlamydial lipopolysaccharide exhibits the same specificity as its natural counterpart. Infect Immun. 1992;60:1314–1321. doi: 10.1128/iai.60.4.1314-1321.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst O, Broer W, Thomas-Oates JE, Mamat U, Brade H. Structural analysis of two oligosaccharide bisphosphates isolated from the lipopolysaccharide of a recombinant strain of Escherichia coli F515 (Re chemotype) expressing the genus-specific epitope of Chlamydia lipopolysaccharide. Eur J Biochem. 1993;214:703–710. doi: 10.1111/j.1432-1033.1993.tb17971.x. [DOI] [PubMed] [Google Scholar]
- Kosma P, Hofinger A, Müller-Loennies S, Brade H. Synthesis of a neoglycoconjugate containing a Chlamydophila psittaci specific branched Kdo trisaccharide epitope. Carbohydr Res. 2009 doi: 10.1016/j.carres.2009.12.015. [DOI] [PubMed] [Google Scholar]
- Lefranc MP, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, Wu Y, Gemrot E, Brochet X, Lane J, Regnier L, Ehrenmann F, Lefranc G, Duroux P. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie R, To R. The role of valency in the selection of anti-carbohydrate single-chain Fvs from phage display libraries. Journal of Immunological Methods. 1998;220:39–49. doi: 10.1016/s0022-1759(98)00143-4. [DOI] [PubMed] [Google Scholar]
- Mamat U, Baumann M, Schmidt G, Brade H. The genus-specific lipopolysaccharide epitope of Chlamydia is assembled in C. psittaci and C. trachomatis by glycosyltransferases of low homology. Mol Microbiol. 1993;10:935–941. doi: 10.1111/j.1365-2958.1993.tb00965.x. [DOI] [PubMed] [Google Scholar]
- Manimala JC, Roach TA, Li Z, Gildersleeve JC. High-throughput carbohydrate microarray profiling of 27 antibodies demonstrates widespread specificity problems. Glycobiology. 2007;17:17C–23C. doi: 10.1093/glycob/cwm047. [DOI] [PubMed] [Google Scholar]
- Morea V, Tramontano A, Rustici M, Chothia C, Lesk AM. Conformations of the third hypervariable region in the VH domain of immunoglobulins. J Mol Biol. 1998;275:269–294. doi: 10.1006/jmbi.1997.1442. [DOI] [PubMed] [Google Scholar]
- Moulder JW. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Loennies S, Grimmecke D, Brade L, Lindner B, Kosma P, Brade H. A novel strategy for the synthesis of neoglycoconjugates from deacylated deep rough lipopolysaccharides. J Endotoxin Res. 2002;8:295–305. doi: 10.1179/096805102125000506. [DOI] [PubMed] [Google Scholar]
- Müller-Loennies S, Gronow S, Brade L, MacKenzie R, Kosma P, Brade H. A monoclonal antibody against a carbohydrate epitope in lipopolysaccharide differentiates Chlamydophila psittaci from Chlamydophila pecorum, Chlamydophila pneumoniae, and Chlamydia trachomatis. Glycobiology. 2006;16:184–196. doi: 10.1093/glycob/cwj055. [DOI] [PubMed] [Google Scholar]
- Müller-Loennies S, MacKenzie CR, Patenaude SI, Evans SV, Kosma P, Brade H, Brade L, Narang S. Characterization of high affinity monoclonal antibodies specific for chlamydial lipopolysaccharide. Glycobiology. 2000;10:121–130. doi: 10.1093/glycob/10.2.121. [DOI] [PubMed] [Google Scholar]
- Nguyen HP, Seto NO, MacKenzie CR, Brade L, Kosma P, Brade H, Evans SV. Germline antibody recognition of distinct carbohydrate epitopes. Nat Struct Biol. 2003;10:1019–1025. doi: 10.1038/nsb1014. [DOI] [PubMed] [Google Scholar]
- Rund S, Lindner B, Brade H, Holst O. Structural analysis of the lipopolysaccharide from Chlamydia trachomatis serotype L2. J Biol Chem. 1999;274:16819–16824. doi: 10.1074/jbc.274.24.16819. [DOI] [PubMed] [Google Scholar]
- Rund S, Lindner B, Brade H, Holst O. Structural analysis of the lipopolysaccharide from Chlamydophila psittaci strain 6BC. Eur J Biochem. 2000;267:5717–5726. doi: 10.1046/j.1432-1327.2000.01635.x. [DOI] [PubMed] [Google Scholar]
- Schoonbroodt S, Steukers M, Viswanathan M, Frans N, Timmermans M, Wehnert A, Nguyen M, Ladner RC, Hoet RM. Engineering antibody heavy chain CDR3 to create a phage display Fab library rich in antibodies that bind charged carbohydrates. J Immunol. 2008;181:6213–6221. doi: 10.4049/jimmunol.181.9.6213. [DOI] [PubMed] [Google Scholar]
- Shirai H, Kidera A, Nakamura H. Structural classification of CDR-H3 in antibodies. FEBS Lett. 1996;399:1–8. doi: 10.1016/s0014-5793(96)01252-5. [DOI] [PubMed] [Google Scholar]
- Shirai H, Kidera A, Nakamura H. H3-rules: identification of CDR-H3 structures in antibodies. FEBS Lett. 1999;455:188–197. doi: 10.1016/s0014-5793(99)00821-2. [DOI] [PubMed] [Google Scholar]