Abstract
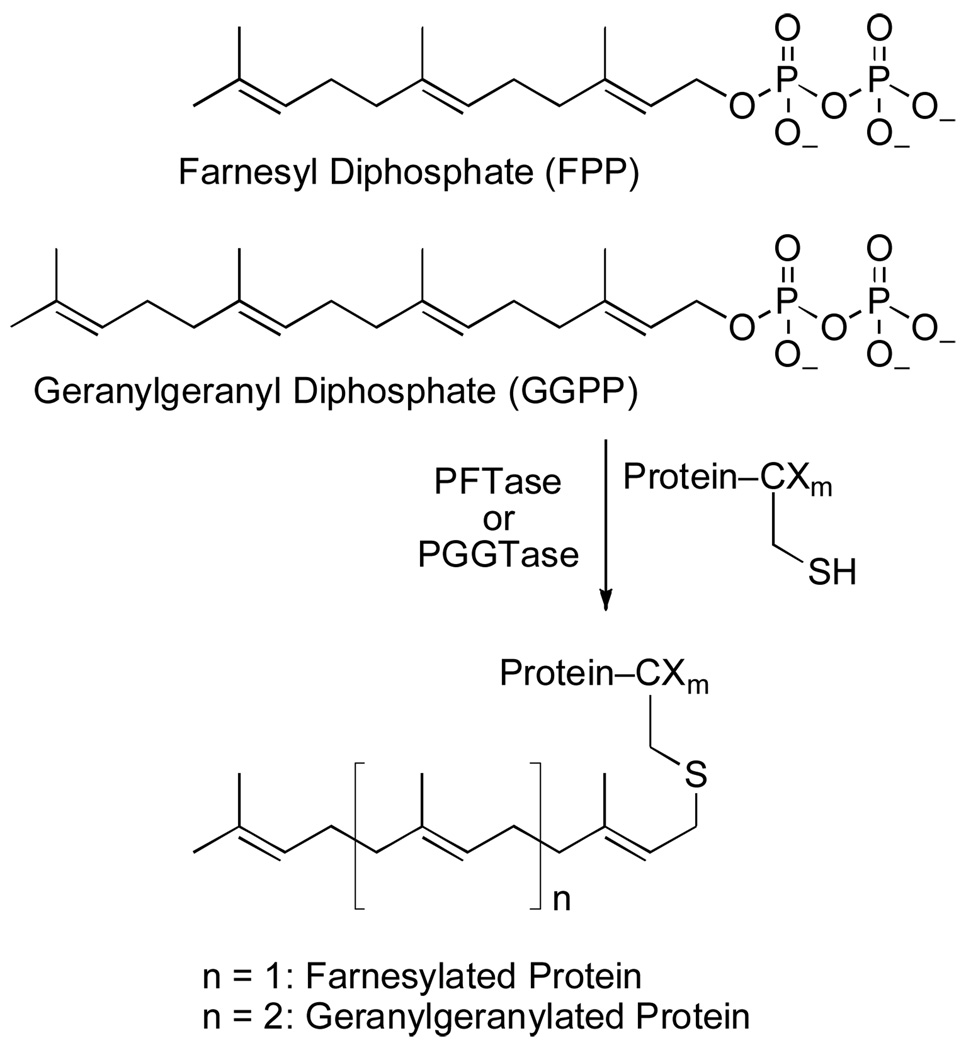
Protein prenyltransferases catalyze the attachment of C15 (farnesyl) and C20 (geranylgeranyl) groups to proteins at specific sequences localized at or near the C-termini of specific proteins. Determination of the specific protein prenyltransferase substrates affected by the inhibition of these enzymes is critical for enhancing knowledge of the mechanism of such potential drugs. Here we investigate the utility of alkyne-containing isoprenoid analogues for chemical proteomics experiments by showing that these compounds readily penetrate mammalian cells in culture and become incorporated into proteins that are normally prenylated. Derivatization via Cu(I) catalyzed Click reaction with a fluorescent azide reagent allows the proteins to be visualized and their relative levels to be analyzed. Simultaneous treatment of cells with these probes and inhibitors of prenylation reveals decreases in the levels of some but not all of the labeled proteins. Two-dimensional electrophoretic separation of these labeled proteins followed by mass spectrometric analysis allowed several labeled proteins to be unambiguously identified. Docking experiments and DFT calculations suggest that the substrate specificity of PFTase may vary depending on whether azide- or alkyne-based isoprenoid analogues are employed. These results demonstrate the utility of alkyne-containing analogues for chemical proteomic applications.
Keywords: chemical proteomics, click reaction, farnesyl diphosphate, prenyltransferase, prenylome, protein prenylation, protein profiling
INTRODUCTION
Protein prenyltransferases catalyze the attachment of C15 (farnesyl) and C20 (geranylgeranyl) groups to proteins at specific sequences localized at or near the C-termini of certain proteins via the reaction shown in Figure 1. Protein farnesyltransferase (PFTase) and protein geranylgeranyltransferase type 1 (PGGTase I) alkylate simple tetrapeptide (CAAX box) substrates while protein geranylgeranyltransferase type 2 (PGGTase II) modifies more cryptic sequences.(1) The inhibition of protein farnesylation has been a target for disease intervention for the past two decades and protein farnesyltransferase inhibitors (FTIs) have been evaluated as therapeutic agents for several medical problems. These include a number of forms of cancer, malaria and related protozoan infections, and certain progerias; protein geranylgeranyltransferase inhibitors (GGTIs) are also in development.(2, 3) Despite copious amounts of research, much still remains unclear about protein prenylation and its inhibition. For example, the driving force behind FTI development for cancer therapy has focused on the oncogenic Ras proteins, since they must be farnesylated to be active.(4) During pre-clinical studies with these inhibitors, antiproliferative and pro-apoptotic activity were observed in cases where oncogenic Ras was not present, suggesting that other downstream effectors contribute to the anti-cancer activity of FTIs.(5) While decreases in the levels of a number of prenylated proteins have been shown to occur upon treatment with FTIs, direct evidence that these species, and not other undiscovered prenylated proteins, are relevant to the physiological effects of FTIs is severely lacking.(6, 7) Determination of the protein farnesyltransferase (PFTase) and/or protein geranylgeranyltransferase (PGGTase) substrates affected by the inhibition of these enzymes is critical for enhancing our knowledge of the mechanism of action of FTIs and GGTIs. Only a fraction of prenylated proteins have been observed experimentally despite the hundreds predicted by bioinformatics approaches.(8, 9) Clearly, comprehensive experimental techniques designed to study the post-translational modification of proteins with isoprenoids by protein prenyltransferases are needed. Such techniques would help provide a better understanding of the mechanism(s) of action of FTIs and GGTIs and could assist in the creation of more potent and selective compounds.
Figure 1.
Reactions catalyzed by protein prenyltransferases. For PFTase and PGGTase-I, Xm is a tripeptide. For PGGTase-II, Xm can be a range of short sequences.
Recently a relatively new technique known as chemical proteomics has been developed that allows post-translationally modified proteins to be selectively labeled or enriched for subsequent analysis.(10, 11) In this strategy, the proteins of interest are first labeled with a small chemical tag at the site of the modification. This can be done by exploiting the cells own machinery to label the proteins in situ with a tagged substrate analogue based on the post-translational modification, or by post-lysis modification by chemical or enzymatic means. The next step involves performing a bioorthogonal chemical ligation reaction with a capture/labeling reagent. A number of such reagents have been created bearing affinity labels (e.g.; biotin, FLAG, etc.), reporter dyes, radiolabels, oligonucleotide tags, and stable isotope tags. The choice of capture chemistry depends on the downstream application with the most common being the “Click” reaction and Staudinger ligation.(12) To date, chemical proteomics have been applied towards the study of a number of post-translational modifications including glycosylation,(13–17) phosphorylation,(18, 19) myristoylation,(20–22) palmitoylation,(21, 23–25) and prenylation.(26–28)
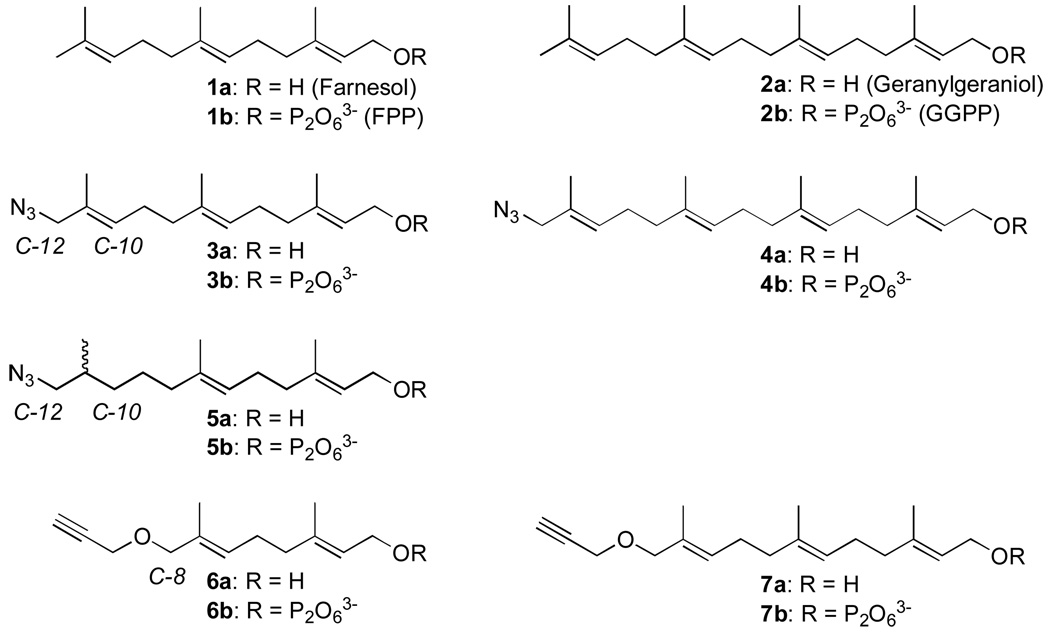
In the prenylation field, Tamanoi, Zhang and coworkers explored the use of farnesyl azide (3a/3b, Figure 2) in proteomics experiments as a surrogate for FPP (1b).(27) Growth of COS cells in the presence of either alcohol 3a or diphosphate 3b resulted in incorporation of these azide-containing analogues into proteins. This was established using a biotinylated phosphine capture reagent that reacted with the azide-labeled proteins via Staudinger ligation chemistry. Subsequent mass spectrometric analysis allowed them to identify a number of farnesylated proteins. In collaboration with Invitrogen, Corp., Tamanoi and coworkers followed up on this work and used an azide-containing analogue of GGPP (4a) to identify a number of geranylgeranylated proteins;(26) Berry et al. extended this approach to labeling in whole animals.(29) Other approaches for studying the “prenylome” including the use of biotinylated substrates(28) and antibodies directed against isoprenoid analogues have also been employed(30).
Figure 2.
Azide- and alkyne-containing isoprenoid analogues of farnesyl diphsophate (FPP) and geranylgeranyl diphosphate (GGPP).
The rapid rate of the Cu(I)-catalyzed click reaction has made it the method of choice for many proteomic profiling protocols. However, as noted by Cravatt and coworkers in related activity-based profiling experiments, background labeling does occur in the click reaction when the alkyne reagent is present in excess.(31) Significantly lower levels of nonspecific reaction occur when the azide partner is employed in high concentration. Thus, for proteomic analysis of prenylated proteins, it would be advantageous to use isoprenoid analogues that incorporate alkyne functional groups so that subsequent labeling could be performed with the more selective azide-containing reagent present in excess.
In 2007, we reported the synthesis of alkyne-containing analogues 6b(32) and 7b(33) and demonstrated that 6b was an alternative substrate for PFTase while 7b was an alternative substrate for both PFTase and PGGTase-I; related alkyne-containing analogues have also been reported by other groups.(34, 35) In light of their potentially greater specificity, we decided to investigate the utility of our alkyne-functionalized analogues for proteomics applications. Here, we explore the use of these probes as reporters of protein prenylation in the presence of various inhibitors in a number of different mammalian cell lines and compare these molecules with the aforementioned azides.
MATERIALS AND METHODS
General
Protease inhibitor cocktail and benzonase were purchased from Sigma Aldrich (St. Louis, MO, USA). PFTase inhibitor L-778,834 (FTI), PGGTase-I inhibitor GGTI-286 (GGTI) and ProteoExtract protein precipitation kits were obtained from Calbiochem (EMD Chemicals, Gibbstown, NJ, USA). TAMRA-azide and TAMRA-alkyne were purchased from Invitrogen (Carlsbad, CA, USA). Detergent compatible protein assay reagents and Tris-HCl SDS-PAGE Protean™ II Ready gels were obtained from Bio-Rad (Hercules, CA, USA). Immobline™ DryStrips and and ampholyte buffer were purchased from GE Healthcare (Piscataway, NJ, USA). 1D gels were visualized using a BioRad FX Molecular Imager. 2D electrophoresis was performed using an Ettan™ IPGphor™ IEF apparatus and the resulting fluorescent spots visualized using a Typhoon 8610 scanner both obtained from GE Healthcare. Fluorescent spots were picked using an Investigator ProPic™ instrument (Genomic Solutions, Ann Arbor, MI, USA). The Paradigm Platinum Peptide Nanotrap precolumn and Magic C18 AQ RP column were purchased from Michrom Bioresources (Auburn, CA, USA). LC-MS/MS analysis was performed using Paradigm 2D capillary LC system (Michrom Bioresources) interfaced with a linear ion trap spectrometer (LTQ, Thermo Scientific, Waltham, MA, USA). For data analysis, Sequest embedded in BioWorks Browser (v 3.3) was obtained from Thermo Scientific and Scaffold (v_2_00_03) was licensed from Proteome Software (Portland, OR, USA). Large scale (1 L) growth of HeLa cell was performed by Biovest International Inc./ NCCC (Minneapolis, MN, USA). Compounds 5a,(36) 6a,(32) 7a and 7b(33) were prepared as previously described.
General cell growth and lysis for in-gel fluorescence experiments
All cell lines except for MCF10A were routinely cultured in DMEM supplemented with 10% FBS; MCF10A cells were grown in DMEM/F-12 supplemented with 5% horse serum, 0.5 ug/mL hydrocortisone, 10 ug/mL insulin, 20 ng/mL EGF (epidermal growth factor), 0.1 ug/mL cholera toxin, 100 units/mL penicillin/streptomycin, 2 m M L-glutamine and 0.5 ug/mL fungizone. For labeling experiments, cell media (40–50% confluence) was supplemented with 25 µM lovastatin, and 50 µM 5a, 6a, 7a or 7b. In some cases, the PFTase inhibitor L-778,834 at 10 µM (FTI) or the PGGTase-I inhibitor GGTI-286 (GGTI) at 5.0 µM were included in the cell media. The cells were allowed to reach 80–90% confluence (~ 24 h) and then washed with PBS. Cells were then suspended in PBS, placed in 1.5 mL microcentrifuge tubes, and pelleted by centrifugation at 2,000 × g, 4°C, for 5 min. After discarding the supernatant, a PBS solution containing 0.10% SDS, 0.20% Triton X-100, protease inhibitor cocktail, Benzonase and 2.4 µM PMSF was added to the cell pellet. After vigorous vortexing, cell lysate was harvested by sonicating 4 times for 10 sec each with 20 sec intervals between sonication cycles. The concentration of protein in the lysate was determined using a detergent compatible protein assay reagent kit.
Comparison of azide- and alkyne-modified isoprenoid analogues
To 100 µg (0.83 mg/mL) of HeLa lysate protein was added 25, 50, or 100 µM TAMRA-azide or TAMRA-alkyne, 50 mM TCEP, and 100 µM TBTA. After vortexing, 1.0 mM CuSO4 was added and the reaction was allowed to proceed at rt for 1 h. Excess reagents were removed by protein preciption with a ProteoExtract protein precipitation kit. Protein pellets were suspended in 1x Laemmli SDS-PAGE loading buffer and sonicated in a water bath for 20 min to insure dissolution. A 12% acrylamide gel was loaded with 15 µg of protein for in-gel fluorescence scanning and visualized on a BioRad FX Molecular Imager.
Analysis of changes in protein prenylation upon exposure to an inhibitor
To the cell lysate (1–2 mg/mL of protein) from one 60 mm dish of cells was added 50 µM TAMRA-azide or TAMRA-alkyne, 50 mM TCEP, and 100 µM TBTA. After vortexing, 1.0 mM CuSO4 was added and the reaction was allowed to proceed at rt for 1 h. Excess reagents were removed by protein preciption with a ProteoExtract protein precipitation kit. Protein pellets (200–300 µg of total protein) were suspended in 1x Laemmli SDS-PAGE loading buffer and sonicated in a water bath for 20 min to insure dissolution. A 12% acrylamide gel was loaded with ~ 10 µg of protein for in-gel fluorescence scanning and visualized with a BioRad FX Molecular Imager.
HeLa cell growth, lysis and labeling for identification of prenylated proteins
Large scale (1 L) cultures of HeLa cells were grown in Joklik’s modified MEM with 5.0% FBS. For metabolic labelling, cells at mid log phase were treated with 25 µM Lovastatin and 50 µM C10-alkyne (6a) or C15-alkyne (7a), and allowed to grow for 24 h. Cells were collected by centrifugation at 2500 × g, followed by two washes in cold PBS (Ca/Mg free). Cell pellets were resuspended in PBS containing 1.0% SDS, protease inhibitor cocktail and benzonase and sonicated on ice for 2 min total (10 sec pulse, 10 sec rest). The concentration of protein in the lysate was determined using detergent compatible protein assay reagents. Proteins in the lysate were precipitated using a TCA/acetone protocol. Briefly, a 100% TCA solution (100 g TCA in 45.4 mL water) was added to the lysate so as to obtain a 10% final concentration of TCA in the solution which was vortexed and stored on ice for 1 h. HPLC grade acetone (4x lysate volume) was then added to the mixture and maintained at −20°C overnight. The lysate was then centrifuged at 10,000 × g for 30 min to obtain a protein pellet. The supernatant was discarded and the pellet was washed twice with acetone and air dried for 5 min. Precipitated proteins were resuspended in PBS containing 0.20% SDS by brief vortexing and sonication. Protein concentration was measured using detergent compatible protein assay reagents. For protein labeling, 3.0 mg (1.9 mg/ml) of HeLa lysate protein was treated with TAMRA-azide, TCEP, and TBTA at final concentrations of 25 µM, 1.0 mM and 100 µM, respectively. After vortexing, CuSO4 (1.0 mM, final concentration) was added and the reaction was allowed to proceed at rt for 1 h. Excess reagents were removed by protein preciption using above mentioned TCA/acetone protocol and the resulting protein pellet was stored at −20 °C prior to subsequent use.
2D gel electrophoresis
Two-dimensional gel electrophoresis was performed at the Center for Mass Spectrometry and Proteomics, University of Minnesota, Twin Cities using a standard procedure.(37) In brief, the protein pellet was resuspended in IPG running buffer (7.0 M urea, 2.0 M thiourea, 2.0 % CHAPS, bromophenol blue, 0.50 % (v/v) ampholyte buffer, 1.0 % n-dodecyl β-D-maltoside and 12 mM DTT). Samples containing 800 µg of protein were rehydrated into 18 cm pH 3–10 Immobline™ DryStrips overnight under low current, and resolved in an Ettan™ IPGphor™ IEF apparatus per manufacturer’s protocol. Strips were then equilibrated in SDS equilibration buffer (50 mM Tris, pH 8.8, 8.0 M urea, 30% glycerol (v/v), 4.0 % SDS, 1.0 % DTT) for 0.5 h, and resolved on 8–16% Tris-HCl SDS-PAGE Protean™ II Ready gels. TAMRA labelled proteins on the gel were visualized with Typhoon 8610 scanner using an excitation wavelength of 532 nm and a 580BP30 emission filter (580 nm). Gel images were imported into a Genomic Solutions® Investigator ProPic™ instrument for robotic excision and robotic trypsin digestion of excised spots.(38) Tryptic peptides were then lyophilized and stored at −20 °C prior to subsequent use.
MS analysis of protein spots
LC-MS analysis was performed using a Michrom Bioresources Paradigm 2D capillary LC system interfaced with a linear ion trap spectrometer using a standard procedure.(37) Lyophilized tryptic peptides disolved in water/CH3CN/formic acid (95:5:0.1) were desalted and concentrated with a Paradigm Platinum Peptide Nanotrap (Michrom Bioresources) precolumn, and eluted onto a Magic C18 AQ RP column at a flow rate of ~ 250 nL/min. A 60 min (10–40% CH3CN) linear gradient was used to separate the peptides. The peptides were ionized with a voltage of 2.0 kV applied distally on the column, and analyzed on the LTQ instrument set to positive polarity and data-dependent acquisition method (one survey MS scan followed by MS/MS (relative collision energy of 35%) on the four most abundant ions detected in the survey scan). Dynamic exclusion was employed for 30 s time intervals.
Database searching and protein identification
The MS/MS data were searched with Sequest embedded in BioWorks Browser (v 3.3, Thermo Scientific) against the database of human, mouse, and rat [NCBI (http://www.ncbi.nlm.nih.gov)] non-redundant protein sequences, in addition to 179 common contaminant proteins (Thermo Scientific), for a total of 263,677 proteins. Fragment ion mass tolerance and parent ion tolerance were set at 1.00 Da. The search parameters were as follows: trypsin digestion, fixed carbamidomethyl modification of cysteine, and variable oxidation of methionine, three missed trypsin cleavage sites allowed. The dta/out files generated by Bioworks were analyzed in Scaffold (v_2_00_03, Proteome Software, Portland, OR, USA) to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at >95.0% probability as specified by the Peptide Prophet algorithm.(39) Protein identifications were accepted if they could be established at >99.0% probability by the Protein Prophet algorithm,(40) and contained at least three identified peptides.
Docking of 5b and 6b in the active sites of PFTase
For modeling compounds 5b and 6b in the active site of PFTase (pdb file 1D8D), docking was performed using Glide (Schrodinger, version 5.5). The PFTase crystal structure was prepared using the default settings in the protein preparation wizard as part of the Maestro 9.0 package. A receptor grid large enough to encompass the entire binding site for compounds 5b and 6b was generated from the prepared PFTase enzyme. A standard precision docking parameter was set and 10,000 ligand poses per docking were run. The 20 top poses with the lowest docking score were then further subjected to post-docking minimization. The conformations with overall lowest energy were chosen for display for each probe.
Electrostatic potential calculations of 1b, 5b and 6b
Structures of 1b, 5b and 6b were generated in Gaussian View and the geometries were optimized at the b3lyp/6-31G(d) level using Gaussian 03 (M.J. Frisch, G.W. Trucks, H.B. Schlegel et al., Gaussian 03, Revision E.01, Gaussian, Inc., Wallingford, CT, 2004). The extended conformation of each molecule was produced by constraining all dihedral angles of the backbone. A map of electrostatic potential and a map of the electron density were generated for each molecule. To generate the molecular electrostatic potential (MEP), the electrostatic potential was plotted on an isodensity surface (MO = 0.02 and density = 0.02) using Gaussian View. The minimum and maximum potentials were set to −0.05 and 0.2, respectively.
RESULTS AND DISCUSSION
Comparison of azide- and alkyne-modified isoprenoid analogues for studying the protein prenylome
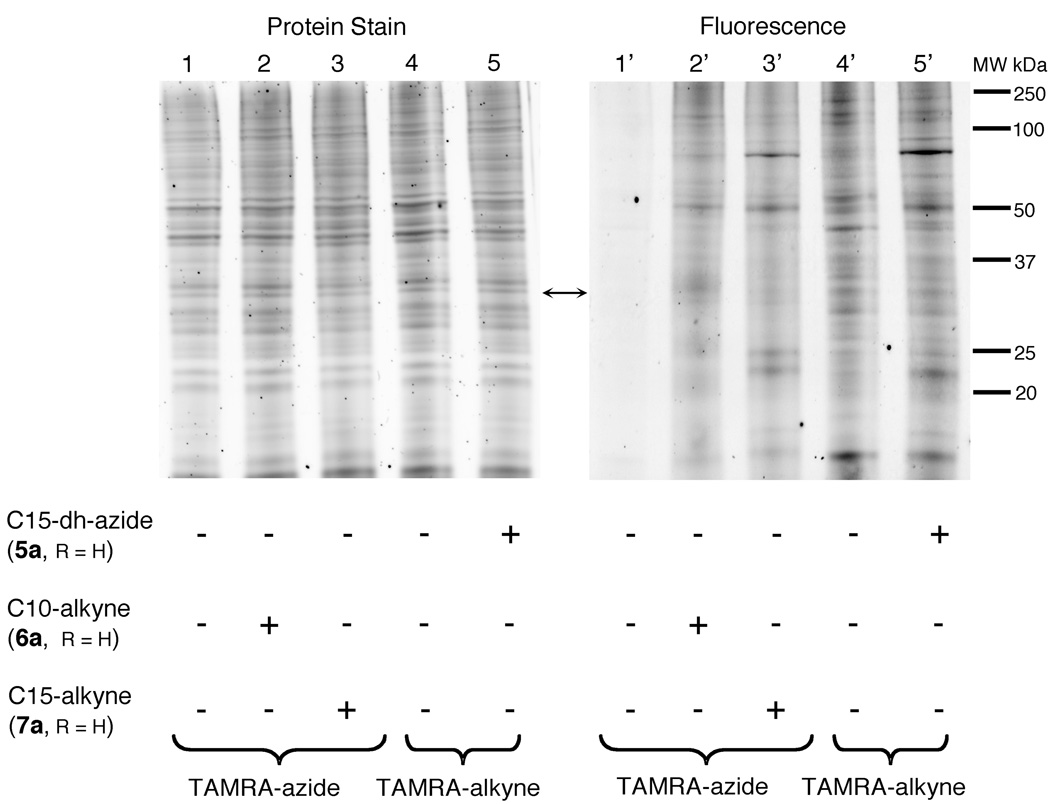
To examine the utility of alkyne-containing isoprenoid analogues, HeLa cells were treated with the alcohol forms (unphosphorylated forms) of the C10-alkyne probe (6a), the C15-alkyne probe (7a) or the C15-dh-azide probe (5a) in the presence of lovastatin to suppress the production of endogenous FPP and GGPP for 24 h. The resulting cells were then lysed and treated with excess azido- or propargyl-labeled tetramethylcarboxyrhodamine (TAMRA) fluorophore via the Cu(I)-catalyzed click reaction. Separation of the proteins was accomplished by SDS-PAGE and the labeled proteins were visualized by in-gel fluorescence. The labeling of specific proteins in the crude cell lysate obtained with compounds 5a, 6a, and 7a is shown in Figure 3. First, it should be noted that significant labeling was observed with all three probes indicating that both the azide- and alkyne-containing isoprenoid analogues are incorporated into proteins. While this has been demonstrated before for related azide-based probes 3a and 3b, this is the first reported example for the incorporation of alkyne-containing isoprenoid analogues in live cells; the fact that these molecules become incorporated indicates that the alcohols must be effectively phosphorylated to the corresponding diphosphates to convert them into substrates for the protein prenyltransferases.
Figure 3.
Analysis of labeled proteins in crude HeLa cell lysates by Cu(I)-catalyzed click chemistry. Lanes 1–5 show proteins stained with Sypro Ruby. Lanes 1’–5’ shown fluorescently labeled proteins. Lanes 1/1’ and 4/4’ contain lysate from cells not treated with isoprenoid analogue. Lanes 2/2’ and 3/3’ contain lysate from cells treated with analogues 6a and 7a respectively. Lane 5/5’ contains lysate from cells treated with analogue 5a. Lanes 1/1’ through 3/3’ were reacted with TAMRA-azide (100 µM) while lanes 4/4’ and 5/5’ were reacted with TAMRA-alkyne (100 µM).
Next, we examined the relative amount of background labeling when lysates were treated with a fluorescent azide or alkyne reagent. Lanes 1 through 3 (Figure 3) contain lysates treated with excess TAMRA-azide, while lanes 4 and 5 (Figure 3) contain lysates treated with excess TAMRA-alkyne. Despite equal amounts of proteins being loaded in each lane, significant background labeling was observed in cell lysate not treated with any isoprenoid analogues and treated with excess TAMRA-alkyne (Lane 4). Remarkably, lysate not treated with isoprenoid analogues and reacted with excess TAMRA-azide shows virtually no background labeling. This reduced background is consistent with previous studies where comparisons between azide- and alkyne-based labeling were performed(31) and serves to highlight the utility of these probes. It should also be noted that some proteins appear to be labeled by both of the alkyne probes while others are labeled by only one. This is probably a reflection of the fact that the C15-alkyne probe is a substrate for both PFTase and PGGTase whereas the C10-alkyne probe is incorporated only by PFTase and is consistent with earlier in vitro work with these compounds.
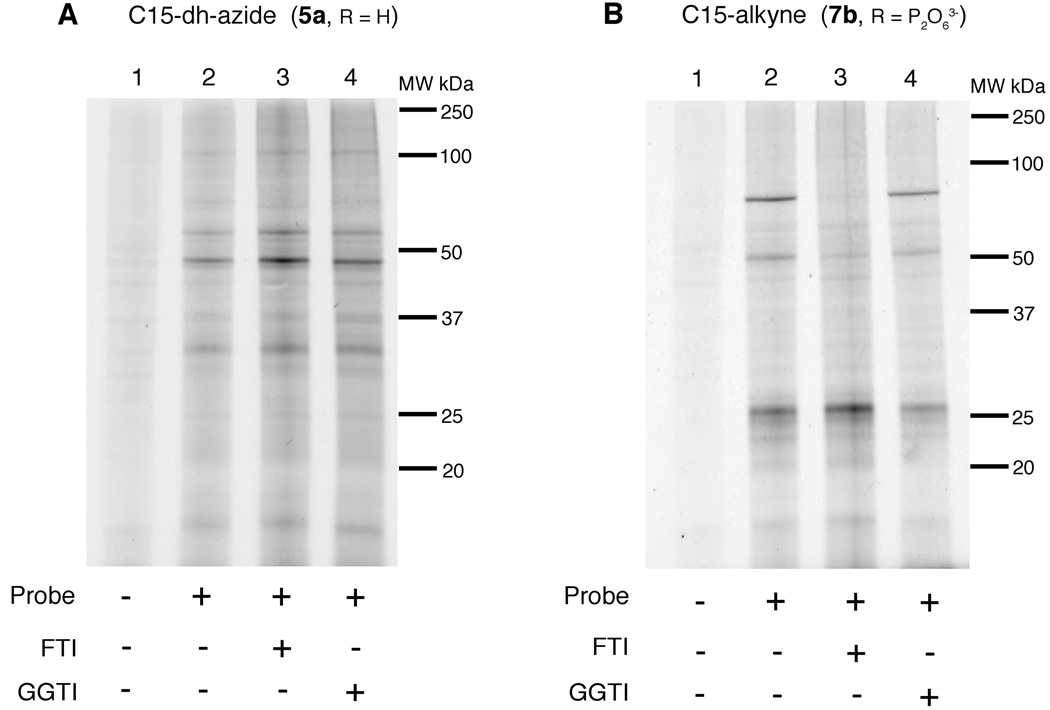
Visualizing prenylated proteins across various cell lines and changes in prenylation status upon exposure to protein prenyltransferase inhibitors
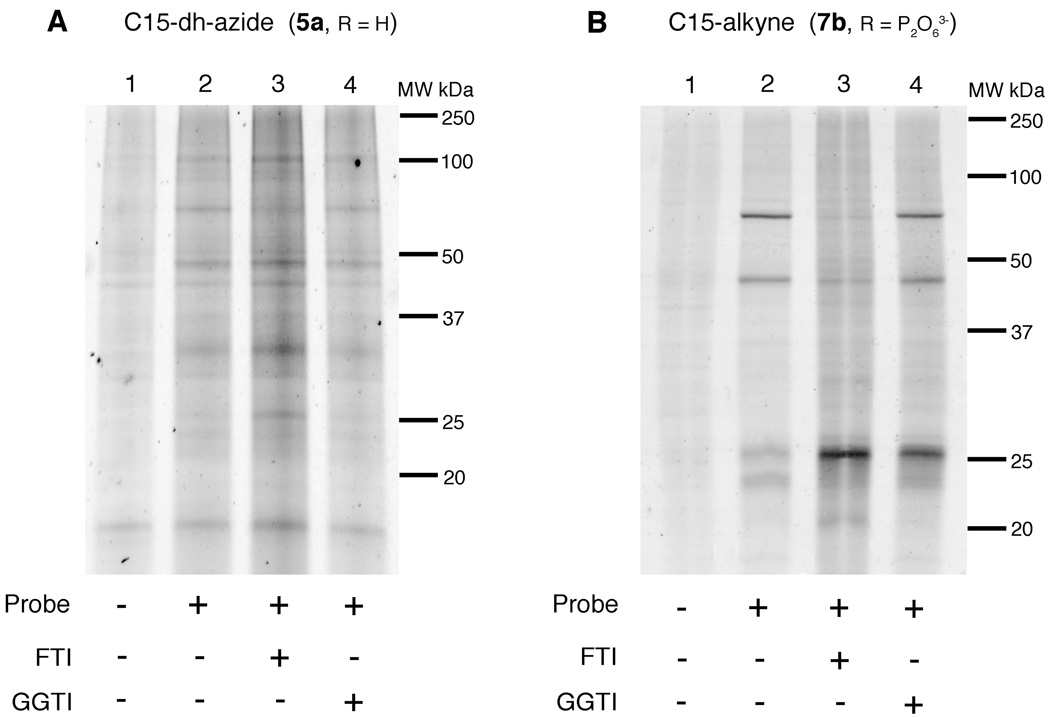
With the utility of the alkyne probes established from experiments described above, we next employed them for the analysis of protein prenylation in a number of cultured mammalian cell lines. Through simultaneous treatment with either an FTI or GGTI, the targets and specificity of these inhibitors could also be observed. HeLa cells, an immortal cell line derived from cervical cancer, were treated with C15-dh-azide probe (5a) or C15-alkyne probe (7b), the lysates were allowed to react with the corresponding TAMRA capture reagent and subjected to in-gel fluorescence analysis (Figure 4). Proteins ranging in molecular weights from 15 to 100 kDa were labeled with 5a (Figure 4A: lane 2). Treatment with the FTI (Figure 4A: lane 3) resulted in diminished labeling of one labeled protein at ~ 70 kDa while treatment with the GGTI (Figure 4A: lane 4) did not affect the labeling pattern. This latter result is not particularly surprising since it is unlikely that 5a is efficiently incorporated into geranylgeranylated proteins via PGGTase. Interestingly, labeled proteins at ~ 25 and 30 kDa appeared to be labeled to a greater extent in the presence of the FTI; the origin of that effect is unclear. In contrast, the C15-alkyne probe 7b manifests a very different labeling pattern with only a few proteins being labeled (Figure 4B: lane 2) from ~ 20–30 kDa and ~ 40–70 kDa. As was seen in HeLa cells treated with probe 5a, little change was observed upon treatment with the GGTI (Figure 4B: lane 4). In contrast, the protein at ~ 70 kDa was diminished and the protein at ~ 30 kDa was enhanced upon treatment with the FTI, as evidenced by the increase in fluorescence (Figure 4B: lane 3); this latter result is similar to what was observed with 5a. It should also be noted that in this experiment, the phosphorylated form (7b) of the C15-alkyne analogue was employed instead of the free alcohol form (7a) used in the intial experiments described first (Figure 3). The fact that the phosphorylated analogue can enter the cells and become incorporated into proteins is consistent with earlier observations with azide-based probe 3b where that phosphorylated compound was shown to enter cells and serve as a protein prenyltransferase substrate.(27)
Figure 4.
In-gel fluorescence analysis of prenylated proteins in HeLa cells. A: Treatment with C15-dh-azide (5a) isoprenoid analogue and reacted with TAMRA-alkyne (50 µM). B: Treatment with C15-alkyne (7b) isoprenoid analogue and reacted with TAMRA-azide (50 µM). Lane 1: no treatment control; Lane 2: treatment with isoprenoid analogue only; Lane 3: treatment with analogue and FTI; Lane 4: treatment with analogue and GGTI.
Next, MCF10A cells, a non-tumorigenic epithelial cell line, were treated with the C15-dh-azide probe (5a) or C15-alkyne probe (7b) and analyzed to see how cancerous and non-cancerous derived cell lines vary in prenylation levels (Figure 5). A significantly different labeling pattern was observed in these cells treated with azide probe 5a (compared to HeLa cells). Two proteins in particular (one just under 50 kDa in molecular weight, the other above) appear to predominate (Figure 5A: lane 2). Interestingly, neither of these proteins, or any of the other proteins in MCF10A cells are affected by treatment with the FTI or the GGTI (Figure 5A: lanes 3 and 4). Treatment with the alkyne analogue 7b showed a pattern very similar to that observed with the same probe in HeLa cells, including the proteins affected by treatment with the FTI (Figure 5B: lane 3).
Figure 5.
In-gel fluorescence analysis of prenylated proteins in MCF10A cells. A: Treatment with C15-dh-azide (5a) isoprenoid analogue and reacted with TAMRA-alkyne (50 µM). B: Treatment with C15-alkyne (7b) isoprenoid analogue and reacted with TAMRA-azide (50 µM). Lane 1: no treatment control; Lane 2: treatment with isoprenoid analogue only; Lane 3: treatment with analogue and FTI; Lane 4: treatment with analogue and GGTI.
While isoprenoids themselves may play a role in Alzheimer’s disease given their elevated levels(41), a recent study points towards the involvement of a yet unknown prenylated protein in Alzheimer’s disease neuropathophysiology.(42) To explore the applicability of the alkyne-containing probes reported here for the study of prenylated proteins in brain cells, astrocytes were treated with C15-dh-azide probe (5a) or C15-alkyne probe (7b). Astrocytes are cells found in the brain and spinal cord that serve a variety of functions.(43) The labeling patterns observed upon treatment with either probe (Figure 6) are similar to those observed in the HeLa cell line (Figure 4) although close inspection reveals subtle differences which undoubtedly reflect the presence of different populations of prenylated proteins in the different cell lines.
Figure 6.
In-gel fluorescence analysis of prenylated proteins in astrocytes. A: Treatment with C15-dh-azide (5a) isoprenoid analogue and reacted with TAMRA-alkyne (50 µM). B: Treatment with C15-alkyne (7b) isoprenoid analogue and reacted with TAMRA-azide (50 µM). Lane 1: no treatment control; Lane 2: treatment with isoprenoid analogue only; Lane 3: treatment with analogue and FTI; Lane 4: treatment with analogue and GGTI.
Upon treatment with the C15-dh-azide (5a), a variety of proteins ranging from ~ 15 to 100 kDa in molecular weight were labeled (Figure 6A, lane 2). While treatment with the GGTI had no effect on the labeling (Figure 6A, lane 4), treatment with the FTI results in significant diminishment in labeling of nearly every protein (Figure 6A, lane 3). This decrease in labeling, however, was not observed in the HeLa cell line, suggesting that prenyltransferases found in astrocytes are particularly sensitive to the FTI. Only proteins from ~ 20–30 kDa and ~ 50–100 kDa were labeled with the C15-alkyne (7b) (Figure 6B, lane 2), as was observed in the other cell lines tested. Additionally, proteins from ~ 50–100 kDa were affected by FTI treatment (Figure 6B, lane 3) while none of the proteins were affected by treatment with the GGTI (Figure 6B, lane 4).
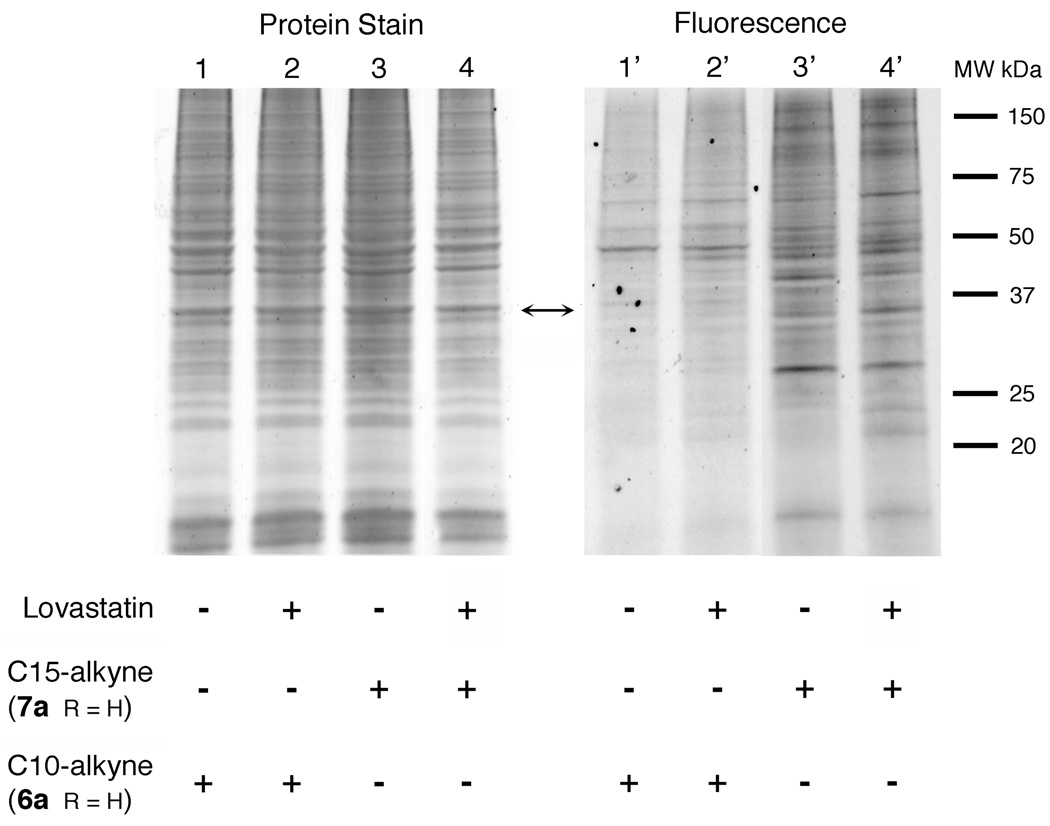
Incorporation of 6a and 7a into prenylated proteins in the absence of lovastatin, an inhibitor of isoprenoid biosynthesis
Proteomic experiments employing isoprenoid analogues have often been performed in the presence of lovastatin, an inhibitor of HMG-CoA reductase. This reduces the level of endogenous FPP and hence increases the incorporation efficiency of the desired analogue. However, in order to use alkyne-based probes 6a and 7a to monitor real changes that occur in the presence of various inhibitors and drugs, it would be preferable to avoid the perturbing effects of simultaneous treatment with lovastatin. Accordingly, the effect of lovastatin on the incorporation of 6a and 7a was evaluated by growing HeLa cells in media containing the alkyne probes in the absence or presence of lovastatin. Cells were cultured, lysed, reacted with TAMRA-azide, analyzed by SDS-PAGE and the results are presented in Figure 7. For probe 6a, comparison of lane 1’ (without lovastatin) with lane 2’ (with lovastatin) shows only a small increase in the labeling pattern. Similar results were observed with 7a (compare lanes 3’ and 4’). In total, these experiments suggest that both 6a and 7a can be efficiently incorporated even in the absence of lovastatin.
Figure 7.
In-gel fluorescence analysis of prenylated proteins in HeLa cells in the absence and presence of lovastatin. Cells were grown in the presence of the C10-alkyne, 6a (lanes 1/1' and 2/2') or the C15-alkyne 7a (lanes 3/3' and 4/4'). Lanes 1–4 show proteins stained with Sypro Ruby. Lanes 1’–4’ shown fluorescently labeled proteins. Lanes 1/1’ and 3/3’ contain samples from cells not grown in the presence of lovastatin. Lanes 2/2’ and 4/4’ contain samples from cells treated with lovastatin. Following lysis, samples were treated with TAMRA-azide (50 µM). to allow the prenylated proteins to be visualized.
Identification of prenylated proteins from HeLa cells via 2D electrophoretic separation and tandem mass spectrometry
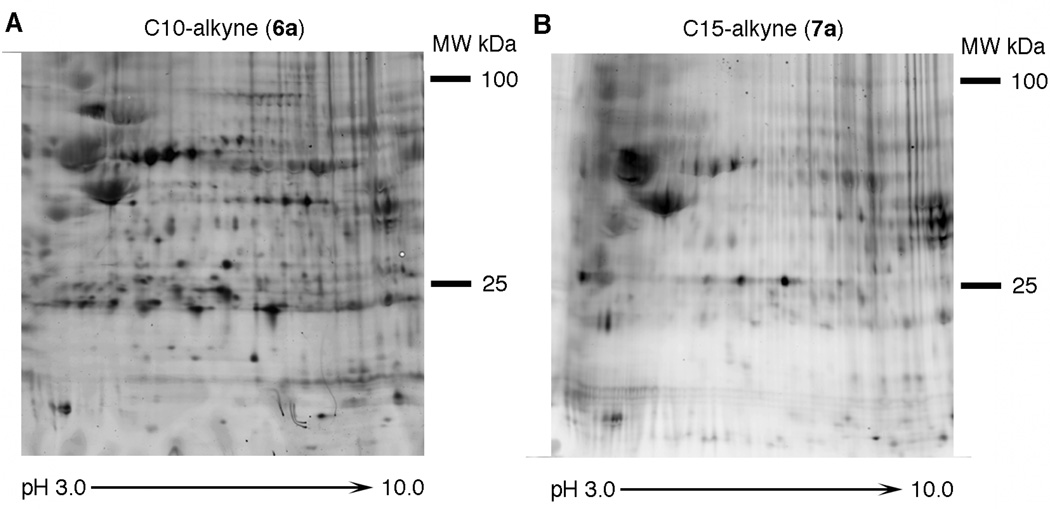
While azide-based probes 3a and 4a have been used to label and identify specific prenylated proteins, to date, alkyne-based isoprenoid analogues have not been employed for this purpose. Such studies provide a broad overview of the protein prenylome and a qualitative look as to which proteins are affected by prenyltransferase inhibitors. Thus, to examine the utility of these molecules for the profiling and identification of specific prenylated polypeptides, HeLa cells were grown in the presence of C10-alkyne 6a and C15-alkyne 7a for 24 h. After cell disruption in the presence of detergent, the resulting lysate was reacted with TAMRA-azide followed by fractionation by 2D electrophoresis and visualization of the labeled prenylated proteins by in-gel fluorescence scanning. Examination of the images from those gels (Figure 8) reveals a large number of labeled proteins of varying intensity.
Figure 8.
In-gel fluorescence analysis of prenylated proteins from HeLa cells after 2D electrophoretic separation: A: 2D gel of labeled proteins obtained from HeLa cells grown in the presence of C10-alkyne (6a). B: 2D gel of labeled proteins obtained from HeLa cells grown in the presence of C15-alkyne (7a).
To identify a subset of the labeled proteins, 20 spots from the 2D gel shown in Figure 8A were excised and subjected to in-gel tryptic digestion. Following concentration and desalting, the resulting peptides were then analyzed by LC-MS/MS to obtain peptide sequence information. From those 20 samples, 7 proteins were identified (Table 1) with a confidence level of >99%. Several additional proteins were identified at lower confidence levels.
Table 1.
Summary of proteins identified via proteomic analysis with alkyne probe 6a.
| Protein | Accession Number |
C-terminal Sequence |
Number of Peptides |
Sequence Coverage |
Previously Identifieda (probe used) |
|---|---|---|---|---|---|
| GNBP | gi 5729850 ref NP_006487.1 | CGLY | 3 | 7% | N |
| Lamin B1 | gi 5031877 ref NP_005564.1 | CAIM | 10 | 19% | Y (3a) |
| Rab 1B | gi 13569962 ref NP_112243.1 | GGCC | 8 | 38% | Y (4a) |
| Rab 2A | gi 4506365 ref NP_002856.1 | GGCC | 3 | 16% | Y (4a) |
| Rab 6A | gi 38679888 ref NP_942599.1 | GCSC | 4 | 24% | Y (4a) |
| Rab 7 | gi 34147513 ref NP_004628.4 | SCSC | 3 | 15% | Y (4a) |
| Annexin A3 | gi 4826643 ref NP_005130.1 | CGGDD | 6 | 19% | Y/N (3a) |
Of the seven proteins identified here, one, GNBP, has not been previously labeled using azide-based probes although it has been determined to be a bona fide PFTase substrate based on in vitro biochemical evaluation.(44) Five of the seven labeled proteins observed here have been previously detected in experiments employing either 3a or 4a. Interestingly, Lamin B1 was labeled with the FPP analogue 3a while the various Rab proteins were all detected with the larger GGPP-based probe 4a. In contrast, in the experiment reported here, all of these proteins were visualized in cells treated with 6a. This may result from the phosphorylated form of the C10-alkyne (6b) acting as an alternative substrate for farnesyl- or geranylgeranyl synthase or PGGTase-II (the enzyme that normally prenylates Rab proteins). The seventh protein identified here was Annexin A3. Interestingly, the closely related protein Annexin A2 was previously identified using the C15-azide probe 3a.(27) However, both of these proteins contain a non-canonical C-terminal sequence that has never been experimentally verified as a substrate for protein prenyltransferases. Moreover, recent results from experiments employing an antibody-based approach for the detection of prenylated proteins suggested that annexin proteins are not substrates for prenylation.(30) Evidently, more work is required to address this issue.
Comparison by computational analysis of the properties of azide- and alkyne containing isoprenoid analogues
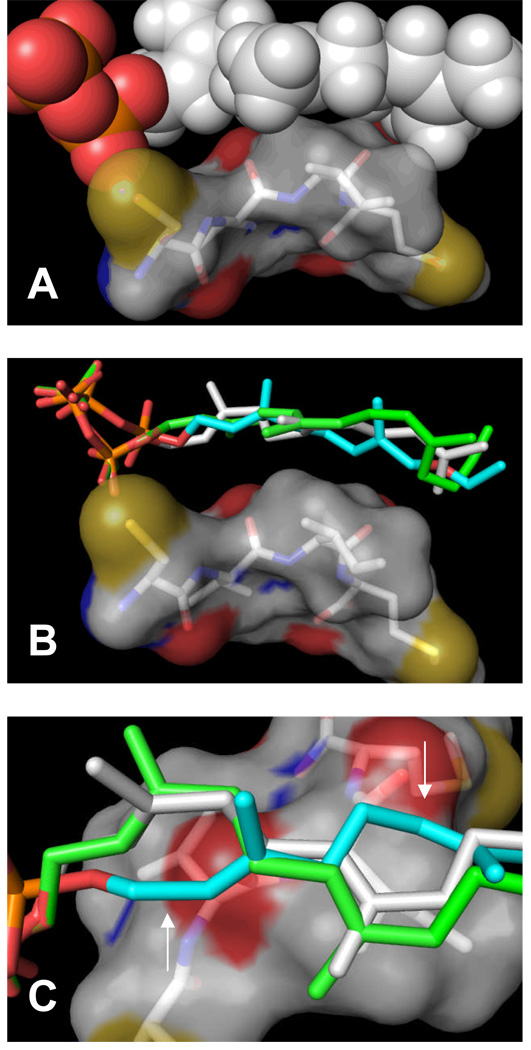
While the results presented above clearly show that azide- and alkyne-based analogues can be used to label many of the same proteins within cells, it does not appear that these probes are interchangeable. For example, comparison of the labeling obtained using 5a with that seen with 6a (compare lanes 2’ and 5’, Figure 3) shows that while both of these probes label many of the same proteins, the relative intensities of the labeled protein bands vary depending on the probe employed. Recently, it has been noted that modifications in prenyl group structure can alter the peptide specificity of PFTase.(45) This occurs because the isoprenoid and peptide substrates extensively contact each other when bound in the enzyme active site as noted in Figure 9A.(46) Accordingly, 5b and 6b were docked into the active site of PFTase. The lowest energy pose for each of these two analogues superimposed on the cystallographically determined structures of FPP and a substrate peptide CVIM are shown in Figures 9B and 9C. The side view (Figure 9B) illustrates that both the azide and alkyne analogues remain in close contact with the peptide substrate in the docked structures. The top view (Figure 9C) provides a clearer picture of how the conformation of alkyne 6b may differ in the active site. Such a difference would alter the contacts between the isoprenoid and peptide that would, in turn, affect peptide substrate specificity. It is interesting to note that in a least one case, crystallographic evidence exists for an isoprenoid analogue in the active site of PFTase that adopts a conformation significantly different from that of FPP.(47)
Figure 9.
Interactions between isoprenoid substrates and a peptide substrate in the active site of PFTase probed by docking experiments. Top (A): Structure of FPP and a peptide substrate bound in the active site of PFTase as determined by X-ray crystallography. Note the significant interface of interaction between the two. Middle (B): Docked structures of 5b and 6b (lowest energy poses) superimposed on the structure of FPP and a peptide substrate bound in the active site of PFTase from panel A. Note how isoprenoid substrates remain in contact with the peptide. Bottom (C): 90° rotation and zoomed view of structures shown in panel B. Major conformational differences between FPP (white), 5b (green) and 6b (aqua) are noted with arrows. Color scheme and representations: In all panels the peptide is shown as a transparent surface with the structure given in a stick representation colored by element. In panel B, FPP is presented in CPK format (C and H: white; P: orange; O: red). In panels B and C, FPP (white), 5b (green) and 6b (aqua) are presented as sticks.
Another important difference between azide and and alkyne analogues can be seen by examining their electrostatics. Accordingly, DFT calculations were employed to calculate molecular electrostatic potential (MEP) maps for FPP, 5b and 6b. CPK representations and MEP maps of the distal regions (beyond the first isoprene unit) for these molecules are presented in Figure 10. Given the differences in heteroatom and functional group content between these three molecules, it is not surprising that their MEP maps differ significantly. One striking difference concerns the region of greatest negative electrostatic potential. In compound 5b, the nitrogen attached directly to C-12 bears the greatest negative charge (except for the diphosphate which is not shown). In contrast, the most negatively charged atom in 6b is the oxygen attached to C-8. These differences in charge distribution significantly affect the interface between the isoprenoid and peptide substrate and may be important for modulating substrate specificity. Finally, it should be noted that while the computational analysis described above was performed with compounds 5b and 6b, it is likely that similar conclusions would be reached if 7b was included in the study. Overall, given that the isoprenoid and peptide substrates interact extensively over a large interface, it is likely that each probe will manifest a distinct pattern of incorporation into the prenylome; that conclusion is consistent with the experimental results reported above.
Figure 10.
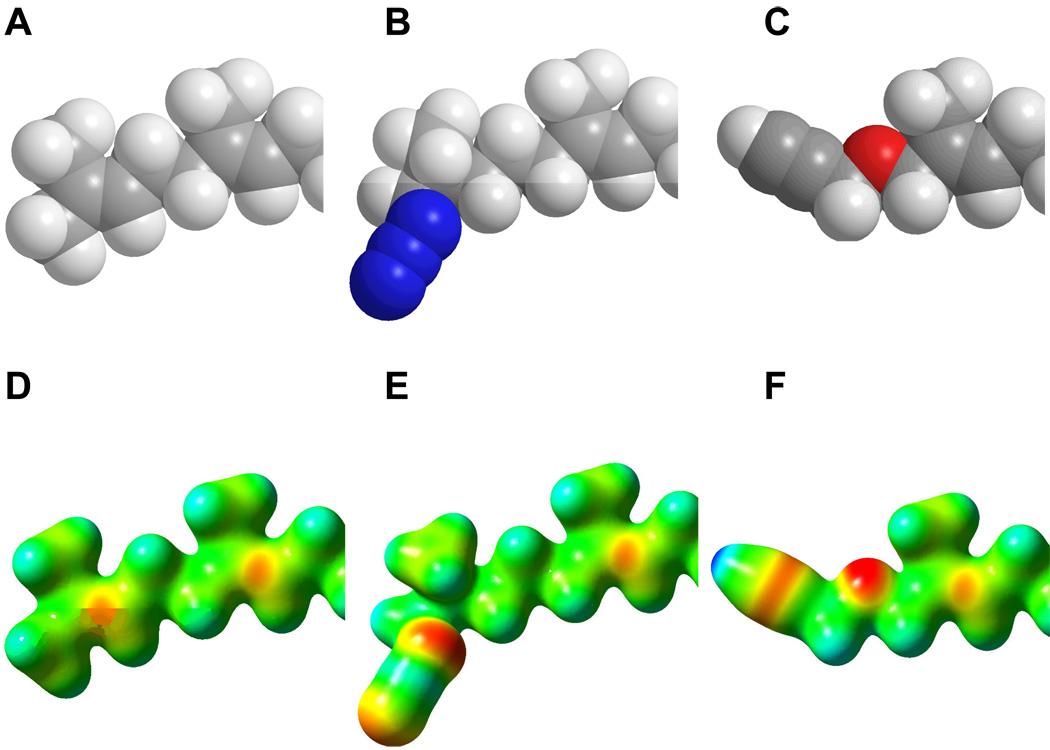
CPK models and molecular electrostatic potential (MEP) maps calculated for FPP (1b), dihydroazide 5b and alkyne 6b. Top: CPK models of (A) FPP (1b); (B) 5b; (C) 6b. Bottom: MEP for (D) FPP (1b); (E) 5b; (F) 6b. For clarity, only cabons distal to C-5 are shown. Carbons 1–4 and the diphosphate groups are not shown since these are the same in all structures. For CPK models, the following colors were used: Carbon (grey), Hydrogen (white), Nitrogen (blue) and Oxygen (red). For MEP maps, potentials were plotted on a surface of constant density (density = 0.02). Electronegative areas are shown in red and electropositive areas are shown in blue.
CONCLUSIONS AND FUTURE DIRECTIONS
In an effort to develop improved probes that can be used for chemical proteomic experiments, alkynecontaining analogues of isoprenoid diphosphates 6 and 7 have been studied here. These analogues readily penetrate mammalian cells in culture and become incorporated into proteins that are normally prenylated. Derivatization via Cu(I) catalyzed Click reaction with a fluorescent azide reagent allows the proteins to be visualized and their relative levels to be analyzed. Simultaneous treatment of cells with these probes and inhibitors of prenylation reveals decreases in the levels of some but not all of the labeled proteins. Two dimensional electrophoretic separation of these labeled proteins followed by mass spectrometric analysis allowed several labeled proteins to be unambiguously identified. Docking experiments and DFT calculations suggest that the sequence specificity of PFTase (the efficiency by which C-terminal CAAX box sequences are prenylated) may vary depending on whether azide- or alkyne-based isoprenoid analogues are employed. These results demonstrate the utility of alkyne-containing analogues for chemical proteomic applications. A variety of experiments that monitor the levels of prenylated proteins in various disease state models are currently underway using these probes.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by a National Institutes of Health Grant GM58442 (M.D.D.), by a National Institutes of Health Predoctoral Training Grant T32-GM08700 (A.J.D.) and by a National Institutes of Health Predoctoral Training Grant T32 GM008347 (J.D.O.). This work was carried out in part using instrumentation from the University of Minnesota Center for Mass Spectrometry and Proteomics, the Minnesota Supercomputer Institute and facilities of the Masonic Cancer Center. We thank Dr. Lorraine Anderson, Todd Markowski, and Bruce Witthuhn of the University of Minnesota Center for Mass Spectrometry for their assistance and technical guidance.
Abbreviations
- BSA
bovine serum albumin
- DFT
density functional theory
- DMEM
Dulbecco's modified Eagle's medium
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- ESI-MS
electrospray ionization mass spectrometry
- FBS
fetal bovine serum
- FPP
farnesyl diphosphate
- FTI
farnesyltransferase inhibitor
- GGPP
geranylgeranyl diphosphate
- GGTI
geranylgeranyltransferase inhibitor
- PBS
phosphate buffered saline
- PGGTase I
protein geranylgeranyltransferase type 1
- PGGTase II
protein geranylgeranyltransferase type 2
- PFTase
protein farnesyl transferase
- SDS PAGE
sodium dodecylsulfate polyacrylamide gel electrophoresis
- TBTA
tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl] amine
- TCA
trichloroacetic acid
- TCEP
tris(2-carboxyethyl)phosphine
REFERENCES
- 1.Zhang FL, Casey PJ. Protein prenylation: Molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 2.Sebti SM, Hamilton AD. Inhibition of Rho GTPases using protein geranylgeranyltransferase I inhibitors. Methods Enzymol. 2000;325:381–383. doi: 10.1016/s0076-6879(00)25459-1. [DOI] [PubMed] [Google Scholar]
- 3.Sebti SM, Hamilton AD. Farnesyltransferase and geranylgeranyltransferase I inhibitors and cancer therapy: lessons from mechanism and bench-to-bedside translational studies. Oncogene. 2000;19:6584–6593. doi: 10.1038/sj.onc.1204146. [DOI] [PubMed] [Google Scholar]
- 4.Crul M, de Klerk GJ, Beijnen JH, Schellens JH. Ras biochemistry and farnesyl transferase inhibitors: a literature survey. Anti-cancer drugs. 2001;12:163–184. doi: 10.1097/00001813-200103000-00001. [DOI] [PubMed] [Google Scholar]
- 5.End DWML, Angibaud P. Farnesyl Protein Transferase Inhibitors: Medicinal Chemistry, Molecular Mechanisms, and Progress in the Clinic. Top Med Chem. 2007;1:133–168. [Google Scholar]
- 6.Prendergast GC. Actin' up: RhoB in cancer and apoptosis. Nat Rev Cancer. 2001;1:162–168. doi: 10.1038/35101096. [DOI] [PubMed] [Google Scholar]
- 7.Sebti SM, Der CJ. Opinion: Searching for the elusive targets of farnesyltransferase inhibitors. Nat Rev Cancer. 2003;3:945–951. doi: 10.1038/nrc1234. [DOI] [PubMed] [Google Scholar]
- 8.Sebti SM. Protein farnesylation: implications for normal physiology, malignant transformation, and cancer therapy. Cancer Cell. 2005;7:297–300. doi: 10.1016/j.ccr.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Maurer-Stroh S, Koranda M, Benetka W, Schneider G, Sirota FL, Eisenhaber F. Towards complete sets of farnesylated and geranylgeranylated proteins. PLoS Comput Biol. 2007;3:e66. doi: 10.1371/journal.pcbi.0030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rix U, Superti-Furga G. Target profiling of small molecules by chemical proteomics. Nat Chem Biol. 2009;5:616–624. doi: 10.1038/nchembio.216. [DOI] [PubMed] [Google Scholar]
- 11.Heal WP, Wickramasinghe SR, Tate EW. Activity based chemical proteomics: profiling proteases as drug targets. Curr Drug Discovery Technol. 2008;5:200–212. doi: 10.2174/157016308785739866. [DOI] [PubMed] [Google Scholar]
- 12.Sletten EM, Bertozzi CR. Bioorthogonal Chemistry: Fishing for Selectivity in a Sea of Functionality. Angewandte Chemie International Edition. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertozzi CR, Kiessling LL. Chemical glycobiology. Science. 2001;291:2357–2364. doi: 10.1126/science.1059820. [DOI] [PubMed] [Google Scholar]
- 14.Gama CI, Hsieh-Wilson LC. Chemical approaches to deciphering the glycosaminoglycan code. Curr Opin Chem Biol. 2005;9:609–619. doi: 10.1016/j.cbpa.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Laughlin ST, Bertozzi CR. Metabolic labeling of glycans with azido sugars and subsequent glycan-profiling and visualization via Staudinger ligation. Nat Protoc. 2007;2:2930–2944. doi: 10.1038/nprot.2007.422. [DOI] [PubMed] [Google Scholar]
- 16.Luchansky SJ, Argade S, Hayes BK, Bertozzi CR. Metabolic functionalization of recombinant glycoproteins. Biochemistry. 2004;43:12358–12366. doi: 10.1021/bi049274f. [DOI] [PubMed] [Google Scholar]
- 17.Sprung R, Nandi A, Chen Y, Kim SC, Barma D, Falck JR, Zhao Y. Tagging-via-substrate strategy for probing O-GlcNAc modified proteins. J Proteome Res. 2005;4:950–957. doi: 10.1021/pr050033j. [DOI] [PubMed] [Google Scholar]
- 18.Warthaka M, Karwowska-Desaulniers P, Pflum MK. Phosphopeptide modification and enrichment by oxidation-reduction condensation. ACS Chem Biol. 2006;1:697–701. doi: 10.1021/cb6003564. [DOI] [PubMed] [Google Scholar]
- 19.Zhou H, Watts JD, Aebersold R. A systematic approach to the analysis of protein phosphorylation. Nat Biotechnol. 2001;19:375–378. doi: 10.1038/86777. [DOI] [PubMed] [Google Scholar]
- 20.Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, Shamir E, Hang HC. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J Am Chem Soc. 2009;131:4967–4975. doi: 10.1021/ja810122f. [DOI] [PubMed] [Google Scholar]
- 21.Hang HC, Geutjes EJ, Grotenbreg G, Pollington AM, Bijlmakers MJ, Ploegh HL. Chemical probes for the rapid detection of Fatty-acylated proteins in Mammalian cells. J Am Chem Soc. 2007;129:2744–2745. doi: 10.1021/ja0685001. [DOI] [PubMed] [Google Scholar]
- 22.Heal WP, Wickramasinghe SR, Bowyer PW, Holder AA, Smith DF, Leatherbarrow RJ, Tate EW. Site-specific N-terminal labelling of proteins in vitro and in vivo using N-myristoyl transferase and bioorthogonal ligation chemistry. Chem Commun. 2008:480–482. doi: 10.1039/b716115h. [DOI] [PubMed] [Google Scholar]
- 23.Kostiuk MA, Corvi MM, Keller BO, Plummer G, Prescher JA, Hangauer MJ, Bertozzi CR, Rajaiah G, Falck JR, Berthiaume LG. Identification of palmitoylated mitochondrial proteins using a bio-orthogonal azido-palmitate analogue. Faseb J. 2008;22:721–732. doi: 10.1096/fj.07-9199com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin BR, Cravatt BF. Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roth AF, Wan J, Green WN, Yates JR, Davis NG. Proteomic identification of palmitoylated proteins. Methods. 2006;40:135–142. doi: 10.1016/j.ymeth.2006.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan LN, Hart C, Guo L, Nyberg T, Davies BS, Fong LG, Young SG, Agnew BJ, Tamanoi F. A novel approach to tag and identify geranylgeranylated proteins. Electrophoresis. 2009;30:3598–3606. doi: 10.1002/elps.200900259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kho Y, Kim SC, Jiang C, Barma D, Kwon SW, Cheng J, Jaunbergs J, Weinbaum C, Tamanoi F, Falck J, Zhao Y. A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc Natl Acad Sci U S A. 2004;101:12479–12484. doi: 10.1073/pnas.0403413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen UT, Guo Z, Delon C, Wu Y, Deraeve C, Franzel B, Bon RS, Blankenfeldt W, Goody RS, Waldmann H, Wolters D, Alexandrov K. Analysis of the eukaryotic prenylome by isoprenoid affinity tagging. Nat Chem Biol. 2009;5:227–235. doi: 10.1038/nchembio.149. [DOI] [PubMed] [Google Scholar]
- 29.Berry AFH, Heal WP, Tarafder AK, Tolmachova T, Baron RA, Seabra MC, Tate EW. Rapid Multilabel Detection of Geranylgeranylated Proteins by Using Bioorthogonal Ligation Chemistry. ChemBioChem. 2010;11:771–773. doi: 10.1002/cbic.201000087. [DOI] [PubMed] [Google Scholar]
- 30.Onono FO, Morgan MA, Spielmann HP, Andres DA, Subramanian T, Ganser A, Reuter CWM. A tagging-via-substrate approach to detect the farnesylated proteome using two-dimensional electrophoresis coupled with Western blotting. Mol Cell Proteomics. 2010;9:742–751. doi: 10.1074/mcp.M900597-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Speers AE, Cravatt BF. Profiling Enzyme Activities In Vivo Using Click Chemistry Methods. Chem Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 32.Duckworth BP, Zhang Z, Hosokawa A, Distefano MD. Selective Labeling of Proteins using Protein Farnesyltransferase. ChemBioChem. 2007;8:98–105. doi: 10.1002/cbic.200600340. [DOI] [PubMed] [Google Scholar]
- 33.Hosokawa A, Wollack JW, Zhang Z, Chen L, Barany G, Distefano MD. Evaluation of an alkyne-containing analogue of farnesyl diphosphate as a dual substrate for protein-prenyltransferases. Int J Peptide Res. 2007;13:345–354. [Google Scholar]
- 34.Gauchet C, Labadie GR, Poulter CD. Regio- and Chemoselective Covalent Immobilization of Proteins through Unnatural Amino Acids. J Am Chem Soc. 2006;128:9274–9275. doi: 10.1021/ja061131o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Labadie GR, Viswanathan R, Poulter CD. Farnesyl Diphosphate Analogues with w-Bioorthogonal Azide and Alkyne Functional Groups for Protein Farnesyl Transferase-Catalyzed Ligation Reactions. J Org Chem. 2007;72:9291–9297. doi: 10.1021/jo7017747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duckworth BP, Xu J, Taton TA, Guo A, Distefano MD. Site-Specific, Covalent Attachment of Proteins to a Solid Surface. Bioconj Chem. 2006;17:967–974. doi: 10.1021/bc060125e. [DOI] [PubMed] [Google Scholar]
- 37.Andersen JD, Boylan KLM, Xue FS, Anderson LB, Witthuhn BA, Markowski TW, Higgins L, Skubitz APN. Identification of candidate biomarkers in ovarian cancer serum by depletion of highly abundant proteins and differential in-gel electrophoresis. Electrophoresis. 2010;31:599–610. doi: 10.1002/elps.200900441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shevchenko A, Wilm M, Vorm O, Mann M. Mass Spectrometric Sequencing of Proteins from Silver-Stained Polyacrylamide Gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 39.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical Statistical Model To Estimate the Accuracy of Peptide Identifications Made by MS/MS and Database Search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 40.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 41.Cole SLV, R Isoprenoids and Alzheimer's disease: A complex relationship. Neurobiol Dis. 2006;22:209–222. doi: 10.1016/j.nbd.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Eckert GP, Hooff GP, Strandjord DM, Igbavboa U, Volmer DA, Muller WE, Wood WG. Regulation of the brain isoprenoids farnesyl- and geranylgeranylpyrophosphate is altered in male Alzheimer patients. Neurobiol Dis. 2009;35:251–257. doi: 10.1016/j.nbd.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fiacco TA, Agulhon C, McCarthy KD. Sorting out astrocyte physiology from pharmacology. Annu Rev Pharmacol Toxicol. 2009;49:151–174. doi: 10.1146/annurev.pharmtox.011008.145602. [DOI] [PubMed] [Google Scholar]
- 44.Hougland JL, Hicks KA, Hartman HL, Kelly RA, Watt TJ, Fierke CA. Identification of Novel Peptide Substrates for Protein Farnesyltransferase Reveals Two Substrate Classes with Distinct Sequence Selectivities. J Mol Biol. 2009;395:176–190. doi: 10.1016/j.jmb.2009.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reigard SA, Zahn TJ, Haworth KB, Hicks KA, Fierke CA, Gibbs RA. Interplay of Isoprenoid and Peptide Substrate Specificity in Protein Farnesyltransferase. Biochemistry. 2005;44:11214–11223. doi: 10.1021/bi050725l. [DOI] [PubMed] [Google Scholar]
- 46.Strickland CL, Windsor WT, Syto R, Wang L, Bond R, Wu Z, Schwartz J, Le HV, Beese LS, Weber PC. Crystal Structure of Farnesyl Protein Transferase Complexed with a CaaX Peptide and Farnesyl Diphosphate Analogue. Biochemistry. 1998;37:16601–16611. doi: 10.1021/bi981197z. [DOI] [PubMed] [Google Scholar]
- 47.Hovlid ML, Edelstein RL, Henry O, Ochocki J, Talbot T, Lopez-Gallego F, Schmidt-Dannert C, Distefano MD. Synthesis and Applications of DATFP-Containing Photoactive Analogues of Farnesyl Diphosphate Containing Modified Linkages for Enhanced Stability. Chem Biol Drug Des. 2010;75:51–67. doi: 10.1111/j.1747-0285.2009.00914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.