Abstract
The bromodomain is an ∼110 amino acid module found in histone acetyltransferases and the ATPase component of certain nucleosome remodelling complexes. We report the crystal structure at 1.9 Å resolution of the Saccharomyces cerevisiae Gcn5p bromodomain complexed with a peptide corresponding to residues 15–29 of histone H4 acetylated at the ζ-N of lysine 16. We show that this bromodomain preferentially binds to peptides containing an N-acetyl lysine residue. Only residues 16–19 of the acetylated peptide interact with the bromodomain. The primary interaction is the N-acetyl lysine binding in a cleft with the specificity provided by the interaction of the amide nitrogen of a conserved asparagine with the oxygen of the acetyl carbonyl group. A network of water-mediated H-bonds with protein main chain carbonyl groups at the base of the cleft contributes to the binding. Additional side chain binding occurs on a shallow depression that is hydrophobic at one end and can accommodate charge interactions at the other. These findings suggest that the Gcn5p bromodomain may discriminate between different acetylated lysine residues depending on the context in which they are displayed.
Keywords: acetylated histone H4/bromodomain structure/histone acetyltransferase Gcn5p/recognition/Saccharomyces cerevisiae
Introduction
The recognition of specific modifications in the N- and C-terminal tails of the nucleosomal histones has been postulated to be one of the key interactions determining the transcriptional competence of chromatin (Strahl and Allis, 2000). One such modification is the acetylation of particular lysine residues in the N-terminal regions of histones H3 and H4. This type of modification has long been correlated with the activation of transcription (reviewed by Turner, 1991; Grunstein, 1997; W.L.Cheung et al., 2000; Kouzarides, 2000; Sterner and Berger, 2000). Histone acetylation is mediated by acetyltransferases including Gcn5p (Brownell et al., 1996), p300/CBP (Bannister and Kouzarides, 1996; Ogryzko et al., 1996), P/CAF (Yang et al., 1996) and TAFII250 (Mizzen et al., 1996). The selectivity of these enzymes depends on both their substrates and their environment within the cell. Free Gcn5p preferentially acetylates Lys14 of free histone H3 and to a lesser extent Lys8 and Lys16 of free histone H4 (Kuo et al., 1996). However, Gcn5p by itself is unable to modify nucleosomal histones (Kuo et al., 1996). Such an ability is conferred by association with other proteins in large macromolecular histone acetylation complexes such as the ADA and SAGA complexes (Grant et al., 1997). In these complexes the pattern of lysine modification by Gcn5p is altered (Grant et al., 1997).
In addition to its histone acetyltransferase (HAT) domain, Gcn5p contains a C-terminal domain that is also found both in other HATs, including P/CAF, p300/CBP and TAFII250, and in the ATPase subunit of certain chromatin remodelling complexes, including RSC, SWI/SNF and their homologues (Haynes et al., 1992; Jeanmougin et al., 1997). This conserved 110 amino acid module is termed the bromodomain and was originally identified as a sequence motif common to the Drosophila brahma and female–sterile homeotic proteins, the yeast SWI2/SNF2 proteins and the human CCG1 protein (Tamkun et al., 1992). The name is derived from brahma by analogy to the chromodomain. Several lines of evidence suggest that the bromodomain may be directly involved in chromatin remodelling, particularly that associated with transcriptional activation. In the absence of Gcn5p, the nuclease-sensitive region of the HIS3 promoter is invaded by nucleosomes and the gene is repressed (Filetici et al., 1998). Efficient activation of HIS3 gene transcription specifically requires the bromodomain of Gcn5p in addition to the HAT domain (Marcus et al., 1994), although the former module is not required for the interaction of Gcn5p with ADA2, a component of the ADA complex. Similarly, Syntichaki et al. (2000) demonstrated that the Gcn5p bromodomain was not required in vivo for Gcn5p-mediated histone acetylation at a synthetic promoter, but was necessary for subsequent Swi2p-dependent nucleosome remodelling and consequent transcriptional activation. At the yeast PHO5 and PHO8 promoters, loss of Gcn5p HAT activity prevents the complete chromatin remodelling associated with sub-maximal and maximal activation, respectively (Gregory et al., 1998, 1999), while no remodelling is observed at PHO8p in the absence of active Swi2 (Gregory et al., 1999). At the HO promoter, the Swi2p-containing SWI–SNF remodelling complex recruits the SAGA complex (Cosma et al., 1999; Krebs et al., 1999) prior to activation. Taken together these observations suggest a model which proposes that transcriptional activation at certain promoters requires the concerted action of both the SAGA and SWI–SNF complexes, both of which contain bromodomains.
This correlation of bromodomain function with chromatin remodelling suggested that this module might target different activities to nucleosomes (Winston and Allis, 1999). Consistent with this finding, three binding studies showed that the bromodomain interacted with the N-terminal regions of histones H3 and H4. However, while Ornaghi et al. (1999) demonstrated that the Gcn5p bromodomain bound specific unmodified sequences of histone H4, both Dhalluin et al. (1999) and Jacobson et al. (2000) showed that the P/CAF bromodomain and the TAFII250 dibromodomain, respectively, exhibited a strong preference for acetylated histone tails. The solution structure of the P/CAF bromodomain (Dhalluin et al., 1999) and the crystal structure of the TAFII250 dibromodomain (Jacobson et al., 2000) revealed that the bromodomain is a conserved four-helix bundle with a pronounced cleft between two loops. Dhalluin et al. (1999) proposed, on the basis of interactions with analogues to N-acetyl lysine, that this cleft was the primary recognition site for the acetylated histone tails, although N-acetyl lysine by itself did not bind to the P/CAF bromodomain.
In this paper we report the crystal structure of a complex between an acetylated peptide from histone H4 and the Gcn5p bromodomain at a resolution of 1.9 Å. We show that the binding site for N-acetyl lysine is a deep cleft containing a number of conserved residues, including at its mouth three tyrosine residues proposed to be involved in histone tail binding from NMR studies (Dhalluin et al., 1999). The structure explains the molecular basis for the selective binding of this modified amino acid. We also show that additional interactions between the peptide and the domain can explain the binding of unmodified histone tails and may provide a means for discriminating between different sequences adjacent to N-acetylated lysines.
Results and discussion
Binding of a peptide from histone H4 depends on N-acetylation of lysine
To test the binding selectivity of the Gcn5p bromodomain we used a 15 amino acid peptide corresponding to residues 15–29 of histone H4 (Figure 1). This sequence was chosen because it contained a lysine residue (Lys16) that can be acetylated in vivo and also because when unacetylated it interacts preferentially with the Gcn5p bromodomain in vitro (Ornaghi et al., 1999).
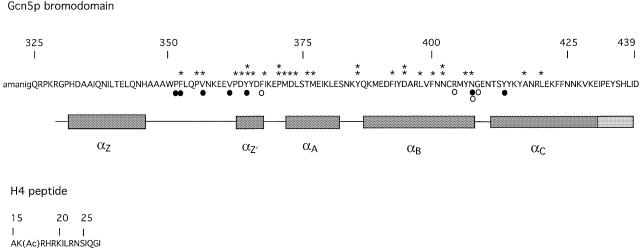
Fig. 1. Sequence of the Gcn5p bromodomain used for crystallization and for NMR spectroscopy. Highly and absolutely conserved residues in the bromodomain are marked by one and two asterisks, respectively. Amino acids encoded by the vector are in lower case. Residues directly contacting the N-acetyl lysine in the peptide are indicated by closed circles and residues contacting other parts of the peptide by open circles. The nomenclature for the secondary structure follows that of Dhalluin et al. (1999). αZ′ is a short α-helix that is present in all bromodomain structures but was previously unnamed. α- and 310-helices are indicated by dark grey and light grey boxes, respectively.
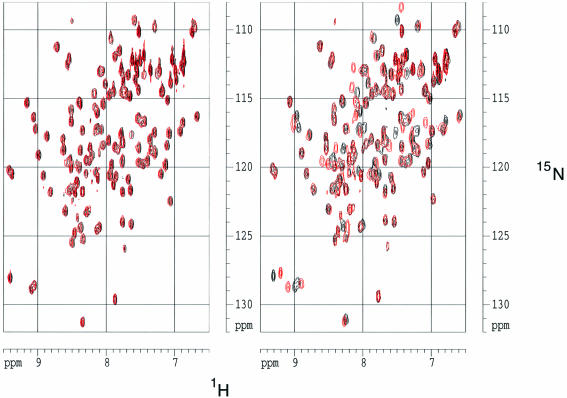
We acquired and assigned the HSQC spectrum of the free bromodomain and then titrated the protein with acetylated and non-acetylated versions of the peptide. The acetylated, but not the non-acetylated, peptide induced a number of distinct chemical shift changes in the resonances of backbone amide groups of the bromodomain (Figure 2). These shift differences gradually increased throughout the titration, which was taken to a maximum acetylated peptide:bromodomain ratio of 5:1. We conclude from these data that the bromodomain has a strong preference for the acetylated, relative to the non-acetylated, peptide. The data also show that the peptide and the bromodomain are in fast exchange between free and bound states on the NMR chemical shift timescale.
Fig. 2. The Gcn5p bromodomain binds specifically to an H4 peptide containing N-acetyl lysine. Superimposed (15N, 1H) HSQC spectra of free bromodomain and bromodomain plus unacetylated peptide (left) and free bromodomain and bromodomain plus acetylated peptide (right). In both cases, the spectrum for the free bromodomain is shown in black and that complexed with the peptide in red.
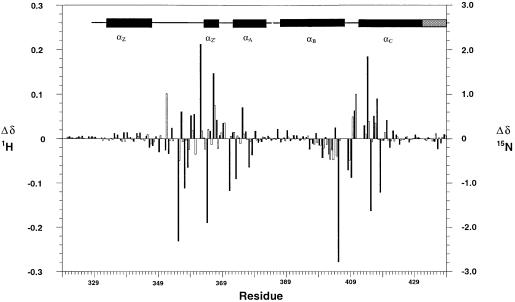
Signals for 109 and 108 backbone amide groups were assigned for the free protein and complex, respectively. There are 114 non-proline residues present in the domain, and the two N-terminal residues gave no backbone amide signals. Other absences were the NH signals of residues Asn407, Gly408 and Ser412 in the free protein and additionally of Tyr413 in the complex, probably reflecting the mobility of the BC loop [in the nomenclature of Dhalluin et al. (1999)]. The residues exhibiting changed chemical shifts on addition of the peptide were concentrated in two regions of sequence particularly including loops ZA and BC and the α-helical regions immediately flanking the BC loop (Figure 3), both spatially located at one end of the molecule (Figure 4D). The number of changed chemical shifts observed in the presence of the H4 15–29 peptide, acetylated at position 16, although generally congruent with those observed by Dhalluin et al. (1999), on addition of an H4 peptide corresponding to residues 1–12 in which Lys8 was acetylated, was significantly greater, suggesting that the H4 15–29 peptide is involved in more extensive interactions. This inference is consistent with the binding studies of Ornaghi et al. (1999), who observed that the Gcn5p bound the unacetylated H4 peptide containing residues 16–34, but not that containing residues 1–16.
Fig. 3. Histogram of chemical shift changes induced on addition of H4 peptide acetylated at Lys16. 1H and 15N changes are denoted by filled and open bars, respectively.
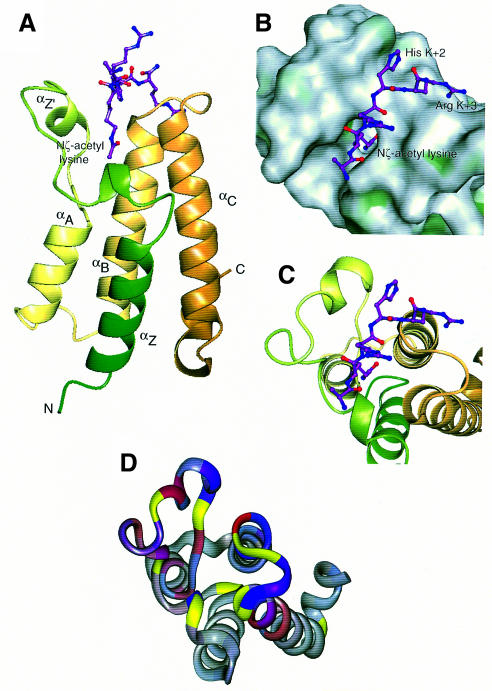
Fig. 4. Overall structure and peptide binding. (A) Ribbon diagram showing the bundle of four main α-helices, Z, A, B and C, with the Nζ-acetyl lysine side chain of the peptide bound in a deep slot at the top of the bundle. The ribbon is coloured from green (N-terminus) to gold (C-terminus). (B) ‘Top’ view of the molecular surface coloured by hydrophobic potential (dark green marks favourable regions for hydrophobic interaction; grey, less favourable; light green, intermediate). The five visible residues of the peptide fit into a shallow groove, with the acetyl lysine side chain in a deep slot. The hydrophobic surface at the base of the pocket formed by Phe352 and Val356, as well as that lining the side of the pocket formed by Pro351, Val 356, Tyr364 and Tyr413, are not easily visible in this view. (C) The same view as (B), showing the underlying helices coloured as in (A). (D) Mapping of backbone NH chemical shift changes onto residues 329–438 of the peptide backbone of the Gcn5p bromodomain in the same orientation as (B) and (C). 1H and 15N changes are indicated in red and blue, respectively. The colours are merged so that the shade reflects the relative contributions of the two shift changes and the intensity reflects the magnitude of the changes. Residues for which no backbone amide groups were assigned are shown in yellow (see text).
Crystal structure of a Gcn5p bromodomain–acetylated H4 peptide complex
The crystal structure of the Gcn5p bromodomain–H4 peptide complex was solved at a resolution of 1.9 Å using phasing from a single mercury derivative. No electron density was found for the first 10 N-terminal residues and the C-terminal residue of the bromodomain construct, and also for the C-terminal 10 residues of the peptide corresponding to histone H4 positions 20–29.
The structure of the bromodomain from Gcn5p was, as expected from previous structures solved (Dhalluin et al., 1999; Jacobson et al., 2000), a four-helical bundle (Figure 4A). A number of conserved, mainly hydrophobic residues, but also some hydrophilic ones including Asn402, are involved in forming and stabilizing the hydrophobic core of the domain. A surface representation shows a narrow cleft with a cavity at the bottom formed by the ZA and BC loops (Figure 4B). Hydrophobic surface potential mapping showed the sides to be hydrophobic in nature, indicating its suitability as a site of hydrophobic contact mediating protein–peptide interactions.
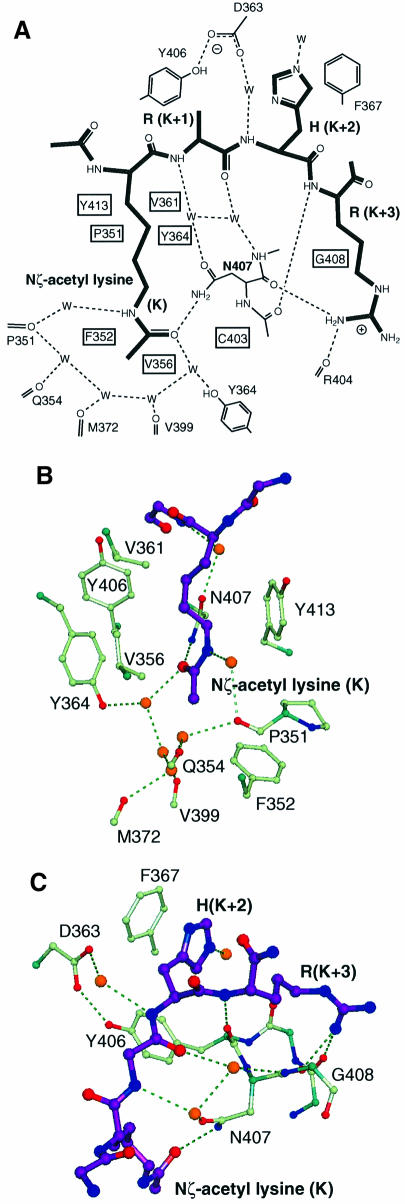
The structure was determined in a complex with a peptide corresponding to residues 15–29 of histone H4. The peptide binds in an extended conformation across the patch of high hydrophobic surface potential with specificity being provided by the interactions of N-acetyl lysine at H4 position 16 and the side chains at positions 18 and 19 (Figures 4B, C and 5A). This ‘peptide-surface’ or ‘pronged plug in socket’ type of interaction between a small folded domain and a linear peptide is common. Such interactions have been described between SH2 or PTB domains and phosphotyrosine-containing peptides (Eck et al., 1993, 1996; Waksman et al., 1993; Zhou et al., 1995), PDZ domains with C-terminal xS/TxV sequences (Doyle et al., 1996), µ2 subunit of AP2 with YxxΦ internalization motifs (Owen and Evans, 1998) and the clathrin N-terminal β-propeller with the LΦxΦD/E clathrin box motifs (ter Haar et al., 2000). The peptide is spatially complementary with the groove in which it lies (Figure 4B), but its backbone makes only one direct H-bond with the protein surface.
The major binding determinant is the acetylated lysine. This sits in the deep cleft, which is accessible to solvent on one side (Figure 4B). The walls of the cleft, which pack against the aliphatic part of the N-acetyl lysine side chain, are composed of Val361 and Tyr364 on one side and Pro351 and Tyr413 on the other. This accounts for its high hydrophobic potential. The interaction that plays the major role in orientating the N-acetyl group such that its carbonyl points towards the hydrophobic interior of the protein is a H-bond formed between the amide nitrogen of Asn407 and the oxygen of the acetyl carbonyl group (Figures 5B, C and 6). The carbonyl group and amide nitrogen from the N-acetyl group also form H-bonds to a number of well-ordered water molecules, which participate in a network of H-bonds between each other and carbonyl groups and side chains of the protein, in particular the carbonyl group of Pro351 and the hydroxyl group of Tyr364 (Figures 5A and 6). The methyl of the N-acetyl group is surrounded at the base of the pocket by the exposed hydrophobic surfaces of Phe352 and Val356. An unacetylated lysine residue that is positively charged would have no compensatory negative charge to interact within this hydrophobic environment and consequently its binding would be considerably less energetically favourable. This would explain both the failure to observe chemical shifts on addition of the unacetylated peptide at a concentration of 5 mM (Figure 2) and also the >20-fold weaker binding measured for the binding of an unacetylated as opposed to an acetylated lysine-containing peptide to the TAFII250 dibromodomain (Jacobson et al., 2000).
Fig. 5. Details of the peptide binding site. (A) Schematic view of interactions, with water molecules represented as W. (B) The N-acetyl lysine slot showing the ring of water molecules around the acetyl group at the base of the slot, and the hydrophobic walls left and right. (C) The binding groove for the (K + 2) and (K + 3) peptide residues lies across the 405–408 loop between αB and αC. His(K + 2) packs against Phe367, and Arg(K + 3) forms hydrogen bonds back to the protein backbone. The peptide backbone forms four hydrogen bonds to the protein, three of them via water molecules.

Fig. 6. The Nζ-acetyl lysine binding site, showing electron density from the final 2mFo – DFc map. Nitrogen atoms are black and oxygen atoms are grey. The acetyl group is surrounded by a ring of water molecules (grey balls) at the base of the slot, and the carbonyl oxygen of the acetyl forms a hydrogen bond to the side chain of Asn407. An unacetylated lysine could not form these hydrogen bonds, and would introduce an unpaired charge into the slot.
Specificity for the sequence surrounding the N-acetyl lysine comes from the binding of side chains at the K + 2 and K + 3 positions, where K defines the position of the acetylated lysine. The side chains at K – 1 and K + 1 make no contacts with the protein, while no density can be seen for any of the 10 residues following K + 3 and these are therefore not likely to be involved in binding to the bromodomain. In this peptide, the K + 2 residue is a histidine which sits in a shallow hydrophobic pocket formed by the side chains of Tyr406 and Phe367 (Figures 4B and 5C). A hydrophobic residue or an arginine at this position would also be expected to form an energetically beneficial interaction and indeed there are other examples, shown in Table I, where a hydrophobic residue, in particular leucine or proline, is found in a corresponding position relative to other acetylatable lysine residues. The K + 3 residue is an arginine and interacts with the protein via two H-bonds to the backbone carbonyl groups of Arg404 and Asn407 (Figure 5C). It is interesting to note that 5 Å away from the guanidinium group of the K + 3 side chain is a glutamate side chain (Glu409). If this were able to form an ionic interaction in vivo with the K + 3 arginine, it would increase the specificity for an arginine residue at this position. However, in the crystal structure there is a crystal contact at this point, in which the side chain of Asn345 from an adjacent molecule interacts with the K + 3 arginine. This would prevent the Glu409 side chain from interacting with the K + 3 arginine. Since it cannot form this interaction in the crystal, this glutamic acid residue is relatively mobile and has poor electron density.
Table I. Sequence context of acetylatable lysines in histones H3 and H4 from S.cerevisiae.
| Histone | Acetylatable lysine | Sequence |
|---|---|---|
| H3 | 9 | RKSTG |
| H3 | 14 | GKAPR |
| H3 | 18 | RKQLA |
| H3 | 23 | SKAAR |
| H4 | 5 | GKGGK |
| H4 | 8 | GKGLG |
| H4 | 12 | GKGGA |
| H4 | 16 | AKRHR |
The table shows the sequences from K – 1 to K + 3, where K is the acetylatable lysine. Residues that could potentially make secondary interactions with the Gcn5p bromodomain are shown in bold.
The identification of two interaction sites reconciles previous apparently disparate observations. Dhalluin et al. (1999) and Jacobson et al. (2000) reported that the bromodomain of P/CAF and the dibromodomain of TAF250, respectively, preferentially bound peptides from histones H3 and H4 that contain N-acetylated lysine residues. In contrast, Ornaghi et al. (1999) found that the bromodomain of Gcn5p selectively bound certain sequences from the N-terminal region of histone H4 when challenged with sequences in which the lysine residues were unmodified. The most likely explanation for these observations is that both Dhalluin et al. (1999) and Jacobson et al. (2000) measured the effect of the primary interaction of N-acetyl lysine with its binding cleft, while the results of Ornaghi et al. (1999) were dependent only on the secondary interaction. This interpretation is consistent with the observation that the strongest binding sequence selected by Ornaghi et al. (1999) contains H4 residues His18 and Arg19, which interact well with the secondary site. Arginine residues, which as discussed above could interact particularly favourably with the secondary site, were important for this binding. By comparison, the sequences following the other acetylatable lysines in histone H4 (Lys5, Lys8, Lys12, as well as Lys9, Lys18 and Lys23 in histone H3; Table II) would be expected to interact less favourably with the secondary site. This suggests that the Gcn5p bromodomain may discriminate between different acetylated lysines, depending on the sequence context in which they are found. Interestingly, the sequences following two of the three preferred substrates for free Gcn5p (Kuo et al., 1996), H4 Lys16 and H3 Lys14, are also those we would predict to interact most strongly with the Gcn5p bromodomain at the K + 2 and K + 3 positions. The hydrophobic part of the secondary binding site (see Figure 4B) is well conserved in other bromodomains, but the region interacting with Arg19 shows more variation, both in charge and hydrophobicity, raising the possibility that different bromodomains might target different acetylated lysine residues as a result of the interaction of the peptide K + 3 residue at this site.
Table II. Statistics on data collection and phasing.
| Native | EMTS | |
|---|---|---|
| Data collectiona | ||
| resolution (Å) (outer bin) | 19–1.87 (1.97) | 19–1.85 (1.95) |
| Rmergeb | 0.073 (0.204) | 0.042 (0.283) |
| Rmeasc | 0.079 (0.220) | 0.052 (0.336) |
| <<I>/σ(<I>)> | 24.5 (8.7) | 29.4 (6.7) |
| completeness (%) | 99.8 (99.5) | 99.1 (95.2) |
| multiplicity | 7.4 (7.0) | 6.7 (5.9) |
| Wilson plot B (Å2) | 19 | 22 |
| MIR phasing | ||
| no. of sites | 1 | |
| Rderivd | 0.078 | |
| Rcullise | 0.92 | |
| phasing power: isomorphous (anomalous)f | 0.73 (0.91) | |
| mean figure of merit | 0.23 | |
| figure of merit after solvent flattening (all data) | 0.80 | |
| Refinement | ||
| R (Rfree)g | 0.185 (0.204) | |
| <B> (Å2) | 20 | |
| Nreflections (Nfree) | 11 944 (573) | |
| Natoms (Nwater) | 1090 (109) | |
| r.m.s.d. bond length (Å) | 0.013 | |
| r.m.s.d. bond angle (°) | 1.3 | |
| no. of Ramachandran violations | 0 | |
aValues in parentheses apply to the high resolution shell.
bRmerge = ΣΣi |Ih – Ihi|/ΣΣi Ih, where Ih is the mean intensity for reflection hi.
cRmeas = Σ √(n/n – 1)Σi |Ih – Ihi|/ΣΣi Ih, the multiplicity weighted Rmerge (Diederichs and Karplus, 1997).
dRderiv = Σ|FPH – FP|/ΣFP
eRcullis = Σ||FPH – FP| – |FHcalc||/Σ|FPH – FP|
fPhasing power = <|FHcalc|/phase-integrated lack of closure>.
gR = Σ|FP – Fcalc|/ΣFP
The asparagine residue corresponding to Asn407 is highly, but not absolutely, conserved in the bromodomain sequences reported so far, but is present in all GCN5 homologues. Where this residue is present we would expect that the interactions in the primary N-acetyl lysine binding site observed in this structure would also be conserved and, therefore, that the specificity for N-acetyl lysine binding should be maintained. So far, there are five examples of sequences homologous to the Gcn5p bromodomain in which the residue corresponding to Asn407 is different: in three of these cases, this residue is a serine, in the other two, a tyrosine. It should be noted that these are both residues that could potentially form H-bonds from their side chains to the N-acetyl lysine carbonyl group. However, the binding of N-acetyl lysine in such a non-asparagine pocket would be of a different strength to that of a pocket containing asparagine. Three of these deviations occur in open reading frames with more than one bromodomain, while the latter two, SP-140 and LYSP-100, are both components of distinct subnuclear structures (Bloch et al., 1996; Dent et al., 1996). These latter two also lack part of the highly hydrophobic lining to the cleft and could thus potentially recognize a different amino acid. We note, however, that neither of these domains has yet been shown functionally or structurally to be a true bromodomain.
In addition to binding to the bromodomain of Gcn5p, the N-terminal regions of the histone tails also bind to the HAT domain. Recent crystal structures of the HAT domain include those of Saccharomyces cerevisiae Gcn5p (Trievel et al., 1999), the related Hat1p (Dutnall et al., 1998), and a complex between the Tetrahymena Gcn5p HAT domain and a histone H3 peptide (Rojas et al., 1999). In these examples, the proposed (Dutnall et al., 1998) and observed (Rojas et al., 1999) interaction of the peptide is considerably more extensive than that with the bromodomain, involving ∼10 amino acids bound in an extended cleft. There are many contacts with the main chain of the peptide, but interestingly, as for the Gcn5p bromodomain, residues corresponding to K + 2 and K + 3 in the H4 peptide make important contacts. In particular, in the histone H3 sequence 14-KGPR-17, Pro16 binds to a hydrophobic surface comprising a phenylalanine and an alanine residue, while Arg17 also makes hydrophobic contacts with a proline residue. The primary acetylation target of Gcn5p is nucleosomal histone H3 rather than histone H4 (Grant et al., 1997). The acetylation of Lys14 of H3 is also correlated with phosphorylation of Ser10 (P.Cheung et al., 2000; Clayton et al., 2000; Lo et al., 2000). It is an open question as to whether this additional modification influences or is influenced by the binding of the bromodomain.
The recognition of N-acetyl lysine by the Gcn5p bromodomain differs from that of ω-acetyl histamine by the P/CAF bromodomain in two important respects. In the P/CAF–ω-acetyl histamine complex, intermolecular NOEs are observed between the side chain of Ala757 and the methyl group of the acetyl group in the amino acid analogue (Dhalluin et al., 1999), implying that these groups are separated by <6 Å. In contrast, the distances between the methyl groups of the corresponding Val361 in Gcn5p and the methyl group of N-acetyl lysine are 7.7 and 8.3 Å, respectively. In addition, the side chain of P/CAF Asn803 is oriented differently from that of the corresponding Asn407 in Gcn5p, such that the amide group of Asn803 is unlikely to form a hydrogen bond with the acetyl group of ω-acetyl histamine. Taken together, these differences suggest that the specificity for N-acetyl lysine involves the active recognition of the acetyl group by Asn407, whereas that for ω-acetyl histamine may rely simply on the exclusion of the unacetylated histamine.
Jacobson et al. (2000) have recently proposed that the dibromodomain of TAFII250 could bind a diacetylated H4 tail when either Lys5 and Lys12 or Lys8 and Lys16 are acetylated. However, we find that when the structure of the Gcn5p–H4 peptide is superimposed on that of the dibromodomain, the straight-line distance between the α-carbons of the two bound N-acetyl lysine residues is 29 Å, rather than the 25 Å estimated by Jacobson et al. (2000). Assuming that there is no substantial rearrangement of the relative positions of the two bromodomains, this distance is too long to accommodate either the Lys5/Lys12 or the Lys8/Lys16 diacetylated peptide, with six and seven intervening residues, respectively. Since the path of the backbone between N-acetyl lysine binding sites cannot be straight, we estimate that the minimum number of intervening residues compatible with the binding of a diacetylated peptide is 10. The dibromodomain could thus bind a diacetylated peptide in which Lys5 and Lys16 were both acetylated without distortion of the protein, although this particular pattern of acetylation may not be physiologically significant. An alternative possibility would be for the dibromodomain to bind simultaneously to two acetylated lysines located on different histone tails.
The primary specificity of the Gcn5p bromodomain for N-acetyl lysine reinforces the argument that a major function of this domain in vivo is to facilitate the coupling between histone acetylation and nucleosome remodelling (Syntichaki et al., 2000), possibly by aiding the release of the acetylated histone tails either from folded nucleosomes or from nucleosome core particles. We note that Arg19 of histone H4 is in close proximity to the bound DNA in the core nucleosome particle (Luger et al., 1997) and binding by the bromodomain could in principle break this contact. An inefficient release of tails from folded oligonucleosomes, and hence incomplete unfolding, could explain the observed reduced levels of acetylation by a SAGA complex containing a Gcn5p component lacking a functional bromodomain (Sterner et al., 1999). It also remains possible that an initial binding of histone tails by the secondary binding site of the bromodomain could facilitate acetylation. Since the bromodomain apparently interacts with a stretch of only four amino acids, it is also conceivable that it could bind proteins other than histones that contain acetylated lysine residues followed by an appropriate sequence.
Materials and methods
Materials
The histone H4 peptide, acetylated at Lys16, used for crystallization was obtained from Peptide Products (Oldham, UK).
Cloning
The DNA sequence containing the bromodomain of S.cerevisiae GCN5 was cloned into pET30a immediately downstream of the encoded enterokinase cleavage site. The recombinant Gcn5p bromodomain obtained from this construct was a 121 amino acid polypeptide corresponding to residues 325–439 of Gcn5p, i.e. to the C-terminal sequence; the first six amino acids were derived from the vector (Figure 1).
Purification of the Gcn5p bromodomain
The Gcn5p bromodomain was purified from 8 l of an induced culture of BL21(DE3) containing pET30a-Gcn5pBr grown in 2× TY medium. After collection, the bacteria were frozen and resuspended in 5 ml lysis buffer (50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 1 mM 2-mercaptoethanol, 10 mM imidazole) per gram of cells. Lysozyme was added to a final concentration of 1 mg/ml and the resuspended cells were incubated on ice for 30 min. Aliquots (30 ml) of the suspended cells were then sonicated for 6× 10 s and the lysate centrifuge at 10 000 r.p.m. for 20 min. The supernatant was saved, the pellet resuspended in an equal volume of lysis buffer and again centrifuged. The supernatants were combined and added to a 50% slurry of Ni-NTA–agarose (Qiagen) (1 ml agarose slurry/6 ml lysate). After mixing for 2 h at 4°C, the mixture was centrifuged at 2000 r.p.m. for 2 min at 4°C and resuspended in an equal volume of wash buffer (50 mM Na2HPO4 pH 8.0, 300 mM NaCl, 1 mM 2-mercaptoethanol, 20 mM imidazole). The resin was then transferred to a column and washed with wash buffer until the eluate contained no more protein. The column was washed with buffer A (50 mM Na2HPO4 pH 7.5, 250 mM NaCl, 1 mM 2-mercaptoethanol) and the resin resuspended in an equal volume of the same buffer. Recombinant enterokinase (Novagen) was added to the resin at a concentration of 1 U/500 µg of protein and the mixture incubated at room temperature overnight with mixing. The mixture was then transferred to a column and the released protein allowed to flow through. Buffer A (0.5 column volumes) was added and the column voided by increasing the air pressure. The eluate was concentrated initially in a Centriprep 3 and finally in a Centricon 3. At this stage, the protein was ∼99% pure and the yield was ∼12 mg/1 l of culture.
The protein for NMR studies was purified by essentially the same procedure, but with the following modifications. For 15N-labelled protein, the culture was grown as described by Churchill et al. (1995) and for 15N,13C-labelled protein the initial culture was grown in Martek 9-CN medium. For protein for NMR studies, 2-mercaptoethanol was omitted from the buffers.
Crystallization and structure determination
For crystallization, the protein was purified further by gel filtration on Superdex S200 in buffer B (20 mM HEPES pH 7.5, 250 mM NaCl, 1 mM dithiothreitol). The protein was concentrated to ∼20 mg/ml and a peptide corresponding to residues 15–29 of histone H4 [AK(Ac)RHRKILRNSIQGI] was added to a final peptide:protein ratio of 5:1. Crystallization was carried out by hanging drop vapour diffusion at 16°C against a reservoir containing 2.2 M ammonium sulfate, 20% (v/v) glycerol, 4 mM dithiothreitol and 100 mM HEPES pH 7.5. Crystals (space group C2221 with one molecule in the asymmetric unit, unit cell 43.7 × 71.9 × 89.2 Å) were obtained over a period of 4 days with final dimensions 0.2 × 0.2 × 0.05 mm in the best cases. X-ray diffraction data (native and derivative) to 1.87 Å resolution were collected at 100 K using a CuKα rotating anode source, images recorded on a 345 mm MAR-research image plate, integrated with MOSFLM (Leslie, 1992) and scaled using CCP4 programs (CCP4, 1994). A single mercury derivative was made by soaking a crystal in cryoprotected buffer containing 1 mM ethylmercury thiosalicylate for 30 min. The single mercury site was determined from difference Pattersons, and heavy atom refinement and phasing were performed with SHARP (de la Fortelle and Bricogne, 1997), followed by solvent flipping and flattening with SOLOMON (Abrahams and Leslie, 1996) (solvent content 47%). The phases from the rather weak derivative (one Hg atom with occupancy ∼0.25) were not good, and the initial solvent-flattened map was not immediately interpretable, but it was improved and traced using the ARP/wARP procedure (Perrakis et al., 1999), which automatically built the model for 93% of the structure. The model was refined with REFMAC5 (Murshudov et al., 1997), and rebuilding and water addition were performed with O (Jones et al., 1991). The final model contains residues 329–438 of the protein (Figure 1), the first five residues of the peptide [AK(Ac)RHR] and 109 water molecules.
The coordinates and structure factors have been deposited in the Protein Data Bank, accession code 1E6I. Hydrophobic interaction potentials were calculated as in Owen et al. (1999) and displayed using Aesop (M.Noble, unpublished).
NMR spectroscopy
Samples of the free protein for NMR spectroscopy contained 0.5–2.0 mM 15N-labelled protein or 15N,13C-labelled protein, 250 mM NaCl, 50 mM sodium phosphate pH 7.5 in 10% 2H2O. Samples of the complex were prepared by adding peptide and following the relative intensity of the 1H signals from the protein and the peptide until a ratio of ∼5:1 peptide:protein was reached.
NMR spectra were recorded on Bruker DMX 600, DRX 500 and Avance 800 spectrometers, equipped with 5 mm triple resonance (1H/15N/13C) probes (600 and 800 MHz). Data were processed using the program XWIN-NMR (Bruker GmbH, Karlsruhe, Germany) and analysed using the programs XWIN-NMR and Felix (Molecular Simulations, San Diego, CA). Heteronuclear experiments included 2D 15N-HSQC (generally acquired with spectral widths of 4000 Hz for 15N and 8012.82 Hz for 1H), 3D HNCA, 3D CBCA(CO)NH, 3D CBCANH, 3D HBHA(CO)NH, 3D 15N NOESY-HSQC and 3D 13C NOESY-HSQC. All spectra were acquired in phase-sensitive mode and frequency discrimination in indirect dimensions was achieved using either TPPI, States-TPPI or echo-antiecho (15N or 13C dimensions with gradient selection). Water suppression was achieved by selective irradiation during the relaxation delay and during the mixing time in NOESY and HSQC-NOESY experiments. Backbone assignments were made using the spectra from both 15N and 13C,15N-labelled protein. 1H, 13C and 15N chemical shifts were referenced following the method described by Wishart et al. (1995), using sodium 3,3,3-trimethylsilylpropionate as internal 1H reference.
Acknowledgments
Acknowledgements
This work was partially supported by the Fondazione Cenci-Bolognetti and by MURST-Cofin University ‘La Sapienza’ 1999. P.O. thanks EMBO for a short-term fellowship. We thank Dr M.E.M.Noble for the hydrophobic potential calculation and Dr J.O.Thomas for careful reading of the manuscript. This project was initiated during the tenure of an EMBO short-term fellowship by A.A.T.
References
- Abrahams J.P. and Leslie,A.G.W. (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D, 52, 30–42. [DOI] [PubMed] [Google Scholar]
- Bannister A.J. and Kouzarides,T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. [DOI] [PubMed] [Google Scholar]
- Bloch D.B., de la Monte,S.M., Guigaouri,P., Filippov,A. and Bloch,K.D. (1996) Identification and characterization of a leukocyte-specific component of the nuclear body. J. Biol. Chem., 271, 29198–29204. [DOI] [PubMed] [Google Scholar]
- Brownell J.E., Zhou,J., Ranalli,T., Kobayashi,R., Edmonson,D.G., Roth,S.Y. and Allis,C.D. (1996) Tetrahymena histone acetyl transferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell, 84, 843–851. [DOI] [PubMed] [Google Scholar]
- Cheung P., Tanner,K.G., Cheung,W.L., Sassone-Corsi,P., Denu,J.M. and Allis,C.D. (2000) Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell, 5, 905–915. [DOI] [PubMed] [Google Scholar]
- Cheung W.L., Briggs,S.D. and Allis,C.D. (2000) Acetylation and chromosomal functions. Curr. Opin. Cell Biol., 12, 326–333. [DOI] [PubMed] [Google Scholar]
- Churchill M.E.A., Jones,D.N.M., Glaser,T., Hefner,H., Searles,M.A. and Travers,A.A. (1995) HMG-D is an architecture-specific protein that binds to DNA containing the dinucleotide TG. EMBO J., 14, 1264–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton A.L., Rose,S., Barratt,M.J. and Mahadevan,L.C. (2000) Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J., 19, 3714–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cosma M.P., Tanaka,T. and Nasmyth,K. (1999) Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell, 97, 299–311. [DOI] [PubMed] [Google Scholar]
- de la Fortelle E. and Bricogne,G. (1997). Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol., 277, 472–494. [DOI] [PubMed] [Google Scholar]
- Dent A.L., Yewdell,J., Puvion-Dutilleul,F., Koken,M.H., de The,H. and Staudt,L.M. (1996) LYSP100-associated nuclear domains (LANDs): description of a new class of subnuclear structures and their relationship to PML nuclear bodies. Blood, 88, 1423–1426. [PubMed] [Google Scholar]
- Dhalluin C., Carlson,J.E., Zeng,L., He,C., Aggarwal,A.K. and Zhou,M.M. (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature, 399, 491–496. [DOI] [PubMed] [Google Scholar]
- Diederichs K. and Karplus,P.A. (1997) Improved R-factors for diffraction data analysis in macromolecular crystallography. Nature Struct. Biol., 4, 269–275. [DOI] [PubMed] [Google Scholar]
- Doyle D.A., Lee,A., Lewis,J., Kim,E., Sheng,M. and Mackinnon,R. (1996) Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell, 85, 1067–1076. [DOI] [PubMed] [Google Scholar]
- Dutnall R.N., Tafrov,S.T., Sternglanz,R. and Ramakrishnan,V. (1998) Structure of the histone acetyltransferase Hat1: a paradigm for the GCN5-related N-acetyltransferase superfamily. Cell, 94, 427–438. [DOI] [PubMed] [Google Scholar]
- Eck M.J., Shoelson,S.E. and Harrison,S.C. (1993) Structure of the regulatory domains of the Src-family tyrosine kinase Lck. Nature, 362, 87–91. [DOI] [PubMed] [Google Scholar]
- Eck M.J., Pluskey,S., Trub,T. and Harrison,S.C. (1996) Spatial constraints on the recognition of phosphoproteins by the tandem SH2 domains of the phosphatase SH-PTP2. Nature, 379, 277–280. [DOI] [PubMed] [Google Scholar]
- Filetici P., Aranda,C., Gonzalez,A. and Ballario,P. (1998) GCN5, a yeast transcriptional co-activator, induces chromatin reconfiguration of HIS3 promoter in vivo. Biochem. Biophys. Res. Commun., 242, 84–87. [DOI] [PubMed] [Google Scholar]
- Grant P.A. et al. (1997) Yeast Gcn5p functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev., 11, 1640–1650. [DOI] [PubMed] [Google Scholar]
- Gregory P.D., Schmid,A., Zavari,M., Lui,L., Berger,S.L. and Hörz,W. (1998) Absence of Gcn5p HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast. Mol. Cell, 1, 495–505. [DOI] [PubMed] [Google Scholar]
- Gregory P.D., Schmid,A., Zavari,M., Munsterkotter,M. and Hörz,W. (1999) Chromatin remodelling at the PHO8 promoter requires SWI–SNF and SAGA at a step subsequent to activator binding. EMBO J., 18, 6407–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. (1997) Histone acetylation in chromatin structure and transcription. Nature, 389, 349–352. [DOI] [PubMed] [Google Scholar]
- Haynes S.R., Dollard,C., Winston,F., Beck,S., Trowsdale,J. and Dawid,I.B. (1992) The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res., 20, 2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson R.H., Ladurner,A.G., King,D.S. and Tjian,R. (2000) Structure and function of a human TAFII250 double bromodomain module. Science, 288, 1422–1425. [DOI] [PubMed] [Google Scholar]
- Jeanmougin F., Wurtz,J.M., Le Douarin,B., Chambon,P. and Losson,R. (1997) The bromodomain revisited. Trends Biochem. Sci., 22, 151–153. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. (2000) Acetylation: a regulatory modification to rival phosphorylation? EMBO J., 19, 1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs J.E., Kuo,M.H., Allis,C.D. and Peterson,C.L. (1999) Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev., 13, 1412–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M.B., Brownell,J.B., Sobel,R.E., Ranalli,T.A., Cook,R.G., Edmondson,D.G., Roth,S.Y. and Allis,C.D. (1996) Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature, 383, 269–272. [DOI] [PubMed] [Google Scholar]
- Leslie A.G.W. (1992) Recent changes to the MOSFLM package for processing film and image plate data. In Joint CCP4 and ESF-EACMB Newsletter on Protein Crystallography, No. 26. Daresbury Laboratory, Warrington, UK. [Google Scholar]
- Lo W.S., Trievel,R.C., Rojas,J.R., Duggan,L., Hsu,J.Y., Allis,C.D., Marmorstein,R. and Berger,S.L. (2000) Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell, 5, 917–926. [DOI] [PubMed] [Google Scholar]
- Luger K., Mader,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- Marcus G.A., Silverman,N., Berger,S.L., Horiuchi,J. and Guarente,L. (1994) Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J., 13, 4807–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzen C.A. et al. (1996) The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell, 87, 1261–1270. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin,A.A. and Dodson,E.J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D, 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Ogryzko V.V., Schiltz,R.L., Russanova,V., Howard,B.H. and Nakatani,Y. (1996) The transcriptional co-activators p300 and CBP are histone acetyltransferases. Cell, 87, 953–959. [DOI] [PubMed] [Google Scholar]
- Ornaghi P., Ballario,P., Lena,A.M., Gonzalez,A. and Filetici,P. (1999) The bromodomain of Gcn5p interacts in vitro with specific residues in the N-terminus of histone H4. J. Mol. Biol., 287, 1–7. [DOI] [PubMed] [Google Scholar]
- Owen D.J. and Evans,P.R. (1998) A structural explanation for the recognition of tyrosine-based endocytotic signals. Science, 282, 1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D.J., Vallis,Y., Noble,M.E., Hunter,J.B., Dafforn,T.R., Evans,P.R. and McMahon,H.T. (1999) A structural explanation for the binding of multiple ligands by the α-adaptin appendage domain. Cell, 97, 805–815. [DOI] [PubMed] [Google Scholar]
- Perrakis A., Morris,R. and Lamzin,V.S. (1999) Automated protein model building combined with iterative structure refinement. Nature Struct. Biol., 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Rojas J.R., Trievel,R.C., Zhou,J., Mo,Y., Li,X., Berger,S.L., Allis,C.D. and Marmorstein,R. (1999) Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature, 401, 93–98. [DOI] [PubMed] [Google Scholar]
- Sterner D.E. and Berger,S.L. (2000) Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev., 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner D.E., Grant,P.A., Roberts,S.M., Duggan,L.J., Belotserkovskaya,R., Pacella,L.A., Winston,F., Workman,J.L. and Berger,S.L. (1999) Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation and TATA-binding protein interaction. Mol. Cell. Biol., 19, 86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Syntichaki P., Topalidou,I. and Thireos,G. (2000) The Gcn5p bromodomain co-ordinates nucleosome remodelling. Nature, 404, 414–417. [DOI] [PubMed] [Google Scholar]
- Tamkun J.W., Deuring,R., Scott,M.P., Kissinger,M., Pattatucci,A.M., Kaufman,T.C. and Kennison,J.A. (1992) brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell, 68, 561–572. [DOI] [PubMed] [Google Scholar]
- ter Haar E., Harrison,S.C. and Kirchhausen,T.K. (2000) Peptide-in-groove interactions link target proteins to the β-propeller of clathrin. Proc. Natl Acad. Sci. USA, 97, 1096–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trievel R.C., Rojas,J.R., Sterner,D.E., Venkataramani,R.N., Wang,L., Zhou,J., Allis,C.D., Berger,S.L. and Marmorstein,R. (1999) Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional co-activator. Proc. Natl Acad. Sci. USA, 96, 8931–8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B.M. (1991) Histone acetylation and control of gene expression. J. Cell Sci., 99, 13–20. [DOI] [PubMed] [Google Scholar]
- Waksman G., Shoelson,S.E., Pant,N., Cowburn,D. and Kuriyan,J. (1993) Binding of a high affinity phosphotyrosyl peptide to the Src SH2 domain: crystal structures of the complexed and peptide-free forms. Cell, 72, 779–790. [DOI] [PubMed] [Google Scholar]
- Winston F. and Allis,C.D. (1999) The bromodomain: a chromatin-targeting module? Nature Struct. Biol., 6, 601–604. [DOI] [PubMed] [Google Scholar]
- Wishart D.S., Bigam,C.G., Yao,J., Abildgaard,F., Dyson,H.J., Oldfield,E., Markley,J.L. and Sykes,B.D. (1995) 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR, 6, 135–141. [DOI] [PubMed] [Google Scholar]
- Yang X.J., Ogryzko,V.V., Nishikawa,J., Howard,B.H. and Nakatani,Y.A. (1996) p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature, 382, 319–324. [DOI] [PubMed] [Google Scholar]
- Zhou M.-M. et al. (1995) Structure and ligand recognition of the phosphotyrosine binding domain of Shc. Nature, 378, 584–592. [DOI] [PubMed] [Google Scholar]