Abstract
Most organisms contain several members of a recently discovered class of DNA polymerases (umuC/dinB superfamily) potentially involved in replication of damaged DNA. In Escherichia coli, only Pol V (umuDC) was known to be essential for base substitution mutagenesis induced by UV light or abasic sites. Here we show that, depending upon the nature of the DNA damage and its sequence context, the two additional SOS-inducible DNA polymerases, Pol II (polB) and Pol IV (dinB), are also involved in error-free and mutagenic translesion synthesis (TLS). For example, bypass of N-2-acetylaminofluorene (AAF) guanine adducts located within the NarI mutation hot spot requires Pol II for –2 frameshifts but Pol V for error-free TLS. On the other hand, error-free and –1 frameshift TLS at a benzo(a)pyrene adduct requires both Pol IV and Pol V. Therefore, in response to the vast diversity of existing DNA damage, the cell uses a pool of ‘translesional’ DNA polymerases in order to bypass the various DNA lesions.
Keywords: chemical carcinogens/DNA polymerases/frameshift mutagenesis/NarI mutation hot spot/slippage mutagenesis
Introduction
A new class of DNA polymerases involved in mutagenesis was discovered recently in bacteria (Bacher-Reuven et al., 1999; Tang et al., 1999; Wagner et al., 1999), yeast (Nelson et al., 1996a,b; Johnson et al., 1999a,b) and mammalian cells (Masutani et al., 1999b). In Escherichia coli, three DNA polymerases, Pol II (polB) (Bonner et al., 1988) and the recently discovered Pol IV (dinB) (Wagner et al., 1999) and Pol V (umuDC) (Bacher-Reuven et al., 1999; Tang et al., 1999), are induced as part of the SOS system, a coordinated cellular response to environmental stress. While the umuDC gene products were known to be required for base substitution mutagenesis triggered by UV light or abasic sites (for a recent review see Smith and Walker, 1998), the roles of Pol II and Pol IV in induced mutagenesis have not yet been established. Here, we show that while Pol II (polB) and Pol V (umuDC) are necessary and sufficient for –2 and –1 frameshift mutagenesis induced by the chemical carcinogen N-2-acetylaminofluorene (AAF), respectively, benzo(a)pyrene (BaP)-induced –1 frameshift mutagenesis requires both Pol IV (dinB) and Pol V. The present work shows that, in response to the vast diversity of existing DNA damage, the cell uses a pool of specialized DNA polymerases in order to perform translesion synthesis (TLS). As exemplified by the human disease xeroderma pigmentosum variant (XPV), a defect in one of these polymerases (Johnson et al., 1999b; Masutani et al., 1999a,b) confers a hypermutator phenotype, leading to increased susceptibility to UV-induced skin cancer by eliciting an alternative, but more mutagenic pathway for the bypass of UV lesions.
Point mutations are formed during DNA replication either as genuine replication errors or as a consequence of the presence of damage in the parental DNA. DNA damage arises from both endogenous sources, such as reactive oxygen species, or exogenous sources such as sunlight and tobacco smoke. By changing the chemical structure of the bases, these agents are often effective blocks to the progression of replicative DNA polymerases. It has now become clear that these blocks are overcome by specialized enzymes (translesional DNA polymerases) that can read through damaged bases in a process known as TLS (Nelson et al., 1996a,b; Bacher-Reuven et al., 1999; Johnson et al., 1999a; Masutani et al., 1999a; Tang et al., 1999). Owing to the presence of the lesion, this process is inevitably less accurate than normal replication and will thus lead to mutations in the newly synthesized strand opposite the damaged base.
Following metabolic activation, chemical carcinogens interact covalently with DNA, forming lesions that may be converted into mutations during the process of replication, thus triggering malignant transformation. Aromatic amines and polycyclic aromatic hydrocarbons form the two largest families of chemicals, with potent carcinogenic effects in animals and humans (Singer and Grunberger, 1983). Aromatic amines such as the heterocyclic food mutagens and 4-aminobiphenyl are found bound to the C8 position of guanine in DNA in human colon and pancreas tissue, where these chemicals are suspected to be involved in tumour formation (Miller, 1970; Kasai et al., 1980). On the other hand, polycyclic aromatic hydrocarbons (PAH) are found in cigarette smoke and combustion material, and are involved in human lung and bladder cancer (Harvey, 1991). While the genetics and biochemistry of mutagenesis by UV light and abasic sites have been investigated in detail (reviewed in Friedberg et al., 1995), relatively little is known about the mechanisms of induction of mutations by these bulky chemical carcinogens.
Results and discussion
Pol II (polB) is responsible for AAF-induced –2 frameshift mutagenesis
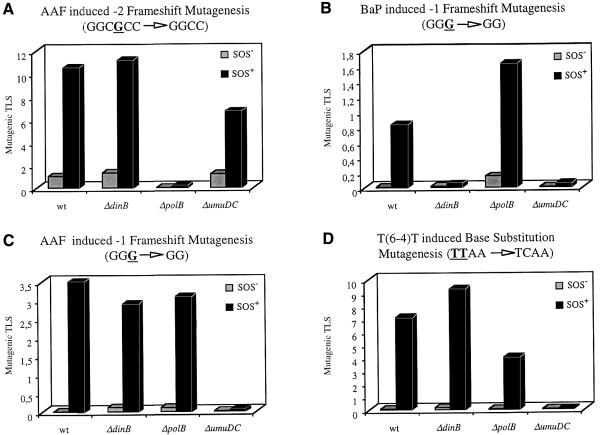
AAF, a representative member of the family of aromatic amides, is known to strongly induce frameshift mutations within short alternating GpC sequences (hot spots) such as the NarI sequence. The loss of a GC dinucleotide within the NarI sequence (GGCGCC→GGCC), which occurs spontaneously at an extremely low frequency (<10–8), is increased up to 107-fold when an AAF residue is bound to the underlined guanine (Koffel-Schwartz et al., 1984; Burnouf et al., 1989; Veaute and Fuchs, 1993; Koffel-Schwartz and Fuchs, 1995; Napolitano et al., 1997). The genetic control of this frameshift mutation pathway was found to be distinct from that leading to base substitutions. Indeed, although both pathways depend upon SOS-controlled functions, the induction of base substitutions requires the combined action of the activated form of RecA (RecA*) and the umuDC gene products (for a recent review see Smith and Walker, 1998), while the –2 frameshift mutation pathway requires neither of these functions, but another, previously unknown SOS function (Koffel-Schwartz et al., 1984; Koffel-Schwartz and Fuchs, 1989; Maenhaut-Michel et al., 1992; Napolitano et al., 1997). Here we demonstrate, using a plasmid construction containing a single AAF adduct within the sequence 5′-GGCGAAFCC-3′, that Pol II, encoded by the SOS-regulated polB gene, is responsible for the induction of –2 frameshift mutations, since the mutation frequency falls to ∼0.05% in the polB deletion strain under both –SOS and +SOS conditions (Table I; Figure 1A). The severe loss of mutagenesis under –SOS conditions in the polB deletion strain (mutagenic TLS = 0.026%) as compared with the wild-type strain (mutagenic TLS ∼1%) suggests that the constitutive level of Pol II expression [∼35 molecules in –SOS cells (Qiu and Goodman, 1997)] mediates the mutagenic response under these conditions. The observed 10-fold increase in mutagenesis upon induction of the SOS response also parallels well the known 7-fold increase in Pol II expression (Qiu and Goodman, 1997). It can be noted that the dinB gene product does not seem to be involved in the –2 frameshift mutation pathway. In contrast, deletion of umuDC partially affects the level of induced mutagenesis obtained in the wild-type strain under +SOS conditions. In conclusion, while Pol II is absolutely required for the –2 frameshift pathway, Pol V, although not essential, appears to facilitate one or several steps in this pathway (Table I; Figure 1A).
Table I. Mutagenic TLS in different DNA polymerase-deficient strains.
| Strains MGZ | UV irradiation (J/m2) | Mutagenic TLS (%) |
|||
|---|---|---|---|---|---|
| GGCGAAFCC | GGGBaP | GGGAAF | T(6–4)T | ||
| wt | 0 | 1.0 (164/15 637)a | <0.007 (0/13 716) | 0.01 (2/20 003) | 0.08 (8/10 067) |
| 50 | 10.5 (532/5066) | 0.83 (195/23 571) | 3.5 (872/25 064) | 7.1 (374/5238) | |
| ΔdinB | 0 | 1.3 (211/15 806) | 0.022 (3/13 855) | 0.12 (16/13 492) | 0.16 (14/8868) |
| 45 | 11.2 (809/7233) | 0.041 (13/31 404) | 2.9 (342/11 782) | 9.3 (1204/12 961) | |
| ΔpolB | 0 | 0.026 (5/19 271) | 0.15 (27/17 917) | 0.105 (25/23 708) | 0.047 (10/21 154) |
| 45 | 0.11 (13/11 765) | 1.63 (866/53 253) | 3.1 (1169/37 369) | 4.0 (309/7732) | |
| ΔumuDC | 0 | 1.2 (281/23 136) | 0.007 (1/14 460) | 0.02 (4/19 655) | 0.046 (6/13 100) |
| 45 | 6.7 (789/11 747) | 0.055 (16/29 035) | 0.046 (7/15 035) | 0.046 (4/8642) | |
aNumber of blue colonies/total number of colonies. The relative errors in the mutagenic TLS values are within 10–20%. These values represent the average of at least three independent experiments.
Fig. 1. Induced mutation frequencies (%) in wild-type and isogenic E.coli strains with deletions in each of the three SOS-inducible DNA polymerases: ΔpolB (Pol II), ΔdinB (Pol IV) and ΔumuDC (Pol V), under –UV and +UV conditions. The nature of the sequence context and the DNA lesion are specified for each panel.
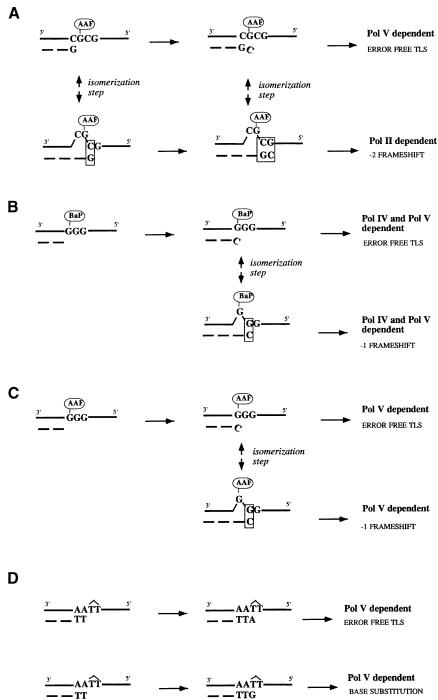
Based on experiments with single-adducted plasmids, the following model for frameshift mutagenesis has been proposed (Lambert et al., 1992; Koffel-Schwartz and Fuchs, 1995; Napolitano et al., 1997). Incorporation of a cytosine residue opposite the G-AAF lesion generates a key replication intermediate, hereafter called the lesion terminus. When elongation proceeds from this intermediate, the replication product is error free (non-slipped TLS, Figure 2A). On the other hand, induction of –2 frameshift mutations involves the formation of a slipped mutagenic intermediate characterized by the presence of one or two correct base pairs at the primer terminus (slipped TLS, Figure 2A). Formation of a slipped intermediate is thermodynamically favoured by the presence of the AAF adduct (Garcia et al., 1993; Milhe et al., 1994, 1996). Our present results suggest that TLS leading to –2 frameshift mutagenesis specifically involves Pol II (Table I; Figure 1A). Thus, polB represents the SOS function, previously named npf, that was found to be required for the –2 frameshift mutation pathway (Koffel-Schwartz et al., 1984; Koffel-Schwartz and Fuchs, 1989; Maenhaut-Michel et al., 1992; Napolitano et al., 1997).
Fig. 2. Potential involvement of the different DNA polymerases in distinct TLS pathways. (A) Induction of –2 frameshift mutations by an AAF adduct located within the NarI sequence context. The non-slipped and slipped TLS pathways are strictly dependent upon Pol V and Pol II, respectively. (B) Induction of –1 frameshift mutations by a BaP adduct located within the sequence GGG. Both the non-slipped and slipped TLS pathways require Pol IV and Pol V. (C) Induction of –1 frameshift mutations by an AAF adduct located within the sequence GGG. Both the non-slipped and slipped replication pathways only require Pol V. (D) Induction of base substitutions by a T(6–4)T dimer. Both replication intermediates leading to error-free or mutagenic bypass strictly require the umuDC gene products (Pol V). Whether Pol V is required at both incorporation and elongation steps remains to be established.
Insertion of C across G-AAF (i.e. error-free TLS) represents >99% of all TLS events (Napolitano et al., 1997). Using a strand segregation analysis assay (Koffel-Schwartz et al., 1996), we have previously shown that error-free TLS requires Pol V (umuDC) (Napolitano et al., 1997; Broschard et al., 1999). Here we confirm this requirement, and in addition show that neither Pol II nor Pol IV is involved in error-free TLS (Table II).
Table II. Error-free TLS in different polymerase-deficient strains.
| Strains MGZ | UV irradiation (J/m2) | Error-free TLS (%) |
||
|---|---|---|---|---|
| GGCGAAFCC | GGGBaP | GGGAAF | ||
| wt | 50 | 17.2 (80/465)a | 8.25 (26/315) | 28 (87/310) |
| ΔdinB | 45 | 23.6 (39/165) | 0.55 (2/315) | 23.6 (71/300) |
| ΔpolB | 45 | 15.2 (71/465) | 9.2 (29/315) | 20.6 (62/300) |
| ΔumuDC | 45 | <0.6 (0/165) | 1.3 (4/315) | <0.3 (0/300) |
aNumber of colonies giving a positive hybridization signal with the target strand probe/total number of colonies analysed.
Therefore, the following model can be proposed (Figure 2A): depending upon the conformation of the replication intermediate (slipped versus non-slipped) TLS proceeds with two distinct DNA polymerases (Pol II or Pol V), suggesting that the replicative DNA polymerase, when blocked in the vicinity of the lesion site, gives up the primer template terminus, which becomes accessible to the pool of translesional DNA polymerases. Whether the process of DNA polymerase switching is highly coordinated with the potential involvement of additional proteins or occurs on a purely competitive basis remains to be established. Although a series of phenotypes related to DNA repair has been described in polB strains (Masker et al., 1973; Escarcellar et al., 1994; Foster et al., 1995; Berardini et al., 1999; Rangarajan et al., 1999), only one study has pointed to the potential role of the polB gene product in the bypass of abasic sites in vivo (Tessman and Kennedy, 1994). However, the present work unambiguously shows the involvement of Pol II in induced mutagenesis justifying it being part of the SOS response. The reason why Pol II, which possesses a robust 3′–5′ exonuclease activity (Cai et al., 1995), is able to elongate the slipped intermediate, while Pol III does not, remains to be understood.
Pol IV (dinB) and Pol V (umuDC) are both involved in BaP bypass
BaP, a representative member of the PAH family, binds covalently to DNA following metabolic conversion to diolepoxides (Harvey, 1991). The major BaP adduct to the N2 position of guanine, (+)-trans-anti-BaP-N2-dG, is known to induce both base substitutions and –1 frameshift mutations within short runs of guanines (Rodriguez and Loechler, 1993). Similar to the mechanism of induction of –2 frameshift mutations, –1 frameshifts also occur during replication through a misaligned primer/terminus intermediate (Lambert et al., 1992; Napolitano et al., 1997). Using a plasmid construction containing a single BaP adduct within sequence 5′-GGGBaP-3′, –1 frameshift mutations are induced ∼100-fold in the wild-type strain upon SOS induction (Table I; Figure 1B). While a similar induction is seen in the polB deletion strain, the inducible mutagenic response is essentially lost in both dinB and umuDC deletion strains (Table I; Figure 1B). In contrast, the absence of the polB gene seems to favour the formation of –1 frameshift mutations under both –SOS and +SOS conditions (Table I). The significance of this observation remains to be discovered.
Insertion of C across G-BaP within the GGG sequence context (i.e. error-free TLS) represents ∼95% of all TLS events (Lenne-Samuel et al., 2000). This error-free pathway is severely decreased in both dinB and umuDC mutant strains (Table II). Although both Pol IV and Pol V appear to be necessary for both error-free and frameshift BaP TLS (Figure 2B), the distinct biochemical steps performed by each of them remain to be established. While the role of the dinB gene product in untargeted mutagenesis is well documented (Brotcorne-Lannoye and Maenhaut-Michel, 1986; Kim et al., 1997), the present results represent the first demonstration of its involvement in induced mutagenesis.
Surprisingly, although AAF also induced –1 frameshifts within the same repetitive sequence (GGG) (Fuchs et al., 1981; Koffel-Schwartz et al., 1984; Lambert et al., 1992), only the umuDC gene product, but neither the dinB nor polB gene product, is necessary for either error-free or frameshift pathways (Tables I and II; Figure 1C). Therefore, the structural distortion of the DNA template induced within the same sequence context by two distinct lesions (BaP versus AAF) necessitates a different combination of polymerases to achieve TLS.
Base substitution mutagenesis was investigated using a single T(6–4)T lesion, which is known to induce 3′-T to C transitions with high efficiency (LeClerc et al., 1991). In all strains tested, base substitution mutagenesis was strongly enhanced upon induction of the SOS response (60- to 100-fold) except in the umuDC deletion strain (Table I; Figure 1D), demonstrating, in agreement with previous results (Szekeres et al., 1996; Becherel and Fuchs, 1999; Tang et al., 2000), that Pol V is involved in the induction of base substitutions (Figure 2D). The partial reduction in the level of induced mutagenesis under +SOS conditions seen in the polB deletion strain compared with the wild-type strain suggests a potential facilitation by Pol II of this TLS pathway. While error-free TLS also depends upon Pol V (Becherel and Fuchs, 1999), the possible involvements of the dinB or polB gene products have not been checked.
Multiple DNA polymerases to bypass all potential DNA lesions?
In conclusion, members of the UmuC/DinB superfamily of DNA polymerases have recently been identified in many prokaryotes and eukaryotes as being involved in TLS and mutagenesis (Gerlach et al., 1999; McDonald et al., 1999; Ogi et al., 1999). It should also be noted that TLS is not confined to polymerases of this superfamily, as Pol ζ in yeast (Nelson et al., 1996b) and Pol II in E.coli belong to the Pol(alpha) family. For those polymerases where biochemical data are available, it was found that they lack proofreading function and exhibit low processivity, leading to a new model for TLS and mutagenesis, referred to as the DNA polymerase switch model (Cordonnier and Fuchs, 1999). Although the lack of proofreading intuitively appears to be essential during TLS in order to avoid futile cycles of incorporation–excision at the lesion terminus, it should be noted that Pol II clearly performs TLS despite its robust 3′–5′ exonuclease activity. In the vicinity of a replication-blocking lesion (targeted mutagenesis) or a difficult to replicate sequence, a less accurate DNA polymerase (translesional DNA polymerase) is temporarily recruited, thus replacing the high fidelity replicative DNA polymerase. After a short patch of less accurate DNA synthesis, high fidelity DNA replication resumes by means of the replicative DNA polymerase. However, as the partners of the translesional DNA polymerase are not yet known, it is not clear whether they are indeed distributive in vivo. In vitro, the β clamp was recently shown to target Pol IV to DNA and to strongly increase its processivity (Wagner et al., in press). It is not yet known how DNA polymerase switching occurs: it may either be a highly coordinated process involving additional proteins or involve a simple competition among the different translesional DNA polymerases that are present.
Why are there multiple DNA polymerases involved in TLS within a single organism? The present work sheds some light on this intriguing question as it suggests that, depending upon the structure of the lesion and the sequence context, a given translesional DNA polymerase or a specific combination of them is necessary to achieve TLS. Additional examples of the involvement of Pol II and Pol IV in the bypass of DNA lesions will be described in the future. For example, it was brought to our attention that Pol II is also involved in the error-free bypass of a benz(a)anthracene adenine adduct (A.G.McNees, M.F.Goodman and R.Stephen Lloyd, personal communication), while Pol IV is essential for the induction of –1 frameshift mutations by the chemical 4-nitroquinoline-N-oxide (S.R.Kim, K.Matsui, M.Yamada, P.Gruz and T.Nohmi personal communication). As more lesions are studied, it is also likely that cases of functional redundancy among the polymerases will be discovered (i.e. a given lesion can be handled efficiently by several polymerases). This appears to be the case in humans where a defective Pol η confers a hypermutator phenotype in XPV patients (Masutani et al., 1999a). In these patients, in the absence of Pol η, TLS may be achieved by an alternative minor, but more mutagenic pathway for the bypass of UV light-induced lesions. The vast diversity of ‘lesions/sequence context’ combinations that are potentially encountered by a replication fork will be dealt with by a limited number of translesional DNA polymerases. It is therefore likely that there will be no general (predictable) model as to which polymerase, or combination thereof, will bypass a given lesion. The fine mechanistics of the bypass of any given lesion will await the determination of the structure of complexes between these DNA polymerases and lesion-containing primer templates.
Materials and methods
Strains
The lacZ-α-complementing allele (lacZΔM15) was introduced into the chromosome of strain MG1655 by P1 transduction using a donor strain with the following genotype (S1521: lacZΔM15, lacY::Tn10), kindly provided by Dr S.Brown (University of Copenhagen). The resulting strain, MGZ, was selected on Luria–Bertani (LB) plates containing 10 µg/ml tetracycline. The presence of the lacZΔM15 allele was veri fied as follows: strain MGZ forms white colonies when plated on X-gal-containing medium (lacZ– genotype) and blue colonies on these same plates when transformed with plasmid pUC8 (α-complementation assay). The previously described null alleles of polB (Escarcellar et al., 1994), dinB (Kim et al., 1997) and umuDC (Woodgate, 1992) were then introduced into strain MGZ by P1 transduction. The presence of the Δ(araD-polB)::Ω allele was selected for on LB agar plates with 20 µg/ml spectinomycin and then confirmed by PCR analysis. The dinB::Tn5 (Kim et al., 1997) allele was selected for on LB agar plates with 20 µg/ml kanamycin and confirmed by PCR analysis. Similarly, the Δ(umuDC)595::cat allele was selected using agar plates with 20 µg/ml chloramphenicol.
Mutagenesis assays
Mutagenesis induced by the different lesions was determined in the various strains following transformation by electroporation (Bio-Rad gene pulser) with the single-adducted plasmids described below. SOS induction was achieved as previously described (Koffel-Schwartz et al., 1996) by prior UV irradiation of the different strains at a UV dose (as specified in Table I) yielding survival levels between 10 and 30%.
AAF-induced –2 frameshift mutagenesis (GGCGCC→GGCC) was assayed using a single-adducted pUC-derived plasmid described previously (Broschard et al., 1999). The single AAF adduct is located within a NarI sequence 5′-GGCGAAFCC-3′ in the N-terminal part of the lacZ′ gene of a pUC-derived plasmid. The construction has a LacZ– phenotype that can be reverted to LacZ+ by a –2 frameshift mutation. Wild-type and mutant transformants are scored on indicator plates containing carbenicillin, X-gal and isopropyl-β-d-thiogalactopyranoside as white and blue colonies, respectively. Frequencies of –2 frameshift mutations were measured in each strain, under both SOS-uninduced (–UV) and SOS-induced (+UV) conditions, as the ratio of blue transformants over the total number of transformants.
BaP-induced –1 frameshift mutagenesis (5′-GGGBP-3′→GG) was assayed using a single-adducted pUC-derived plasmid described previously (Koffel-Schwartz et al., 1996; Lenne-Samuel et al., 2000). The single BaP adduct is located as indicated in the N-terminal part of the lacZ′ gene of a pUC-derived plasmid (Koffel-Schwartz et al., 1996; Lenne-Samuel et al., 2000). The construction has a LacZ– phenotype that can be reverted to LacZ+ by a –1 frameshift mutation. Frequencies of –1 frameshift mutations were measured on indicator plates as described above.
AAF-induced –1 frameshift mutagenesis (5′-GGGAAF-3′→GG) was assayed using the single-adducted pUC-derived plasmid described previously (Koffel-Schwartz et al., 1996). Minus 1 frameshift mutations are scored as described above for the BaP adduct.
T(6–4)T-induced base substitution mutagenesis was assayed using a single-adducted pUC-derived plasmid described previously (Becherel and Fuchs, 1999). The T(6–4)T lesion was engineered such that its 3′ T is part of an in-frame TAA ochre codon in the lacZ′ gene. Insertion of the correct base (i.e. A) opposite the 3′ T of the lesion maintains the stop codon. Any base substitution involving the 3′ T of the TT photoproduct will suppress the stop codon and yield a lacZ+ colony. Frequencies of base substitution mutations were determined on indicator plates as indicated for the AAF lesion.
Strand segregation analysis
Among the non-mutant colonies (i.e. white colonies), the fraction of colonies resulting from error-free TLS was determined using the colony hybridization assay developed previously (Koffel-Schwartz et al., 1996). Briefly, all plasmid constructions (Koffel-Schwartz et al., 1996; Broschard et al., 1999; Lenne-Samuel et al., 2000) contain a short sequence heterology located across from the adduct site. The adduct-containing strand is called the target strand, while the other strand is referred to as the marker strand. Using oligonucleotides that hybridize specifically with the target strand of the heteroduplex, it is possible to determine the fraction of colonies that have undergone error-free TLS. The majority of colonies do not hybridize with the target strand probe but only with the marker strand, thus representing colonies where plasmid replication occurred via damage avoidance (Koffel-Schwartz et al., 1996; Broschard et al., 1999; Lenne-Samuel et al., 2000).
Acknowledgments
Acknowledgements
This work was partially supported by a Human Frontier Research Grant (RG0351/1998-M).
References
- Bacher-Reuven N., Arad,G., Maor-Shoshani,A. and Livneh,Z. (1999) The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA and SSB and specialized for translesion replication. J. Biol. Chem., 274, 31763–31766. [DOI] [PubMed] [Google Scholar]
- Becherel O.J. and Fuchs,R.P.P. (1999) SOS mutagenesis results from up-regulation of translesion synthesis. J. Mol. Biol., 294, 299–306. [DOI] [PubMed] [Google Scholar]
- Berardini M., Foster,P.L. and Loechler,E.L. (1999) DNA polymerase II (polB) is involved in a new DNA repair pathway for DNA interstrand cross-links in Escherichia coli. J. Bacteriol., 181, 2878–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner C.A., Randall,S.K., Rayssiguier,C., Radman,M., Eritja,R., Kaplan,B.E., McEntee,K. and Goodman,M.F. (1988) Purification and characterization of an inducible Escherichia coli DNA polymerase capable of insertion and bypass at abasic lesions in DNA. J. Biol. Chem., 263, 18946–18952. [PubMed] [Google Scholar]
- Broschard T.H., Koffel-Schwartz,N. and Fuchs,R.P.P. (1999) Sequence-dependent modulation of frameshift mutagenesis at NarI-derived mutation hot spots. J. Mol. Biol., 288, 191–199. [DOI] [PubMed] [Google Scholar]
- Brotcorne-Lannoye A. and Maenhaut-Michel,G. (1986) Role of RecA protein in untargeted UV mutagenesis of bacteriophage λ: evidence for the requirement of the dinB gene. Proc. Natl Acad. Sci. USA, 83, 3904–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnouf D., Koehl,P. and Fuchs,R.P.P. (1989) Single adduct mutagenesis: strong effect of the position of a single acetylaminofluorene adduct within a mutation hot spot. Proc. Natl Acad. Sci. USA, 86, 4147–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Yu,H., McEntee,K., Kunkel,T.A. and Goodman,M.F. (1995) Purification and properties of wild-type and exonuclease-deficient DNA polymerase II from E.coli. J. Biol. Chem., 270, 15327–15335. [DOI] [PubMed] [Google Scholar]
- Cordonnier A.M. and Fuchs,R.P.P. (1999) Replication of damaged DNA: molecular defect in xeroderma pigmentosum variant cells. Mutat. Res., 435, 111–119. [DOI] [PubMed] [Google Scholar]
- Escarcellar M., Hicks,J., Gudmundsson,G., Trump,G., Touati,D., Lovett,S., Foster,P., McEntee,K. and Goodman,M.F. (1994) Involvement of E.coli DNA polymerase II in response to oxidative damage and adaptive mutation. J. Bacteriol., 176, 6221–6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P.L., Gudmundsson,G., Trimarchi,J.M., Cai,H. and Goodman, M.F. (1995) Proofreading-defective DNA polymerase II increases adaptive mutation in Escherichia coli. Proc. Natl Acad. Sci. USA, 92, 7951–7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. American Society for Microbiology, Washington, DC. [Google Scholar]
- Fuchs R.P.P., Schwartz,N. and Daune,M.P. (1981) Hot spots of frameshift mutations induced by the ultimate carcinogen N-acetoxy-N-2-acetylaminofluorene. Nature, 294, 657–659. [DOI] [PubMed] [Google Scholar]
- Garcia A., Lambert,I.B. and Fuchs,R.P.P. (1993) DNA adduct induced stabilization of slipped frameshift intermediates within repetitive sequences: implications for mutagenesis. Proc. Natl Acad. Sci. USA, 90, 5989–5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach V.L., Aravind,L., Gotway,G., Schulz,R.A., Koonin,E.V. and Friedberg,E.C. (1999) Human and mouse homologs of E.coli DinB (DNA polymerase IV), members of the UmuC/DinB superfamily. Proc. Natl Acad. Sci. USA, 96, 11922–11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey R.G. (1991) Polycyclic Aromatic Hydrocarbons: Chemistry and Carcinogenicity. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Johnson R.E., Prakash,S. and Prakash,L. (1999a) Efficient bypass of a thymine–thymine dimer by yeast DNA polymerase, Pol η. Science, 283, 1001–1004. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Kondratick,C.M., Prakash,S. and Prakash,L. (1999b) hRAD30 mutations in the variant form of xeroderma pigmentosum. Science, 285, 263–265. [DOI] [PubMed] [Google Scholar]
- Kasai H. et al. (1980) Potent novel mutagens produced by broiling fish under normal conditions. Proc. Jpn Acad., 56B, 278–283. [Google Scholar]
- Kim S.R., Maenhaut-Michel,G., Yamada,M., Yamamoro,Y., Matsui,K., Sofuni,T., Nohmi,T. and Ohmori,H. (1997) Multiple pathways for SOS-induced mutagenesis in E.coli: an overexpression of dinB/P results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc. Natl Acad. Sci. USA, 94, 13792–13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffel-Schwartz N. and Fuchs,R.P.P. (1989) Genetic control of AAF-induced mutagenesis at alternating GC sequences: an additional role for RecA. Mol. Gen. Genet., 215, 306–311. [DOI] [PubMed] [Google Scholar]
- Koffel-Schwartz N. and Fuchs,R.P.P. (1995) Sequence determinants for –2 frameshift mutagenesis at NarI-derived hot spots. J. Mol. Biol., 252, 507–513. [DOI] [PubMed] [Google Scholar]
- Koffel-Schwartz N., Verdier,J.M., Bichara,M., Freund,A.M., Daune,M.P. and Fuchs,R.P.P. (1984) Carcinogen induced mutation spectrum in wild type, uvrA and umuC strains in E.coli: strain specificity and mutation prone sequences. J. Mol. Biol., 177, 33–51. [DOI] [PubMed] [Google Scholar]
- Koffel-Schwartz N., Coin,F., Veaute,X. and Fuchs,R.P.P. (1996) Cellular strategies for accommodating replication-hindering adducts in DNA: Control by the SOS response in E.coli. Proc. Natl Acad. Sci. USA, 93, 7805–7810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert I.B., Napolitano,R.L. and Fuchs,R.P.P. (1992) Carcinogen-induced frameshift mutagenesis in repetitive sequences. Proc. Natl Acad. Sci. USA, 89, 1310–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClerc J.E., Borden,A. and Lawrence,C.W. (1991) The thymine–thymine pyrimidine–pyrimidone(6–4) ultraviolet light photoproduct is highly mutagenic and specifically induces 3′ thymine-to-cytosine transitions in Escherichia coli. Proc. Natl Acad. Sci. USA, 88, 9685–9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenne-Samuel N.H., Janel-Bintz,R., Kolbanovskiy,A., Geacintov,N.E. and Fuchs,R.P.P. (2000) Distinct genetic processing of a single benzo(a)pyrene adduct into frameshift or base substitution mutations during translesion synthesis in E.coli. Mol. Microbiol., in press. [DOI] [PubMed] [Google Scholar]
- Maenhaut-Michel G., Janel-Bintz,R. and Fuchs,R.P.P. (1992) A umuDC-independent SOS pathway for frameshift mutagenesis. Mol. Gen. Genet., 235, 373–380. [DOI] [PubMed] [Google Scholar]
- Masker W., Hanawalt,P. and Shizuya,H. (1973) Role of DNA polymerase II in repair replication in Escherichia coli. Nature New Biol., 244, 242–243. [DOI] [PubMed] [Google Scholar]
- Masutani C., Araki,M., Yamada,A., Kusumoto,R., Nogimori,T., Maekawa,T., Iwai,S. and Hanaoka,F. (1999a) Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J., 18, 3491–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C. et al. (1999b) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature, 399, 700–704. [DOI] [PubMed] [Google Scholar]
- McDonald J.P., Rapic-Otrin,V., Epstein,J.A., Broughton,B.C., Wang,X., Lehmann,A.R., Wolgemuth,D.J. and Woodgate,R. (1999) Novel human and mouse homologs of S. cerevisiae DNA polymerase η. Genomics, 60, 20–30. [DOI] [PubMed] [Google Scholar]
- Milhe C., Dhalluin,C., Fuchs,R.P.P. and Lefevre,J.-F. (1994) NMR evidence of the stabilization by the carcinogen N-2-acetylaminofluorene of a frameshift mutation intermediate. Nucleic Acids Res., 22, 4646–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milhe C., Fuchs,R.P.P. and Lefevre,J.-F. (1996) NMR data show that the carcinogen N-2-acetylaminofluorene stabilizes an intermediate of –2 frameshift mutagenesis in a region of high mutation frequency. Eur. J. Biochem., 235, 120–127. [DOI] [PubMed] [Google Scholar]
- Miller J.A. (1970) Carcinogenesis by chemicals: an overview. Cancer Res., 30, 559–576. [PubMed] [Google Scholar]
- Napolitano R.L., Lambert,I.B. and Fuchs,R.P.P. (1997) SOS factors involved in translesion synthesis. Proc. Natl Acad. Sci. USA, 94, 5733–5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J.R., Lawrence,C.W. and Hinkle,D.C. (1996a) Deoxycytidyl transferase activity of yeast REV1 protein. Nature, 382, 729–731. [DOI] [PubMed] [Google Scholar]
- Nelson J.R., Lawrence,C.W. and Hinkle,D.C. (1996b) Thymine–thymine dimer bypass by yeast polymerase ζ. Science, 272, 1646–1649. [DOI] [PubMed] [Google Scholar]
- Ogi T., Kato,T.,Jr, Kato,T. and Ohmori,H. (1999) Mutation enhancement by DINB1, a mammalian homologue of the E.coli mutagenesis protein DinB. Genes Cells, 4, 607–618. [DOI] [PubMed] [Google Scholar]
- Qiu Z. and Goodman,M.F. (1997) The E.coli polB locus is identical to dinA, the structural gene for DNA polymerase II. Characterization of Pol II purified from a polB mutant. J. Biol. Chem., 272, 8611–8617. [DOI] [PubMed] [Google Scholar]
- Rangarajan S., Woodgate,R. and Goodman,M.F. (1999) A phenotype for enigmatic DNA polymerase II: a pivotal role for polII in replication restart in UV-irradiated E.coli. Proc. Natl Acad. Sci. USA, 96, 9224–9229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez H. and Loechler,E.L. (1993) Mutagenesis by the (+)-anti-diolepoxide of B(a)P: What controls mutation specificity. Biochemistry, 32, 1759–1769. [DOI] [PubMed] [Google Scholar]
- Singer B. and Grunberger,D. (1983) Molecular Biology of Mutagens and Carcinogens. Plenum Press, New York, NY. [Google Scholar]
- Smith B.T. and Walker,G.C. (1998) Mutagenesis and more: umuDC and the E.coli SOS response. Genetics, 148, 1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres E.S., Woodgate,R. and Lawrence,C.W. (1996) Substitution of mucAB or rumAB for umuDC alters the relative frequencies of two classes of mutations induced by a site-specific T–T cyclobutane dimer and the efficiency of translesion DNA synthesis. J. Bacteriol., 178, 2559–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M., Shen,X., Frank,E.G., O’Donnell,M., Woodgate,R. and Goodman,M.F. (1999) UmuD′2C is an error-prone DNA polymerase, E. coli polV. Proc. Natl Acad. Sci. USA, 96, 8919–8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M., Pham,P., Shen,X., Taylor,J.S., O’Donnell,M., Woodgate,R. and Goodman,M.F. (2000) Roles of E.coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature, 404, 1014–1018. [DOI] [PubMed] [Google Scholar]
- Tessman I. and Kennedy,M.A. (1994) DNA polymerase II of Escherichia coli in the bypass of abasic sites in vivo. Genetics, 136, 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute X. and Fuchs,R.P.P. (1993) Greater susceptibility to mutations in lagging strand of DNA replication in Escherichia coli than in leading strand. Science, 261, 598–600. [DOI] [PubMed] [Google Scholar]
- Wagner J., Gruz,P., Kim,S.-R., Yamada,M., Matsui,K., Fuchs,R.P.P. and Nohmi,T. (1999) The dinB gene encodes a novel E.coli DNA polymerase, DNA PolIV, involved in mutagenesis. Mol. Cell, 4, 281–286. [DOI] [PubMed] [Google Scholar]
- Wagner J., Fujii,S., Gruz,P., Nohmi,T. and Fuchs,R.P.P. (2000) The β clamp targets DNA Pol IV to DNA and strongly increases its processivity. EMBO rep., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgate R. (1992) Construction of a umuDC operon substitution mutation in E.coli. Mutat. Res., 281, 221–225. [DOI] [PubMed] [Google Scholar]