Abstract
In Saccharomyces cerevisiae, snoRNAs are encoded by independent genes and within introns. Despite this heterogenous organization, snoRNA biosynthesis relies on a common theme: entry sites for 5′–3′ and 3′–5′ exonucleases are created on precursor molecules allowing the release of mature snoRNAs. In independently transcribed snoRNAs, such entry sites are often produced by the Rnt1p endonuclease. In many cases, cleavage sites are absent in the 3′ portion of the pre-snoRNAs, suggesting that processing starts from the 3′ end of the primary transcript. Here we show that cleavage/polyadenylation sites driving efficient polyadenylation, such as CYC1, prevent production of mature and functional snoRNPs. With these sites, snoRNA accumulation is restored only if polyadenylation activity is inhibited. Analysis of sequences downstream of snoRNA-coding units and the use of strains carrying mutations in RNA polymerase II (polII) cleavage/polyadenylation activities allowed us to establish that formation of snoRNA mature 3′ ends requires only the cleavage activity of the polII 3′-processing machinery. These data indicate that, in vivo, uncoupling of cleavage and polyadenylation is necessary for an essential cellular biosynthesis.
Keywords: 3′ end formation/polyadenylation/processing/snoRNA/yeast
Introduction
SnoRNAs represent a complex family of RNA molecules localized in the nucleolus where they participate in different steps of ribosome biogenesis. More then 100 different species have been identified in higher eukaryotes and in yeast; few snoRNAs are required for processing of the pre-rRNA, whereas most of them guide site-specific 2′-O-ribose methylation or pseudouridylation of rRNAs (reviewed in Maxwell and Fournier, 1995; Smith and Steitz, 1997; Tollervey and Kiss, 1997; Lafontaine and Tollervey, 1998). These modifications are directed by two distinct classes of snoRNAs defined on the basis of conserved sequences and structural elements (Bachellerie et al., 1995; Balakin et al., 1996; Ganot et al., 1997a). Methylation guide snoRNAs are defined by the presence of the conserved boxes C and D and by a terminal stem (Kiss-László et al., 1996; Nicoloso et al., 1996), while pseudouridylation guide snoRNAs belong to the box H/ACA class, defined by evolutionarily conserved H/ACA motifs together with the conserved ‘hairpin–hinge– hairpin–tail’ secondary structure (Ganot et al., 1997b; Ni et al., 1997). Besides their special functions, snoRNAs have very characteristic and variegated genomic organizations (Weinstein and Steitz, 1999). In vertebrates, the majority are encoded in introns of protein-coding genes; few exceptions to this general rule are represented by the independently transcribed U3, U8 and U13 snoRNAs (Maxwell and Fournier, 1995). In contrast, in plants, snoRNAs are encoded by polycistronic clusters (Leader, 1994, 1997). A different organization is found in yeast, in which most snoRNAs are encoded in independent monocistronic or polycistronic transcripts and only a minority are encoded in introns (Maxwell and Fournier 1995; Chanfreau et al., 1998a,b). The diversity in the genomic organization of snoRNA genes raises the issue of how many different biosynthetic mechanisms exist and how many different factors are involved in snoRNA production.
Vertebrate intron-encoded snoRNAs are produced by two different pathways: one is splicing dependent and utilizes the spliced-debranched lariat, while the second is splicing independent and utilizes the pre-mRNA cleaved endonucleolytically. In both cases, pre-snoRNA molecules are produced which are then converted by exonucleolytic trimming to mature species (Tycowski et al., 1993; Caffarelli et al., 1994, 1996; Kiss and Filipowicz, 1995; Watkins et al., 1996). The mature 5′ and 3′ termini of the snoRNA are determined by snoRNA-associated proteins which provide barriers against further exonucleolytic trimming (Caffarelli et al., 1996). Both pathways have been identified for the intron-encoded snoRNAs in yeast (Ooi et al., 1998; Villa et al., 1998, 2000). For independently transcribed units, whether polycistronic or monocistronic, the release of individual and mature species involves both endonucleolytic cleavage and exonucleolytic trimming (Chanfreau et al., 1998a,b; Allmang et al., 1999; Van Hoof et al., 2000). Besides the diversity of snoRNA gene organization (introns or independently transcribed units), a unifying rule can be derived: in all cases, the crucial step for snoRNA processing is the production of entry sites for exonucleases in the precursor molecule, followed by exo-trimming to produce a snoRNA with mature 5′ and 3′ ends.
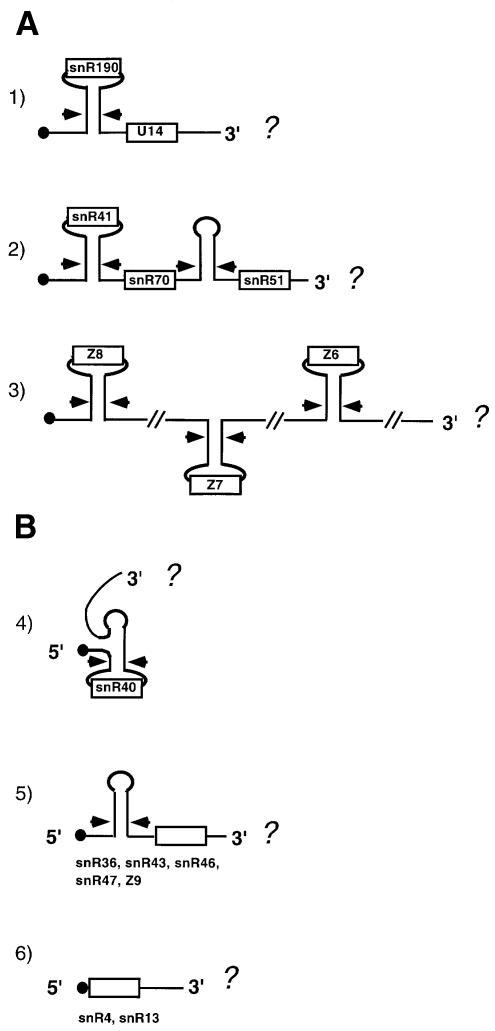
Figure 1 summarizes the different prototypes of independently transcribed C/D box snoRNA units. It appears that dicistronic or polycistronic units possess Rnt1 cleavage sites (arrows) located so as to allow the release of individual snoRNAs from the precursor molecule and the liberation from the cap structure, which protects the 5′ end. In the case of monocistronic units, the 5′ end of the snoRNA either corresponds to the 5′ end of the primary transcript, in this case the snoRNA is capped [Figure 1 (6)], or it is produced by 5′–3′ trimming from an Rnt1p-processing site [Figure 1 (4 and 5)]. From the available data, it appears that many poly- and monocistronic units do not have Rnt1p-processing sites in their 3′ portion, raising the question of how the entry site for trimming from the 3′end is produced and, more generally, how the 3′ end of snoRNA precursors is formed. Even if these genes are transcribed by RNA polymerase II (polII) it is not known whether they possess canonical cleavage/polyadenylation sites. Several papers have shown that, in exosome mutant strains, several snRNAs and snoRNAs accumulate as 3′-extended polyadenylated species, suggesting that the exosome is required to process these 3′-extended transcripts (Allmang et al., 1999; Van Hoof et al., 2000). So far, it has not yet been defined whether these polyadenylated molecules represent physiological biosynthetic intermediates or whether they identify products directed to the discard pathway. In this study, we show that canonical polyadenylation signals prevent C/D box snoRNA accumulation, indicating that this reaction interferes with snoRNA processing. Instead, 3′ end formation of independently transcribed snoRNA relies on sequences that uniquely require the cleavage activity and not the polyadenylation activity of the polII 3′ end-processing apparatus.
Fig. 1. Schematic representation of the different prototypes of independently transcribed snoRNA-coding units. The boxes represent the snoRNA-coding regions, the stems and arrows indicate the Rnt1p substrates and cleavage sites, respectively, and the dots represent the cap structure. Poly- and monocistronic units are schematically represented in (A) and (B), respectively. In all cases studied so far, the 5′ cap is either present in the mature snoRNA (6) or it is removed by Rnt1p cleavage (1–5) (Chanfreau et al., 1998).
Results
snoRNAs encoded in different contexts but common themes
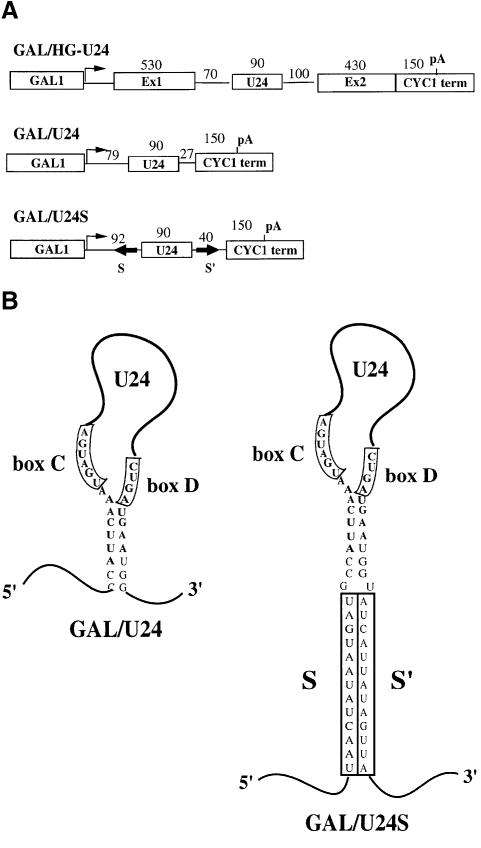
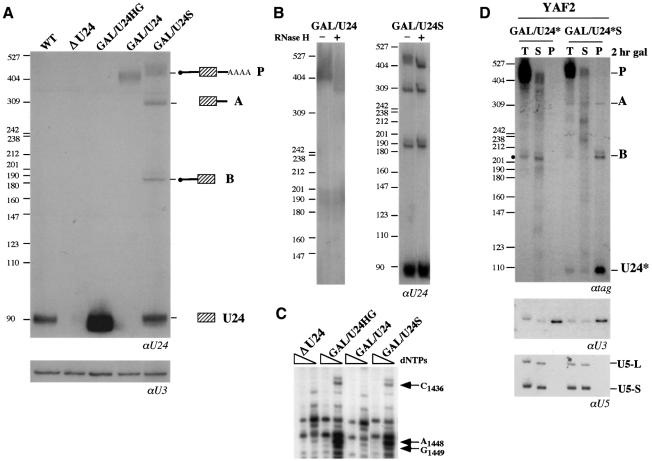
In order to analyze the role of the genomic context on the biosynthesis and processing of independently transcribed snoRNAs, we have produced a test construct in which the U24 snoRNA was cloned between the GAL1 promoter and the CYC1 cleavage/polyadenylation site of vector p413GAL1 (Mumberg et al., 1994), giving rise to plasmid GAL/U24 (Figure 2). The CYC1 cleavage/polyadenylation site is a very well characterized sequence that directs efficient 3′ end processing and polyadenylation (Guo and Sherman, 1996) mediated by the CFI and CPF complexes together with the poly(A) polymerase (Pap1p) (Keller and Minvielle-Sebastia, 1997; Birse et al., 1998; Ohnacker et al., 2000). The U24 snoRNA transcribed from this construct can only be produced by trimming from the ends of the primary transcript since no processing sites are present in the flanking regions. GAL/U24S is a derivative of GAL/U24: it contains two inverted repeats (S and S′), flanking the snoRNA-coding region, which form a 13 nucleotide external stem (Figure 2A and B). This structure was described to enhance snoRNA biosynthesis by bringing together the C and D boxes and, in turn, activating endonucleolytic cleavage (Villa et al., 2000). As a control, the entire transcriptional unit of the U24 host gene (Qu et al., 1995) was cloned inside the same vector backbone producing GAL/HG-U24 (Figure 2A). The three constructs were transformed into the YAF2 strain, which is depleted of the endogenous U24 snoRNA-coding sequence (strain ΔU24) (Kiss-László et al., 1996) and which expresses a ‘tagged’ Nop56 protein (see Materials and methods). In this strain, it is possible to determine whether the U24 snoRNAs expressed from the transformed plasmids are functional, by analyzing the methylation state of the 25S rRNA at positions C1436, A1448 and G1449 (Kiss-László et al., 1996); in addition, since Nop56p is a component of the C/D box snoRNP (Gautier et al., 1997; Lafontaine and Tollervey, 2000), it is possible to analyze which of the U24-containing molecules are associated with C/D box-specific complexes. Northern blot analysis showed that no mature U24 snoRNA was produced from the GAL/U24 construct; instead, a molecule longer than 400 nucleotides accumulated (Figure 3A). RNase H treatment with oligo(dT) indicated that this molecule was polyadenylated (Figure 3B). The analysis of the methylation state of rRNA and immunoprecipitation experiments of the RNA associated with the ‘tagged’ Nop56 protein showed that this RNA species was not able either to restore rRNA methylation (Figure 3C, lanes GAL/U24) or to assemble into C/D box-specific complexes (Figure 3D, lanes GAL/U24*).
Fig. 2. Schematic representation of the GAL/CYC1 constructs containing the U24 snoRNA-coding sequences. (A) The different boxes indicate the promoter (GAL1), the cleavage/polyadenylation site (CYC1 term), the BEL1 exons (Ex1 and Ex2) and the snoRNA-coding region (U24). Numbers indicate the size of the different portions of the transcripts, and the divergent arrows (S and S′) represent the inverted repeats forming a 13 nucleotide external stem. GAL/HG-U24 contains the U24/EST genomic coding region (Qu et al., 1995) cloned between the GAL and CYC1 sequences of plasmid p413GAL1 (Mumberg et al., 1994). GAL/U24 contains the U24 coding region flanked by 79 and 27 nucleotides of the 5′-untranslated region of GAL1 and of vector sequences, respectively; GAL/U24S contains, in addition, the S/S′ inverted repeats flanking the intron-encoded U18 snoRNA (Villa et al., 2000). (B) Schematic representation of the pre-snoRNAs produced by the GAL/U24 and GAL/U24S constructs. The conserved C and D boxes are indicated together with the terminal stem; the nucleotides present in the mature snoRNA are represented in bold. The external stem is represented by the S and S′ boxes.
Fig. 3. Processing of the different U24 constructs. (A) Northern blot analysis of 5 µg of total RNA extracted from exponentially grown cells of strain YAF2 (see Materials and methods) either untransformed (lane ΔU24) or transformed with GAL/U24HG (lane GAL/U24HG), GAL/U24 (lane GAL/U24) or GAL/U24S (lane GAL/U24S) plasmids. RNA from strain CH1462 was also loaded as control (lane WT). The probe was a 32P-labeled anti-U24 oligonucleotide. The different products of the reaction and the molecular weight markers (MspI-digested pBR322) are indicated at the sides. The lower panel shows a control hybridization with a U3-specific probe. (B) A 10 µg aliquot of total RNA, extracted from strain YAF2 transformed with GAL/U24 (lane GAL/U24) and GAL/U24S (lane GAL/U24S) plasmids, was incubated in the absence (–) or presence (+) of RNase H and oligo(dT) and analyzed by northern blotting with the U24 probe. (C) Primer extension mapping of the ribose methylations in strain YAF2 either untransformed (lanes ΔU24) or transformed with GAL/U24HG (lanes GAL/U24HG), GAL/U24 (lanes GAL/U24) or GAL/U24S (lanes GAL/U24S) plasmids. A 32P-labeled primer, complementary to nucleotides 1458–1477 of yeast 25S rRNA, was annealed to each RNA sample and extended with AMV reverse transcriptase in the presence of decreasing concentrations of dNTPs (Maden et al., 1995; Kiss-László et al., 1996). The arrows indicate the positions of the modified nucleotides. (D) Tagged (*) versions of the different U24 constructs were utilized in this experiment. Strain YAF2, containing GAL/U24* or GAL/U24*S plasmids, was induced for 2 h in galactose, cells were lysed and immunoprecipitated with IgG beads. RNA was extracted from pellets (P) and supernatants (S) and run on a 6% polyacrylamide–urea gel in parallel with RNA extracted from non-immunoprecipitated extracts (T). Northern blot analysis was performed with a 32P-labeled tag-specific oligonucleotide. The different products of the reaction are indicated at the side by letters, as in (A). The band indicated by a dot represents a degradation product. The lower panels show control hybridizations with U3 snoRNA- and U5 snRNA-specific probes.
The analysis of the GAL/U24S construct indicated that the 13 nucleotide external stem was able to resume the synthesis of U24 at a considerable level (Figure 3A) and to restore methylation of rRNA at the correct sites (Figure 3C, lanes GAL/U24S). Also in this case, a precursor molecule corresponding in size to a full-length polyadenylated molecule was visualized (band P). RNase H experiments with oligo(dT) have proven that this RNA species is indeed polyadenylated (Figure 3B). GAL-U24S produced two additional molecules (A and B). Reverse transcriptase analysis (not shown) and RNase H experiments (Figure 3B) indicated that the A species extends from the 5′ end of U24 to the 3′ end of the transcript, deprived of the poly(A) tail, while B RNA extends from the 5′ end of the precursor to the 3′ end of U24. It is not clear at the moment how A molecules are deprived of the poly(A) tail. Analogous processing intermediates have been described already in studies of U18 snoRNA processing and were shown to represent products stabilized by the binding of snoRNP-specific factors (Villa et al., 1998, 2000). Immuno precipitation experiments indicated that these species, as well as mature U24 snoRNA, are assembled into Nop56p-containing snoRNPs; on the contrary, the polyadenylated species did not bind C/D box-specific factor (Figure 3D, lanes GAL/U24*S), demonstrating that Nop56p associates only with non-polyadenylated molecules.
For immunoprecipitations and for the other experiments of this study, we utilized constructs with a tagged U24 snoRNA (U24*). The tag allows the snoRNA produced by the transforming plasmids to be distinguished from the endogenous U24. The tag utilized was shown previously not to affect snoRNP formation (Villa et al., 2000) or U24 processing and stability (unpublished data). In order to study the relationship between polyadenylation and snoRNA accumulation in more detail, we tested the GAL/U24 construct in strains carrying the rna14-1 and pap1-5 mutant alleles. At the non-permissive temperature, rna14-1 blocks the cleavage step while pap1-5 drastically reduces the polyadenylation reaction (Minvielle-Sebastia et al., 1994). In both strains, a slight recovery of U24* snoRNA accumulation was observed (Figure 4A). Since the production of mature U24* snoRNA from the GAL/U24* construct can occur exclusively by exonucleolytic trimming, entry sites for such activitites should have been formed. In the rna14-1 strain, very low levels of transcripts accumulated due to the lack of efficient 3′ end formation; nevertheless, random transcription termination in the 3′ downstream sequences should have provided 3′ end substrates for 3′–5′ trimming (Allmang et al., 1999; Van Hoof et al., 2000). In the pap1-5 strain, since the cleavage reaction is unaffected, higher levels of transcripts were produced; in addition, inefficient polyadenylation should have provided more substrate for 3′–5′ exonucleases. In both strains, mature 5′ ends of the snoRNA were also produced, indicating that removal of the cap structure had occurred. Nevertheless, the accumulation of the B intermediate, extending from the 5′ end of the precursor to the 3′ end of U24, indicated that cap removal is a limiting step in the final maturation of this type of construct.
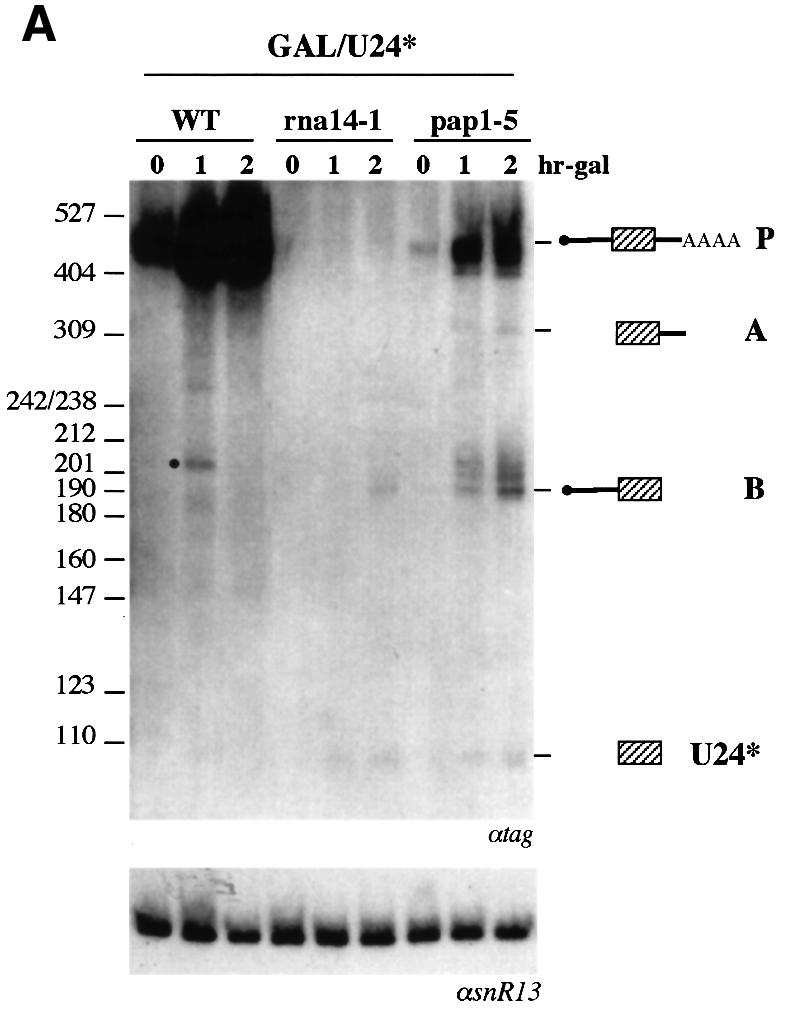
Fig. 4. Analysis of U24 processing in different mutant strains. After 1 h shift at the non-permissive temperature (time 0), cultures were induced with galactose for the indicated times. Northern blot analysis of RNA was performed with the CH1462 strain (WT) or the strains carrying the mutant alleles rna14-1, pap1-5, rnt1Δ, rat1-1 and nup145N transformed with GAL/U24* (A) or GAL/U24*S (B) plasmids. Both blots were hybridized with a 32P-labeled tag-specific oligonucleotide. Size markers (MspI-digested pBR322) and schematic representations of the different processing products are indicated at the sides. The lower panels show control hybridizations with an snR13 snoRNA-specific probe.
The same mutant strains were utilized for testing the activity of the GAL/U24*S construct. The absence of (lanes rna14-1) or limited (lanes pap1-5) polyadenylation strongly enhanced the release of mature U24* snoRNA as well as of the A and B intermediates (Figure 4B), once again indicating that poly(A) tails also interfere with snoRNA release in the presence of a processing site. In the strain carrying the nup145N mutant allele (lanes nup145N), which blocks cytoplasmic transport of mRNA (Fabre et al., 1994), the levels of mature U24* snoRNA were also increased. This result correlated the retention of precursor molecules in the nucleus with the enhancement of U24 release, suggesting that the major inhibitory effect of polyadenylation could be the rapid export of the poly(A)+ pre-snoRNA molecules to the cytoplasm (Huang and Carmichael, 1996) where no snoRNA processing occurs (Caffarelli et al., 1996).
The behavior of GAL/U24*S in the strain carrying the rnt1Δ (Chanfreau et al., 1997) and rat1-1 (Amberg et al., 1992) mutations was also analyzed. The first strain showed that the external stem of U24S is susceptible to Rnt1p activity: in fact, a strong decrease of mature U24* was observed as well as of intermediate molecules (lanes rnt1Δ), better visualized in longer exposures (not shown). These and other data (C.Giorgi, unpublished observations) indicated that the stem utilized, even if it does not appear as a canonical substrate of Rnt1p (Chanfreau et al., 2000), is nevertheless cleaved by this enzyme. In this strain, the residual production of U24* must be accounted for by exonucleolytic trimming from the pre-snoRNA termini (see Discussion). The rat1-1 mutation, in which the nuclear 5′–3′ exonuclease Rat1p was inactivated at 37°C, produced an increase in the accumulation of all the processing products. Since Rat1p has been implicated in the nuclear turnover of the RNA (Bousquet-Antonelli et al., 2000; Villa et al., 2000), we must assume that a considerable proportion of the processing products undergo rapid degradation in the nucleus unless they are assembled into snoRNP particles, indicating competition between turnover of the RNA and assembly into functional particles. The production of mature 5′ ends of U24* snoRNA in the rat1-1 strain is very likely to be due to some degree of complementation by the cytoplasmic Xrn1p activity (Bousquet-Antonelli et al., 2000).
Accumulation of snR4, snR13 and snR40 snoRNAs is also inhibited by 3′ polyadenylation sites
The effect of the presence of poly(A) tails on the biosynthesis of snR4, snR13 and snR40 snoRNAs was also tested (see Figure 1). The U24 coding region of plasmid GAL/U24 was replaced with that of the three snoRNAs, giving plasmids GAL/snR4*, GAL/snR13* and GAL/snR40* (see schematic representation of Figure 5). The transcripts originated from all three constructs contained a tag (*) inside the snoRNA-coding region in order to distinguish them from the corresponding endogenous snoRNAs. After galactose induction, large amounts of transcripts accumulated (Figure 5). All three constructs had sizes compatible with polyadenylated pre-snoRNAs species. No detectable amounts of mature snoRNAs were visualized, or of the A and B type of intermediates, again indicating that polyadenylation was interfering with snoRNA processing and that sequences internal to the snoRNA-coding region were not sufficient for directing efficient maturation.
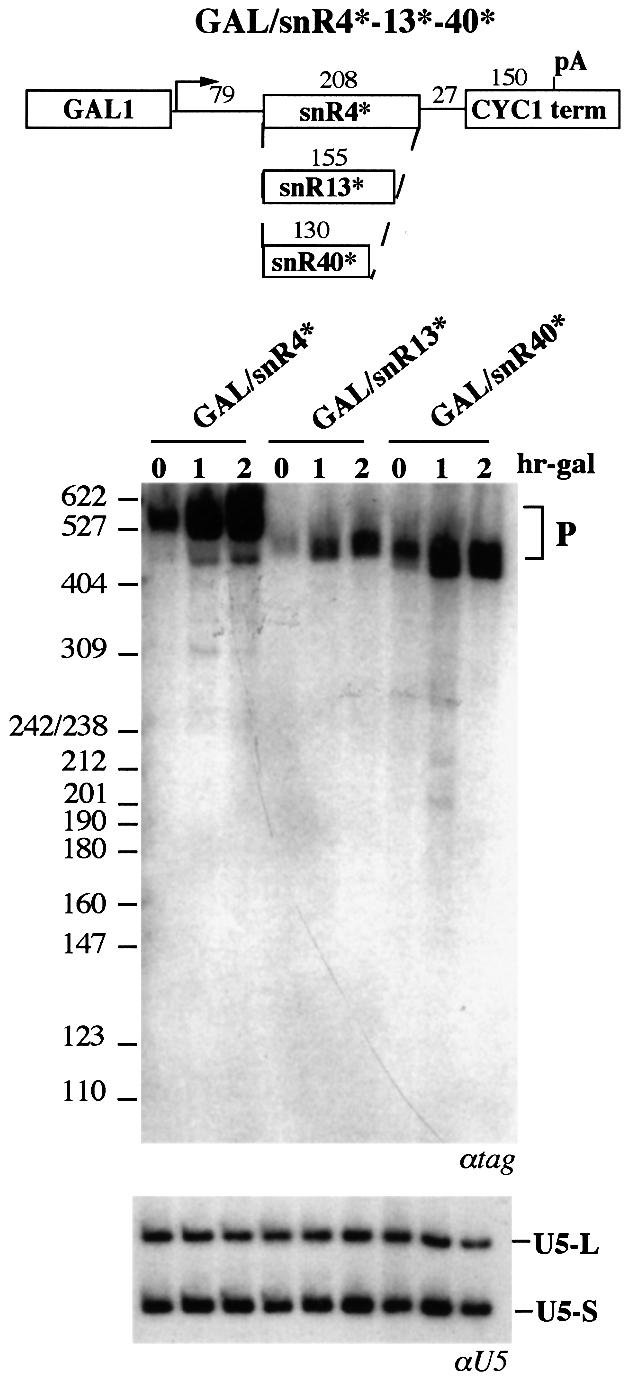
Fig. 5. Processing activity of monocistronic snoRNAs under a polyadenylation-dependent site. The schematic representation of the constructs utilized is indicated above. The boxes indicate the GAL1 promoter, the CYC1 cleavage/polyadenylation site and the sequences coding for the mature snoRNAs, snR4, snR13 and snR40. The snoRNA-coding regions are flanked by 79 and 27 nucleotides of the 5′-untranslated region of GAL1 and of vector sequences, respectively. All three snoRNAs possess a six or seven nucleotide terminal stem. All these constructs contain the same tag sequence (*) in the snoRNA-coding region. Middle panel: northern blot analysis of RNA, extracted at different times after galactose induction (indicated in hours above the lanes), from the CH1462 strain transformed with GAL/snR4* (lanes GAL/snR4*), GAL/snR13* (lanes GAL/snR13*) and GAL/snR40* (lanes GAL/snR40*) plasmids. The blot was hybridized with a 32P-labeled tag-specific oligonucleotide. Size markers (MspI-digested pBR322) are indicated at the side. The lower panel shows a control hybridization with a U5 snRNA-specific probe.
Characterization of 3′ downstream regions of independently transcribed snoRNAs
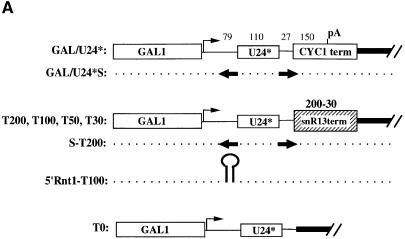
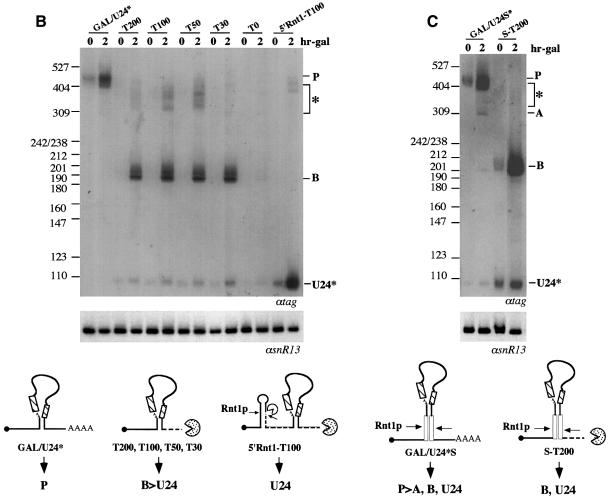
The CYC1 cleavage/polyadenylation site of the GAL/U24* plasmid was substituted with different portions of the downstream region of the snR13 snoRNA gene. The first clone tested, plasmid T200 (see Figure 6A), contained 200 nucleotides of this region (from position +20 to +220 with respect to the 3′ end of the mature snoRNA, named ‘T200’). This construct was transformed into the CH1462 strain and the accumulation of U24* snoRNA was analyzed in comparison with that of the construct containing the CYC1 cleavage/polyadenylation site. The presence of the ‘T200’ region strongly increased the accumulation of B molecules and mature snoRNA (Figure 6B). These results indicated that the ‘T200’ region induced efficient production of entry sites for 3′–5′ trimming. Progressive shortening to 100, 50 and 30 nucleotides (constructs T100, T50 and T30, respectively) produced levels of intermediate and mature molecules similar to those of T200 (Figure 6B), indicating that 30 nucleotides are still sufficient for directing efficient 3′ end formation. The higher level of accumulation of B molecules in comparison with mature U24 is due to the presence of the cap structure which hampers the formation of entry sites for 5′–3′ trimming. Efficient conversion into mature 5′ ends was obtained when an Rnt1p-dependent stem was cloned between the 5′ end of the precursor and the 5′ end of the snoRNA (construct 5′Rnt1-T100, see Figure 6A), as it occurs in most snoRNA-coding units (see Figure 1). With this construct, all the B product was converted efficiently into mature U24 snoRNA (Figure 6B). As control, a construct deprived of the snR13 3′ region was derived (construct T0, see Figure 6A). T0 produced almost undetectable amounts of mature and B molecules (Figure 6B).
Fig. 6. Characterization of 3′ downstream regions of independently transcribed snoRNAs. (A) Schematic representation of the constructs utilized. The constructs are the same as those shown in Figure 2 up to the CYC region; in the T-type constructs (T200, T100, T50 and T30), the CYC1 cleavage/polyadenylation site was substituted with 200, 100, 50 and 30 nucleotides present in the 3′-flanking region of the snR13 coding sequence (shaded box). Construct 5′stem-T100 contains an Rnt1p-processing site upstream of the U24 coding sequence. The S-T200 plasmid contain the S/S′ external stem as in construct GAL/U24*S. The T0 construct has no snR13 terminator region. Vector sequences are represented by a thick black line. (B and C) Northern blot analysis of RNA, extracted at different times after galactose induction from the CH1462 strain transformed with the plasmids indicated above the lanes. Molecules indicated by the asterisk are polyadenylated products. The lower panels show control hybridizations with an snR13-specific probe. At the bottom of the figure, the schematic representation of the products obtained with the different constructs is indicated. Dotted lines indicate trimming from the available free ends. Rnt1p cleavage sites are indicated by arrows, and pacmen represent 5′–3′ and 3′–5′ exonucleases. Removal of the cap structure, followed by Rnt1p digestion, also occurs but with a very low efficiency (not indicated in the figure).
The ‘T200’ region was also cloned in place of the CYC1 cleavage/polyadenylation site of GAL/U24*S, producing the S-T200 construct (see Figure 6A). Also in this case, high levels of B molecules were produced, indicating that the snR13 3′-non-coding region strongly enhanced 3′ processing even in the presence of an Rnt1p site (Figure 6C). Due to the Rnt1p cleavage present in the 5′ portion of the transcript, high levels of mature U24 also accumulated.
The T-constructs also produced bands of larger size (indicated in the figures by asterisks). These molecules are polyadenylated products (see experiment of Figure 7) and represent only a minority of the overall amount of transcripts. It is possible that under conditions of massive transcription, such as those obtained with the GAL1 promoter, these precursors represent molecules directed to the discard pathway (Allmang et al., 1999; Bousquet-Antonelli et al., 2000; Van Hoof et al., 2000).

Fig. 7. (A) Plasmids T100 and T0 were transformed into strain W303-1B (WT) and into strains carrying the rna14-1, rna15-2, pap1-5 and pfs2-1 mutant alleles (corresponding lanes); after 1 h shift at the non-permissive temperature (time 0), the cultures were induced with galactose for 2 h (time 2 h). RNA was extracted and run on a 6% polyacrylamide gel. Blots were hybridized with a 32P-labeled tag-specific oligonucleotide. Size markers (MspI-digested pBR322) and the different processing products are indicated at the sides. The lower panel shows a control hybridization with an snR13-specific probe. (B) A 10 µg aliquot of total RNA, extracted from the CH1462 strain (lane WT) and from the rat1-1 xrn1Δ double mutant strain (lane rat1-1 xrn1Δ) grown for 2 h at the restrictive temperature, was elongated with reverse transcriptase in the presence of the R13ter200-b primer, which is complementary to a region 200 nucleotides downstream of the snR13 coding region. The sequence around the stop site is shown at the side, while the diagrammatic representation of the cut-off products is shown below together with an indication of the region contained in the T30 construct.
In order to understand the relationship between polyadenylation and processing, the T100 construct was transformed into strains carrying mutations affecting the cleavage or polyadenylation reactions. In strains carrying the rna14-1 and rna15-2 mutant alleles, which affect the cleavage activity of the CFIA complex (Keller and Minvielle-Sebastia, 1997), both the B and mature molecules disappeared, while in the pap1-5 strain they accumulated at even higher levels than in the wild-type (Figure 7). Since it has been shown that components of the CPF complex are also involved in the cleavage reaction (Ohnacker et al., 2000), we tested one mutant, pfs2-1, which was shown to affect cleavage activity in vitro. In the strain carrying this mutation, the accumulation of the processed products was also affected, even if to a lesser extent than in CFIA mutants (Figure 7A).
The direct evidence for the occurrence of endonucleolytic cleavage is the identification of 3′ cut-off products. In wild-type cells, these intermediates are degraded rapidly, whereas in a strain carrying mutations in the 5′–3′ exonucleases Rat1p and Xrn1p (strain rat1-1, xrn1Δ) they have a longer half-life. In order to map the cleavage site of the polII 3′-processing apparatus on the endogenous snR13 transcriptional unit, primer extension analysis, on RNA extracted from the double mutant strain, was carried out with a downstream oligonucleotide. The results indicated that a strong stop occurred at a specific UAAU sequence (Figure 7B). This region maps some 25 nucleotides downstream of the snoRNA-coding region and is present in the T30 construct. From these data, we could conclude that a major cleavage site for the polII 3′-processing machinery is present in the T30 construct and that it represents a physiological substrate.
In order to analyze whether a region with similar features is present in other snoRNA genes, 500 nucleotides downstream of the snR47 gene were cloned in GAL/U24* in place of the CYC1 cleavage/polyadenylation site (GAL/U24*-47T). This construct also belonged to the snR13 3′-non-coding region, producing B molecules and mature snoRNA (Figure 8).
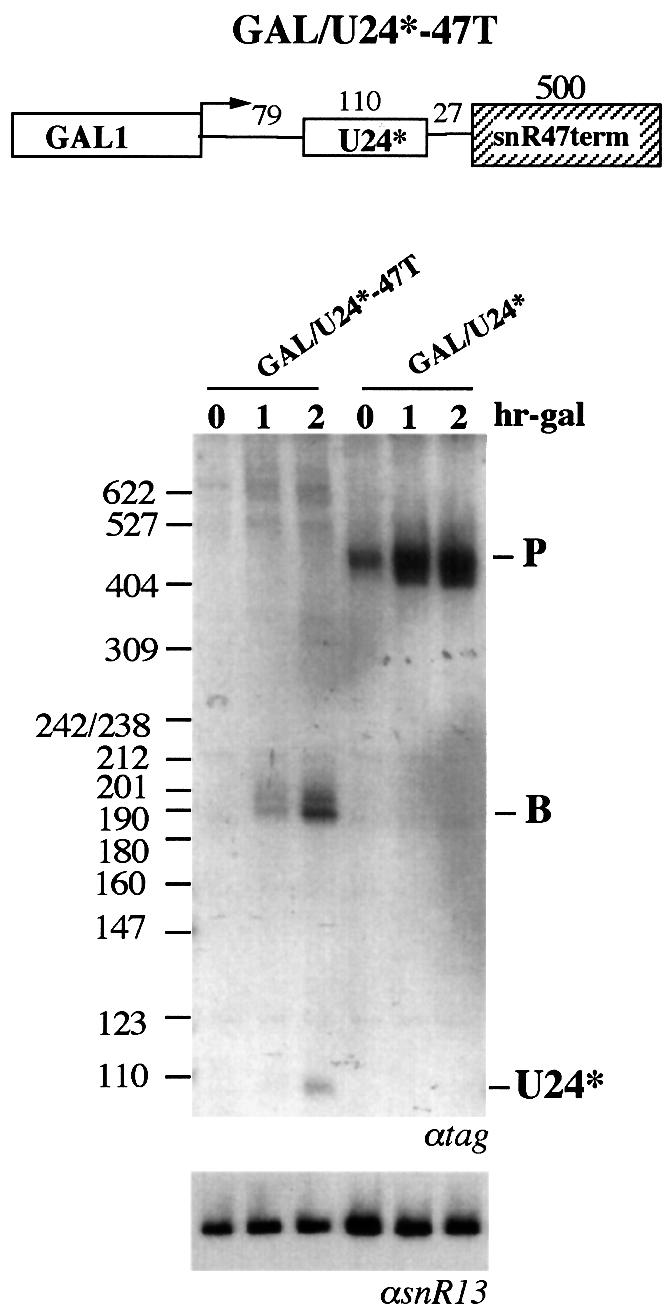
Fig. 8. Analysis of the snR47 terminator. The schematic representation of construct GAL/U24*-47T is shown above. Plasmid was transformed into strain CH1462 (lanes GAL/U24*-47T); after 1 h shift at the non-permissive temperature (time 0), the cultures were induced with galactose for 1 and 2 h. RNA was extracted and run on a 6% polyacrylamide gel in parallel with RNA extracted from cells transformed with GAL/U24* (lanes GAL/U24*). Blots were hybridized with a 32P-labeled tag-specific oligonucleotide. MspI-digested pBR322 and the different processing products are indicated at the sides. The lower panel shows a control hybridization with an snR13-specific probe.
Discussion
Independently transcribed C/D box snoRNAs are a typical feature of yeast: monocistronic, dicistronic and polycistronic units have been described (Figure 1). In most cases, the common theme for C/D box snoRNA biosynthesis is the presence of Rnt1p-dependent processing sites that punctuate the precursor molecules and allow the liberation of individual snoRNAs (Chanfreau et al., 1998b). In the 5′ portion, Rnt1p cleavage eliminates the cap structure, creating entry sites for the Rat1p 5′–3′ exonuclease (Ooi et al., 1998; Petfalski et al., 1998), while in the 3′ portion the endonuclease creates entry sites for 3′–5′ trimming performed by the exosome (Allmang et al., 1999; Van Hoof et al., 2000). Nevertheless, many of the snoRNA transcriptional units do not possess Rnt1p-processing sites in the 3′ region (see Figure 1); in addition, nothing is known about 3′ end formation of the pre-snoRNA molecules. In this study, we have analyzed the 3′-non-coding regions compatible with C/D box snoRNA production. We found that a cleavage/polyadenylation cassette derived from the CYC1 gene is strongly detrimental to snoRNA accumulation: in constructs that do not have Rnt1p sites in either the 5′- or 3′-flanking regions and rely only on exonucleolytic trimming for maturation, the presence of the poly(A) tail totally prevents accumulation of a functional and mature snoRNA; in addition, no snoRNP-specific proteins were found associated with these transcripts. Cells defective in the polyadenylation activity, such as those carrying the pap1-5 mutation, displayed efficient maturation of the 3′ ends and partial recovery of mature snoRNA accumulation, indicating that it is indeed the presence of the poly(A) tail that interferes with snoRNA maturation. Since the type of construct analyzed contained 5′ trailer sequences, which could be processed only by exonucleolytic trimming from the 5′ end, we concluded that, even if with only very low efficiency, removal of the cap structure can occur in the nucleus (see below for discussion; Bousquet-Antonelli, 2000).
The insertion of a 13 nucleotide stem formed by inverted repeats in the flanking regions of the snoRNA counteracts, in part, the inhibitory effect of the polyadenylation signal: processing products and mature snoRNA are produced and are able to assemble into functional C/D box snoRNPs. The stem utilized, essential for processing U18 snoRNA from its host intron, was described previously as playing a dual role: (i) to direct endonucleolytic cleavage of the pre-snoRNA molecule and (ii) to facilitate snoRNP assembly by bringing together the C and D boxes (Villa et al., 2000). Even if this stem is not a canonical substrate for Rnt1p cleavage (Chanfreau et al., 2000), experiments in this study indicate that this structure is indeed cleaved by the yeast homolog of the bacterial RNase III: in fact, in the absence of such activity, processing intermediates completely disappear. Neverthe less, in the rnt1Δ strain, trace amounts of mature snoRNA are detected. Recently, a nuclear discard pathway able to degrade newly synthesized pre-mRNA molecules, possibly capped and polyadenylated, by exonucleolytic trimming has been shown to operate in yeast (Bousquet-Antonelli et al., 2000; C.Presutti, personal communication). In accordance with this, the residual snoRNA accumulation, previously interpreted as Rnt1p insensitivity (Villa et al., 2000), can now be explained by its biosynthesis through the discard pathway by exonucleolytic trimming after cap and poly(A) tails have been removed.
The inactivation of the polyadenylation activity enhanced snoRNA accumulation even in the presence of an Rnt1p-processing site. SnoRNA processing also increased in the strain carrying the nup145N mutant allele, which affects the nuclear–cytoplasmic transport of poly(A)+ RNA. These results suggested that part of the inhibitory effect of the poly(A) tail could be due to the rapid cytoplasmic export of poly(A)+ pre-snoRNA molecules, in agreement with data indicating that polyadenylation drives the RNA very rapidly towards the cytoplasmic export pathway (Huang et al., 1996). If such molecules are forced to be retained in the nucleus, a large pool of precursors will then be available for snoRNA biosynthesis. The use of the rat1-1 strain, in which the nuclear turnover of precursor RNA is decreased due to the inactivation of the nuclear 5′–3′ exonuclease, also increased snoRNA accumulation. This is again consistent with the existence of a nuclear turnover involving 5′–3′ (as well as 3′–5′) exonucleases (Bousquet-Antonelli et al., 2000): when these functions are inactivated, snoRNP assembly is favored and increased levels of snoRNAs are produced through both the endo- and exonucleolytic pathways. The stability of precursor RNA molecules inside the nucleus would then play an important role in gene expression, the levels of mature RNA species being affected by the competition between degradation and other nuclear events such as processing and transport.
The CYC1 cleavage/polyadenylation site was also inserted downstream of other snoRNAs, which are transcribed as monocistronic units, such as those coding for snR4, snR13 and snR40; in all cases, polyadenylated precursors accumulated without any release of processing products or mature snoRNAs. This indicated that also on other substrates, sequences internal to the snoRNAs are not sufficient to drive the processing reaction in the context of a polyadenylated precursor.
Polyadenylated pre-snoRNAs have been found in strains depleted of different subunits of the exosome (Allmang et al., 1999; Van Hoof et al., 2000); the results presented here suggest that these species are unlikely to be converted into functional snoRNAs, but rather they would represent dead-end products possibly directed to the nuclear discard pathway, in analogy with the situation described in prokaryotes (Xu and Cohen, 1995; Sarkar, 1997; Li et al., 1998). Alternatively, polyadenylated species could be exported rapidly to the cytoplasm where they could follow a cytoplasmic degradative pathway.
Since polyadenylation signals were found not to be appropriate for snoRNA synthesis and to identify the regions required for efficient 3′ end formation, we analyzed the sequences present in the 3′ portion of different snoRNA-coding units. Sequences downstream of the snR13 and snR47 coding units, not containing any Rnt1p-processing site (Chanfreau et al., 2000), were found to direct efficient accumulation of snoRNAs with mature 3′ ends, indicating that entry sites for 3′–5′ trimming activity were produced efficiently.
Two major complexes have been described to be involved in polII-dependent 3′ processing in yeast: CFI and CPF (Keller and Minvielle, 1997; Ohnacker et al., 2000). In canonical cleavage/polyadenylation sites, such as CYC1, these complexes direct cleavage and allow polyadenylation by recruiting the Pap1p enzyme. A more in-depth analysis of the snR13 3′-non-coding region revealed that its function is affected by mutations in CFI (rna14-1 and rna15-2) and, even if to a minor extent, in CPF (pfs2-1). Vice versa, mutations affecting the polyadenylation reaction did not affect the accumulation of snoRNA with mature 3′ ends. These data allowed us to conclude that efficient formation of snoRNA 3′ ends requires only the cleavage activity of the polII 3′-processing machinery. A specific cleavage site was identified in the endogenous snR13 coding unit utilizing the double mutant strain for the 5′–3′ exonucleases Rat1p and Xrn1p. This site is localized in a proximal region downstream of the snoRNA-coding region and is present in the constructs tested, in particular, in the shortest one (T30), which was still able to direct efficient 3′ processing. These data confirmed that the region identified is involved in 3′ end formation also in the chromosomal context.
Recently, it has been demonstrated that only factors involved in the pre-mRNA cleavage reaction play a role in directing termination of polII transcription in vivo and that polyadenylation is not required for termination (Birse et al., 1998). C/D box snoRNA genes would then represent examples of transcriptional units in which uncoupling between the two activities could play an important regulatory role: while for pre-mRNAs, which have to be protected from exonucleolytic trimming and to be exported rapidly to the cytoplasm, the 3′-processing apparatus should ensure efficient polyadenylation, for nuclear-localized transcripts, which require 3′ end maturation, such as snoRNAs and possibly snRNAs, only the cleavage reaction would be required. Since the activity of the polII-dependent 3′-processing machinery is expected to be modulated depending on the specific transcript, it will be interesting to analyze whether and how the architecture of the interactions of the cleavage and polyadenylation complexes changes on different substrates. It is possible that different cis-acting elements might be involved in distinguishing between substrates which are to be polyadenylated and those which are just to be cleaved.
Materials and methods
Strains and media
Growth and handling of S.cerevisiae were by standard techniques. Yeast strains used are listed in Table I. Epitope TAP tagging of Nop56p (YAF2 strain) was performed as described in Rigaut et al. (1999). Yeast strains were transformed with the appropiate plasmids as described in Villa et al. (1998) and grown in the appropriate selective media.
Table I. Strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| CH1462 | MATα, ade2, ade3, leu2, ura3, his3, kan1 | Kranz and Holm (1990) |
| W303-1B | MATa, ura3-1, trp1-1, ade2-1, leu2-3, 112, his3-11, 15 | Amrani et al. (1997) |
| LM88 | MATa, ura3-1, trp1-1, ade2-1, leu2-3, 112, his3-11, 15, rna14-1 | Minvielle-Sebastia et al. (1994) |
| rna15-2 | MATa, ura3-1, trp1-1, ade2-1, leu2-3, 112, his3-11, 15, rna15-2 | Amrani et al. (1997) |
| Ypap1-5 | MATa, ura3-1, trp1-1, ade2-1, leu2-3, 112, his3-11, 15, PAP1::LEU2 [pA pap1-5] | Minvielle-Sebastia et al. (1994) |
| MO17 | ura3-1, Δtrp1, ade2-1, leu 2-3, 112, his3-11, 15, pfs2::TRP1, pFL36-pfs2-1 | Ohnacker et al. (2000) |
| SAE-52/1 | MATα, ura3-52, leu2-3, 112, trp1, lys2, his3, pep4, prb1, prc1, rnt1::HIS3 | Chanfreau et al. (1997) |
| D162 | MATα, ura3-52,leu2-Δ, his3-Δ200, rat1-1 | Amberg et al. (1992) |
| FYEF9 | MATα, ura3-Δ851, trp1Δ63, leu2Δ1, Δnup145 [ARS-CEN-LEU2-ProtA-NUP145N] | Teixeira et al. (1997) |
| ΔU24 | MATα, trp1. Δ, his3. Δ, ura3.52, lys2.801, ade2.101, URA3::U24 | Kiss-László et al. (1998) |
| YAF2 | MATα, trp1. Δ, his3. Δ, ura3.52, lys2.801, ade2.101, URA3::U24, NOP56::TAP::TRP1 | this study |
| D174 | MATa, ade2-1, xrn1::URA3, rat1-1 | Henry et al. (1994) |
For induction experiments, galactose at a final concentration of 2% was added directly to cultures grown in non-repressive medium containg 2% raffinose and 0.08% glucose. The strains containing conditional temperature-sensitive alleles, rat1-1, rna14-1, rna15-2, pap1-5, pfs2-1 and nup145N, were grown at 23°C, up to mid-log phase and then were shifted to 37°C for 1 h prior to galactose induction.
Plasmid construction
Genomic U24 sequences (from position –4 to +102, with respect to the beginning of the U24 coding region) were amplified by PCR using oligonucleotides U24a and U24b (see below) and then cloned into the BamHI and EcoRI sites of the Bluescript plasmid to produce pBS-U24. The introduction of the TAG sequence between the D′ and C′ boxes of U24 (pBS-U24tag) was achieved by inverse PCR with oligonucleotides U24tag-a and U24tag-b. Subsequently, the genomic and tagged U24 (U24*) were iserted into the BamHI–EcoRI sites of plasmids p413GAL1 and p414GAL1 (Mumberg et al., 1994) to give plasmids GAL/U24 and GAL/U24*. The introduction of the extra stem element was achieved by PCR on pBS-U24 or pBS-U24tag using oligonucleotides U24Sa and U24Sb. The U24S was inserted into the BamHI–EcoRI sites of plasmids p413GAL1 and p414GAL1 to give constructs GAL/U24S and GAL/U24*S. Part of the BEL1 gene (from position +1 to +1233, with respect to the ATG codon) was amplified by PCR using oligonucleotides U24HGa and U24HGb and then cloned into the BamHI and EcoRI sites of the pGAL413 plasmid to produce construct GAL/U24HG. Plasmids containing the genomic version of snR4 (from position +1 to +188), snR13 (from +1 to +135) and snR40 (from –6 to +103) were obtained as described for U24 with oligonucleotides snR4a, snR4b, snR13a, snR13b, snR40a and snR40b for cloning of genomic snoRNAs, and oligonucleotides snR4tag-a, snR4tag-b, snR13tag-a, snR13tag-b, snR40tag-a and snR40tag-b for snoRNA tagging. Plasmids GAL/U24* and GAL/U24*S were used as starting material to generate plasmids containing the snR13 or snR47 terminators. The CYC1 cleavage/polyadenylation site was removed by XhoI–KpnI digestion and replaced by PCR products obtained with oligonucleotides R13ter-a and R13ter200-b (construct T200); R13ter-a and R13ter100-b (construct T100); R13ter-a and R13ter50-b (construct T50); R13ter-a and R13ter30-b (construct T30); and R47ter-a and R47ter-b (construct GAL/U24*-47T). For constructs T0 and S-T0, the CYC1 cleavage/polyadenylation site was removed and, after conversion to blunt ends, the plasmid was religated. The introduction of the Rnt1p cleavage site (obtained from the 3′ ETS of the 35S pre-rRNA; Kufel et al., 1999) upstream of the U24 coding sequence (Rnt1-T100 plasmid) was accomplished by inverse PCR with oligonucleotides 3′ ETSa and 3′ ETSb.
Oligonucleotides
The sequences of the oligonucleotides used for the different cloning steps are (5′–3′): U24a, CGGGATCCATTCAAATGATGTAAT; U24b, CGGAATTCCATTCATCAGAGATCT; U24tag-a, GACCTACGGAGACTTCATCGTTTATACAAT; U24tag-b, CGTCAGGCGACTTACTCTGTATGGTTAATAG; U24Sa, CGGGATCCTAACTATAATGA TGCCATTCAAATGATGTAATAA; U24Sb, CGGAATTCTAACTATAATGATACCATTCATCAGAGATCTTGGT; U24HGa, GGGGATCCATGGCATCTAACGAAGTTTTAG; U24HGb, GGGAATTCTTAG TTAGCAGTCATAAC; snR4a, GCGGATCCACTATTACAGTCGATGAGGA; snR4b, GCGAATTCTACAGCCTCAGTTATTAATAG; snR4tag-a, GCAGTCCGCTGTTTGTGTAAGATGCTTTGGT; snR4tag-b, CTGGATGCCGTTTGTGAAAAACGCCCCGGAT; snR13a, GCGGATCCAGGAAGTTTTTTCCTTTTTAT; snR13b, GCGAATTCGGAAGTTAAAAGGTCAGATAA; snR13tag-a, GCAGTCCGCTTT TTGCCAAATCAGTAACGGT; snR13tag-b, CTGGATGCCGGCCAAACAGCAACTCGAGCCA; snR40a, GCGGATCCAAAATGGTAAATGACGAGAAA; snR40b, GCGAATTCAAAATGGATCAGA AATTTGGG; snR40tag-a, GCAGTCCGCTAAGTACTCAGGTGTCCTATGA; snR40tag-b, CTGGATGCCGGTGGCATCCATGTTCAGACTG; R13ter-a, CCGCTCGAGTAATCCTTCTTACATTGTA; R13ter 200-b, GGGGTACCCCTGCAAAGTATACGATAAA; R13ter100-b, GGGGTACCCCCAACGTACTAACATCTTT; R13ter50-b, GGGGTA CCGAAAATTTTACGCATTATATA; R13ter30-b, GGGGTACCGCAGCGCTACGATACAAT; R47ter-a, CCGCTCGAGGATCACTCGCCAAGAGAATG; R47ter-b, GGGGTACCGATCACTAACAACG ATATAT; 3′ ETSa, GAGTCGGGGCCTAGTTTAGAGAGAAGTAGACTCATCCATTCAAATGATGTAATAACA; and 3′ ETSb, CTGCCAGTACCCACTTAGAAAGAAATAAAAAAGGATCCACTAGTTCTAGAGCGGCC.
The oligonucleotides used for RNA analyses by northern hybridization or primer extension were (5′–3′): αtag, TGCGGACTGCCTGGATCGCG; αU24, TCAGAGATCTTGGTGATAAT; αU5, CCTGTTTCTATGGAGACAACACCCGGATGGTTCTG; αU3, GAAGAGTCA AAGAGTGAC; αsnR13, TTCCACACCGTTACTGATTT; and α25S, GCTTCAGTCTTCAAAGTTCTC.
RNA analysis
RNA was extracted by the hot-phenol method as previously described (Schmitt et al., 1990). RNA concentrations were calibrated by absorbance at 260 mn and normalized by hybridization with U5-, U3- and snR13-specific oligonucleotides. For RNase H analysis, 10 µg of total RNA was hybridized with oligo(dT) (24 µM) and incubated with 0.4 U of RNase H (Pharmacia) in 80 mM Tris–HCl (pH 7.5), 8 mM MgCl2, 2 mM dithiothreitol (DTT), 60 µg/ml bovine serum albumin (BSA), 8% glycerol for 20 min at 37°C. For northern blot analysis, typically 5 µg of total RNA were run on 6% polyacrylamide–7 M urea gels and electrotransferred at 4°C to Amershan Hybond-N+ filters in 0.5× TBE buffer for 16 h at 10 V. Hybridizations and primer extensions were carried out as previously described (Villa et al., 1998). Oligonucleotides (10 pmol) were routinely 5′ end labeled with 30 µCi of [γ-32P]ATP.
Mapping of 2′-O-ribose-methylated nucleotides in yeast 25S rRNA was performed by primer extension analysis in the presence of a low concentration of dNTPs as previously described (Maden et al., 1995; Kiss-László et al., 1998).
Yeast extract and immunoprecipitation experiments were performed as previously described (Ganot et al., 1997a; Lafontaine and Tollervey, 2000).
Acknowledgments
Acknowledgements
We are particularly grateful to Walter Keller, Bernhard Dichtl, Carlo Presutti, Manny Ares, Tamas Kiss and Bertrand Séraphin for kindly providing strains and plasmids; we wish to thank Massimo Arceci and Genesio Ricci for skillful technical help. A.F. was the recipient of a fellowship from the CNR ‘Target Project on Biotechnology’. This work was partially supported by grants from ‘MURST-CNR Biotechnology Program L.95/95’, from PRIN 40% of MURST and from CNR ‘Target Project on Biotechnology’.
References
- Allmang C., Kufel,J., Chanfreau,G., Mitchell,P., Petfalski,E. and Tollervey,D. (1999) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J., 19, 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg D.C., Goldstein,A.L. and Cole,C.N. (1992) Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev., 6, 1173–1189. [DOI] [PubMed] [Google Scholar]
- Amrani N., Minet,M., Le Gouar,M., Lacroute,F. and Wyers,F. (1997) Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol. Cell. Biol., 17, 3694–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachellerie J.-P., Michot,B., Nicoloso,M., Balakin,A., Ni,J. and Fournier,M.J. (1995) Antisense snoRNAs: a family of nucleolar RNAs with long complementarities to rRNA. Trends Biochem. Sci., 20, 261–264. [DOI] [PubMed] [Google Scholar]
- Balakin A.G., Smith,L. and Fournier,M.J. (1996) The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell, 86, 823–834. [DOI] [PubMed] [Google Scholar]
- Birse C.E., Minvielle-Sebastia,L., Lee,B.A., Keller,W. and Proudfoot,N.J. (1998) Coupling termination of transcription to messenger RNA maturation in yeast. Science, 280, 298–301. [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli C., Presutti,C. and Tollervey,D. (2000) Identification of a regulated turnover pathway for nuclear pre-mRNA. Cell, 102, 765–775. [DOI] [PubMed] [Google Scholar]
- Caffarelli E., Arese,M., Santoro,B., Fragapane,P. and Bozzoni,I. (1994) In vitro study of the processing of the intron encoded U16 snoRNA in X.laevis.Mol. Cell. Biol., 14, 2966–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarelli E., De Gregorio,E., Fatica,A., Prislei,S., Fragapane,P. and Bozzoni,I. (1996) Processing of intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J., 15, 1121–1131. [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G., Abou Elela,S., Ares,M.,Jr and Guthrie,C. (1997) Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev., 11, 2741–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G., Legrain,P. and Jacquier,A. (1998a) Yeast RNase III as a key processing enzyme in small nucleolar RNA metabolism. J. Mol. Biol., 284, 975–988. [DOI] [PubMed] [Google Scholar]
- Chanfreau G., Rotondo,G., Legrain,P. and Jacquier,A. (1998b) Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J., 17, 3726–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G., Buckle,M. and Jacquier,A. (2000) Recognition of a conserved class of RNA tetraloops by Saccharomyces cerevisiae RNase III. Proc. Natl Acad. Sci. USA, 97, 3142–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre E., Boelens,W.C., Wimmer,C., Mattaj,I.W. and Hurt,E.C. (1994) Nup145p is required for nuclear export of mRNA and binds homopolymeric RNA in vitro via a novel conserved motif. Cell, 78, 275–289. [DOI] [PubMed] [Google Scholar]
- Ganot J.-P., Caizergues-Ferrer,M. and Kiss,T. (1997a) The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev., 11, 941–956. [DOI] [PubMed] [Google Scholar]
- Ganot J.-P., Bortolin,M.-L. and Kiss,T. (1997b) Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell, 89, 799–809. [DOI] [PubMed] [Google Scholar]
- Gautier T., Berges,T., Tollervey,D. and Hurt,E.C. (1997) Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol., 17, 7088–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z. and Sherman,F. (1996) 3′-end-forming signals of yeast mRNA. Trends Biochem. Sci., 21, 477–481. [DOI] [PubMed] [Google Scholar]
- Henry Y., Wood,H., Morrissey,J.P., Petfalsky,E., Kearsey,S. and Tollervey,D. (1994) The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J., 13, 2452–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. and Carmichael,G.G. (1996) Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol., 16, 1534–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W. and Minvielle-Sebastia,L. (1997) A comparison of the mammalian and yeast pre-mRNA 3′-end processing. Curr. Opin. Cell Biol., 9, 329–336. [DOI] [PubMed] [Google Scholar]
- Kiss T. and Filipowicz,W. (1995) Exonucleolytic processing of small nucleolar RNAs from pre mRNA introns. Genes Dev., 9, 1411–1424. [DOI] [PubMed] [Google Scholar]
- Kiss-László Z., Henry,Y., Bachellerie,J.P., Caizergues-Ferrer,M. and Kiss,T. (1996) Site-specific ribose methylation of preribosmal RNA: a novel function for small nucleolar RNAs. Cell, 85, 1077–1088. [DOI] [PubMed] [Google Scholar]
- Kiss-László Z., Henry,Y. and Kiss,T. (1998) Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose metylation of pre-rRNA. EMBO J., 17, 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz J.E. and Holm,C. (1990) Cloning by function: an alternative approach for identifying homologs of genes from other organisms. Proc. Natl Acad. Sci. USA, 87, 6629–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel J., Dichtl,B. and Tollervey,D. (1999) Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA, 5, 909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D.L.J. and Tollervey,D. (1998) Birth of snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochem. Sci., 23, 383–388. [DOI] [PubMed] [Google Scholar]
- Lafontaine D.L. and Tollervey,D. (2000) Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell. Biol., 20, 2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader D.J., Sanders,J.F., Waugh,R., Shaw,P.J. and Brown,J.W.S. (1994) Molecular characterization of plant U14 small nucleolar RNA genes: closely linked genes are transcribed as polycistronic U14 transcript. Nucleic Acids Res., 22, 5196–5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader D.J., Clark,G.P., Watters,J., Beven,A.F., Shaw,P.J. and Brown,J.W.S. (1997) Clusters of multiple different small nucleolar RNA genes in plants are expressed as processed from polycistronic pre-snoRNAs. EMBO J., 16, 5742–5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Pandit,S. and Deutscher,M.P. (1998) Polyadenylation of stable RNA precursor in vivo. Proc. Natl Acad. Sci. USA, 95, 12158–12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B.E.H., Corbett,M.E., Heeney,P.A., Pugh,K. and Ajuh,P.M. (1995) Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie, 77, 22–29. [DOI] [PubMed] [Google Scholar]
- Maxwell E.S. and Fournier,M.J. (1995) The small nucleolar RNAs. Annu. Rev. Biochem., 64, 897–934. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Preker,P.J. and Keller,W. (1994) RNA 14 and RNA 15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science, 266, 1702–1705. [DOI] [PubMed] [Google Scholar]
- Mumberg D., Muller,R. and Funk,M. (1994) Regulatable promoters of S.cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res., 22, 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J., Tien,A.L. and Fournier,M.J. (1997) Small nucleolar RNAs direct site-specific synthesis of pseudouridines in ribosomal RNA. Cell, 89, 565–573. [DOI] [PubMed] [Google Scholar]
- Nicoloso M., Qu,L.H., Michot,B. and Bachellerie,J.P. (1996) Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation of rRNAs. J. Mol. Biol., 260, 178–195. [DOI] [PubMed] [Google Scholar]
- Ohnacker M., Barabino,S., Preker,P.J. and Keller,W. (2000) The WD-repeat protein Pfs2p bridges two essential factors within the yeast pre-mRNA 3′-end-processing complex. EMBO J., 19, 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi S.L., Samarsky,D.A., Fournier,M.J. and Boeke,J.D. (1998) Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: intron length effects and activity of a precursor snoRNA. RNA, 4, 1096–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petfalski E., Dandekar,T., Henry,Y. and Tollervey,D. (1998) Processing of the precursor to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol., 18, 1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L.-H. et al. (1999) Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol. Cell. Biol., 19, 1144–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko,A., Rutz,B., Wilm,M., Mann,M. and Séraphin,B. (1999) A generic purification method for protein complex characterization and proteome exploration. Nature Biotechnol., 17, 1030–1032. [DOI] [PubMed] [Google Scholar]
- Sarkar N. (1997) Polyadenylation of mRNA in prokaryotes. Annu. Rev. Biochem., 66, 173–197. [DOI] [PubMed] [Google Scholar]
- Schmitt M.E., Brown,T.A. and Trumpower,B.L. (1990) A rapid simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res., 18, 3091–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.M. and Steitz,J.A. (1997) Sno storm in the nucleolus: new roles for myriad small RNPs. Cell, 89, 669–672. [DOI] [PubMed] [Google Scholar]
- Teixeira M.T., Siniossoglou,S., Podtelejnikov,S., Benichou,J.C., Mann,M., Dujion,B., Hurt,E. and Fabre,E. (1997) Two fuctionally distinct domains generated by in vivo cleavage of Nup145p: a novel biogenesis pathway for nucleoporins. EMBO J., 16, 5086–5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D. and Kiss,T. (1997) Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol., 9, 337–342. [DOI] [PubMed] [Google Scholar]
- Tycowski K.T., Shu,M.D. and Steitz,J.A. (1993) A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S3. Genes Dev., 7, 1176–1190. [DOI] [PubMed] [Google Scholar]
- Van Hoof A., Lennertz,P. and Parker,R. (2000) Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol., 20, 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa T., Ceradini,F., Presutti,C. and Bozzoni,I. (1998) Processing of the intron-encoded U18 small nucleolar RNA in the yeast Saccharomyces cerevisiae relies on both exo- and endonucleolytic activities. Mol. Cell. Biol., 18, 3376–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa T., Ceradini,F. and Bozzoni,I. (2000) Identification of a novel element required for processing of intron-encoded box C/D small nucleolar RNAs in Saccharomyces cerevisiae. Mol. Cell. Biol., 20, 1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins N.J., Leverette,D., Xia,L., Andrew,M.T. and Maxwell,E.S. (1996) Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide boxes C and D. RNA, 4, 118–133. [PMC free article] [PubMed] [Google Scholar]
- Weinstein L.B. and Steitz,J.A. (1999) Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol., 11, 378–384. [DOI] [PubMed] [Google Scholar]
- Xu F. and Cohen,S.N. (1995) RNA degradation in Escherichia coli regulated by 3′ adenylation and 5′ phosphorylation. Nature, 374, 180–183. [DOI] [PubMed] [Google Scholar]