Abstract
Transgenic mice expressing a T-cell-specific dominant interfering allele (MEnT) of the c-Myb transcription factor have a pronounced block in CD4–CD8– (DN) development. In this study we show that differentiation of DN MEnT thymocytes is blocked due to the failure of cells to enter the cell cycle following β-selection, the process by which productive rearrangement of the T-cell receptor (TCR) β-chain permits maturation of cells into CD4+CD8+ (DP) thymocytes. c-myb mRNA continues to be expressed in DN cells in mice lacking a functional pre-TCR signalling pathway, implying that its transcriptional regulation is independent of the signalling events regulating β-selection. It is also expressed in the absence of cytokine signalling. However, we show that c-Myb protein is required for the function in β-selection of its known upstream activator, the serine/threonine kinase Pim1: MEnT expression inhibits the cell cycle in Pim1 transgenic DN thymocytes and prevents Pim1-mediated rescue of a RAG1–/– developmental block. Super activation of c-Myb by Pim1 may therefore be required for β-selection.
Keywords: β-selection/cell cycle/c-Myb/Pim1/T cell
Introduction
Elucidating the mechanism by which immature naive thymocytes differentiate, mature and expand to produce a full peripheral T-cell repertoire is one of the central problems of T-cell immunology. β-selection, one of the earliest control points in this process, occurs in the most immature thymocyte compartment, defined as double negative (DN) for the CD4 and CD8 co-receptors. Progression of thymocytes through the DN stage can be monitored by the expression of CD25 and CD44 (reviewed in Shortman and Wu, 1996). The four stages of DN development, in increasing level of maturity, are known as DN1 (CD44+CD25–), DN2 (CD44+CD25+), DN3 (CD44–CD25+) and DN4 (CD44–CD25–). β-selection, which occurs in DN3 cells, is a major developmental checkpoint and follows productive RAG-mediated rearrangement of the T-cell receptor (TCR) β-chain locus (reviewed in Levelt and Eichmann, 1995; Fehling and von Boehmer, 1997). Functional TCR β-chain associates with a surrogate TCR α-chain, pTα, and CD3 to form the pre-TCR complex, which is expressed at the cell surface (reviewed in Malissen and Malissen, 1996). Formation of the pre-TCR, showing that productive rearrangement has occurred, triggers a signalling cascade involving the non-receptor tyrosine kinases Lck/Fyn and ZAP-70/Syk (Groves et al., 1996; van Oers et al., 1996; Cheng et al., 1997; Sugawara et al., 1998) and the MAPK pathway (Crompton et al., 1996; Michie et al., 1999), which leads to proliferation and differentiation through the DN4 stage and, ultimately, to the generation of CD4+CD8+ (DP) thymocytes.
β-selection is known to be required for the DN to DP transition, as in the absence of RAG proteins (Mombaerts et al., 1992b; Shinkai et al., 1992), TCRβ (Mombaerts et al., 1992a), pTα (Fehling et al., 1995), components of the CD3 complex (Love et al., 1993; Malissen et al., 1993, 1995; Ohno et al., 1993; DeJarnette et al., 1998; Haks et al., 1998; Wang et al., 1998), ZAP-70/syk (Cheng et al., 1997; Sugawara et al., 1998) or Lck (van Oers et al., 1996) developing thymocytes are blocked, to varying degrees, during DN3 progression. It has been proposed that pre-TCR signalling is both necessary and sufficient for differentiation from DN to DP. Extracellular signals, notably the cytokine IL-7 and stem cell factor (SCF), also have a defined role in DN thymopoiesis (Godfrey et al., 1992; Peschon et al., 1994; Rodewald et al., 1995); however, it is thought that they are required for expansion and survival, but not for progression to the DP stage (DiSanto et al., 1995; Rodewald et al., 1997).
Recent work has defined a number of transcription factors able to regulate β-selection. Thymocytes from mice in which the genes for both the HMG family members TCF-1 and LEF-1 are deleted show impaired DN3 progression and a complete block in development at the subsequent CD8+ immature single positive (ISP) stage (Okamura et al., 1998). These defects can be rescued in fetal thymic organ culture by CD3 ligation, thought to mimic pre-TCR signalling, suggesting that knockout of TCF/LEF does not affect events downstream of the pre-TCR. Mutation of the bHLH gene Hes1, which lies on the Notch signalling pathway, results in a severe reduction in proliferation both pre- and post-β-selection (Tomita et al., 1999). Knockout of HEB, an E-box binding transcription activator, also causes a DN3/ISP block (Barndt et al., 1999), as does overexpression of the zinc finger protein Gfi-1, a putative transcriptional repressor. In the case of Gfi-1 this is due to the impaired ability of DN3 cells to enter the cell cycle (Schmidt et al., 1998b). Conversely, overexpression of the zinc finger-containing transcription factor Egr-1 can rescue the DN3 block seen in Rag2-deficient mice (Miyazaki, 1997), as can homologous knockout of Ikaros (Winandy et al., 1999) and p53, whose loss is thought to overcome a DNA damage checkpoint at this stage (Nacht and Jacks, 1998; Haks et al., 1999).
We are interested in the role played by the transcription factor c-Myb in T-cell development. c-Myb, like its close relatives, A- and B-Myb, has been implicated in the control of growth and differentiation in a number of cell types (reviewed in Weston, 1998). c-Myb is expressed almost exclusively in haematopoietic cells and is absolutely required for correct development of the blood; the c-Myb knockout mouse has severely impaired haematopoiesis and, consequently, dies at E15 (Mucenski et al., 1991). Generation of homozygous null c-myb/rag1 chimeric mice has shown that c-myb is also required for maturation beyond the DN1 stage of thymopoiesis (Allen et al., 1999).
To study Myb function later on in T-cell development, we developed a method of inhibiting Myb activity using an active repressor, which comprises the Myb DNA binding domain fused to the Drosophila Engrailed repressor domain (Badiani et al., 1994). This protein, termed MEnT, has proved to be a powerful tool, as it can be expressed at low levels approximating to those of the endogenous c-Myb protein, obviating problems associated with overexpression, and selectively targets and switches off expression of Myb-regulated genes (Taylor et al., 1996; White and Weston, 2000). When expressed at the same level as endogenous c-Myb protein during T-cell development in mice, MEnT causes a marked impairment of the DN to DP transition and T-cell numbers thereafter are greatly reduced (Badiani et al., 1994), due to the increased susceptibility of DP and SP T cells to apoptosis (K.Weston, unpublished results; Taylor et al., 1996). In this paper we have investigated the cause of the DN block. MEnT DN thymocytes turn out to be defective in a process downstream of β-selection; MEnT DN3 thymocytes cannot mature properly into the DN4 compartment and this block cannot be overcome either by mimicking pre-TCR signalling or by preventing apoptosis. Analysis of the cell cycle of both DN3 and DN4 cells in MEnT mice shows that it is substantially impaired. We show that A- and B-myb are not expressed during β-selection, allowing us to ascribe the MEnT phenotype entirely to loss of c-Myb activity. c-myb expression is not regulated by pre-TCR signalling or by the IL-7 receptor, as expression is unchanged in rag1 and γc knockout thymocytes. However, we show, using a genetic approach, that c-Myb lies downstream of the serine/threonine kinase Pim1, which itself can be regulated by many mitogens and cytokines. Our results implicate c-Myb as an important component of the nuclear events regulating proliferation following β-selection.
Results
MEnT blocks DN thymopoiesis at the DN3 stage
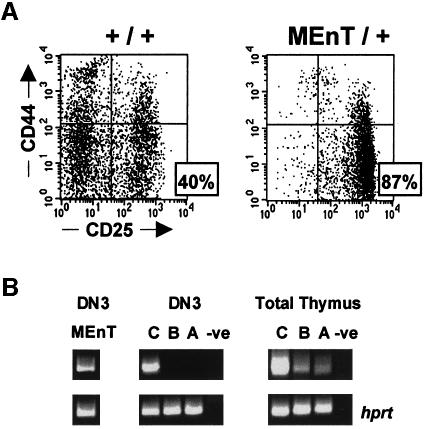
Mice expressing dominant interfering Myb (MEnT) show a partial developmental block in thymopoiesis, with impaired DN to DP progression (Badiani et al., 1994). To define further the MEnT-induced block, we studied DN thymopoiesis in MEnT mice by flow cytometry. Total thymocytes were stained with a cocktail of biotinylated antibodies (anti-CD3ε, anti-CD4, anti-CD8, anti-TCRβ, anti-B220, anti-Gr-1, anti-Ter-119 and anti-Mac-1) against mature and non-T cells, followed by addition of streptavidin–CyChrome; the unwanted mature and non-T cells could then be gated from the analysis population and the remaining DN thymocytes examined for expression of CD44 and CD25 cell surface markers. Figure 1A is a representative plot of such an experiment and shows that thymocyte development in MEnT mice is partially blocked at the DN3 (CD44–CD25+) stage. Analysis of a number of mice (Table I) shows that on average 40% of DN thymocytes are found at the DN3 stage in non-transgenic animals, whereas in the MEnT/+ mouse this figure rises to 87%. The real cell numbers in the DN compartments emphasize the DN3 block in MEnT mice. MEnT mice show an ∼2-fold reduction in total DN cellularity (Table I). In the DN3 compartment MEnT cell numbers are higher than non-transgenic, reflecting transgene-induced DN3 accumulation. However, by the DN4 (CD44–CD25–) stage there is a 26-fold decrease in MEnT thymic cellularity relative to non-transgenic controls. These data show that functional Myb protein is required for progression beyond the DN3 stage in thymopoiesis.
Fig. 1. DN thymocytes expressing dominant-negative Myb are blocked at DN3. (A) Total thymocytes were prepared from non-transgenic and MEnT littermates and stained with a cocktail of biotinylated antibodies against mature thymocyte and non-T-cell markers, followed by streptavidin–CyChrome. DN thymocytes were identified by gating on the CyChrome-negative population. Shown are representative plots of gated DN cells stained with anti-CD44–PE and anti-CD25–FITC. Numerical data from all mice are shown in Table I. (B) RNA from non-transgenic total thymus and from non-transgenic sorted DN3 cells was examined by RT–PCR for expression of c-myb, B-myb and A-myb. Expression of the MEnT transgene in DN3 cells was confirmed by RT–PCR of RNA from sorted MEnT thymocytes. Control reactions in the absence of a cDNA template are shown in the lanes marked –ve.
Table I. Cell number (× 10–4) and percentage in each DN compartment.
| Line | n | All DN | DN1 | DN2 | DN3 | DN4 | %DN1 | %DN2 | %DN3 | %DN4 |
|---|---|---|---|---|---|---|---|---|---|---|
| +/+ | 10 | 438 ± 136 | 43 ± 21 | 120 ± 44 | 174 ± 52 | 104 ± 51 | 9.8 | 27.4 | 39.7 | 23.7 |
| MEnT/+ | 8 | 238 ± 99 | 2 ± 1 | 27 ± 11 | 206 ± 90 | 4 ± 2 | 0.8 | 11.3 | 86.6 | 1.7 |
| LckF505/+ | 4 | 483 ± 235 | 13 ± 10 | 43 ± 22 | 165 ± 46 | 262 ± 206 | 2.7 | 8.9 | 34.2 | 54.2 |
| LckF505/+:MEnT+ | 4 | 316 ± 54 | 2 ± 2 | 33 ± 22 | 279 ± 32 | 2 ± 1 | 0.6 | 10.4 | 88.3 | 0.6 |
| Bcl-2/+ | 8 | 2328 ± 699 | 151 ± 82 | 336 ± 149 | 1195 ± 457 | 646 ± 168 | 6.5 | 14.4 | 51.3 | 27.8 |
| Bcl-2/+:MEnT/+ | 8 | 882 ± 233 | 11 ± 6 | 88 ± 21 | 724 ± 192 | 58 ± 32 | 1.3 | 10.0 | 82.1 | 6.6 |
| Pim1/+ | 3 | 230 ± 60 | 42 ± 3 | 23 ± 3 | 62 ± 22 | 106 ± 35 | 18.7 | 9.9 | 26.2 | 45.2 |
| Pim1/+:MEnT/+ | 5 | 410 ± 180 | 15 ± 5 | 38 ± 10 | 350 ± 185 | 9 ± 4 | 4.1 | 11.0 | 82.0 | 2.8 |
| RAG1–/– | 3 | 286 ± 78 | 3 ± 2 | 3 ± 1.6 | 278 ± 75 | 1.3 ± 0.9 | 1.1 | 1.1 | 97.4 | 0.4 |
c-myb, but not A- or B-myb, is expressed in DN3 thymocytes
The mechanism of action of our dominant-negative Myb mutant, MEnT, is to displace wild-type Myb protein from its target sites on DNA. As the three Myb proteins A-, B- and c-Myb all recognize the same consensus sequence, MEnT could be inhibiting target genes regulated by any or all of them. To determine which Myb proteins were expressed during DN development, we flow sorted normal thymocytes and ran RT–PCRs on RNA prepared from DN3 cells and total thymocytes. Figure 1B shows that A- and B-myb expression is detectable in total thymic RNA, but not at any appreciable level in sorted DN3 cells, where only c-myb is expressed. We also looked at RNA levels of our transgenic MEnT construct, by flow sorting MEnT DN3 thymocytes, and saw good expression in DN3 cells (Figure 1B). Therefore, during the DN3 stage MEnT is likely to be affecting only genes regulated by c-Myb.
Rearrangement of the TCR β-chain locus occurs in thymocytes expressing MEnT
As cells progress through DN3, RAG-mediated rearrangement of the TCR β-chain locus occurs and productive rearrangement leads to proliferation and differentiation (reviewed in Fehling and von Boehmer, 1997). To examine whether MEnT thymocytes are blocked in DN3 because they cannot rearrange their TCR β-chains and initiate β-selection, cells were assayed by PCR for VDJ recombination events. Figure 2A shows that amplification with primers specific for Vβ8-Jβ2, Vβ5-Jβ2 and Dβ2-Jβ2 events yielded several products, indicating that TCRβ rearrangement occurs in MEnT thymocytes. No rearrangement products could be amplified from control non-transgenic tail DNA. The ability of MEnT thymocytes to rearrange the TCR β-chain implies that the effect of the transgene is downstream of this event.
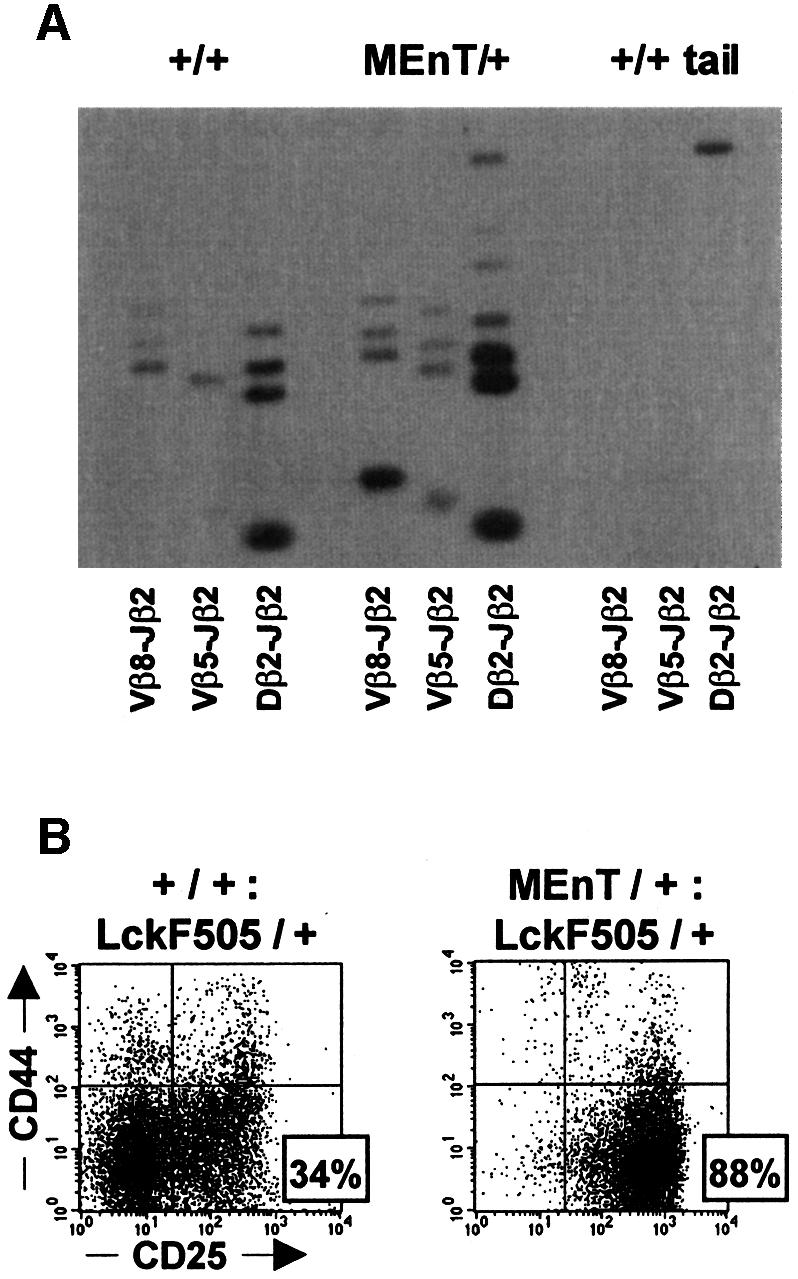
Fig. 2. TCRβ rearrangement occurs in MEnT mice. (A) TCRβ chain rearrangements occur in thymocytes expressing dominant-negative Myb. DNA purified from non-transgenic and MEnT total thymocytes and from non-transgenic tail was amplified by PCR, using primers specific for Vβ8Jβ2, Vβ5Jβ2 and Dβ2Jβ2 recombination events. The amplified products were electrophoresed and Southern blotted to nitrocellulose. TCRβ chain rearrangements were detected by hybridization of a radiolabelled probe specific for the PCR products of recombination events. (B) Expression of activated Lck is insufficient to relieve the MEnT-induced DN3 block. Total thymocytes were prepared from +/+:LckF505/+ and MEnT/+:LckF505/+ littermates, and mature and non-T cells excluded from the analysis, as described in the legend to Figure 1. A representative plot is shown; data from several mice are given in Table I.
Expression of activated Lck fails to relieve the MEnT-induced DN3 block
From the previous experiment we cannot exclude the possibility that assembly of the pre-TCR is actually being affected in cells blocked in DN3 and that the TCRβ rearrangement detected occurred in cells somehow escaping from the MEnT block. Therefore, we investigated whether the MEnT-induced block could be rescued by overexpression of an activated form of the Lck tyrosine kinase. Lck is both necessary and sufficient for correct signalling from the pre-TCR, and its overexpression is able to rescue thymopoiesis in rag1–/–, pTα–/– or CD3γ–/– mice, all of which are unable to assemble the pre-TCR (Mombaerts et al., 1994; Fehling et al., 1997; Jacobs et al., 1999). Interestingly, a previous report has suggested that lck may be a target gene for c-Myb (McCracken et al., 1994). MEnT mice were crossed with LckF505 mice, which overexpress an activated form of Lck in the thymus (Abraham et al., 1991). Expression of the activated construct is under the control of a promoter in which no c-Myb binding sites are present. DN thymocytes from MEnT/+:LckF505/+ mice were gated and analysed for CD44 and CD25 expression by flow cytometry, as before. Figure 2B shows that the LckF505/+ mouse has an over-representation of DN4 cells, due to elevated Lck levels mimicking pre-TCR signalling (Fehling et al., 1997). The MEnT transgene reverses this phenotype (Figure 2B), with 88.3% of DN cells blocked at DN3 compared with 34.2% in the LckF505 mouse (Table I). As a consequence of the DN3 block, the MEnT/+:LckF505/+ DN4 percentage is reduced to 0.6%. Table I shows that, as would be predicted from the DN phenotypes, the LckF505/+ DN4 cell number is ∼130-fold greater than that of MEnT/+:LckF505/+, whereas DN3 cellularity is slightly less. The failure of Lck to relieve the MEnT DN3 block and restore DN cell number suggests that the lck gene is not a c-Myb target during DN development. Furthermore, as activated Lck can rescue cells defective in components of the pre-TCR (see above), the MEnT-induced block to β-selection is unlikely to be affecting the pre-TCR complex itself. To investigate this further, we attempted to overcome the MEnT-induced block by CD3 ligation in E17.5 fetal thymic organ cultures, but saw no change in the number of cells blocked in DN3 relative to untreated MEnT controls (data not shown). Crossing MEnT mice with mice transgenic for the H-Y TCR (Huesmann et al., 1991) also failed to rescue the block (data not shown).
The MEnT-induced DN3 block is not due to increased apoptosis
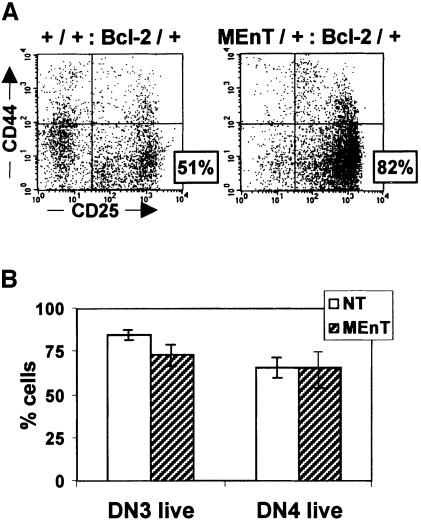
Previously we and others have demonstrated that Myb can protect against apoptosis (Frampton et al., 1996; Taylor et al., 1996) and that this is due, at least in part, to direct regulation of the bcl-2 promoter. We reasoned that the apparent DN3 block in MEnT thymopoiesis might be due to cells progressing into the DN4 compartment and then dying due to an increased susceptibility to apoptosis brought about by down-regulation of bcl-2. To assess whether this was true, MEnT mice were crossed with a line overexpressing Bcl-2 in their T-cell lineage (EµBcl-2-25) (Strasser et al., 1990). MEnT/+:Bcl-2/+ DN cells were gated and analysed for CD44 and CD25 expression, as before. Bcl-2 mice have a slight over-representation of DN3 cells (Figure 3A; 51.3%) compared with non-transgenic mice (Figure 1A; 32%), possibly because elevated levels of Bcl-2 are slowing pre-TCR-induced cell cycle entry (Brady et al., 1996; O’Reilly et al., 1996). However, in the MEnT/+:Bcl-2/+ double transgenic 82% of DN cells are found in the DN3 compartment. The MEnT/+:Bcl-2/+ DN cell numbers (Table I) confirm these percentages. Therefore, the DN3 block in MEnT thymocytes is maintained even in the presence of exogenous Bcl-2, suggesting that down-regulation of bcl-2 is unlikely to be the cause of the MEnT phenotype.
Fig. 3. MEnT does not induce increased apoptosis following β-selection. (A) Bcl-2 overexpression cannot rescue the MEnT DN3 block. Total thymocytes were prepared from +/+:Bcl-2/+ and MEnT/+:Bcl-2/+ littermates, and mature and non-T cells excluded from the analysis as described in the legend to Figure 1. A representative plot is shown; data from several mice are given in Table I. (B) DN3 and DN4 populations were gated and live cells identified by exclusion of 7-AAD and negative staining with annexin V–FITC. Between three and eight mice were assayed for each histogram bar. The mean and standard deviation are plotted.
To test directly whether expression of MEnT leads to increased apoptosis following β-selection, gated DN3 and DN4 cells were analysed for apoptotic events by annexin V–FITC and 7-AAD staining. Although there was a small but significant (P = 0.002) reduction in the percentage of live cells in the MEnT DN3 compartment (+/+, 85%; MEnT/+, 73%; Figure 3B), a comparison of non-transgenic and MEnT DN4 thymocytes revealed a near identical percentage of live cells (+/+, 66%; MEnT/+, 65%; Figure 3B). In addition to annexin V analysis, RNA from pooled DN3 and DN4 thymocytes from non-transgenic and MEnT mice was analysed for expression of bclw, bfl1, bclx, bak, bax, bcl-2 and bad; no differences were found (data not shown). Taken together, these data suggest that following β-selection the MEnT phenotype is unlikely to be caused by increased apoptosis.
The MEnT-induced DN3 block results from reduced cell cycling
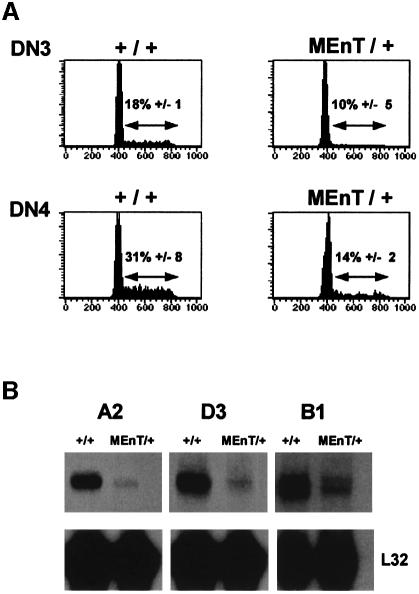
During the late DN3 and DN4 stages, a massive proliferative burst is initiated following β-selection. Thymocyte number is greatly increased as cells undergo up to 10 rounds of division and progress to the DP compartment (Penit et al., 1995). The loss of thymic cellularity in MEnT mice suggested that proliferation of late DN cells might be impaired. To examine this, we studied the cell cycle status of gated DN3 and DN4 MEnT cells by saponin permeabilization and 7-AAD staining. While on average 18% of non-transgenic DN3 cells were in S/G2/M, only 10% of MEnT DN3 cells had progressed from G0/G1 (Figure 4A). In DN4 cells an average of 31% of non-transgenic thymocytes were in S/G2/M, com pared with only 14% in MEnT thymocytes (Figure 4A). In support of the cell cycle data, forward scatter analysis (Penit et al., 1995; Hoffman et al., 1996) showed reduced blasting in DN3 and DN4 MEnT thymocytes (data not shown).
Fig. 4. DN3 and DN4 MEnT thymocytes show reduced cell cycle entry. (A) To determine the cell cycle status of gated DN3 and DN4 populations, cells were permeabilized in 0.03% saponin in the presence of 25 µg/ml 7-AAD prior to flow cytometry. A representative plot is shown, together with the statistical analysis of several mice (+/+, n = 4; MEnT/+, n = 4). (B) MEnT DN3 and DN4 thymocytes show reduced expression of cyclins. Sorted DN3/DN4 non-transgenic and MEnT RNA (10 µg) was analysed by RNase protection assay for cyclin expression levels. The results shown are representative of three experiments.
To confirm that the cell cycle status of late DN MEnT thymocytes was affected, sorted DN3 and DN4 cells were pooled and analysed for cyclin expression by RNase protection assay (Figure 4B). There was a marked reduction in expression of cyclins A2 and D3 relative to wild type; both of these cyclins are required for G1/S progression (reviewed in Sherr, 1993). Cyclin B1, expressed during G2/M (reviewed in King et al., 1994) was also reduced in MEnT cells. Expression of cyclins C, D1, D2, A1 and B2 was either comparable or undetectable (data not shown).
The significant reduction in the percentage of cycling cells provides a good explanation for the partial MEnT DN3 block and probably underlies the dramatic loss of total thymic cellularity; in the MEnT thymus, if cell cycling is reduced ∼2-fold during each of the 10 rounds of division following β-selection, the total number of DP thymocytes generated would be greatly reduced.
γc, the pre-TCR and Lck/Fyn are not required for expression of c-myb
As c-Myb protein appears essential for correct progression through the DN compartment, we were interested to determine whether the signalling pathways involved in DN development regulate c-myb expression. To test whether the IL-7 receptor, which signals using the common cytokine gamma chain γc, the pre-TCR or the tyrosine kinases Lck and Fyn are required for c-myb expression, total DN RNA was isolated from γc– (DiSanto et al., 1995) and lck–/–/fyn–/– (Molina et al., 1992; Stein et al., 1992) thymuses and DN3 RNA was purified from non-transgenic and rag1–/– (Spanopoulou et al., 1994) mice. Interestingly, expression of c-myb could be detected by RT–PCR in all of the sorted subsets (Figure 5). The absence of genomic DNA was confirmed by control PCR of samples prior to reverse transcription (data not shown). Although the DN block in rag1–/– and lck–/–/fyn–/– mice is complete, γc– mice have a small number of mature thymocytes, which could theoretically contaminate the DN RNA. We therefore performed RT–PCR using RNA from rag1–/–:γc– animals, which have a total block, and also saw c-myb expression (data not shown). Therefore, c-myb expression is not regulated by IL-7 signalling, pre-TCR assembly or Lck/Fyn signalling, suggesting that it is either constitutive or lies downstream of a separate, as yet unidentified, pathway.
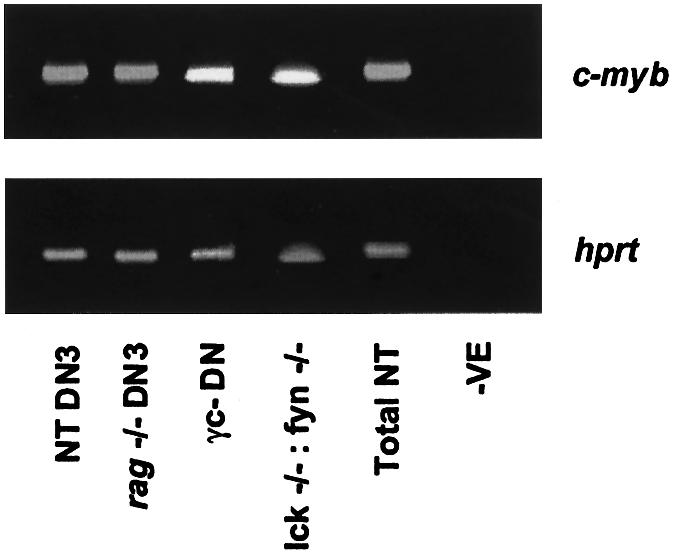
Fig. 5. γc, the pre-TCR and Lck and Fyn tyrosine kinases are not required for expression of c-myb. DNase-treated RNA from sorted cells was reverse transcribed and assayed by PCR for expression of hprt and c-myb. Control reactions in the absence of a cDNA template are shown in the lanes marked –ve.
Pim1 signals proliferation via c-Myb during β-selection
If c-myb expression is not regulated by the signals permitting progression following β-selection, we reasoned that perhaps a post-translational event might be of importance. Recently, the serine/threonine kinase Pim1 has been shown to potentiate the transcription activation function of c-Myb, at least in part by phosphorylation of the c-Myb co-activator p100 (Leverson et al., 1998). Pim1 also plays an important role in proliferative signalling during β-selection (Schmidt et al., 1998a), causing an increase in the number of cycling cells, and is able to rescue the DN3 block in rag–/– thymocytes, allowing development of DP cells (Jacobs et al., 1999). To assess whether c-Myb was required for Pim1 function during β-selection, we first crossed MEnT/+ with pim1/+ (van Lohuizen et al., 1989) mice and analysed DN cells as before. A representative plot (Figure 6A) shows that pim1/+ mice have a higher than normal percentage (49%) of DN4 cells, probably due to the transgene promoting differentiation through β-selection. However, this phenotype is reversed in pim1/+:MEnT/+ mice, with 82% of DN cells blocked at the DN3 stage of development (Figure 6A; Table I). In addition, cell cycle is severely reduced in both DN3 and DN4 pim1/+:MEnT/+ thymocytes (Figure 6B).
Fig. 6. Pim1 signals to c-Myb during β-selection. (A) Expression of MEnT blocks thymopoiesis at DN3 in Pim1 transgenic mice. Total thymocytes were prepared from pim1/+ and pim1/+:MEnT/+: littermates, and mature and non-T cells excluded from the analysis, as described in the legend to Figure 1. A representative plot is shown; data from several mice are given in Table I. (B) Cell cycle is inhibited in pim1/+:MEnT/+ DN3 and DN4 thymocytes. To determine the cell cycle status of gated DN3 and DN4 populations, cells were permeabilized in 0.03% saponin in the presence of 25 µg/ml 7-AAD prior to flow cytometry. A representative plot of three experiments is shown. (C) MEnT reverses the rescue of the RAG–/– phenotype by Pim1. Total thymocytes were prepared from rag1–/–, rag1–/–:pim1/+ and rag1–/–:pim1/+:MEnT/+ littermates aged 9 weeks. Cells were stained with anti-CD8–PE and anti-CD4–CyChrome antibodies.
To examine whether MEnT was capable of reversing the Pim1-mediated rescue of the DN3 block in rag–/– thymocytes, MEnT/+ mice were crossed with pim1/+:rag1–/– mice and cells examined for cell surface expression of CD4 and CD8 co-receptors. While Pim1 achieved a significant rescue of the rag1–/– phenotype (Figure 6C), the presence of MEnT completely abrogated this effect. Taken together with the known biochemical relationship between c-Myb and pim1, these data imply that c-Myb protein lies downstream of pim1 during β-selection and is required for its functionality.
Discussion
This study has examined the role of c-Myb in DN thymopoiesis. Cells expressing dominant interfering Myb show a partial block in development at DN3. This block appears not to affect TCRβ rearrangement and cannot be rescued by co-expression of activated Lck, placing the function of c-Myb after TCRβ rearrangement and early pre-TCR signalling. Loss of c-Myb function does not result in a significant increase in apoptosis during DN development and the DN3 block cannot be rescued by expression of the Myb target gene bcl-2. However, MEnT thymocytes do show a marked decrease in cell cycling following β-selection, implying an important role for c-Myb in regulation of thymocyte expansion during the DN:DP transition. Analysis of thymocytes from various knockout mice shows that the pre-TCR and the IL-7R are not required for expression of c-myb. However, c-Myb activity is necessary for Pim1 function during β-selection, implying that regulation of c-Myb superactivation is required for β-selection to occur.
We noted that when we calculated cell numbers at all four stages of DN development, MEnT mice appeared to have reduced DN1 and DN2 compartments (Table I) and therefore MEnT might be affecting stages both pre- and post-β-selection. To determine whether MEnT was having an earlier effect on DN development, we performed cell cycle and apoptosis analyses on DN2 thymocytes, but found no difference between MEnT and normal mice (data not shown). Analysis of DN subsets from rag1–/– mice showed that in these animals the DN1 and DN2 compartments were also greatly reduced (Table I), suggesting that a feedback control mechanism caused by defective β-selection exists. These data are supported by the results of others, which indicate that blocks to pre-TCR signalling by loss of Rag or CD3ε (Malissen et al., 1995), pTα (DiSanto et al., 1999) or Lck and Fyn (van Oers et al., 1996) reduce the DN1 and DN2 subsets. Therefore, the MEnT phenotype is no different from other mutations solely affecting β-selection.
Myb proteins have been implicated in control of the cell cycle and/or as anti-apoptotic factors in a number of cell types. Our finding that c-Myb is regulating the cell cycle rather than apoptosis tallies with other studies which suggest that Myb proteins have different effects in different circumstances. For example, Frampton et al. (1996) showed that switching off v-Myb could down-regulate bcl-2 and cause apoptosis in chicken myeloblasts, but not in multipotent progenitors, and Hogg et al. (1997), using a similar approach in immature murine myeloid cells, found that cells differentiated but did not die when v-Myb was inactivated. An inducible dominant-negative Myb causes G1 arrest but not cell death in cytotoxic T-cell lines (Lyon and Watson, 1996), but regulates cell cycle arrest, differentiation and apoptosis in the multipotent haematopoietic cell line FDCP-Mix (White and Weston, 2000).
Although we and others have identified bcl-2 as a c-Myb target gene (Frampton et al., 1996; Taylor et al., 1996; Grassilli et al., 1999), we found in this study that bcl-2 expression is unchanged relative to wild-type levels in MEnT DN3 and DN4 cells, indicating that c-Myb is not its primary regulator at this stage of thymopoiesis. However, we observed that cyclins A2, D3 and B1 were down-regulated in MEnT DN thymocytes. We do not think that transcription of these cyclins is directly regulated by c-Myb, as we were unable to find Myb binding sites in their promoters. Rather, their reduced levels are an indication of the dearth of MEnT thymocytes in cycle. A recent report suggested that c-Myb may be able to regulate the cyclin A1 promoter in humans (Muller et al., 1999), but we were unable to detect changes in murine cyclin A1 expression in our system.
We were intrigued to find that c-myb expression in sorted DN3 thymocytes appears to be completely independent of the presence of a functional IL-7R or the pre-TCR. Either c-myb expression is not regulated during β-selection or a novel signalling pathway maintains normal levels of c-myb. Little is known about how the c-myb promoter is regulated during haematopoiesis. One system, the proliferative response of mature T cells to antigenic challenge, has been studied and here c-myb is induced in response to IL-2 (Stern and Smith, 1986; Thompson et al., 1986). Recently we have shown that activation of phosphoinositide 3-kinase (PI3K) by the IL-2 receptor results in c-myb expression, via a signalling cascade terminating at the transcription factor E2F (A.Lauder and K.Weston, submitted for publication). We tested whether incubation of sorted DN thymocytes with a chemical inhibitor of PI3K, LY294002, could down-regulate c-myb, but found that expression levels were unchanged (data not shown), implying that the regulation we see in peripheral T cells is not occurring in DN thymocytes.
Our data showing that MEnT can block the action of Pim1 during β-selection suggests the possibility that c-Myb protein, rather than the c-myb gene, may be the primary regulatory target during DN development. Pim1 has recently been shown by Jacobs et al. (1999) to reconstitute thymic cellularity in IL-7 and γc mutant mice and to rescue maturation in Rag knockout animals. These authors propose that Pim1, which is commonly induced in response to many mitogens and cytokines (reviewed in Domen et al., 1993), is an effector of the IL-7 pathway, which is thought to be required to support the proliferative burst following β-selection (DiSanto et al., 1999). Taken together, the genetic link we have established here between Pim1 and c-Myb, the phenotype of MEnT mice, in which β-selection but not subsequent proliferation occurs, and the biochemical data demonstrating superactivation of c-Myb by Pim1 (Leverson et al., 1998) strongly suggest that c-Myb is on the same pathway as Pim1 and can be regulated by extracellular signalling events.
Overexpression of another transcription factor, Gfi-1, has recently been shown to cause a partial DN3 block during thymopoiesis, due to an inhibitory effect on the cell cycle (Schmidt et al., 1998b). Gfi-1 has been suggested to function as a sequence-specific transcriptional repressor (Grimes et al., 1996; Zweidler-Mckay et al., 1996). Presumably, post-β-selection Gfi-1 and MEnT both repress genes required for proliferation, implying an antagonistic relationship between Gfi-1 and c-Myb in normal DN cells. There is an interesting potential link between Gfi-1 and c-Myb, as expression of Pim1 is able to rescue the Gfi-1-induced DN3 block during thymopoiesis (Schmidt et al., 1998b). Therefore, rescue of the Gfi-1 phenotype could involve up-regulation of c-Myb activity leading to enhanced expression of the subset of genes controlled by c-Myb. It will be of interest to determine whether overexpression of a myb transgene can rescue the Gfi-1 phenotype.
In summary, the data presented here demonstrate an essential role for c-Myb in the proliferative response of DN3 cells to β-selection and place c-Myb at the bottom of a signalling cascade initiated by extracellular events activating Pim1. What might lie downstream of c-Myb? Only a handful of bona fide Myb-regulated genes have been defined and none of those described can fully account for the effects we see in DN cells. We are currently attempting to identify the c-Myb target genes that are crucial for β-selection.
Materials and methods
Immunostaining
All antibodies were supplied by Pharmingen and used at a dilution of 1:100 in phosphate-buffered saline (PBS) containing 1.0% bovine serum albumin, unless otherwise indicated. All antibody incubations were performed at 4°C for 30 min. DN thymocytes were identified by gating mature cells from the analysis population. Total thymocytes were stained with a cocktail of biotinylated antibodies: anti-CD3e (clone 145-2C11), anti-CD4 (clone H129.19), anti-CD8 (clone 53-6.7), anti-TCRβ (clone H57-597), anti-B220 (clone RA3-6B2), anti-Gr-1 (clone RB6-8C5), anti-Ter-119 and anti-Mac-1 (clone M1/70, 1:200 dilution). Bound antibody was revealed by staining with streptavidin–CyChrome (1:50 dilution). Non-specific CyChrome binding was prevented by pre-incubating with Fc block [anti-CD16/CD32 (Fcγ III/II receptor), 1:50 dilution] for 10 min at 4°C. The DN populations were revealed by including anti-CD25–FITC (clone 7D4, 1:50 dilution) and anti-CD44–PE (clone IM7) with the CyChrome incubation. Dead cells were excluded on the basis of forward and side scatter and TO-PRO 3 iodide (Molecular Probes) uptake. CD4 and CD8 co-receptors were visualized by staining with anti-CD4– CyChrome (clone H129.19) and anti-CD8–PE (clone 53-6.7). Flow cytometry was done on FACScan and FACSCalibur analysers (Becton Dickinson) and analysed using the CELLQuest program.
Apoptosis of DN thymocytes
Total thymocytes were stained with a cocktail of biotinylated antibodies, as described in the Immunostaining section. When analysing DN3 cells, anti-CD44–biotin was included in the cocktail; to identify DN4 cells, anti-CD44–biotin and anti-CD25–biotin (1:50 dilution) were included. Bound antibody was revealed by staining with streptavidin–APC (1:50 dilution). DN3 cells were identified from the APC-negative population by staining with anti-CD25–PE (1:50 dilution), while mature cells were located in the APC-positive gate. DN4 cells were visualized by staining with anti-HSA–PE (clone M1/69, 1:400 dilution). Apoptotic cells were revealed by staining with annexin V–FITC (Nexins Research) and 7-AAD, following the supplier’s protocol. 7-AAD was included at a final concentration of 1 µg/ml. Apoptotic profiles were visualized by flow cytometry on the FACSCalibur analyser.
Cell cycle of DN thymocytes
Total thymocytes, from mice aged 4–8 weeks, were stained with a cocktail of biotinylated antibodies, as described above. When analysing DN3 cells, anti-CD44–biotin was included in the cocktail; to identify DN4 cells, anti-CD44–biotin and anti-CD25–biotin (1:50 dilution) were included. Bound antibody was revealed by staining with avidin–PE (Biogenesis, 1:50 dilution). DN3 cells were identified from the PE-negative population by staining with anti-CD25–FITC (1:50 dilution), while DN4 cells were visualized by staining with anti-CD45.2–FITC (clone 104). After surface staining, cells were permeabilized in 0.03% saponin, 25 µg/ml 7-AAD, 0.1% sodium azide in PBS for 1 h at 37°C.
TCR β-chain rearrangements
PCR was used to assay for VDJ recombination of the TCRβ locus exactly as described in Clements et al. (1998).
Mice
Stock C57BL/10 mice were obtained from the MRC facility. METCD2.5 (Badiani et al., 1994), EµBcl-2-25 (Strasser et al., 1990), rag1–/– (Spanopoulou et al., 1994), EµPim1 (van Lohuizen et al., 1989), PLGF.A-2964 Lck (Abraham et al., 1991) and γc– (DiSanto et al., 1995) lines were as described. Thymocytes from lck–/–:fyn–/– mice were provided by R.Zamoyska. All lines, except EµBcl-2-25, were maintained on a C57 BL/10 inbred background. Mice were maintained under SPF conditions in the facility at the ICR and used at age 4–8 weeks, unless otherwise stated.
RNase protection assay
Total thymocytes were purified from 24 mice, aged 4–8 weeks, and stained with a cocktail of biotinylated antibodies specific for markers of mature and non-T cells, as described above. To remove DN1 and DN2 cells, anti-CD44–biotin was included in the cocktail. Cells were then incubated with streptavidin-coated microbeads (Miltenyi), according to the supplier’s instructions. DN3 and DN4 cells were purified by MACS column (Miltenyi) depletion of mature, non-T, DN1 and DN2 cells, again according to the supplier’s instructions. Purities of >95% were confirmed by flow cytometry. The RNase protection assay was performed using the m-Cyc-1 mouse cyclin Multi-Probe Template Set (Pharmingen), following the supplier’s protocol.
RT–PCR of RNA from sorted thymocyte subsets
Total thymocytes were prepared from non-transgenic, MEnT, rag1–/–, lck –/–:fyn –/– and γc– mice, aged 4–8 weeks. To purify non-transgenic and MEnT DN3 cells, thymocytes were stained with anti-CD25–FITC, washed in PBS and treated with anti-FITC microbeads (Miltenyi), according to the supplier’s instructions. CD25-positive cells were purified by MACS column (Miltenyi) positive selection, again according to the supplier’s instructions. Purified thymocytes were then additionally stained with anti-CD44–PE and DN3 cells sorted by FACS. DN3 rag1–/– thymocytes were purified by FACS sorting of anti-CD25–FITC and anti-CD44–PE stained total thymocytes. DN γc– thymocytes were purified by staining with anti-CD45.2–FITC and gating mature cells from the sort population by including anti-CD4–PE, anti-CD8–PE and anti-TCRβ–PE antibodies. Purities of >97% were confirmed by flow cytometry.
RNA was purified from cells using RNeasy mini columns (Qiagen). DNA was removed during the preparation using the RNase-free DNase set recommended by Qiagen. RNA was reverse transcribed with SuperScript II (Gibco BRL) extension of random hexamer (Pharmacia Biotech) primed RNA. cDNA was amplified using a GeneAmp 9700 machine (PE Applied Biosystems) under the following conditions: 1 cycle of 94°C for 2 min; 35 cycles of 94°C for 20 s, 55°C for 30 s, 72°C for 45 s; 1 cycle of 72°C for 7 min; a 4°C hold. cDNA was amplified using the following primer sets: c-myb, 5′-TGCTGCCCGGACGGACTGATAATG and 5′-CCCGGGGTAGCTGCAAGTGTGGTT; A-myb, 5′-ATGGCGAAGAGGTCGCGCAGTGAG and 5′-AGAACGATTTTGAAG ATGACTA; B-myb, 5′-GATCTGGATGAGTTACACTAC and 5′-CCAGCGGTTACCCAGGAC; hprt, 5′-GGGGGCTATAAGTTCTTTGC and 5′-TCCAACACTTCGAGAGGTCC; MEnT, 5′-CCATGGAAGCCGTCATTAAGAACCGG and 5′-CGCATGGTGGAATTCCAGTGGTTCTTG.
Acknowledgments
Acknowledgements
We thank D.Cantrell, M.Owen and R.Zamoyska for gifts of mice and R.Zamoyska for lck–/–:fyn–/– thymocytes. J.Clements generously donated reagents and protocols for examining TCRβ locus rearrangements. Primers for B-myb and c-myb RT–PCR were kindly provided by R.Watson and R.Zamoyska. We are indebted to Rose Zamoyska and Albert Basson for helpful discussions and J.Miller, D.Bird and I.Titley for technical assistance. This work was supported by the Cancer Research Campaign.
References
- Abraham K.M., Levin,S.D., Marth,J.D., Forbush,K.A. and Perlmutter,R.M. (1991) Delayed thymocyte development induced by augmented expression of p56lck. J. Exp. Med., 173, 1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen R.D., Bender,T.P. and Siu,G. (1999) c-Myb is essential for early T cell development. Genes Dev., 13, 1073–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badiani P., Corbella,P., Kioussis,D., Marvel,J. and Weston,K. (1994) Dominant interfering alleles define a role for c-Myb in T cell development. Genes Dev., 8, 770–782. [DOI] [PubMed] [Google Scholar]
- Barndt R., Dai,M.F. and Zhuang,Y. (1999) A novel role for HEB downstream or parallel to the pre-TCR signaling pathway during αβ thymopoiesis. J. Immunol., 163, 3331–3343. [PubMed] [Google Scholar]
- Brady H.J.M., Gil-Gomez,G., Kirberg,J. and Berns,A.J.M. (1996) Baxα perturbs T cell development and affects cell cycle entry of T cells. EMBO J., 15, 6991–7001. [PMC free article] [PubMed] [Google Scholar]
- Cheng A.M., Negishi,I., Anderson,S.J., Chan,A.C., Bolen,J., Loh,D.Y. and Pawson,T. (1997) The Syk and ZAP-70 SH2-containing tyrosine kinases are implicated in pre-T cell receptor signaling. Proc. Natl Acad. Sci. USA, 94, 9797–9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements J.L., Yang,B., Ross-Barta,S.E., Eliason,S.L., Hrstka,R.F., Williamson,R.A. and Koretzky,G.A. (1998) Requirement for the leukocyte-specific adapter protein SLP-76 for normal T cell development. Science, 281, 416–419. [DOI] [PubMed] [Google Scholar]
- Crompton T.J., Gilmour,K.C. and Owen,M.J. (1996) The MAP kinase pathway controls differentiation from double-negative to double-positive thymocyte. Cell, 86, 243–251. [DOI] [PubMed] [Google Scholar]
- DeJarnette J.B., Sommers,C.L., Huang,K., Woodside,K.J., Emmons,R., Katz,K., Shores,E.W. and Love,P.E. (1998) Specific requirement for CD3epsilon in T cell development. Proc. Natl Acad. Sci. USA, 95, 14909–14914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiSanto J.P., Muller,W., Guy-Grand,D., Fischer,A. and Rajewsky,K. (1995) Lymphoid development in mice with a targeted deletion of the interleukin 2 receptor γ chain. Proc. Natl Acad. Sci. USA, 92, 377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiSanto J.P., Aifantis,I., Rosmaraki,E., Garcia,C., Feinberg,J., Fehling,H.J., Fischer,A., von Boehmer,H. and Rocha,B. (1999) The common cytokine receptor gamma chain and the pre-T cell receptor provide independent but critically overlapping signals in early α/β T cell development. J. Exp. Med., 189, 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domen J., van der Lugt,N.M., Laird,P.W., Saris,C.J. and Berns,A. (1993) Analysis of Pim1 function in mutant mice. Leukemia, 7, S108–S112. [PubMed] [Google Scholar]
- Fehling H.J. and von Boehmer,H. (1997) Early αβ T cell development in the thymus of normal and genetically altered mice. Curr. Opin. Immunol., 9, 263–275. [DOI] [PubMed] [Google Scholar]
- Fehling H.J., Krotkova,A., Saint-Ruf,C. and von Boehmer,H. (1995) Crucial role of the pre-T cell receptor α gene in development of αβ but not of γδ cells. Nature, 375, 795–798. [DOI] [PubMed] [Google Scholar]
- Fehling H.J., Iritani,B.M., Krotkova,A., Forbush,K.A., Laplace,C., Perlmutter,R.M. and von Boehmer,H. (1997) Restoration of thymopoiesis in pTα–/– mice by anti-CD3ε antibody treatment or with transgenes encoding activated Lck or tailless pTα. Immunity, 6, 703–714. [DOI] [PubMed] [Google Scholar]
- Frampton J., Ramqvist,T. and Graf,T. (1996) v-Myb of E26 leukemia virus up-regulates bcl-2 and suppresses apoptosis in myeloid cells. Genes Dev., 10, 2720–2731. [DOI] [PubMed] [Google Scholar]
- Godfrey D.I., Zlotnik,A. and Suda,T. (1992) Phenotypic and functional characterization of c-kit expression during intrathymic T cell development. J. Immunol., 149, 2281–2285. [PubMed] [Google Scholar]
- Grassilli E., Salomoni,P., Perrotti,D., Franceschi,C. and Calabretta,B. (1999) Resistance to apoptosis in CTLL-2 cells overexpressing B-Myb is associated with B-Myb-dependent bcl-2 induction. Cancer Res., 59, 2451–2456. [PubMed] [Google Scholar]
- Grimes H.L., Chan,T.O., Zweidler-McKay,P.A., Tong,B. and Tsichlis,P.N. (1996) The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol. Cell. Biol., 16, 6263–6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves T., Smiley,P., Cooke,M.P., Forbush,K., Perlmutter,R.M. and Guidos,C.J. (1996) Fyn can partially substitute for Lck in T-lymphocyte development. Immunity, 5, 417–428. [DOI] [PubMed] [Google Scholar]
- Haks M.C., Krimpenfort,P., Borst,J. and Kruisbeek,A.M. (1998) The CD3γ chain is essential for development of both the TCRαβ and TCRγδ lineages. EMBO J., 17, 1871–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haks M.C., Krimpenfort,P., van den Brakel,J.H. and Kruisbeek,A.M. (1999) Pre-TCR signaling and inactivation of p53 induces crucial cell survival pathways in pre-T cells. Immunity, 11, 91–101. [DOI] [PubMed] [Google Scholar]
- Hoffman E.S., Passoni,L., Crompton,T., Leu,T.M., Schatz,D.G., Koff,A., Owen,M.J. and Hayday,A.C. (1996) Productive T-cell receptor β-chain gene rearrangement: coincident regulation of cell cycle and clonality during development in vivo. Genes Dev., 10, 948–962. [DOI] [PubMed] [Google Scholar]
- Hogg A., Schirm,S., Nakagoshi,H., Bartley,P., Ishii,S., Bishop,J.M. and Gonda,T.J. (1997) Inactivation of a c-Myb/estrogen receptor fusion protein in transformed primary cells leads to granulocyte/macrophage differentiation and down regulation of c-kit but not c-myc or cdc2. Oncogene, 15, 2885–2898. [DOI] [PubMed] [Google Scholar]
- Huesmann M., Scott,B., Kisielow,P. and von Boehmer,H. (1991) Kinetics and efficacy of positive selection in the thymus of normal and T cell receptor transgenic mice. Cell, 66, 533–540. [DOI] [PubMed] [Google Scholar]
- Jacobs H., Krimpenfort,P., Haks,M., Allen,J., Blom,B., Demolliere,C., Kruisbeek,A., Spits,H. and Berns,A. (1999) PIM1 reconstitutes thymus cellularity in interleukin 7- and common γ chain-mutant mice and permits thymocyte maturation in Rag– but not CD3γ-deficient mice. J. Exp. Med., 190, 1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R.W., Jackson,P.K. and Kirschner,M.W. (1994) Mitosis in transition. Cell, 79, 563–571. [DOI] [PubMed] [Google Scholar]
- Levelt C.N. and Eichmann,K. (1995) Receptors and signals in early thymic selection. Immunity, 3, 667–672. [DOI] [PubMed] [Google Scholar]
- Leverson J.D., Koskinen,P.J., Orrico,F.C., Rainio,E.M., Jalkanen,K.J., Dash,A.B., Eisenman,R.N. and Ness,S.A. (1998) Pim1 kinase and p100 cooperate to enhance c-Myb activity. Mol. Cell, 2, 417–425. [DOI] [PubMed] [Google Scholar]
- Love P.E., Shores,E.W., Johnson,M.D., Tremblay,M.L., Lee,E.J., Grinberg,A., Huang,S.P., Singer,A. and Westphal,H. (1993) T cell development in mice that lack the ζ chain of the T cell antigen receptor complex. Science, 261, 918–921. [DOI] [PubMed] [Google Scholar]
- Lyon J. and Watson,R. (1996) Interference of Myb transactivation activity by a conditional dominant negative protein: functional interference in a cytotoxic T-cell line results in G1 arrest. Gene, 182, 123–128. [DOI] [PubMed] [Google Scholar]
- Malissen B. and Malissen,M. (1996) Functions of TCR and pre-TCR subunits: lessons from gene ablation. Curr. Opin. Immunol., 8, 383–393. [DOI] [PubMed] [Google Scholar]
- Malissen M. et al. (1993) T cell development in mice lacking the CD3-ζ/η gene. EMBO J., 12, 4347–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malissen M., Gillet,A., Ardouin,L., Bouvier,G., Trucy,J., Ferrier,P., Vivier,E. and Malissen,B. (1995) Altered T cell development in mice with a targeted mutation of the CD3-ε gene. EMBO J., 14, 4641–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S., Leung,S., Bosselut,R., Ghysdael,J. and Miyamoto,N.G. (1994) Myb and Ets related transcription factors are required for activity of the human lck type I promoter. Oncogene, 9, 3609–3615. [PubMed] [Google Scholar]
- Michie A.M., Trop,S., Wiest,D.L. and Zuniga-Pflucker,J.C. (1999) Extracellular signal-regulated kinase (ERK) activation by the pre-T cell receptor in developing thymocytes in vivo. J. Exp. Med., 190, 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T. (1997) Two distinct steps during thymocyte maturation from CD4–CD8– to CD4+CD8+ distinguished in the early growth response (Egr)-1 transgenic mice with a recombinase-activating gene-deficient background. J. Exp. Med., 186, 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina T.J. et al. (1992) Profound block in thymocyte development in mice lacking p56lck. Nature, 357, 161–164. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. et al. (1992a) Mutations in T-cell antigen receptor genes α and β block thymocyte development at different stages. Nature, 360, 225–231. [DOI] [PubMed] [Google Scholar]
- Mombaerts P., Iacomini,J., Johnson,R.S., Herrup,K., Tonegawa,S. and Papaioannou,V.E. (1992b) RAG-1 deficient mice have no mature B and T lymphocytes. Cell, 68, 869–877. [DOI] [PubMed] [Google Scholar]
- Mombaerts P., Anderson,S.J., Perlmutter,R.M., Mak,T.W. and Tonegawa,S. (1994) An activated lck transgene promotes thymocyte development in RAG-1 mutant mice. Immunity, 1, 261–267. [DOI] [PubMed] [Google Scholar]
- Mucenski M.L., McLain,K., Kier,A.B., Swerdlow,S.H., Schreiner,C.M., Miller,T.A., Pietryga,D.W., Scott,W.J.,Jr and Potter,S.S. (1991) A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell, 65, 677–689. [DOI] [PubMed] [Google Scholar]
- Muller C., Yang,R., Idos,G., Tidow,N., Diederichs,S., Koch,O.M., Verbeek,W., Bender,T.P. and Koeffler,H.P. (1999) c-myb transactivates the human cyclin A1 promoter and induces cyclin A1 gene expression. Blood, 94, 4255–4262. [PubMed] [Google Scholar]
- Nacht M. and Jacks,T. (1998) V(D)J recombination is not required for the development of lymphoma in p53-deficient mice. Cell Growth Differ., 9, 131–138. [PubMed] [Google Scholar]
- Ohno H., Aoe,T., Taki,S., Kitamura,D., Ishida,Y., Rajewsky,K. and Saito,T. (1993) Developmental and functional impairment of T cells in mice lacking CD3 ζ chains. EMBO J., 12, 4357–4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura R.M., Sigvardsson,M., Galceran,J., Verbeek,S., Clevers,H. and Grosschedl,R. (1998) Redundant regulation of T cell differentiation and TCRα gene expression by the transcription factors LEF-1 and TCF-1. Immunity, 8, 11–20. [DOI] [PubMed] [Google Scholar]
- O’Reilly L.A., Huang,D.C.S. and Strasser,A. (1996) The cell death inhibitor Bcl-2 and its homologues influence control of cell cycle entry. EMBO J., 15, 6979–6990. [PMC free article] [PubMed] [Google Scholar]
- Penit C., Lucas,B. and Vasseur,F. (1995) Cell expansion and growth arrest phases during the transition from precursor (CD4–8–) to immature (CD4+8+) thymocytes in normal and genetically modified mice. J. Immunol., 154, 5103–5113. [PubMed] [Google Scholar]
- Peschon J.J. et al. (1994) Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J. Exp. Med., 180, 1955–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodewald H.R., Kretzschmar,K., Swat,W. and Takeda,S. (1995) Intrathymically expressed c-kit ligand (stem cell factor) is a major factor driving expansion of very immature thymocytes in vivo. Immunity, 3, 313–319. [DOI] [PubMed] [Google Scholar]
- Rodewald H.R., Ogawa,M., Haller,C., Waskow,C. and DiSanto,J.P. (1997) Pro-thymocyte expansion by c-kit and the common cytokine receptor γ chain is essential for repertoire formation. Immunity, 6, 265–272. [DOI] [PubMed] [Google Scholar]
- Schmidt T., Karsunky,H., Gau,E., Zevnik,B., Elsasser,H.P. and Moroy,T. (1998a) Zinc finger protein GFI-1 has low oncogenic potential but cooperates strongly with pim and myc genes in T-cell lymphomagenesis. Oncogene, 17, 2661–2667. [DOI] [PubMed] [Google Scholar]
- Schmidt T., Karsunky,H., Rodel,B., Zevnik,B., Elsasser,H.P. and Moroy,T. (1998b) Evidence implicating Gfi-1 and Pim1 in pre-T-cell differentiation steps associated with β-selection. EMBO J., 17, 5349–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C.J. (1993) Mammalian G1 cyclins. Cell, 73, 1059–1065. [DOI] [PubMed] [Google Scholar]
- Shinkai Y. et al. (1992) RAG-2 deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell, 68, 855–867. [DOI] [PubMed] [Google Scholar]
- Shortman K. and Wu,L. (1996) Early T lymphocyte progenitors. Annu. Rev. Immunol., 14, 29–47. [DOI] [PubMed] [Google Scholar]
- Spanopoulou E. et al. (1994) Functional immunoglobulin transgenes guide ordered B-cell differentiation in Rag-1-deficient mice. Genes Dev., 8, 1030–1042. [DOI] [PubMed] [Google Scholar]
- Stein P.L., Lee,H., Rich,S. and Spriano,P. (1992) pp59fyn mutant mice display differential signalling in thymocytes and peripheral T cells. Cell, 70, 741–750. [DOI] [PubMed] [Google Scholar]
- Stern J.B. and Smith,K.A. (1986) Interleukin-2 induction of T-cell G1 progression and c-myb progression. Science, 233, 203–206. [DOI] [PubMed] [Google Scholar]
- Strasser A., Harris,A.W., Vaux,D.L., Webb,E., Bath,M.L., Adams,J.M. and Cory,S. (1990) Abnormalities of the immune system induced by dysregulated bcl-2 expression in transgenic mice. Curr. Top. Microbiol. Immunol., 166, 175–181. [DOI] [PubMed] [Google Scholar]
- Sugawara T., Di Bartolo,V., Miyazaki,T., Nakauchi,H., Acuto,O. and Takahama,Y. (1998) An improved retroviral gene transfer technique demonstrates inhibition of CD4–CD8– thymocyte development by kinase-inactive ZAP-70. J. Immunol., 161, 2888–2894. [PubMed] [Google Scholar]
- Taylor D., Badiani,P. and Weston,K. (1996) A dominant interfering Myb mutant causes apoptosis in T cells. Genes Dev., 10, 2732–2744. [DOI] [PubMed] [Google Scholar]
- Thompson C.B., Challoner,P.B., Neiman,P.E. and Groudine,M. (1986) Expression of the c-myb proto-oncogene during cellular proliferation. Nature, 319, 374–380. [DOI] [PubMed] [Google Scholar]
- Tomita K., Hattori,M., Nakamura,E., Nakanishi,S., Minato,N. and Kageyama,R. (1999) The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes Dev., 13, 1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lohuizen M., Verbeek,S., Krimpenfort,P., Domen,J., Saris,C., Radaszkiewicz,T. and Berns,A. (1989) Predisposition to lymphoma genesis in pim1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell, 56, 673–682. [DOI] [PubMed] [Google Scholar]
- van Oers N.S., Lowin-Kropf,B., Finlay,D., Connolly,K. and Weiss,A. (1996) αβ T cell development is abolished in mice lacking both Lck and Fyn protein tyrosine kinases. Immunity, 5, 429–436. [DOI] [PubMed] [Google Scholar]
- Wang B., Wang,N., Salio,M., Sharpe,A., Allen,D., She,J. and Terhorst,C. (1998) Essential and partially overlapping role of CD3γ and CD3δ for development of αβ and γδ T lymphocytes. J. Exp. Med., 188, 1375–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston K. (1998) Myb proteins in life, death and differentiation. Curr. Opin. Genet. Dev., 8, 76–81. [DOI] [PubMed] [Google Scholar]
- White J.R. and Weston,K. (2000) Myb is required for self-renewal in a model system of early hematopoiesis. Oncogene, 19, 1196–1205. [DOI] [PubMed] [Google Scholar]
- Winandy S., Wu,L., Wang,J.H. and Georgopoulos,K. (1999) Pre-T cell receptor (TCR) and TCR-controlled checkpoints in T cell differentiation are set by Ikaros. J. Exp. Med., 190, 1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweidler-Mckay P.A., Grimes,H.L., Flubacher,M.M. and Tsichlis,P.N. (1996) Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol. Cell. Biol., 16, 4024–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]