Abstract
The interactions of biological macromolecules with water are fundamental to their structure, dynamics and function. Historically, characterization of the location and residence times of hydration waters of proteins in solution has been quite difficult. Confinement within the nanoscale interior of a reverse micelle slows water dynamics, allowing detection of global protein-water interactions using nuclear magnetic resonance techniques. Complications that normally arise from hydrogen exchange and long-range dipolar coupling are overcome by the nature of the reverse micelle medium. Characterization of the hydration of ubiquitin demonstrates that encapsulation within a reverse micelle allows detection of dozens of hydration waters. Comparison of nuclear Overhauser effects obtained in the laboratory and rotating frames indicate a considerable range of hydration water dynamics is present on the protein surface. In addition, an unprecedented clustering of different hydration dynamic classes of sites is evident.
The interactions of biological macromolecules with solvating water are fundamental to their structure, dynamics, and function1. Historically, comprehensive site-resolved experimental insight into the behavior of solvent near protein surfaces has been notoriously difficult to obtain in solution. Some years ago, Wüthrich and coworkers employed multi-dimensional solution NMR to observe site-resolved dipolar magnetization exchange between protein and water molecules having relatively long-lived interactions2 and have provided insight into various properties of water intimately involved in the structure of proteins3. In principle, the same NMR-based methodology could also allow access to a comprehensive site-resolved characterization of the interaction of water across the surface of a protein. Unfortunately this approach has been confounded by the extremely short residence times of hydration water, by complications arising from hydrogen exchange phenomena and by a potential ambiguity in the distinction between long-range contributions from water molecules in the bulk from those of the hydration waters of interest.
It is now well established that the motion of water molecules is somewhat slowed as a result of interaction with the protein surface4. A variety of methods have shown that the 1–2 layers of water molecules nearest the macromolecular surface are motionally slowed though estimates vary between several-fold to perhaps two orders of magnitude5–8. Regardless, hydration water-protein interactions generally remain sufficiently short-lived to quench the intermolecular dipolar magnetization exchange necessary to detect them using the nuclear Overhauser effect in the laboratory (NOE) or the rotating frame (ROE)9. The situation is also considerably clouded by the inherent presence of exchange of hydrogens of the solvent with labile sites on the protein2,3,9. Furthermore, a more recent theoretical analysis also predicts dominant contributions from long-range intermolecular dipolar couplings10. This effect arises primarily from two geometric sources that combine to reduce the effective distance dependence of the NOE and ROE from the familiar inverse sixth-power dependence to a more slowly varying simple inverse dependence10–12. These restrictions and complications have largely frustrated the exploration of the protein-water interface using high-resolution NMR methods5,13. Here we take advantage of several favorable aspects of reverse micelle encapsulation to circumvent these limitations, enabling site-resolved measurement of the hydration water dynamics for human ubiquitin.
RESULTS
Encapsulation enables detection of protein hydration water
Reverse micelles are monodisperse ensembles of surfactant-encapsulated nanoscale water pools that result from spontaneous organization of a mixture of appropriate surfactants, water, and nonpolar solvent such as the alkanes (Fig. 1). The size of the water pool can be tightly controlled by varying the molar ratio of water to surfactant (water loading or Wo). Proteins can be encapsulated within the water pool of reverse micelles reproducibly and with high structural fidelity and at concentrations amenable to high-resolution solution NMR methods14,15. When prepared in low viscosity solvents, the full arsenal of solution multi-dimensional heteronuclear NMR techniques becomes accessible16,17. Reverse micelles have served as a powerful tool for NMR-based studies of protein and nucleic acid structure and biophysics under a range of conditions14,16,18–23.
Figure 1.
Schematic of ubiquitin in an AOT reverse micelle. The surfactant, water pool, and protein are shown to-scale with a single water molecule pictured, also to-scale, for comparison. Nonpolar alkane solvent, which surrounds the reverse micelle particle, is not shown. The diameter of the RM is based on measurements of the rotational correlation time of ubiquitin in AOT RMs at comparable water loading to that used here14. This value agrees closely with estimates of the RM dimensions from simple geometric predictions. The outer dark blue ring approximates the layer of solvent which is dynamically restricted by interactions with the surfactant headgroups24, while the light blue region corresponds to the central water pool which hydrates the protein.
For water loadings such as those used here (W0 ~ 10), the reverse micelle contains approximately one thousand water molecules on average24 and the re-orientational dynamics of encapsulated water is slowed relative to bulk water by approximately one order of magnitude25,26. This suggested that the combined effects of the protein surface and nanoscale confinement could slow the water dynamics to allow efficient detection of hydration water-protein cross-relaxation. At the same time, if reorganization of water contributes to the rate limiting step, hydrogen exchange chemistry in the reverse micelle should also be somewhat slowed, potentially minimizing complications arising from hydrogen-exchange. Finally, reverse micelles also provide an appropriate experimental condition to directly assess the contributions from long-range intermolecular dipolar interactions.
We have employed human ubiquitin to test these ideas. Ubiquitin is a small 76 residue protein comprised of a five stranded mixed beta sheet, a long alpha helix and a short 310 helix27. It engages in an extensive set of protein-protein interactions28. We first examined the hydration of ubiquitin in free aqueous solution at pH 5 and 25 °C using three-dimensional 15N-resolved NOESY29 and ROESY30 experiments (Fig. 2a,b). Only a few amide-water weak cross peaks were observed, which is generally indicative of very short residence times for protein hydration water in bulk aqueous solution3. At this pH and temperature, the exchange of even non-hydrogen bonded amide hydrogens with solvent is generally too slow to impact these experiments. However, every amide hydrogen-water cross peak observed in the free solution spectra involves an amide hydrogen that is within the distance detectable by the NOE (see below) of a labile side chain hydrogen. Chemical exchange of side chain hydrogens with solvent water followed by intramolecular NOE produces cross peaks at the water resonance that are indistinguishable from direct protein-water NOEs2,3. It is likely that many, if not all, of the amide hydrogen – water NOE correlations seen in bulk solution are the result of such exchange-relayed interactions5.
Figure 2.
1H-15N planes at the water 1H resonance of (a) three-dimensional NOESY 15N-HSQC and (b) three-dimensional ROESY 15N-HSQC of uniformly 15N-labeled ubiquitin in aqueous solution. In these spectra, the water resonance is at 4.8 p.p.m. Dipolar interactions between amide hydrogen and alpha hydrogen (nearly) degenerate with the water resonance are also indicated. Also shown are 1H-15N planes at the water 1H resonance of (c) three-dimensional NOESY 15N-HSQC and (d) three-dimensional ROESY 15N-HSQC of uniformly 1H-15N,2H-12C-labeled ubiquitin encapsulated in AOT reverse micelles at W0 = 9.0. In these spectra, the water resonance is at 4.3 p.p.m. For all panels, positive (black) and negative (red) sign of cross peak intensity defines dipolar (positive NOE, negative ROE) versus chemical exchange peaks (positive NOE, positive ROE). The cross peaks between amide NH and the water resonance are labeled by residue. All of the cross peaks observed in the free solution aqueous spectra arise from amide hydrogens that are within NOE distance of labile side chain hydrogens and are ascribed to hydrogen exchange. In contrast, for encapsulated ubiquitin, only the amide hydrogen of Glu-51 (emphasized with an asterisk) is near enough to a rapidly exchanging hydrogen (the hydroxyl of Tyr-59) to have a contribution to the NOE cross peak intensity of greater than two percent. Spectra were obtained at pH 5 and 25 °C (aqueous) or 20 (RM) °C at 500 MHz (1H) using a cryogenically cooled probe.
We have previously demonstrated that the structure of ubiquitin is unperturbed by appropriate encapsulation in reverse micelles composed of bis-2-ethylhexyl sulfosuccinate (AOT)14. In contrast to the paucity of amide hydrogen – water interactions revealed by the NOESY spectrum of ubiquitin in free solution, the protein encapsulated in AOT reverse micelles displays literally dozens of protein-water interactions under similar temperature and pH conditions (Fig. 2c,d). Dipolar exchange is identified by the opposite phase of the cross peaks in the orthogonal frames (shown here as positive NOE, negative ROE), while direct hydrogen exchange with solvent gives rise to peaks of identical phase (shown as positive in both spectra)2,3. Calibration of the NOE obtained at the 40 ms mixing time used here placed it in the linear regime and corresponds to a maximum NOE-detected distance of approximately 4.3 Å. More than half of detected amide hydrogen - water cross peaks involve amides that are over 4.3 Å from any labile side chain hydrogen, as indicated by of the ensemble of structures determined for encapsulated ubiquitin (PDB 1G6J)14. These can therefore be unequivocally assigned to direct NOE interactions between hydration water and the protein. In addition, the hydrogen exchange chemistry catalyzed by hydroxide ion within the reverse micelle water pool is effectively slowed by two orders of magnitude as indicated by the pH dependence of the onset of line broadening due to amide hydrogen exchange (Supplementary Fig. 1). It should be noted that the pKa values of side chain hydroxyls and primary amines are low enough that general base catalysis by buffer or even the head groups of AOT is also a potential concern. However, the fact that lysine side chain ζ amine groups are observed (Fig. 2) and that most side chain hydroxyls could be identified by direct NOE (Supplementary Fig. 2) indicates that minimal exchange with solvent is present. Given the relative concentrations of protein and water, this restricts the hydrogen exchange rates to the 1 s−1 or slower rate regime. The single labile side chain hydrogen whose chemical exchange may be faster than this rate is the hydroxyl of Tyr-59, which is anticipated to have a considerably lower pKa. However, contributions from hydrogen exchange-mediated pathways are sufficiently minimized to allow quantitative interpretation of the vast majority of the NOE and ROE cross peak intensities.
Interpretation of the NOE/ROE ratio (see below) as directly representative of local hydration dynamics also requires that the contributions from long-range dipolar couplings be negligible. NOESY spectra of highly deuterated ubiquitin lack any measurable cross peaks to organic solvent hydrogens, demonstrating that long-range interactions beyond the exterior of the reverse micelle are minimal. It is this long-range contribution in bulk aqueous solution that has been argued to provide the vast majority of the NOE intensity to the water resonance10. It is also of note that only a handful of amide hydrogens show NOEs with hydrogens of the surfactant headgroup. With the exception of Gln 47 and Gln 49, these are located in the disordered C-terminal tail comprised of residues 73–76. The grouping of such interactions suggests that they arise from local close approach of the protein to the surfactant shell rather than general long-range dipolar exchange contributions. It is important to note that these regions of the surface are not dehydrated (Fig. 2). The spatial geometry of the reverse micelle and encapsulated protein also cause the number of water molecules to increase roughly as the square of the distance from a solvent-exposed protein hydrogen in contrast to the cubic dependence seen in bulk solution (Supplementary Fig. 3). These fundamental differences in the physical parameters of the reverse micelle system versus bulk solution combine to largely eliminate contamination of short-range dipolar interactions between protein and hydration water by contribution from long-range couplings to solvent.
In summary, the residence times of water on the surface of an encapsulated ubiquitin molecule are sufficiently increased, the effective rates of specific and general base catalysis of hydrogen exchange sufficiently decreased, and the potential contribution by long-range coupling to bulk solvent sufficiently reduced to allow confident and comprehensive site-resolved measurements of dipolar cross-relaxation between hydration waters and amide hydrogens of the protein. This enables the first site-resolved analysis of relative hydration water mobilities across an entire protein surface.
The dynamical character of the protein-water interface
A quantitative measure of hydration dynamics can be obtained from the ratio of the NOE to ROE cross-relaxation rates, σNOE and σROE2,3. Here ratios of the intensities of the water cross peaks are used (Supplementary Tables 1 and 2). In the slow correlation time limit (ωτ ≫ 1), a rigidly bound water molecule would result in a NOE/ROE ratio of −0.5 (ref. 2,3). We find that a minority of the resolved protein-water correlations have ratios at this limit. Previous relaxation studies indicate that the effective macromolecular tumbling time for these reverse micelle particles dissolved in liquid pentane is about 10 ns16. The majority of detected hydration sites showed a range of NOE/ROE values between 0 and −0.5 indicating the presence of substantial motion of the hydration water on the surface of the protein and/or a somewhat shorter residence time3,31. In previous studies of protein-water cross relaxation, it was unclear to what degree indirect magnetization exchange contributed to NOE/ROE ratios more positive than −0.5. As measurements made in the reverse micelle are generally uncomplicated by indirect exchange effects, we conclude that the range of observed NOE/ROE ratios arise from a distribution of hydration water dynamics that reflect both local geometrical factors and the residence times of the individual water molecules on the surface of ubiquitin31.
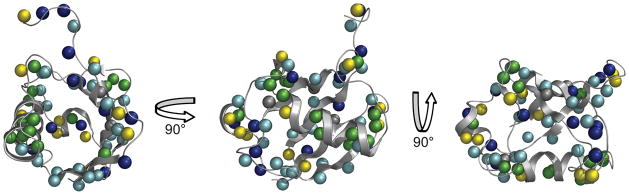
Approximately three fourths of the amide hydrogens within the NOE-detection distance limit (4.3 Å) of solvent showed measurable cross peaks to water. No cross peaks to water from buried amide hydrogens were detected, confirming the absence of spin-diffusion or appreciable penetration of water into the interior of the protein. A detailed view of the relative hydration dynamics of ubiquitin is shown in Figure 3 where the amide hydrogens within NOE distance of the molecular surface are represented as spheres and are color-coded according to dynamic classes. Sites that have NOE/ROE ratios at the bound limit, representing the slowest regions of hydration dynamics, are colored dark blue. Sites with ratios between −0.47 and −0.3 (relatively slow or spatially restricted hydration water) or between −0.3 and 0 (relatively fast or spatially less restricted hydration water) are colored cyan and yellow, respectively. Sites that show no cross peaks to water and are therefore representative of the fastest hydration dynamics are colored green. A clear grouping of very slow and spatially restricted hydration is evident along the C-terminal tail of the protein (top panel), and a cluster of very fast hydration sites is evident along the surface of the alpha helix (center panel). Clusters of intermediate dynamics are also visible along the mixed beta sheet (bottom panel). These groupings illustrate the relative hydration dynamics essentially across the surface of the entire protein. Importantly, a minority of solvent-exposed sites lacked detectable cross-relaxation to water. These sites, again clustered along the solvent-exposed ridge of the alpha helix, also did not show measurable cross peaks to surfactant hydrogens suggesting that long-lived interactions with the inner surface of the reverse micelle shell are absent and that the fastest and least spatially restricted hydration dynamics occur at these amide sites.
Figure 3.
Ubiquitin hydration dynamics map. All amide hydrogens within NOE distance of solvent are shown as spheres. Sites are color coded as follows: dark blue – bound hydration sites (NOE/ROE ≤ −0.47), cyan – relatively slow hydration (−0.47 > NOE/ROE < −0.3), yellow – relatively fast hydration (−0.3 ≤ NOE/ROE), green – fast hydration (solvent-exposed, no water cross peaks). The dark blue hydration class corresponds to sites where water within NOE distance (4.3 Å) of the amide hydrogen has a residence time on the order of the correlation time of the protein (~10 ns) or longer. Green sites are those within NOE distance of solvent, yet show no cross peaks to water. These represent the fastest sites of hydration dynamics. Clustering of hydration dynamic classes is evident. A cluster of greatly restricted hydration sites (dark blue and cyan) is seen in the C-terminal region (top panel), while the sites of fastest hydration dynamics group along the solvent-exposed ridge of the alpha helix (center panel). Clusters of intermediate hydration dynamics are also visible along the mixed beta sheet (cyan grouping) and near the 310 helix (yellow grouping). The differential localization of these hydration dynamic classes illustrates the relative hydration dynamics across the ubiquitin structure. Images were created using PyMOL39.
DISCUSSION
Until now, experimental efforts to develop a site resolved understanding of the hydration layer around proteins have relied largely on crystallographic data. Because most protein crystal structures are heavily hydrated and contain complex ensembles of solvent, the general view seems to be that the environment in protein crystals should be representative of the crowded conditions within the cell and that therefore crystallographic water locations should be representative of the native hydration behavior of the protein. Extensive analysis of crystallographic waters has been combined with simulation data in several systems to propose a picture of the molecular nature of the hydration layer32. The present case of ubiquitin serves as a cautionary example. Two crystal structures for wild type human ubiquitin are available (PDB codes 1UBQ and 1UBI)27,33. Only about half of the defined waters in the two crystal structures were conserved to within 1 Å between the structures even though the proteins were crystallized in the same space group under almost identical conditions. Furthermore, there is little correlation between the waters detected by solution NMR and the locations and occupancies of the crystallographic waters in these crystal structures (Supplementary Fig. 4). To illustrate the degree of correspondence between the long-lived hydration waters detected by NMR to the locations of the defined waters common to both crystal structures of ubiquitin, a color-coded ribbon diagram of 1UBQ is shown in Figure 4. Orange backbone corresponds to amides that are buried deeply enough to be outside NOE detection distance from solvent. No NOEs to water involving these amide hydrogens were seen. Regions of the backbone that have solvent-accessible amide hydrogens but did not show cross peaks to water are colored green. As mentioned above, these sites must have only short-lived interactions with hydration water. Portions of the backbone that have NMR-detected hydration water and also have water common to both crystal structures that are within NOE distance are colored blue, as are the corresponding waters. Crystallographic waters that do not have long-lived counterparts in the NMR hydration data are colored green. Finally, yellow regions correspond to those amide hydrogens that have NMR-detected hydration water but have no counterpart in the waters common to both crystal structures. Interestingly, only ~60% of the crystallographic waters that are common to both crystal structures appear within the 4.3 Å NOE detection distance of sites where NMR-detected hydration waters are long-lived. In addition, more than half of the NMR-detected long-lived protein-water interactions involve amide hydrogens that are outside NOE distance from the nearest conserved crystallographic water molecule. Additionally, we have found no correlation between the NOE/ROE values for the NMR-detected sites of long-lived hydration and the location of the nearest conserved crystallographic water. In the case of ubiquitin, there seems to be limited correlation between the locations of the defined water molecules in the two crystal structures and the NMR-detected hydration waters of encapsulated ubiquitin. Indeed, magnetic relaxation dispersion experiments have suggested one very long-lived water molecule is present in ubiquitin, which on the basis of the crystal structure (1UBQ) was identified with a water molecule (water 28) localized near the amide NH of Leu 43 (ref. 34). Though this water is illuminated by NOEs in encapsulated ubiquitin, its residence time is short enough to cause complete chemical shift averaging with the bulk water resonance. In contrast, encapsulated ubiquitin shows a very long lived water molecule that is localized to a relatively deep surface pocket and is highlighted by short distance NOEs between the amide hydrogens of Val 69 and Lys 6 and a resonance with a chemical shift (5.4 p.p.m.) resolved from the water resonance (4.32 p.p.m.) (Supplementary Fig. 2). These amide hydrogens are not within the NOE detection distance of any hydroxyl group and we therefore identify this water, which is not resolved in the crystal structure, as the long-lived water molecule suggested by the magnetic resonance dispersion experiments. Interestingly, this water resides in a pocket that is lined with hydrophobic side chains, which may account for the lack of ordered crystallographic water at this site35.
Figure 4.
Comparison of crystallographic and solution NMR-detected hydration of human ubiquitin. Two views of the backbone ribbon of a crystallographic structure of human ubiquitin (PDB code 1UBQ) are shown. Waters that are common between the two crystal structures (1UBQ and 1UBI) are shown as spheres. The waters are colored according to whether the nearest amide hydrogen does (blue) or does not (green) show NOE and ROE correlations with the water resonance. Buried and unresolved portions of the backbone are colored orange and gray, respectively. Portions of the backbone which are solvent-exposed but do not show NOE and ROE cross peaks to water are colored green, indicating that waters residing at these sites have too short a residence time to be detected. Backbone amide hydrogens that show NOE and ROE cross peaks to water are colored blue if they are within the NOE detection distance of a conserved crystallographic water or yellow if they are not. Images were created using PyMOL39.
Protein solvation and nanoscale confinement
In recent years, there has been an increasing appreciation of the inherent complexity of the cellular context in which proteins must function36. It is now well known that cells are packed with macromolecular surfaces that confine proteins to nanometer-scale volumes. Confinement on this length scale can potentially alter a number of fundamental properties of proteins36 and creates an intracellular hydration environment that is potentially highly heterogeneous and distinct from bulk water1. Reverse micelle encapsulation provides a convenient way to undertake detailed studies of the effects of confinement on protein hydration, folding and stability and dynamics at the atomic-scale using solution NMR methods20,37,38. Here we have found a previously unseen range of hydration behavior across the surface of the protein. This represents an exciting alternate view for the ongoing effort to unravel the interplay between proteins and solvent that is so vital to living systems.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
Supplementary Material
Acknowledgments
We are grateful to K. Valentine, J. Gledhill and J. Dogan for helpful discussion and to B. Halle for comments on an early draft of this manuscript. Supported by a grant from the National Science Foundation (MCB-0842814). N.V.N. is the recipient of an NIH postdoctoral fellowship (GM 087099).
Footnotes
AUTHOR CONTRIBUTIONS
A.J.W. designed and initiated the study. M.S.P. carried out preliminary NMR experiments. N.V.N. optimized the NMR experiments, collected and analyzed the data. A.J.W. and N.V.N. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
A.J.W. declares a competing financial interest as a Member of Daedalus Innovations, LLC, a manufacturer of high pressure and reverse micelle NMR apparatus.
References
- 1.Ball P. Water as an active constituent in cell biology. Chem Rev. 2008;108:74–108. doi: 10.1021/cr068037a. [DOI] [PubMed] [Google Scholar]
- 2.Otting G, Liepinsh E, Wuthrich K. Protein hydration in aqueous solution. Science. 1991;254:974–80. doi: 10.1126/science.1948083. [DOI] [PubMed] [Google Scholar]
- 3.Otting G. NMR studies of water bound to biological molecules. Prog Nucl Magn Reson Spectrosc. 1997;31:259–285. [Google Scholar]
- 4.Bagchi B. Water dynamics in the hydration layer around proteins and micelles. Chem Rev. 2005;105:3197–3219. doi: 10.1021/cr020661+. [DOI] [PubMed] [Google Scholar]
- 5.Halle B. Protein hydration dynamics in solution: a critical survey. Philos Trans R Soc London, Ser B. 2004;359:1207–1224. doi: 10.1098/rstb.2004.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pal SK, Zewail AH. Dynamics of water in biological recognition. Chem Rev. 2004;104:2099–2123. doi: 10.1021/cr020689l. [DOI] [PubMed] [Google Scholar]
- 7.Zaccai G. The effect of water on protein dynamics. Philos Trans R Soc London, Ser B. 2004;359:1269–1275. doi: 10.1098/rstb.2004.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattea C, Qvist J, Halle B. Dynamics at the protein-water interface from O-17 spin relaxation in deeply supercooled solutions. Biophys J. 2008;95:2951–2963. doi: 10.1529/biophysj.108.135194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gruschus JM, Ferretti JA. Quantitative measurement of water diffusion lifetimes at a protein/DNA interface by NMR. J Biomol NMR. 2001;20:111–126. doi: 10.1023/a:1011266703693. [DOI] [PubMed] [Google Scholar]
- 10.Halle B. Cross-relaxation between macromolecular and solvent spins: The role of long-range dipole couplings. J Chem Phys. 2003;119:12372–12385. [Google Scholar]
- 11.Frezzato D, Rastrelli F, Bagno A. Nuclear spin relaxation driven by intermolecular dipolar interactions: the role of solute-solvent pair correlations in the modeling of spectral density functions. J Phys Chem B. 2006;110:5676–5689. doi: 10.1021/jp0560157. [DOI] [PubMed] [Google Scholar]
- 12.Jeener J, Vlassenbroek A, Broekaert P. Unified derivation of the dipolar field and relaxation terms in the Bloch-Redfield equations of liquid NMR. J Chem Phys. 1995;103:1309–1332. [Google Scholar]
- 13.Modig K, Liepinsh E, Otting G, Halle B. Dynamics of protein and peptide hydration. J Am Chem Soc. 2004;126:102–114. doi: 10.1021/ja038325d. [DOI] [PubMed] [Google Scholar]
- 14.Babu CR, Flynn PF, Wand AJ. Validation of protein structure from preparations of encapsulated proteins dissolved in low viscosity fluids. J Am Chem Soc. 2001;123:2691–2692. doi: 10.1021/ja005766d. [DOI] [PubMed] [Google Scholar]
- 15.Valentine KG, et al. Magnetic susceptibility-induced alignment of proteins in reverse micelles. J Am Chem Soc. 2006;128:15930–1. doi: 10.1021/ja061438n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wand AJ, Ehrhardt MR, Flynn PF. High-resolution NMR of encapsulated proteins dissolved in low-viscosity fluids. Proc Natl Acad Sci U S A. 1998;95:15299–15302. doi: 10.1073/pnas.95.26.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peterson RW, Lefebvre BG, Wand AJ. High-resolution NMR studies of encapsulated proteins in liquid ethane. J Am Chem Soc. 2005;127:10176–10177. doi: 10.1021/ja0526517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson KF, Gierasch LM. Conformation of a peptide solubilizate in a reversed micelle water pool. J Am Chem Soc. 1984;106:3648–52. [Google Scholar]
- 19.Babu CR, Hilser VJ, Wand AJ. Direct access to the cooperative substructure of protein and the protein ensemble via cold denaturation. Nature Struct Mol Biol. 2004;11:352–357. doi: 10.1038/nsmb739. [DOI] [PubMed] [Google Scholar]
- 20.Peterson RW, Anbalagan K, Tommos C, Wand AJ. Forced folding and structural analysis of metastable proteins. J Am Chem Soc. 2004;126:9498–9. doi: 10.1021/ja047900q. [DOI] [PubMed] [Google Scholar]
- 21.Kielec JM, Valentine KG, Babu CR, Wand AJ. Reverse micelles in integral membrane protein structural biology by solution NMR spectroscopy. Structure. 2009;17:345–351. doi: 10.1016/j.str.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Workman H, Flynn PF. Stabilization of RNA oligomers through reverse micelle encapsulation. J Am Chem Soc. 2009;131:3806–7. doi: 10.1021/ja8084753. [DOI] [PubMed] [Google Scholar]
- 23.Valentine KG, et al. Reverse micelle encapsulation of membrane-anchored proteins for solution NMR studies. Structure. 2010;18:9–16. doi: 10.1016/j.str.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piletic IR, Moilanen DE, Spry DB, Levinger NE, Fayer MD. Testing the core/shell model of nanoconfined water in reverse micelles using linear and nonlinear IR spectroscopy. J Phys Chem A. 2006;110:4985–99. doi: 10.1021/jp061065c. [DOI] [PubMed] [Google Scholar]
- 25.Park S, Moilanen DE, Fayer MD. Water dynamics - The effects of ions and nanoconfinement. J Phys Chem B. 2008;112:5279–5290. doi: 10.1021/jp7121856. [DOI] [PubMed] [Google Scholar]
- 26.Fenn EE, Wong DB, Fayer MD. Water dynamics at neutral and ionic interfaces. Proc Natl Acad Sci U S A. 2009;106:15243–15248. doi: 10.1073/pnas.0907875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8. ANG resolution. J Mol Biol. 1987;194:531–44. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 28.Hershko A, Ciechanover A. The ubiquitin system. Ann Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 29.Marion D, Kay LE, Sparks SW, Torchia DA, Bax A. 3-dimensional heteronuclear NMR of 15N-labeled proteins. J Am Chem Soc. 1989;111:1515–1517. [Google Scholar]
- 30.Clore GM, Bax A, Wingfield PT, Gronenborn AM. Identification and localization of bound internal water in the solution structure of interleukin 1 beta by heteronuclear three-dimensional 1H rotating-frame Overhauser 15N-1H multiple quantum coherence NMR spectroscopy. Biochemistry. 1990;29:5671–6. doi: 10.1021/bi00476a004. [DOI] [PubMed] [Google Scholar]
- 31.Brueschweiler R, Wright PE. Water self-diffusion model for protein-water NMR cross relaxation. Chem Phys Lett. 1994;229:75–81. [Google Scholar]
- 32.Nakasako M. Water-protein interactions from high-resolution protein crystallography. Philos Trans R Soc London, Ser B. 2004;359:1191–1206. doi: 10.1098/rstb.2004.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexeev D, et al. Synthetic, structural and biological studies of the ubiquitin system: chemically synthesized and native ubiquitin fold into identical three-dimensional structures. Biochem J. 1994;299:159–63. doi: 10.1042/bj2990159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Persson E, Halle B. Nanosecond to microsecond protein dynamics probed by magnetic relaxation dispersion of buried water molecules. J Am Chem Soc. 2008;130:1774–1787. doi: 10.1021/ja0775873. [DOI] [PubMed] [Google Scholar]
- 35.Ernst JA, Clubb RT, Zhou HX, Gronenborn AM, Clore GM. Demonstration of positionally disordered water within a protein hydrophobic cavity by NMR. Science. 1995;267:1813–7. doi: 10.1126/science.7892604. [DOI] [PubMed] [Google Scholar]
- 36.Zhou HX, Rivas G, Minton AP. Macromolecular crowding and confinement: Biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukherjee S, Chowdhury P, Gai F. Tuning the cooperativity of the helix-coil transition by aqueous reverse micelles. J Phys Chem B. 2006;110:11615–11619. doi: 10.1021/jp062362k. [DOI] [PubMed] [Google Scholar]
- 38.Simorellis AK, Flynn PF. Fast local backbone dynamics of encapsulated ubiquitin. J Am Chem Soc. 2006;128:9580–1. doi: 10.1021/ja061705p. [DOI] [PubMed] [Google Scholar]
- 39.DeLano WL. The PyMOL molecular graphics system. DeLano Scientific LLC; Palo Alto, California, USA: 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.