Abstract
Acetylation of core histone N-terminal tails influences chromatin condensation and transcription. To examine how the SIN3–RPD3 deacetylase complex contributes to these events in vivo, we examined binding of SIN3 and RPD3 to Drosophila salivary gland polytene chromosomes. The binding patterns of SIN3 and RPD3 were highly coincident, suggesting that the SIN3–RPD3 complex is the most abundant chromatin-bound RPD3 complex in salivary gland cells. SIN3– RPD3 binding was restricted to less condensed, hypoacetylated euchromatic interbands and was absent from moderately condensed, hyperacetylated euchromatic bands and highly condensed, differentially acetylated centric heterochromatin. Consistent with its demonstrated role in transcriptional repression, SIN3–RPD3 did not co-localize with RNA polymer ase II. Chromatin binding of the complex, mediated by SMRTER, decreased upon ecdysone-induced transcriptional activation but was restored when transcription was reduced. These results implicate SIN3–RPD3 in maintaining histone acetylation levels or patterns within less condensed chromatin domains and suggest that SIN3–RPD3 activity is required, in the absence of an activation signal, to repress transcription of particular genes within transcriptionally active chromatin domains.
Keywords: chromatin/deacetylation/RPD3/SIN3/transcription
Introduction
Transcription in eukaryotic cells takes place on a chromatin template composed of DNA packaged by core histones (Wolffe, 1998). Although chromatin is held together tightly through protein–protein and protein– DNA interactions, its structure is dynamic. Structural constraints on DNA imposed by histones can be altered by chromatin remodeling complexes such as SWI/SNF family members or by a variety of core histone post-translational modifications, including acetylation of evolutionarily conserved lysine residues in the N-terminal tails of histones H3 and H4 (Grunstein, 1997; Wu, 1997; Spencer and Davie, 1999).
One model to explain how histone acetylation status affects gene expression proposes that the extent of chromatin condensation is directly correlated to the level of histone acetylation. Accordingly, hyperacetylation reduces the affinity of histone tails for DNA, resulting in less compact chromatin and increased accessibility of transcription factors to DNA (Lee et al., 1993; Vettese-Dadey et al., 1996; Hansen et al., 1998; Tse et al., 1998). A second, not necessarily mutually exclusive, model proposes that specific patterns of acetylation within histone tails dictate chromatin compaction and transcriptional activity (Turner, 1998; Strahl and Allis, 2000). It is well established that particular chromatin domains exhibit distinct patterns of histone acetylation. In Drosophila, antibodies against specific acetylated lysine (K) residues in histone H4 reveal that transcriptionally repressed centric heterochromatin is enriched in acetylated K12, while the transcriptionally hyperactive X chromosome in males is enriched in acetylated K16 (Turner et al., 1992). In yeast, the transcriptionally silenced mating type loci are enriched in acetylated K12 of H4 (Braunstein et al., 1996). Thus, specific acetylation patterns, perhaps combined with additional histone modifications, could serve as signals for binding of non-histone chromatin-binding factors that affect transcriptional activity (Strahl and Allis, 2000). It is also possible that specific acetylation patterns could serve as signals for recruitment of chromatin remodeling machines and/or heterochromatic factors (e.g. HP1) that modulate higher order chromatin packaging (Hansen et al., 1998; Strahl and Allis, 2000).
The acetylation status of histones is determined by the relative activities of multiprotein complexes that possess acetylase or deacetylase activities. Mutations in the yeast RPD3 deacetylase lead to a global increase in acetylation of H3 (K14 and K9/18) and H4 (K5, K12 and K16) (Rundlett et al., 1996). RPD3 (HDAC 1 and 2 in mammals) is a component of a 2 MDa complex that also contains SIN3, RbAp46/48, and SIN3-associated polypeptides 18 (SAP18) and 30 (SAP30) (Ayer, 1999). The SIN3–RPD3 complex does not directly bind DNA, but instead is targeted to particular genes by interactions with sequence-specific DNA-binding proteins (such as Mad, p53 and UME6) or nuclear hormone receptor-specific corepressors (such as N-CoR, SMRT and SMRTER) (Pazin and Kadonaga, 1997; Knoepfler and Eisenman, 1999; Murphy et al., 1999; Tsai et al., 1999). Repression of UME6-regulated genes in yeast is associated with localized SIN3- and RPD3-dependent deacetylation of H3 and H4 (preferentially K5 and K12) (Kadosh and Struhl, 1998; Rundlett et al., 1998).
SIN3-mediated repression is not completely RPD3 dependent. Mutations in SIN3 that prohibit RPD3 binding do not abolish transcriptional repression, suggesting that SIN3 has intrinsic repressor activity or may associate with other deacetylases (Laherty et al., 1997; Wong and Privalsky, 1998). Similarly, RPD3 functions independently of SIN3. RPD3 is a component of the Mi-2–NURD complex, which genetically interacts with the Drosophila repressor Hunchback (Kehle et al., 1998; Ayer, 1999). In addition, RPD3 directly interacts with corepressors, such as Drosophila Groucho (Chen et al., 1999). It is critical to note that the SIN3–RPD3 complex may not function exclusively in transcriptional repression. Studies in yeast have shown that the SIN3–RPD3 complex is required for maximal repression of uninduced genes and activation of induced genes (Vidal and Gaber, 1991; Vidal et al., 1991; Wang et al., 1994). Furthermore, the yeast SIN3–RPD3 complex reduces variegated transcriptional silencing of genes placed in close proximity to telomeres as well as stable silencing of genes at HM mating and rDNA loci (Vannier et al., 1996; Sun and Hampsey, 1999). In Drosophila, the role of RPD3 in regulating transcriptional silencing of euchromatic genes placed adjacent to heterochromatin by chromosomal rearrangements [position– effect variegation (PEV)] is less clear (Wakimoto, 1998). Mutations in RPD3 have been reported to enhance, suppress or not affect PEV (De Rubertis et al., 1996; Chen et al., 1999; Mannervik and Levine, 1999; Mottus et al., 2000). Mottus et al. (2000) propose that this discrepancy is due, in part, to the redundant activity of other deacetylases in RPD3 mutant flies. Drosophila encodes four other deacetylases with sequence similarity to RPD3 (Adams et al., 2000).
To understand how the Drosophila SIN3–RPD3 complex regulates transcription in vivo, we have asked whether there is a correlation between chromatin binding of the SIN3–RPD3 complex and chromatin condensation, histone acetylation status or transcriptional activity. An ideal system to address such relationships is polytene chromosomes prepared from wild-type Drosophila salivary gland cells. First, differences in chromatin condensation along polytene chromosomes can be visualized using a light microscope (Ashburner, 1989). Secondly, antibodies can be used to define the location of chromatin-associated proteins and have been used to map positions of histones, including various forms of acetylated histones and RNA polymerase II (pol II), which serves as a marker for active transcription (Jamrich et al., 1977; Turner et al., 1992; Weeks et al., 1993). Thirdly, polytene chromosomes reflect the transcriptional activity and factor-binding properties associated with chromatin of diploid interphase cells (Hill et al., 1987). This study uncovers properties of the SIN3–RPD3 complex that are critical to understanding how and to what extent it regulates transcription in vivo.
Results
Specificity of antibodies against SIN3 and RPD3
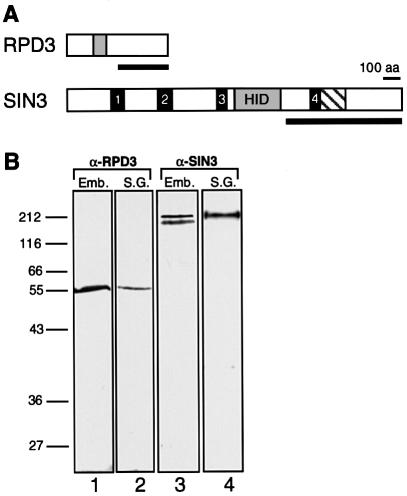
Antibodies specific for Drosophila RPD3 or SIN3 proteins were raised against recombinant proteins containing regions of the proteins that are divergent in primary sequence from their respective mammalian homologs (Figure 1A). The RPD3 antibody recognized a single protein of ∼56 kDa on western blots of Drosophila embryo and salivary gland extracts, consistent with the predicted size of 58 kDa (De Rubertis et al., 1996) (Figure 1B). The SIN3 antibody recognized two bands of ∼200 and 220 kDa in embryo extracts, but only a single 220 kDa band in salivary gland extracts (Figure 1B). Three Drosophila SIN3 isoforms are predicted, based on alternatively spliced cDNA clones; two differ by only 35 amino acids and are predicted to co-migrate at ∼190 kDa, while the third has an additional 330 C-terminal amino acids and is predicted to be 220 kDa (Neufeld et al., 1998; Pennetta and Pauli, 1998). Whole-mount immunostaining of Drosophila embryos revealed that SIN3 and RPD3 are present in all nuclei of the ovary, pre- and post-blastoderm embryos, and larval salivary glands (data not shown; Pennetta and Pauli, 1998; Chen et al., 1999). This is consistent with a general requirement for these proteins throughout Drosophila development. In fact, SIN3 and RPD3 may play more roles in Drosophila than they do in yeast, since they are essential for viability of flies but not yeast (Vidal et al., 1990; De Rubertis et al., 1996; Neufeld et al., 1998; Mottus et al., 2000).
Fig. 1. SIN3 and RPD3 polyclonal antibodies are highly specific. (A) Schematic diagrams of SIN3 and RPD3 proteins. Solid bars indicate regions used as antigens for generating polyclonal antibodies. The RPD3 region does not include the deacetylase domain, indicated by a shaded box. The SIN3 region contains paired-amphipathic helix (PAH) 4, indicated by a solid box, and a conserved region of undefined function, indicated by a hatched box, but does not include PAH1–3 or the histone deacetylase interaction domain (HID). (B) Western blots of total protein extracted from 0–12 h Drosophila embryos (Emb.) and from larval salivary glands (S.G.) were probed with purified RPD3 antibody (lanes 1 and 2) or SIN3 antibody (lanes 3 and 4). The positions of protein molecular weight size markers are indicated on the left.
The SIN3–RPD3 complex binds less-condensed interbands and is absent from more-condensed euchromatic bands and heterochromatin
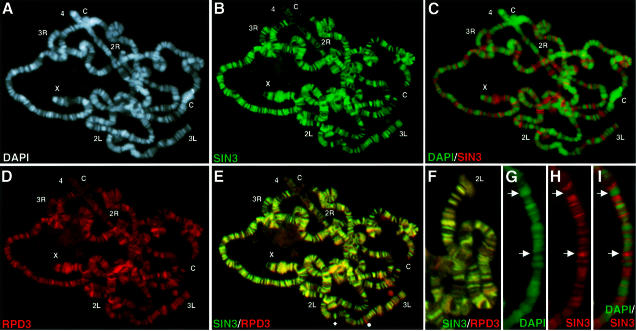
To identify loci associated with SIN3 and RPD3 in vivo, third instar larval salivary gland polytene chromosomes were stained with the antibodies described above. Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize the DNA (Figure 2A). DAPI staining is brightest in the condensed banded regions of euchromatin and in the constitutively condensed α-heterochromatin that comprises proximal regions of the six chromosome arms adjacent to the centromere (Ashburner, 1989; Gatti and Pimpinelli, 1992; Urata et al., 1995). β-heterochromatin, which connects α-heterochromatin to the euchromatic arms and is dispersed throughout chromosome 4, also stains brightly with DAPI. In salivary gland nuclei, the centric heterochromatin of all four chromosomes coalesces to form the chromocenter (‘C’ in Figure 2). DAPI staining is weak or absent in interbands and puffs. Chromatin condensation levels inferred from DAPI staining intensity are supported by atomic force microscopy, which reveals that interbands contain chromatin fibers that range from 11 to 300 nm, while bands contain predominantly 700 nm fibers (de Grauw et al., 1998).
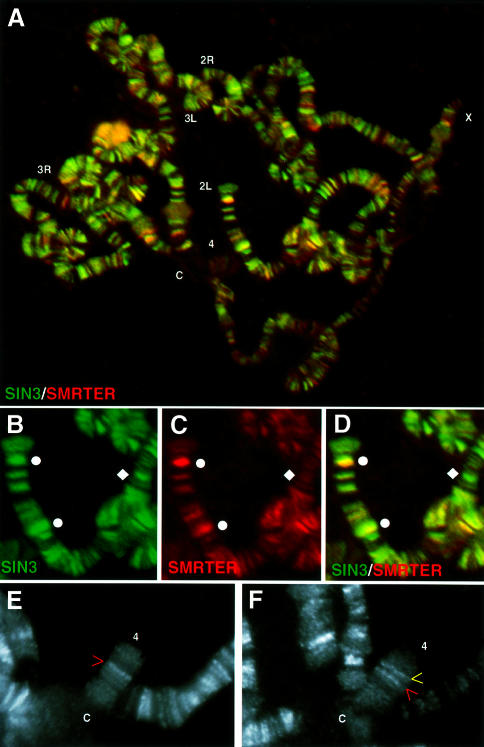
Fig. 2. SIN3 and RPD3 co-localize throughout euchromatin but are absent from heterochromatin. (A–E) A single polytene chromosome spread stained for both SIN3 and RPD3 and counterstained with DAPI. In (E), the diamond and sphere indicate loci that stain predominantly for SIN3 and RPD3, respectively. (F) Higher magnification image of a spread co-stained for SIN3 and RPD3. (G–I) Higher magnification images of another spread stained for SIN3 and counterstained with DAPI. Arrows highlight the non-overlapping pattern of SIN3 and DAPI. Co-localization of two antibodies appears as yellow fluorescence. Chromosome arms (X, 2L, 2R, 3L, 3R and 4) are indicated at the tip, and the chromocenter is indicated by ‘C’. In (A–E), the chromocenter is broken into two pieces. Antibodies used for staining are indicated at the bottom of each panel. The color of the lettering matches the color of the fluorescence.
Genes are present at different densities in each of these chromatin domains. Heterochromatin comprises approximately one-third of the total DNA in a diploid cell and is gene poor (Weiler and Wakimoto, 1995; Adams et al., 2000). α-heterochromatin is thought to lack genes and β-heterochromatin contains ∼40 genes. Euchromatic bands and interbands comprise approximately two-thirds of the total DNA and contain the balance of the RNA pol II genes. A limited number of studies comparing the distribution of genes in bands with that in interbands have found that some band–interband junctions have a high gene density and some bands have few genes per unit length (Hall et al., 1983; Spierer et al., 1983; Friedman et al., 1991). Detailed analysis of the Notch gene indicates that the transcriptional regulatory region maps to an interband, while the protein-coding region maps to the adjacent band (Rykowski et al., 1988). However, on a genome-wide basis, the question of gene density in bands versus interbands remains open.
SIN3 and RPD3 localized to portions of interbands throughout the genome (Figure 2B–D and H and I). The staining patterns of antibodies against SIN3 and RPD3 did not overlap the DAPI pattern, with the exception of some telomeres, and to varying degrees of intensity appeared to include most regions weakly stained by DAPI. Identical results were observed in single chromosome spreads stained for both SIN3 and RPD3. No staining above background was observed with pre-immune sera (data not shown). These results argue that SIN3 and RPD3 function in Drosophila at loci with a particular degree of chromatin condensation.
While SIN3 and RPD3 co-localized at almost all sites along the chromosome arms, the binding patterns were not identical (Figure 2E and F). The vast majority of interbands stained equally for SIN3 and RPD3 (yellow), but some are enriched for SIN3 (green) or RPD3 (red). If the intensity of fluorescence is a reflection of the number of binding sites for SIN3 and RPD3, as has been shown in the case of heat shock factor (Shopland et al., 1995), then these data suggest that SIN3 and RPD3 function together at most sites, as a SIN3–RPD3 complex, and independently at a minority of loci, perhaps as components of other complexes.
The SIN3–RPD3 complex was not present in centric heterochromatin, along most of chromosome 4, at some telomeres or in euchromatic bands (Figure 2A–F). Within constitutively condensed heterochromatic and telomeric regions, SIN3 and RPD3 were observed in a small band at the extreme tips of chromosome arms 2R and X as well as in a broad band at the tips of 2L and 3L (Figure 2A–F). Drosophila telomeres contain heterochromatic repeats and HeT-A and TART genes, which are actively transcribed during Drosophila development; it is not known whether the genes are active in salivary gland cells (Weiler and Wakimoto, 1995; Pardue et al., 1996). Therefore, the SIN3–RPD3 complex may perform similar functions at telomeres and interbands, both of which contain genes that are not constitutively silenced.
The SIN3–RPD3 complex is associated with chromatin containing hypoacetylated histones
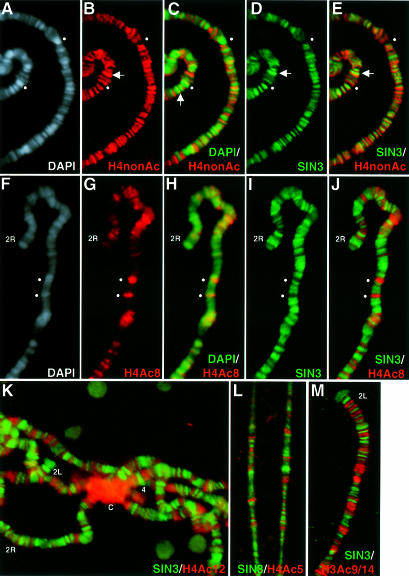
To determine the level of histone acetylation at loci bound by the SIN3–RPD3 complex, polytene chromosomes were stained with antibodies against both SIN3 and specific isoforms of histones H3 or H4, including non-acetylated H4 (H4nonAc), di-acetylated H3 and monoacetylated H4. The specificity of each of these antibodies has previously been demonstrated (Turner et al., 1989; Boggs et al., 1996; Taplick et al., 1998). This approach was used by Turner et al. (1992) to show that isoforms of H4 acetylated at K5, K8, K12 or K16 have distinct distribution patterns. In accord with these data, we found that antibodies against H4nonAc, H4 acetylated at lysine 5 (H4Ac5), H4Ac8, H4Ac12 or H3Ac9/14 stained subsets of bands, interbands and band–interband junctions (Figure 3). As previously reported using Chironomus polytene chromosomes (Turner et al., 1990), H4nonAc localized principally to interbands and was also commonly found at band– interband junctions (vertical arrow in Figure 3C). Finally, acetylated H3 and H4 predominantly localized to euchromatic bands that stain strongly with DAPI (Figure 3F–H and data not shown).
Fig. 3. Chromosomal sites of SIN3 binding and histone hyperacetylation are mutually exclusive. (A–E) A section of a single polytene chromosome spread stained for both H4nonAc and SIN3. Horizontal arrows indicate a region with strong H4nonAc and SIN3 binding. The vertical arrow indicates binding of H4nonAc at the band–interband junction. (F–J) A section of a single polytene chromosome spread stained for both SIN3 and H4Ac8. In (A–J), spheres indicate loci that stain strongly with DAPI and α-H4Ac8, but do not stain with α-H4nonAc or α-SIN3. (K–M) Polytene chromosome spreads co-stained for SIN3 and H4Ac12, H4Ac5 or H3Ac9/14, respectively. Co-localization of two antibodies appears as yellow fluorescence. Chromosome arms (2L, 2R and 4) are indicated at the tip and the chromocenter is indicated by ‘C’ in (K). Antibodies used for staining are indicated at the bottom of each panel. The color of the lettering matches the color of the fluorescence.
Polytene chromosome spreads stained for both SIN3 and specific acetylated histone H3 or H4 isoforms revealed that sites of major SIN3 and H4nonAc binding are very similar, although not entirely overlapping, as might be predicted from their positions relative to DAPI staining. Note that while the binding patterns of SIN3 and H4nonAc are very similar, the extent of co-localization does not match the high degree of SIN3 and RPD3 co-localization (Figure 3E, compare with Figure 2E and F). Often, the region bound by SIN3 was slightly shifted relative to H4nonAc, such that only a portion of the bands overlapped (horizontal arrow in Figure 3B, D and E). In striking contrast, strong staining with anti-SIN3 antibodies and strong staining with antibodies against acetylated histone were mutually exclusive (Figure 3H–M). Although there were some locations of overlap, the relative intensity of staining was not similar. Taken together, these data indicate that SIN3 is associated with loci that contain unacetylated or weakly acetylated histones and is not associated with loci that contain histones hyperacetylated at any of the lysine positions examined. Presumably, the same is true for RPD3, since the patterns of SIN3 and RPD3 binding were highly coincident (Figure 2).
The SIN3–RPD3 complex is not associated with actively transcribed loci
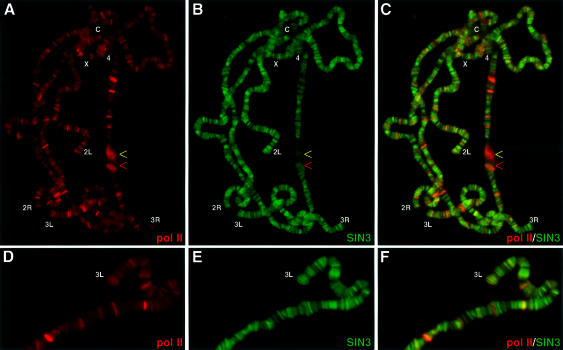
Previous studies have shown that active transcription is concentrated in the less-condensed interband and puff regions of polytene chromosomes (Zhimulev and Belyaeva, 1975; Visa et al., 1991). Furthermore, ribonucleoprotein particles and both the paused and elongating forms of RNA pol II have been localized to these same regions (Jamrich et al., 1977; Mott and Hill, 1986; Weeks et al., 1993). To examine directly the relationship between the SIN3–RPD3 complex and active transcription, polytene chomosomes were stained for both SIN3 and RNA pol II. The antibody against RNA pol II is directed against subunit IIc and recognizes both paused and elongating forms of the polymerase (Skantar and Greenleaf, 1995). As presented in Figure 4, the bulk of SIN3 and RNA pol II bound distinct regions of euchromatic interbands. There were numerous unequal bands (green or red) and very few equal intensity bands (yellow). Thus, while the SIN3– RPD3 complex was bound at loci containing protein-coding genes, the majority of the complex did not bind regions that are actively transcribed by RNA pol II. These data support the model that the SIN3–RPD3 complex is involved in transcriptional repression in vivo.
Fig. 4. SIN3 and RNA pol II do not co-localize on polytene chromosomes. (A–C) A single polytene chromosome spread stained for SIN3 and RNA pol IIc. (D–F) Higher magnification images of a different spread co-stained for SIN3 and RNA pol IIc. Co-localization of two antibodies appears as yellow fluorescence. Chromosome arms (X, 2L, 2R, 3L, 3R and 4) are indicated at the tip; the chromocenter is indicated by a C. The red arrow indicates the puff at 74EF and the yellow arrow indicates the puff at 75B, as in Figure 6. Antibodies used for staining are indicated at the bottom of each panel. The color of the lettering matches the color of the fluorescence.
The dynamic pattern of SIN3–RPD3 complex binding to ecdysone-regulated loci suggests that it functions to repress uninduced genes
To address the question of whether binding of the SIN3–RPD3 complex is abrogated when a gene becomes transcriptionally active, we focused on ecdysone-inducible genes, which comprise interband chromatin at 74EF and 75B. In Drosophila, pulses of the steroid hormone ecdysone control gene expression patterns, which trigger the metamorphosis from larva to adult (Ashburner, 1972; Thummel, 1996). The beginning of the ecdysone response is marked by chromatin decondensation at 74EF and 75B during puff stage 2 [PS2, as designated by Ashburner (1972)]. Puffing largely parallels transcriptional activation of the E74 and E75 genes, except for E74A transcripts, which remain abundant until PS11 while the 74EF puff regresses between PS8 and PS10. These morphological and molecular events are directed by the nuclear hormone receptors, ecdysone receptor (EcR) and ultraspiracle (USP), the Drosophila retinoid X receptor (RXR) homolog (Yao et al., 1992, 1993; Thomas et al., 1993). EcR–USP heterodimers regulate transcription in an ecdysone-dependent manner, repressing target genes in the absence of ecdysone and activating them in its presence (Yao et al., 1992, 1993; Thomas et al., 1993; Tsai et al., 1999). Tsai et al. (1999) have shown that the corepressor SMRTER (a protein with sequence similarity to the mammalian nuclear hormone receptor corepressor SMRT) mediates repression by interacting with EcR and SIN3. These studies predict that SMRTER and SIN3 should co-localize on polytene chromosomes, and that SMRTER and SIN3 chromatin-binding levels may change during the ecdysone pulse.
Co-staining of polytene chromosomes showed that SIN3 and SMRTER binding patterns largely overlapped throughout the genome (Figure 5A–D). Sites of strong staining with anti-SMRTER antibodies are sites of strong staining with anti-SIN3 antibodies. Thus, it appears that a major mechanism for targeting SIN3 to specific chromosomal loci is through association with SMRTER. However, there were sites of strong staining with anti-SIN3 antibody that were weakly stained or not stained by anti-SMRTER antibody, implying that SIN3 is targeted by other corepressors or DNA-binding proteins.
Fig. 5. SIN3 binds steroid hormone-regulated loci. (A) A single polytene chromosome spread stained for both SIN3 and SMRTER. (B–D) Higher magnification images of a section of the spread in (A). Spheres indicate loci strongly stained for both SIN3 and SMRTER (yellow); a diamond indicates a locus stained for SIN3 but not SMRTER (green). Co-localization of two antibodies appears as yellow fluorescence. Chromosome arms (X, 2L, 2R, 3L, 3R and 4) are indicated at the tip and the chromocenter is indicated by a C. Antibodies used for staining are indicated at the bottom of each panel. The color of the lettering matches the color of the fluorescence. (E and F) High magnification images of chromosome 4 of spreads stained with SIN3 prepared from flies of a wild-type (E) or an Sgs-4 transgenic (F) line. The transgenic line has a fluorescent signal at the P-element insertion site at 102D3-5 (yellow arrow) in addition to the single strong band present in the wild-type line (red arrow).
To confirm that the polytene staining assay accurately reflects SIN3 association with individual genes that are developmentally regulated, we asked whether a new SIN3-binding site is present in a fly strain containing a salivary gland secretion protein-4 (Sgs-4) transgene (Lehmann and Korge, 1996). Sgs-4 is one of the best studied of the Sgs genes that are induced by ecdysone in the middle of the third larval instar and are downregulated at the end of the third larval instar. The Sgs-4 P-element transgene is composed of four copies of the Sgs-4 regulatory region (–567 to –151). This region contains sites that bind the EcR–USP heterodimer in vitro (Lehmann and Korge, 1995). Figure 5E and F reveals that in this strain a new fluorescent signal was detected at the P-element integration site, 102D3-5 on the fourth chromosome, in late third instar larval salivary glands. Thus, SIN3 binding is detected on a single gene at a time in development when the gene is transcriptionally repressed.
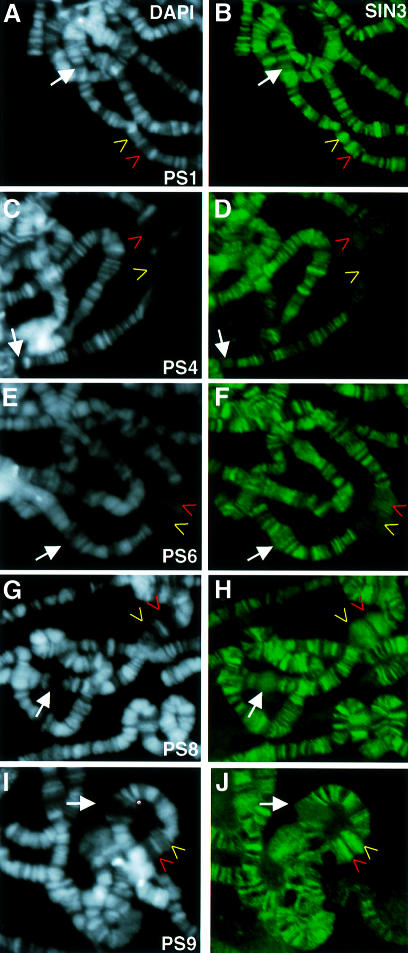
Furthermore, binding of SIN3 to polytene chromosomes changes as a gene progresses through the transcription cycle from a transcriptionally repressed to an active state and then back to a repressed state. This is evident upon analysis of SIN3 binding at 74EF and 75B from PS1 to PS9, during which time the transcription cycle is correlated with the formation and regression of puffs (Ashburner, 1972; Thummel, 1996; Richards, 1997). At PS1, SIN3 moderately bound unpuffed 74EF and 75B (Figure 6A and B), but binding was absent or weak at PS4 (Figure 6C and D) and up through PS6 (Figure 6E and F) when puffing was apparent. Strong USP staining persisted during PS4–PS6, supporting the idea that USP is required for transcriptional activation at these loci (data not shown). USP and RNA pol II staining of PS4–PS6 puffs also argues that the lack of SIN3 staining at these stages is not due to an inability to detect proteins bound at highly decondensed loci (Figure 4A and C). At PS8, SIN3 moderately bound the regressing puffs (Figure 6G and H) and by PS9 strongly bound the almost completely regressed puffs (Figure 6I and J). Similar analysis revealed that RPD3 bound 74EF and 75B with kinetics indistinguishable from those of SIN3 (data not shown). Thus, high levels of SIN3–RPD3 complex binding correlate with transcriptional repression and low levels with transcriptional activation.
Fig. 6. SIN3 binding changes during the transcription cycle. (A, C, E, G and I) Images of DAPI-stained polytene chromosome spreads. (B, D, F, H and J) Images of corresponding polytene chromosome spreads stained with SIN3 antibody. Spreads are arranged according to developmental timing, from PS1 (A and B) to PS9 (I and J). In each panel, the red arrow indicates the puff at 74EF, the yellow arrow the puff at 75B and the white arrow the puff at 78D. The degree of puffing at 78D relative to that at 74EF and 75B was one of several criteria used to determine the puff stage, as designated by Ashburner (1972). Panels are shown at the same magnification.
Acetylated H3 or H4 staining was rarely observed in the transcriptionally active 74EF and 75B puffs (data not shown and Turner et al., 1992). As suggested by recent studies in yeast and Drosophila, this may be due to the transient or localized nature of histone acetylation during gene activation (Cavalli and Paro, 1999; Krebs et al., 1999).
Discussion
The determination of SIN3 and RPD3 chromosome-binding patterns in wild-type interphase cells has provided a framework for understanding how these proteins affect chromatin condensation, histone acetylation and transcription in vivo.
The SIN3–RPD3 complex is the predominant form of chromatin-bound SIN3 and RPD3
The binding patterns of SIN3 and RPD3 on polytene chromosomes were highly coincident, suggesting that the SIN3–RPD3 complex is the major form of chromatin-bound SIN3 and RPD3 in salivary gland cells. The extent of co-localization was unexpected given that the Mi-2– NURD complex is the major form of RPD3 present in nuclear extracts prepared from metazoan cells (Wade et al., 1998, 1999; Xue et al., 1998; Zhang et al., 1998). This inconsistency may reflect organismal differences or differences between salivary gland cells that are not dividing and cells that are prepared for rapid division (Xenopus laevis eggs) or are actively dividing (mammalian tissue culture cells). Alternatively, the chromatin-bound SIN3–RPD3 complex may be under-represented in nuclear extracts because it is resistant to extraction from nuclei. To date, the constellation of SIN3- and RPD3-containing complexes has not been characterized in Drosophila. However, Drosophila does encode homologs of all of the Mi-2–NURD subunits (Kehle et al., 1998; Wade et al., 1999; E.Ballestar, L.A.Pile, D.A.Wassarman, A.P.Wolffe and P.A.Wade, manuscript in preparation). Staining of polytene chromosomes with an antibody to the dMBD-like subunit of the Mi-2–NURD complex indicates that this complex is bound at only a small number of loci and that these loci do not overlap significantly with those bound by SIN3 (E.Ballestar, L.A.Pile, D.A.Wassarman, A.P.Wolffe and P.A.Wade, manuscript in preparation). Thus, in interphase salivary gland cells, chromatin-bound RPD3 is primarily a component of the SIN3–RPD3 complex.
The SIN3–RPD3 complex regulates transcription within decondensed chromatin domains
The SIN3–RPD3 complex is bound at most if not all euchromatic interbands and is absent from euchromatic bands and constitutive heterochromatin at the chromocenter. Since the SIN3–RPD3 complex functions as a transcriptional repressor and antagonizes transcriptional silencing of genes within and adjacent to heterochromatic domains, it might have been expected to localize to more-condensed chromatin domains, such as euchromatic bands, as well as regions that are transcriptionally silenced, such as centric heterochromatin. This is clearly not the case and consequently eliminates models proposing that the SIN3–RPD3 complex represses transcription by maintaining chromatin in a condensed state or antagonizes PEV by binding heterochromatic domains and preventing them from spreading. Instead, the localization pattern argues that the SIN3–RPD3 complex functions to repress transcription within decondensed regions of the genome.
The localization pattern of the SIN3–RPD3 complex provides additional evidence that it functions to repress transcription in vivo. First, the SIN3–RPD3 complex did not co-localize with transcriptionally active or paused forms of RNA pol II or the TAF250 subunit of the general transcription factor TFIID (data not shown). This indicates that the SIN3–RPD3 complex may produce histone acetylation levels or patterns that are inhibitory to RNA pol II transcription. Secondly, the SIN3–RPD3 complex dissociates from specific loci upon transcriptional activation triggered by a pulse of ecdysone and reassociates coincident with a reduction in transcription levels. This indicates that the SIN3–RPD3 complex functions as a transcriptional repressor at sites where it is bound and is not just stored at these sites. Furthermore, this indicates that the SIN3–RPD3 complex probably does not function as a transcriptional activator and that reduced levels of activation in yeast sin3 or rpd3 mutants may be an indirect effect (Vidal and Gaber, 1991; Vidal et al., 1991; Wang et al., 1994).
The localization pattern also provides a rough indication of the extent to which the SIN3–RPD3 complex functions in interphase cells. The complex localized to portions (maybe as much as half) of almost all interbands. The amount of DNA in interbands has been estimated to be between 4 and 26% of total DNA (Beermann, 1972; Laird, 1980; Kress et al., 1985). Assuming that (i) acetylated histones and possibly other chromatin-associated proteins are the in vivo targets of the RPD3 deacetylase, (ii) the SIN3–RPD3 complex is functional at sites where it binds and (iii) gene density is equivalent in bands and interbands (see Results), the SIN3–RPD3 complex probably regulates 2–13% of Drosophila genes. This estimation is in line with the finding that the expression of ∼2% of genes is changed in response to treatment of mammalian tissue culture cells with histone deacetylase inhibitors (Van Lint et al., 1996).
The SIN3–RPD3 complex is bound to chromatin domains with hypoacetylated histones
Co-localization of the SIN3–RPD3 complex with unacetylated but not acetylated histones suggests that it functions to maintain histones in a hypoacetylated state. In vitro, yeast RPD3 can deacetylate all lysine residues in the tails of histones H3 and H4, and in vivo, mutations in rpd3 result in increased acetylation of 6 of the 10 lysine residues (Rundlett et al., 1996; Kuo and Allis, 1998). Thus, co-localization of the SIN3–RPD3 complex with hypoacetylated histones suggests that RPD3 also displays broad substrate specificity in vivo in Drosophila.
The assay can only detect general changes in acetylation patterns and is not meant to be used as a quantitative measure. For example, the assay is not sensitive enough to detect changes in histone acetylation at ecdysone-induced puffs, which presumably occur after dissociation of the SIN3–RPD3 complex (Figure 3 and data not shown). The inability to detect these changes may be due to transient acetylation, as occurs in other systems but which was not captured in any of the single time point ‘snap-shots’ of fixed polytene chromosome spreads (Cavalli and Paro, 1999; Krebs et al., 1999). On the other hand, if acetylation is stable for some period of time but is localized to promoter regions of these genes, as has been demonstrated for some genes, then the antibody staining signal may be too weak to detect (Kadosh and Struhl, 1998; Rundlett et al., 1998; Burgess et al., 1999).
The lack of SIN3–RPD3 complex binding at centric heterochromatin, which is hypoacetylated relative to euchromatin at all lysine residues in histones H3 and H4 except for K8 (data not shown) and K12 of H4, indicates that other deacetylases function within this domain. The SIR2 deacetylase is an excellent candidate, since it is required for silencing in yeast and there are five SIR2 homologs in Drosophila (Adams et al., 2000; Imai et al., 2000). Similarly, non-acetylated H4 is localized to some interband regions that are not bound by the SIN3–RPD3 complex, suggesting that other members of the RPD3 family may regulate histone deacetylation in interbands.
The SIN3–RPD3 complex may generate a transcriptionally repressed but inducible chromatin configuration
The SIN3–RPD3 complex is found at less condensed, hypoacetylated and transcriptionally inactive regions of the genome. The complex does not bind DNA, but, rather, is targeted to specific promoters through protein–protein interactions with DNA-binding factors such as Mad, nuclear hormone receptors and CBF1/RPK-Jκ (Pazin and Kadonaga, 1997; Kao et al., 1998). Each of these factors binds the transcriptional regulatory region of genes and provides a mechanism for them to be switched from an inactive to active state by an activation signal. Localization of these inducible genes to decondensed regions of the genome may allow them to respond quickly to an activation signal. The role of the SIN3–RPD3 complex may be to modify the pattern of histone acetylation at these genes, resulting in a transcriptionally repressive local nucleosomal environment. Our hypothesis regarding the activity of the complex is in accord with the supposition that ‘genes that must be readily induced would be designed to respond to appropriate transcription factors through a mechanism in which nucleosomes are easily disrupted’ (Felsenfeld, 1996). In conclusion, the activity of the SIN3–RPD3 complex is not to aid in the maintenance of repressive chromatin domains (e.g. silenced domains), but rather to inactivate the transcription of genes that are located in less condensed chromatin and therefore require active repression.
Materials and methods
Antibody production
Rabbit polyclonal antibodies were generated against recombinant proteins containing a portion of RPD3 (amino acids 273–522) or SIN3 (amino acids 1328–1746) (De Rubertis et al., 1996; Neufeld et al., 1998). The IgG fraction of each serum was prepared using the Econo-Pac serum IgG purification kit (Bio-Rad). Purified antibodies against SIN3 were directly conjugated to fluorescein using a fluorescein labeling kit (Boehringer Mannheim).
Western blot analysis
Western blot analysis was performed using standard protocols (Sambrook et al., 1989). To prepare embryo extracts, ∼100 µl of 0–12 h w1118 embryos were homogenized in 400 µl of 2× Laemmli sample buffer (Bio-Rad). To prepare salivary gland extracts, glands were dissected from third instar larvae or white pre-pupae in phosphate-buffered saline (PBS) and homogenized in 2× Laemmli sample buffer. Extracts were separated by 10% SDS–PAGE, transferred to Immobilon-P polyvinylidene fluoride (PVDF) membrane (Millipore), probed with purified IgG antibodies against RPD3 or SIN3 (1:500) followed by donkey anti-rabbit horseradish peroxidase IgG (1:3000) (Amersham Pharmacia Biotech) and detected using enhanced chemiluminescence reagents (Amersham Pharmacia Biotech). The SIN3 isoform, described by Pennetta and Pauli (1998) as potentially ovary specific, was detected in embryos (Figure 1B). Control blots probed with pre-immune SIN3 and RPD3 sera, at concentrations equivalent to those indicated above, showed no signal.
Polytene chromosome staining
Polytene chromosome squashes and staining were performed as previously described (Zink and Paro, 1989; Westwood et al., 1991) on Canton-S flies except for the experiment shown in Figure 5F, which was performed on Sgs-4 transgenic flies line T4, 7-6 (kindly provided by G.Korge). Briefly, salivary glands were dissected in PBS and treated by one of two methods. In the first method, glands were placed in fixative containing 3.7% formaldehyde, 1% Triton X-100 and PBS for 30 s and then transferred to fixative containing 3.7% formaldehyde, 45% acetic acid for 1 min before squashing. In the second method, glands were placed directly in fixative containing 3.7% formaldehyde, 45% acetic acid for 1 min before squashing. We were unable to detect differences in staining patterns of spreads prepared by the two methods, but those prepared by the second method tended to have better morphology.
For spreads stained with the histone antibodies, salivary glands were dissected in PBS containing 5 mM sodium butyrate as described in Turner et al. (1992). As the antibodies against RPD3, SIN3, SMRTER (kindly provided by R.Evans), non-acetylated histone H4 (Serotec) and acetylated lysine (Upstate Biotechnology) were all raised in rabbits, co-staining was performed using a SIN3 antibody that was directly conjugated to fluorescein. The sequence of staining was as follows: (i) anti-RPD3 (1:20), anti-H4nonAc (1:50), anti-H4Ac5 (1:20), anti-H4Ac8 (1:20), anti-H4Ac12 (1:20), anti-H3Ac9/14 (1:75) or anti-SMRTER (1:100) primary; (ii) Alexa 594 goat anti-rabbit IgG (1:400) secondary (Molecular Probes); (iii) fluorescein anti-SIN3 (1:10). The SIN3/RNA pol IIc spreads were simultaneously stained with anti-SIN3 (1:100) and goat anti-RNA pol IIc (1:100) (gAP-αD1 kindly provided by A.Greenleaf) primary antibodies followed by Alexa 594-conjugated donkey anti-goat IgG (1:400) (Molecular Probes) and fluorescein isothiocyanate-conjugated donkey anti-rabbit IgG (1:100) (Jackson Laboratories). DNA was visualized with DAPI (1:1000). Control spreads stained with pre-immune SIN3 and RPD3 sera, at concentrations equivalent to those indicated above, showed no staining (data not shown). Each staining experiment was performed several times. Figures 2–6 show representative spreads.
Acknowledgments
Acknowledgements
We thank Ron Evans, Arno Greenleaf, Gunter Korge and Tatiana Kozlova for providing reagents, Todd Laverty for analysis of polytene chromosomes, Frances Lee for characterizing the SIN3 and RPD3 antibodies, Mary Lilly for assistance with microscopy, Paul Wade and Eric Baehrecke for providing thoughtful advice throughout the course of these experiments, and Norikazu Aoyagi, Sonja Ghidelli, Dmitry Guschin, Rohinton Kamakaka, Fyodor Urnov, Paul Wade and Alan Wolffe for comments that greatly improved the manuscript. This work was supported by the Intramural Program in the National Institute of Child Health and Human Development.
References
- Adams M.D. et al. (2000) The genome sequence of Drosophila melanogaster.Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- Ashburner M. (1972) Puffing patterns in Drosophila melanogaster and related species. In Beermann,W., Reinert,J. and Urspriun,H (eds), Results and Problems in Cell Differentiation, Vol. 4. Springer-Verlag, New York, NY, pp. 102–151. [DOI] [PubMed] [Google Scholar]
- Ashburner M. (1989) Drosophila—A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 33–71. [Google Scholar]
- Ayer D.E. (1999) Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol., 9, 193–198. [DOI] [PubMed] [Google Scholar]
- Beermann W. (1972) Chromomeres and genes. In Beermann,W., Reinert,J. and Urspriun,H (eds), Results and Problems in Cell Differentiation, Vol. 4. Springer-Verlag, New York, NY, pp. 1–34. [DOI] [PubMed] [Google Scholar]
- Boggs B.A., Connors,B., Sobel,R.E., Chinault,A.C. and Allis,C.D. (1996) Reduced levels of histone H3 acetylation on the inactive X chromosome in human females. Chromosoma, 105, 303–309. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Sobel,R.E., Allis,C.D., Turner,B.M. and Broach,J.R. (1996) Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol., 16, 4349–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S.M., Ajimura,M. and Kleckner,N. (1999) GCN5-dependent histone H3 acetylation and RPD3-dependent histone H4 deacetylation have distinct, opposing effects on IME2 transcription, during meiosis and during vegetative growth, in budding yeast. Proc. Natl Acad. Sci. USA, 96, 6835–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G. and Paro,R. (1999) Epigenetic inheritance of active chromatin after removal of the main transactivator. Science, 286, 955–958. [DOI] [PubMed] [Google Scholar]
- Chen G., Fernandez,J., Mische,S. and Courey,A.J. (1999) A functional interaction between the histone deacetylase RPD3 and the corepressor Groucho in Drosophila development. Genes Dev., 13, 2218–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Grauw C.J., Avogadro,A., van den Heuvel,D.J., van den Werf,K.O., Otto,C., Kraan,Y., van Hulst,N.F. and Greve,J. (1998) Chromatin structure in bands and interbands of polytene chromosomes imaged by atomic force microscopy. J. Struct. Biol., 121, 2–8. [DOI] [PubMed] [Google Scholar]
- De Rubertis F., Kadosh,D., Henchoz,S., Pauli,D., Reuter,G., Struhl,K. and Spierer,P. (1996) The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature, 384, 589–591. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. (1996) Chromatin unfolds. Cell, 86, 13–19. [DOI] [PubMed] [Google Scholar]
- Friedman T.B., Owens,K.N., Burnett,J.B., Saura,A.O. and Wallrath,L.L. (1991) The faint band/interband region 28C2 to 28C4-5(–) of the Drosophila melanogaster salivary gland polytene chromosomes is rich in transcripts. Mol. Gen. Genet., 226, 81–87. [DOI] [PubMed] [Google Scholar]
- Gatti M. and Pimpinelli,S. (1992) Functional elements in Drosophila melanogaster heterochromatin. Annu. Rev. Genet., 26, 239–275. [DOI] [PubMed] [Google Scholar]
- Grunstein M. (1997) Histone acetylation in chromatin structure and transcription. Nature, 389, 349–352. [DOI] [PubMed] [Google Scholar]
- Hall L.M., Mason,P.J. and Spierer,P. (1983) Transcripts, genes and bands in 315,000 base-pairs of Drosophila DNA. J. Mol. Biol., 169, 83–96. [DOI] [PubMed] [Google Scholar]
- Hansen J.C., Tse,C. and Wolffe,A.P. (1998) Structure and function of the core histone N-termini: more than meets the eye. Biochemistry, 37, 17637–17641. [DOI] [PubMed] [Google Scholar]
- Hill R.J., Mott,M.R. and Steffensen,D.M. (1987) The preparation of polytene chromosomes for localization of nucleic acid sequences, proteins, and chromatin conformation. Int. Rev. Cytol., 108, 61–118. [DOI] [PubMed] [Google Scholar]
- Imai S., Armstrong,C.M., Keiberlein,M. and Guarente,L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature, 403, 795–800. [DOI] [PubMed] [Google Scholar]
- Jamrich M., Greenleaf,A.L. and Bautz,E.K.F. (1977) Localization of RNA polymerase in polytene chromosomes of Drosophila melanogaster.Proc. Natl Acad. Sci. USA, 74, 2079–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D. and Struhl,K. (1998) Targeted recruitment of the SIN3–RPD3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol., 18, 5121–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao H.-Y., Ordentlich,P., Koyano-Nakagawa,N., Tang,Z., Downes,M., Kinter,C.R., Evans,R.M. and Kadesch,T. (1998) A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev., 12, 2269–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehle J., Beuchle,D., Treuheit,S., Christen,B., Kennison,J.A., Bienz,M. and Muller,J. (1998) dMi-2, a Hunchback-interacting protein that functions in polycomb repression. Science, 282, 1897–1900. [DOI] [PubMed] [Google Scholar]
- Knoepfler P.S. and Eisenman,R.N. (1999) Sin meets NuRD and other tails of repression. Cell, 99, 447–450. [DOI] [PubMed] [Google Scholar]
- Krebs J.E., Kou,M.-H., Allis,C.D. and Peterson,C.L. (1999) Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev., 13, 1412–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress H., Meyerowitz,E.M. and Davidson,N. (1985) High resolution mapping of in situ hybridized biotinylated DNA to surface-spread Drosophila polytene chromosomes. Chromosoma, 93, 113–122. [DOI] [PubMed] [Google Scholar]
- Kuo M.-H. and Allis,C.D. (1998) Roles of histone acetyltransferases and deacetylases in gene regulation. BioEssays, 20, 615–626. [DOI] [PubMed] [Google Scholar]
- Laherty C.D., Yang,W.M., Sun,J.M., Davie,J.R., Seto,E. and Eisenman,R.N. (1997) Histone deacetylases associated with the mSIN3 corepressor mediate Mad transcription repression. Cell, 89, 349–356. [DOI] [PubMed] [Google Scholar]
- Laird C.D. (1980) Structural paradox of polytene chromosomes. Cell, 22, 869–874. [DOI] [PubMed] [Google Scholar]
- Lee D.Y., Hayes,J.J., Pruss,D. and Wolffe,A.P. (1993) A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell, 72, 73–84. [DOI] [PubMed] [Google Scholar]
- Lehmann M. and Korge,G. (1995) Ecdysone regulation of the Drosophila Sgs-4 gene is mediated by the synergistic action of ecdysone receptor and SEBP 3. EMBO J., 14, 716–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M. and Korge,G. (1996) The forkhead product directly specifies the tissue-specific hormone responsiveness of the Drosophila Sgs-4 gene. EMBO J., 15, 4825–4834. [PMC free article] [PubMed] [Google Scholar]
- Mannervik M. and Levine,M. (1999) The Rpd3 histone deacetylase is required for segmentation of the Drosophila embryo. Proc. Natl Acad. Sci. USA, 96, 6797–6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott M.R. and Hill,R.J. (1986) The ultrastructural morphology of native salivary gland chromosomes of Drosophila melanogaster: the band–interband question. Chromosoma, 94, 403–411. [DOI] [PubMed] [Google Scholar]
- Mottus R., Sobel,R.E. and Grigliatti,T.A. (2000) Mutational analysis of a histone deacetylase in Drosophila melanogaster: missense mutations suppress gene silencing associated with position effect variegation. Genetics, 154, 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M., Ahn,J., Walker,K.K., Hoffman,W.H., Evans,R.M., Levine,A.J. and George,D.L. (1999) Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev., 13, 2490–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld T.P., Tang,A.H. and Rubin,G.M. (1998) A genetic screen to identify components of the sina signaling pathway in Drosophila eye development. Genetics, 148, 277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue M.L., Danilevskaya,O.N., Lowenhaupt,K., Slot,F. and Traverse,K.L. (1996) Drosophila telomeres: new views on chromosome evolution. Trends Genet., 12, 48–51. [DOI] [PubMed] [Google Scholar]
- Pazin M.J. and Kadonaga,J.T. (1997) What’s up and down with histone deacetylation and transcription. Cell, 89, 325–328. [DOI] [PubMed] [Google Scholar]
- Pennetta G. and Pauli,D. (1998) The Drosophila sin3 gene encodes a widely distributed factor essential for embryonic viability. Dev. Genes Evol., 208, 531–536. [DOI] [PubMed] [Google Scholar]
- Richards G. (1997) The ecdysone regulatory cascades in Drosophila. Adv. Dev. Biol., 5, 81–135. [Google Scholar]
- Rundlett S.E., Carmen,A.A., Kobayashi,R., Bavykin,S., Turner,B.M. and Grunstein,M. (1996) Hda1 and Rpd3 are members of distinct histone deacetylase complexes that regulate silencing and transcription. Proc. Natl Acad. Sci. USA, 93, 14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlett S.E., Carmen,A.A., Suka,N., Turner,B.M. and Grunstein,M. (1998) Transcription repression by Ume6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature, 392, 831–835. [DOI] [PubMed] [Google Scholar]
- Rykowski M.C., Parmelee,S.J., Agard,D.A. and Sedat,J.W. (1988) Precise determination of the molecular limits of a polytene chromosome band: regulatory sequences for the Notch gene are in the interband. Cell, 54, 461–472. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Shopland L.S., Hirayoshi,K., Fernandes,M. and Lis,J.T. (1995) HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites. Genes Dev., 9, 2756–2769. [DOI] [PubMed] [Google Scholar]
- Skantar A.M. and Greenleaf,A.L. (1995) Identifying a transcription factor interaction site on RNA polymerase II. Gene Expr., 5, 49–69. [PMC free article] [PubMed] [Google Scholar]
- Spencer V.A. and Davie,J.R. (1999) Role of covalent modifications of histones in regulating gene expression. Gene, 240, 1–12. [DOI] [PubMed] [Google Scholar]
- Spierer P., Spierer,A., Bender,W. and Hogness,D.S. (1983) Molecular mapping of genetic and chromomeric units in Drosophila melanogaster.J. Mol. Biol., 168, 35–50. [DOI] [PubMed] [Google Scholar]
- Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Sun Z.-W. and Hampsey,M. (1999) A general requirement for the SIN3–RPD3 histone deacetylase complex in regulating silencing in Saccharomyces cerevisiae. Genetics, 152, 921–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taplick J., Kurtev,V., Lagger,G. and Seiser,C. (1998) Histone H4 acetylation during interleukin-2 stimulation of mouse T cells. FEBS Lett., 436, 349–352. [DOI] [PubMed] [Google Scholar]
- Thomas H.E., Stunnenberg,H.G. and Stewart,A.F. (1993) Heterodimer ization of the Drosophila ecdysone receptor with retinoid X receptor and ultraspiracle. Nature, 362, 471–475. [DOI] [PubMed] [Google Scholar]
- Thummel C.S. (1996) Flies on steroids—Drosophila metamorphosis and the mechanisms of steroid hormone action. Trends Genet., 12, 306–310. [DOI] [PubMed] [Google Scholar]
- Tsai C.-C., Kao,H.-Y., Yao,T.-P., McKeown,M. and Evans,R.M. (1999) SMRTER, a Drosophila nuclear receptor coregulator, reveals that EcR-mediated repression is critical for development. Mol. Cell, 4, 175–186. [DOI] [PubMed] [Google Scholar]
- Tse C., Sera,T., Wolffe,A.P. and Hansen,J.C. (1998) Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol., 18, 4629–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B.M. (1998) Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell Mol. Life Sci., 54, 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B.M., O’Neill,L.P. and Allan,I.M. (1989) Histone H4 acetylation in human cells. Frequency of acetylation at different sites defined by immunolabeling with site-specific antibodies. FEBS Lett., 253, 141–145. [DOI] [PubMed] [Google Scholar]
- Turner B.M., Franchi,L. and Wallace,H. (1990) Islands of acetylated histone H4 in polytene chromosomes and their relationship to chromatin packaging and transcriptional activity. J. Cell Sci., 96, 335–346. [DOI] [PubMed] [Google Scholar]
- Turner B.M., Birley,A.J. and Lavender,J. (1992) Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell, 69, 375–384. [DOI] [PubMed] [Google Scholar]
- Urata Y., Parmelee,S.J., Agard,D.A. and Sedat,J.W. (1995) A three-dimensional structural dissection of Drosophila polytene chromo somes. J. Cell Biol., 131, 279–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lint C., Emiliani,S. and Verdin,E. (1996) The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr., 5, 245–253. [PMC free article] [PubMed] [Google Scholar]
- Vannier D., Balderes,D. and Shore,D. (1996) Evidence that the transcriptional regulators SIN3 and RPD3, and a novel gene (SDS3) with similar functions, are involved in transcriptional silencing in S. cerevisiae. Genetics, 144, 1343–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vettese-Dadey M., Grant,P.A., Hebbes,T.R., Crane-Robinson,C., Allis,C.D. and Workman,J.L. (1996) Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J., 15, 2508–2518. [PMC free article] [PubMed] [Google Scholar]
- Vidal M. and Gaber,R.F. (1991) RPD3 encodes a second factor required to achieve maximal positive and negative transcriptional states in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 6317–6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M., Buckley,A.M., Hilger,F. and Gaber,R.F. (1990) Direct selection for mutants with increased K+ transport in Saccharomyces cerevisiae. Genetics, 125, 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M., Strich,R., Esposito,R.E. and Gaber,R.F. (1991) RPD1 (SIN3/UME4) is required for maximal activation and repression of diverse yeast genes. Mol. Cell. Biol., 11, 6306–6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N., Diez,J.L. and Santa-Cruz,M.C. (1991) Comparative analysis of four parameters involved in puffing activity along chromosome arm 2L of D. melanogaster. Biol. Cell, 73, 71–78. [DOI] [PubMed] [Google Scholar]
- Wade P.A., Jones,P.L., Vermaak,D. and Wolffe,A.P. (1998) A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol., 8, 843–846. [DOI] [PubMed] [Google Scholar]
- Wade P.A., Gegonne,A., Jones,P.L., Ballestar,E., Aubry,F. and Wolffe,A.P. (1999) Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nature Genet., 23, 62–66. [DOI] [PubMed] [Google Scholar]
- Wakimoto B.T. (1998) Beyond the nucleosome: epigenetic aspects of position–effect variegation in Drosophila. Cell, 93, 321–324. [DOI] [PubMed] [Google Scholar]
- Wang H., Reynolds-Hager,L. and Stillman,D.J. (1994) Genetic interactions between SIN3 mutations and the Saccharomyces cerevisiae transcriptional activators encoded by MCM1, STE12, and SWI1. Mol. Gen. Genet., 245, 675–685. [DOI] [PubMed] [Google Scholar]
- Weeks J.R., Hardin,S.E., Shen,J., Lee,J.M. and Greenleaf,A.L. (1993) Locus-specific variation in phosphorylation state of RNA polymerase II in vivo: correlations with gene activity and transcript processing. Genes Dev., 7, 2329–2344. [DOI] [PubMed] [Google Scholar]
- Weiler K.S. and Wakimoto,B.T. (1995) Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet., 29, 577–605. [DOI] [PubMed] [Google Scholar]
- Westwood J.T., Clos,J. and Wu,C. (1991) Stress-induced oligomer ization and chromosomal relocalization of heat-shock factor. Nature, 353, 822–827. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P. (1998) Chromatin Structure and Function. Academic Press, San Diego, CA, pp. 7–41. [Google Scholar]
- Wong C.W. and Privalsky,M.L. (1998) Transcriptional repression by the SMRT–mSin3 corepressor: multiple interactions, multiple mechanisms, and a potential role for TFIIB. Mol. Cell. Biol., 18, 5500–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. (1997) Chromatin remodeling and the control of gene expression. J. Biol. Chem., 272, 28171–28174. [DOI] [PubMed] [Google Scholar]
- Xue Y., Wong,J., Moreno,G.T., Young,M.K., Cote,J. and Wang,W. (1998) NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell, 2, 851–861. [DOI] [PubMed] [Google Scholar]
- Yao T.-P., Segraves,W.A., Oro,A.E., McKeown,M. and Evans,R.M. (1992) Drosophila ultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell, 71, 63–72. [DOI] [PubMed] [Google Scholar]
- Yao T.-P., Forman,B.M., Jiang,Z., Cherbas,L., Chen,J.-D., McKeown,M., Cherbas,P. and Evans,R.M. (1993) Functional ecdysone receptor is the product of EcR and Ultraspiricle genes. Nature, 366, 476–479. [DOI] [PubMed] [Google Scholar]
- Zhang Y., LeRoy,G., Seelig,H.P., Lane,W.S. and Reinberg,D. (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell, 95, 279–289. [DOI] [PubMed] [Google Scholar]
- Zhimulev I.F. and Belyaeva,E.S. (1975) 3H-uridine labelling patterns in the Drosophila melanogaster salivary gland chromosomes X, 2R, and 3L. Chromosoma, 49, 219–231. [Google Scholar]
- Zink B. and Paro,R. (1989) In vivo binding pattern of a trans-regulator of homoeotic genes in Drosophila melanogaster. Nature, 337, 468–471. [DOI] [PubMed] [Google Scholar]