Abstract
The G-protein coupled receptor (GPCR) second extracellular loop (E2) is known to play an important role in receptor structure and function. The brain cannabinoid (CB1) receptor is unique in that it lacks the inter-loop E2 disulfide linkage to the transmembrane (TM) helical bundle, a characteristic of many GPCRs. Recent mutation studies of the CB1 receptor, however, suggest the presence of an alternative intra-loop disulfide bond between two E2 Cys residues. Considering the oxidation state of these Cys residues, we determine the molecular structures of the 17-residue E2 in the dithiol form (E2dithiol) and in the disulfide form (E2disulfide) of the CB1 receptor in a fully hydrated 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayer, employing a combination of simulated annealing (SA) and molecular dynamics (MD) simulation approaches. We characterize the CB1 receptor models with these two E2 forms, CB1(E2dithiol) and CB1(E2disulfide), by analyzing interaction energy, contact number, core crevice and cross-correlation. The results show that the distinct E2 structures interact differently with the TM helical bundle and uniquely modify the TM helical topology, suggesting that E2 plays a critical role in stabilizing receptor structure, regulating ligand binding, and ultimately modulating receptor activation. Further studies on the role of E2 of the CB1 receptor are warranted; particularly comparisons of the ligand-bound form with the present ligand-free form.
Keywords: G-protein coupled receptor (GPCR), second extracellular loop (E2), simulated annealing (SA), molecular dynamics (MD), transmembrane (TM), helical topology, toggle switch
INTRODUCTION
Brain cannabinoid (CB1) receptors are G-protein coupled receptors (GPCRs)1 and belong to the rhodopsin-like subfamily.2 Members of this integral membrane protein (IMP) family are characterized by seven transmembrane (TM) helices (TMH) (H1-H7) connected by three intracellular loops (I1-I3) and three extracellular loops (E1-E3). Located in the highly polar environment of the membrane-water interfacial region of a lipid bilayer,3 the GPCR loops are not restricted by the membrane4 and are known to be important for stabilizing the multi-spanning receptor,5–7 most likely in a sequence-specific manner.8 In particular, E2, as part of the binding pocket, is known to play a central role in integrating ligand binding into receptor activation.7,9–13
Even within the rhodopsin family of GPCRs, E2 structures vary. The X-ray structure of rhodopsin14 shows that E2, containing a β-sheet lid, is deeply inserted into the receptor TM helical core and completely covers the binding pocket thereby restricting solvent access. In contrast, the X-ray structures of the β-adrenergic receptors (βARs)15,16 and the adenosine A2A receptor (AA2R)17 show that E2, containing a short α-helical segment, is displaced away from the receptor core and partially covers the binding pocket thereby allowing solvent access. Further, it has been shown that E2 can achieve distinct conformations upon agonist and antagonist binding for modulating receptor function.18,19
Several studies on the cannabinoid receptors20,21 demonstrate that E2 is important for ligand binding and receptor function. However, with little structural information available the structural determination of the 17-residue E2 of the CB1 receptor is quite challenging due to its conformational flexibility,22 location in the membrane-water interface, and conformational sensitivity to its anchoring positions at the TM helical ends. This challenge is partially ameliorated by several popular secondary prediction programs, including PSIPRED,23 JPRED324 and APSSP2,25 that correctly predict the presence of the E2 helical segment found in the X-ray structures of βARs15,16 and suggest the presence of a helical segment within E2 in the CB1 receptor (Table I).
Table I.
Secondary structure prediction of E2 of β1AR, β2AR, AA2R, and the CB1 receptor by various prediction programs
| Prediction method | Sequence and secondary structuref | |||
|---|---|---|---|---|
| β1AR | β2AR | |||
| Start | End | Start | End | |
| 181 | 204 | 173 | 196 | |
| WWRDEDPQALKCYQDPGCCDFVTN | WYRATHQEAINCYAEETCCDFFTN | |||
| PSIPREDa | ||||
| JPRED3bc | ||||
| APSSP2d | ||||
| X-raye | ||||
| AA2R | CB1 | |||
| Start | End | Start | End | |
| 143 | 173 | 255 | 271 | |
| WNNCGQPKEGKNHSQGCGEGQVACLFEDVVP | WNCEKLQSVCSDIFPHI | |||
| PSIPREDa | ||||
| JPRED3bc | ||||
| APSSP2d | ||||
| X-raye | ||||
Ref. 23.
Ref. 24.
Based on a consensus from several methods, including DSC, PHD, NNSSP, PREDATOR, ZPRED, and MULPRED.
Ref. 25.
The E2 secondary structures, assigned by DSSP,78 are taken from the X-ray β1AR,16 β2AR15 and AA2R.17
Estimated secondary structure codes: H, α-helix (shaded); E, extended (β-sheet); C, coil; and T, turn.
Of interest, the CB1 receptor is unique in that it lacks the inter-loop disulfide bond between E2 and H3, common to many GPCRs, which is known to be important for maintaining the correct receptor structure and function26–28 through the coupling between the extracellular loop and the TM helical core domains.29 The absence of the inter-loop disulfide bond in the CB1 receptor, due to the absence of the H3 Cys residue, makes E2 less inserted into the receptor core region and the TM helical bundle more closely packed thereby limiting water access to the crevice.30 Recent mutation studies of the CB1 receptor, however, suggest the presence of an alternative intra-loop disulfide linkage between two E2 Cys residues.21,31
In the present study, we determine E2 structure of the CB1 receptor by employing a combination of simulated annealing (SA) and molecular dynamics (MD) simulation approaches. We use the recently developed homology model of the CB1 receptor TM helical bundle,32 which provides the precise boundaries of the TMH, to aggressively explore the E2 conformations in detail, and the predicted E2 secondary structure (Table I) as a molecular constraint to reduce the conformational space. Considering the reduced state and the oxidized state of E2 (i.e., E2 with two Cys residues in the dithiol form (E2dithiol) and in the disulfide form (E2disulfide)) separately, we determine the CB1 receptor structures with these two E2 conformations, named CB1(E2dithiol) and CB1(E2disulfide), in a fully hydrated 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayer. The results show that the distinct E2 structures, interacted differently with the receptor bundle, uniquely modify the TM helical topology: E2dithiol, with two helical turns, isolated from TMH, which interferes less with the packing of the TM helical bundle, leading to the attachment of H5 to H3; and E2disulfide, with one helical turn and a cyclic ring enclosed by the disulfide linkage, closely packed to TMH, which interacts more with the TM helical bundle, leading to the detachment of H5 from H3. The results of the present studies suggest the critical role of E2 in stabilizing the receptor, regulating ligand binding by allowing or preventing access to the crevice, and ultimately modulating receptor function. In the present manuscript, a numbering, similar to the Ballesteros-Weinstein numbering is used33; TM helical residues and the loop position for every loop residue are indicated by superscription.
MATERIALS AND METHODS
MD simulations
For the CB1 receptor system necessary for exploring the E2 conformation, we used the recently developed homology model of the CB1 receptor in a fully hydrated POPC bilayer,32 which resulted in a system size of 80 Å × 98 Å × 98 Å. All simulations of the CB1 receptor in a POPC bilayer were performed by the NAMD simulation package (http://www.ks.uiuc.edu/Research/namd/)34 using CHARMM22 force field parameters with the CMAP correction for the ϕ/ψ angles35,36 for the protein and the TIP3 water model,37,38 and CHARMM32 force field parameters for the lipids.39 The topology definitions and the parameters for the palmitoylated Cys residue (CYP), including the parameters around the bond connecting C415 of the CB1 receptor and the carbonyl carbon of the palmitoyl moiety, as used in the literature,40 were found in the NAMD ParameterTopologyRepository site (http://www.ks.uiuc.edu/Research/namd/wiki/index.cgi?ParameterTopologyRepository). The temperature was maintained at 310 K through the use of Langevin dynamics41 with a damping coefficient of 1/ps. The pressure was maintained at 1 atm by using the Nosé-Hoover method42 with the modifications as described in the NAMD User’s Guide (http://www.ks.uiuc.edu/Research/namd/2.6/ug/). The van der Waals interactions were switched at 10 Å and zero smoothly at 12 Å. Electrostatic interactions were treated using the PME method.43 A pair list for calculating the van der Waals and electrostatic interactions was set to 13.5 Å and updated every ten steps. A multiple timestepping integration scheme, the impulse-based Verlet-I (r-RESPA) method,44 was used to efficiently compute full electrostatics. The timestep size for integration of each step of the simulation was 1 fs.
Conformational searches of the CB1 receptor E2
Two separate conformational searches of the CB1 receptor E2 in a POPC bilayer were performed: one of E2dithiol; and the other of E2disulfide. Employing an SA approach similar to the protocol by Fiser et al.45, we sampled the conformation of E2dithiol (i.e., W255E2-I271E2) by constraining the predicted consensus sequence (C257E2 – C264E2) (Table I) to be an ideal α-helix by applying torsional constraints (ϕ = −57 degrees and ψ = −47 degrees) using adaptive biasing force46 as implemented in NAMD.47 To save CPU time, a rectangular box (40 Å × 40 Å × 35 Å), large enough to accommodate any change associated with the conformational change of the targeted E2, was defined and only atoms inside the box were simulated while the atoms outside the box were held fixed. Further, the backbone atoms of E1 and E3 as well as the TM helical residues other than the side chain atoms within one helical turn from the extracellular boundaries were also held fixed. The simulation was performed in the constant volume (NVT) ensemble.
The SA cycle consisted of the following steps: energy minimization using 1,000 steps; heating to 2,510 K in 15 ps; MD simulation at 2,510 K for 20 ps; cooling down gradually to 310 K within 60 ps; MD simulation at 310 K for 20 ps; and then energy minimization using 2,000 steps. By repeating the SA cycle, a total of 166 energy-minimized E2 conformations were collected, for which a hierarchical (pairwise average linkage) cluster analysis, using MaxCluster (http://www.sbg.bio.ic.ac.uk/~maxcluster/index.html), was performed resulting in 16 clusters with a maximum inter-cluster distance of 0.7. By visual inspection, seven clusters that showed similar positions for the E2 helical segment and a cluster in which the E2 helical segment stayed far off the helical bundle were eliminated. For the remaining eight clusters, representative conformations were selected for further exploration: the number of the conformation in each cluster was 5, 4, 2, 1, 1, 1, 1, and 1, respectively, which in part reflected their relative abundance in the cluster. The number of the structure at each step of the conformational search procedure is summarized in Table II.
Table II.
The number of the structure at each step of the conformational search procedure for the determination of CB1(E2dithiol) and CB1(E2dithiol)
| i. step 1 | ii. step 2 | iii. step 3 | iv. step 4 | |||
|---|---|---|---|---|---|---|
| SA no. of conf. |
cluster analysis no. of cluster |
MD no. of conf. (time) |
further MD no. of conf. (time) |
|||
| sampled | obtained | retained | explored | explored | ||
| E2dithiol | 166 | 16 | 8 | CB1(E2dithiol) | 16a (5 ns) |
4 (A1, A2, B1 & A3) (8 – 78 ns) |
| E2disulfide | 115 | 28 | 11 | CB1(E2disulfide) | 12b (10 ns) |
4 (B2, B3, B4 & B5) (0 – 78 ns) |
A total of 16 different E2dithiol conformations in 8 clusters (5, 4, 2, 1, 1, 1, 1, and 1) were used to derive 16 different CB1(E2dithiol) models.
A total of 12 different E2disulfide conformations in 11 clusters (2, 1, 1, 1, 1, 1, 1, 1, 1, 1, and 1) were used to derive 12 different CB1(E2disulfide) models.
The CB1 receptor model32 whose E2 was replaced by each of these sixteen E2 conformations was embedded in a POPC bilayer and subjected to a 5 ns to 20 ns MD simulation in the constant pressure (NPT) ensemble without any constraint to relieve any poor geometry on the E2 anchor regions connecting H4 and H5 and to examine the stability of the E2 helical segment. Any of the resulting E2 conformations for which the helical segment became unstructured or stayed far off the helical bundle was dropped from further examination. The E2 C-terminal residues of the remaining four distinct conformations were then remodeled by ModLoop (http://modbase.compbio.ucsf.edu/modloop/modloop.html).45 The resulting structures were subjected to an additional MD simulation and the validity of the modeled E2 structures was examined by WHAT IF (http://swift.cmbi.kun.nl/whatif/).48
The conformational search protocol for E2disulfide was almost the same as for E2dithiol. For this conformational search, two E2 Cys residues were connected to form a disulfide bond. By the fold recognition program GenTHREADER (http://bioinf.cs.ucl.ac.uk/psipred/),49 a suitable fold containing the disulfide-enclosed cyclic segment (i.e., C257E2-C264E2) and the α-helical segment (C257E2 – C260E2) was identified from the X-ray structure of the human NOTCH2 (PDB code: 2OO4).50 During the SA simulation, this fold was constrained by using adaptive biasing force46 as implemented in NAMD.47 A total of 115 energy-minimized E2 conformations were collected from the SA simulations during which the disulfide-enclosed cyclic segment was constrained. These 115 conformations were then grouped into twenty eight clusters. Seventeen clusters were then eliminated due to the disulfide-enclosed cyclic segment either straying too far off of the helical bundle or occupying too similar a position. For the remaining eleven clusters, representative conformations were selected for further exploration: the number of the conformation in each cluster was 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, and 1, respectively (Table II). The CB1 receptor model32 whose E2 was replaced by each of these twelve E2 conformations was embedded in a POPC bilayer and further refined by MD simulation as described for E2dithiol.
Data analysis
The MD analysis software g_correlation (http://www.mpibpc.mpg.de/home/grubmueller/downloads/GeneralizedCorrelations/index.html)51 was used to create cross-correlation matrices from ~ 12,000 coordinates recorded every 2 ps from the last 24 ns MD simulations of CB1(E2dithiol) and CB1(E2disulfide), respectively. Both the least square fit and the non-linear generalized correlation were calculated using the Cα atoms of the receptor residues. To minimize any loss of the detail of interest from the full protein, the N-terminal end residues before H1 and the C-terminal end residues after H7 were not considered in this analysis.
Aromatic stacking was defined as any two aromatic residues (i.e., Phe, Tyr, Trp, or His) for which the distance between the two aromatic ring centroids was less than 8 Å. To determine the concentration of stacking within a range along the z-axis, the z-coordinate of one of the residues participating in the interaction was used; the remaining residue’s coordinates were not considered.
The binding core crevice diameter was calculated using the HOLE program (http://hole.biop.ox.ac.uk/hole), developed to measure the pore radius of ion channels.52 Sampling was done every 0.5 Å along the z-axis and a midpoint (i.e., c-point) between H2, H3, and H7 that was dynamically determined for every coordinate was used to define the core crevice at the extracellular region.
Circular dichroism spectroscopy of the CB1 receptor E2 peptide
E2disulfide and E2dithiol of the peptide WNCEKLQSVCSDIFPHI were custom synthesized (> 95 %) by ProImmune (Oxford, UK) and used without further purification. The peptides were dissolved in phosphate buffered saline (PBS, 10 mM phosphate, pH 7.4) at a concentration of 0.1 mg/ml (50 µM). Far UV circular dichroism (CD) spectra were collected on a JASCO J815 spectrophotometer at ambient temperature using 0.1 cm cuvette. Fifty acquisitions were made for each spectrum. Deconvolution of the recorded spectra was accomplished using the CONTIN L analysis program with reference set 3, provided by Dichroweb (http://dichroweb.cryst.bbk.ac.uk).53
RESULTS
Conformational analysis of the CB1 receptor with two distinct E2
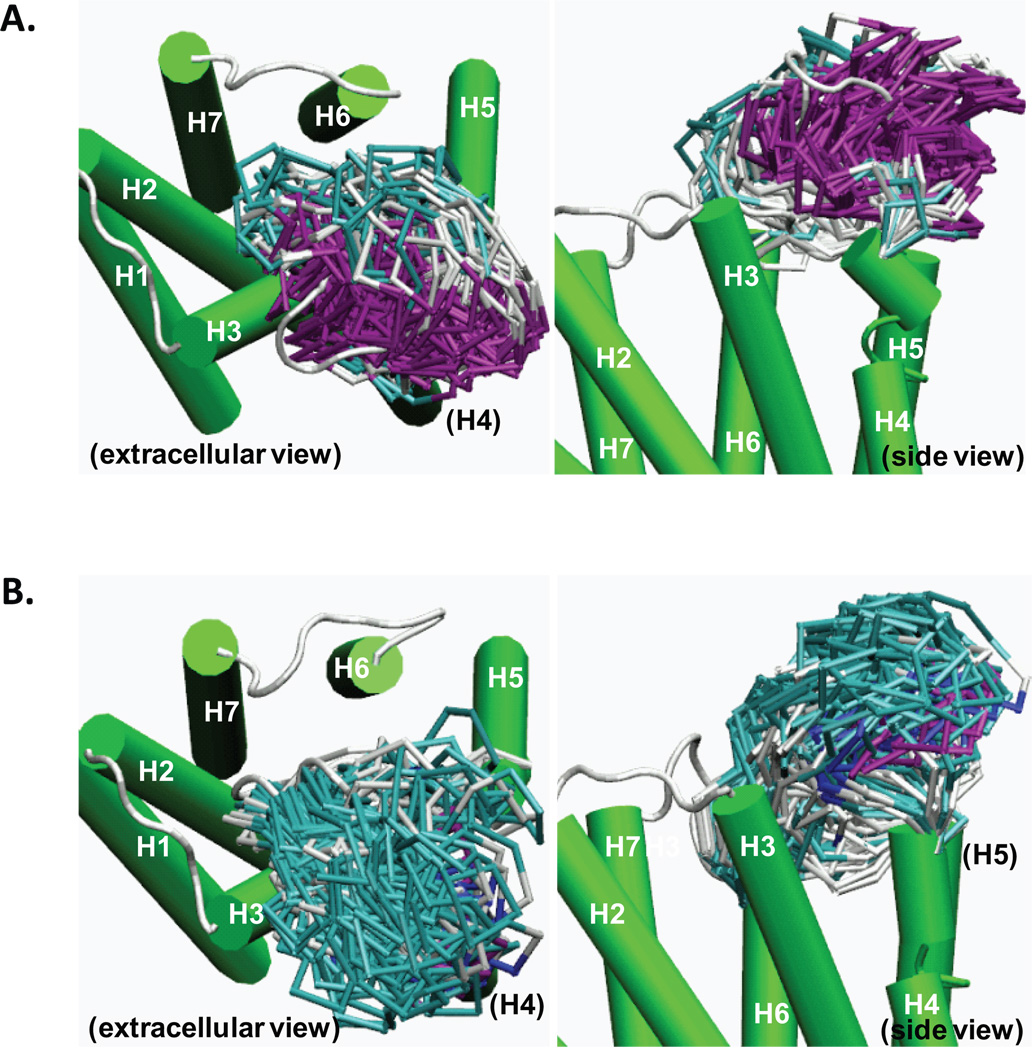
The sampled 166 conformations of E2dithiol and 115 conformations of E2disulfide occupied similar extracellular regions formed by H3/H4/H5, leaving the region near H2/H6/H7 relatively unoccupied (Fig. 1). After clustering analyses and the ensuing MD simulations, the final eight, four from each E2 form, that contained an α-helical segment, as predicted (Table I), were obtained. These E2 conformations were classified into two distinct conformational classes with respect to the spatial orientation of the α-helical segment within E2: in class A the α-helical segment is parallel to the membrane surface, as seen in the X-ray structures of βARs,15,16 while in class B it is perpendicular to the membrane. Class A represents the majority of the high helicity E2dithiol having two helical turns, while class B represents the majority of the low helicity E2disulfide having only one helical turn.
FIGURE 1.
Sampled conformations of E2 of the CB1 receptor in a POPC bilayer by the SA simulation. A. Superposition of 166 conformations of E2dithiol, with extracellular top view and side view. B. Superposition of 115 conformations of E2disulfide, with extracellular top view and side view. Only Cα atoms of E2 are shown for clarity. The TM helical bundle is represented in green cartoon, while the extracellular loops are colored according to the secondary structure: α-helix, purple; 310 helix, blue; turn, cyan; and coil, white.
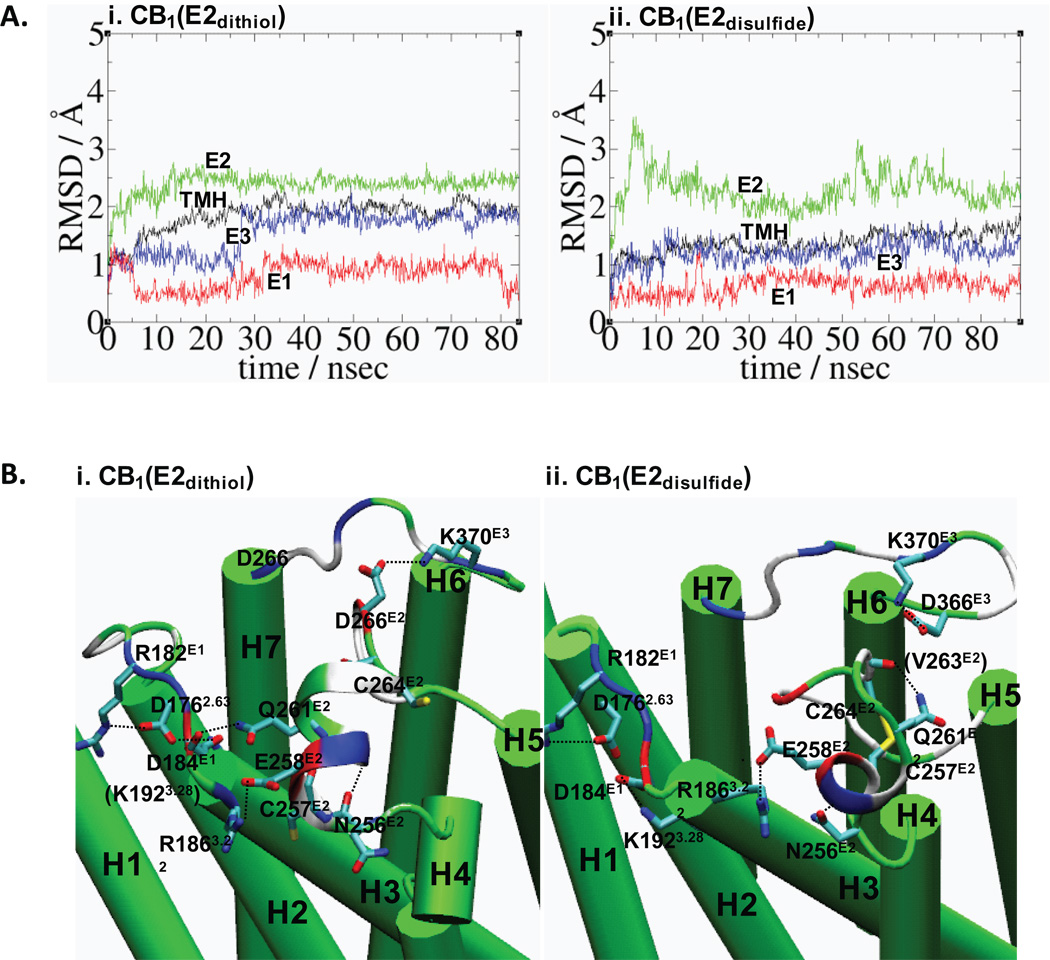
Conformation A1 of E2dithiol and conformation B3 of E2disulfide were chosen as the best conformations of each E2 form based upon receptor stability and the degree of molecular interactions with the other regions of the receptor (data shown in Fig. S1 & Table SI). The CB1 receptor models with these two E2 conformations (i.e., CB1(E2dithiol) and CB1(E2disulfide)) were subjected to long MD simulations for further examination. The rest of our studies are discussed based upon the MD simulation results of these two CB1 receptor models. The root mean-square deviations (RMSDs) show that the TM helical bundle of CB1(E2dithiol) quickly converges in 20 ns of the simulation, while the TM helical bundle of CB1(E2disulfide) is slow to converge even at the end of the simulation (Fig. 2A). It appears that large fluctuations of E2 in CB1(E2disulfide) throughout the simulation cause the rest of the receptor to fluctuate accordingly. The helical backbone RMSDs of CB1(E2dithiol) are 0.5 Å higher than those of CB1(E2disulfide) due to the large outward movement of H1 at the extracellular region from the helical bundle. Differences in structural convergence shown in CB1(E2dithiol) and CB1(E2disulfide) indicate distinct E2 structures play a role in altering the receptor stability.
FIGURE 2.
MD simulations of CB1(E2dithiol) conformation A1 for a total of 83 ns duration and CB1(E2disulfide) conformation B3 for a total of 88 ns duration. A. The RMSDs for CB1(E2dithiol) (i) and for CB1(E2disulfide) (ii), calculated by RMS fitting to the initial coordinates with respect to the backbone heavy atoms of the receptor TM helical residues: TMH, black; E1, red; E2, green; and E3, blue. The TM helical and extracellular loop boundaries of CB1(E2dithiol) are defined as follows: H1, P1131.29-H1431.59; H2, P1512.38-D1762.63; H3, R1863.22-H2193.55; H4, R2304.39-L2534.62; H5, I2715.35-H3025.66; H6, M3376.29-F3686.60; H7, K3767.32-Y3977.53; E1, F177E1-D184E1; E2, G254E2-H270E2 and E3, G369E3-I375E3. Similarly, the TM helical boundaries of CB1(E2disulfide) are defined as follows: H1, P1131.29-H1431.59; H2, P1512.38-V1792.66; H3, R1863.22-H2193.55; H4, R2304.39-G2544.63; H5, D2725.36-R3115.75; H6, M3376.29-F3686.57; H7, T3777.33-Y3977.53; E1, F180E1-S185E1; E2, W255E2-I271E2 and E3, D366E3-K376E3 B. The extracellular loop region in CB1(E2dithiol) (i) and CB1(E2disulfide) (ii) at the end of the MD simulation. The extracellular loop residues (colored according to the atom type) participating in salt bridges and E2 H-bonds (by dotted lines) are shown (see Table III). Hydrogen atoms as well as some side chains are omitted for clarity. The E2 α-helical segment is represented in ribbon, while all other residues are in cartoon. TMH are colored in green while the extracellular loops are colored according to the residue type: hydrophobic, white; hydrophilic, green; positively charged, blue; and negatively charged, red.
Structural features of two distinct E2 conformations
A close examination of E2 in CB1(E2dithiol) and CB1(E2disulfide) exhibits a combination of stabilizing interactions, including hydrophobic interactions, salt bridges, H-bonding interactions, and aromatic stacking interactions (Fig. 2B). E2 in CB1(E2dithiol) contains a helical segment (i.e., C257E2-V263E2), as seen in the X-ray structures of βARs,15,16 in an amphipathic alignment: the polar face, exposed to water, is stabilized by a salt bridge by E258E2/R1863.22 and a H-bond by Q261E2/D184E1, while the non-polar face, buried in the receptor core, is stabilized by forming a hydrophobic cluster with the E2 non-polar residues (i.e., W255E2, C257E2, L260E2, C264E2, F268E2 and P269E2). In CB1(E2dithiol), several salt bridges, including R182E1/D1762.63/K1923.28/D184E1, R1863.22/E258E2 and D266E2/K370E3, exist at the receptor extracellular region (Fig. 2Bi and Table III). Additionally, F268E2 of E2 in CB1(E2dithiol) is involved in a network of aromatic stacking at the receptor extracellular region (see below).
Table III.
Molecular interactions in CB1(E2dithiol) and CB1(E2dithiol) at the receptor extracellular region
| CB1(E2dithiol) | CB1(E2disulfide) | |
|---|---|---|
| salt bridgea | D1762.63/R182E1 | D1762.63/R182E1 |
| D1762.63/K1923.28 | K183E1/D266E2 | |
| D184E1/K1923.28 | D184E1/K1923.28 | |
| R1863.22/E258E2 | R1863.22/E258E2 | |
| D266E2/K370E3 | D3666.58/K370E3 | |
| H-bondingb | D184E1 O(back)/Q261E2 N(side) | N256E2 O(side)/K259E2 N(back) |
| N256E2 O(back)/L260E2 N(back) | N256E2 O(back)/L260E2 N(back) | |
| C257E2 O(back)/Q261E2 N(back) | C257E2 O(back)/Q261E2 N(back) | |
| V263E2 O(back)/S265E2 N(back) | E258E2 O(side)/R1863.22 N(side) | |
| aromatic stackingc | F1742.61/ F1772.64 | F1772.64/F1893.25 |
| F1742.61/ H1782.65 | F1772.64/F3797.35 | |
| F1742.61/ F3817.37 | H1782.65/H1812.68 | |
| F1772.64/ F268E2 | H1782.65/F3817.37 | |
| F1772.64/F3797.35 | F1802.67/H1812.68 | |
| H1782.65/ F3817.37 | F1893.25/F268E2 | |
| F1893.25/ F268E2 | F1893.25/F3797.35 | |
| F268E2/Y2755.39 | F268E2/H270E2 | |
| F268E2/F3797.35 | F268E2/Y2755.39 | |
| Y3656.57/F3686.60 | F268E2/F3797.35 | |
| H270E2/Y2755.39 | ||
| Y3656.57/F3686.60 | ||
Estimated by measuring the distance between the side chain O atom of a negatively charged residue and the side chain N atom of a positively charged residue with the cut-off distance of 3.20 Å.
side, side chain; back, backbone
Estimated by measuring the centroid to centroid distance between any two aromatic residues (Phe, Tyr, Trp, or His) with the cut-off distance of 8.0 Å.
E2 in CB1(E2disulfide) contains a short helical segment (i.e., C257E2-L260E2) due to the geometric constraint by the disulfide bond that interferes with α-helix formation. Salt bridges by D266E2/K183E1 and E258E2/R1863.22 contribute to E2 and receptor stabilization and the cyclic ring enclosed by the disulfide linkage is stabilized by several H-bonds: N256E2/K259E2, N256E2/L260E2, C257E2/Q261E2 and E258E2/R1863.22 (Fig. 2Bii and Table III). Two intra-loop hydrophobic clusters (i.e., one: W255E2, L260E2, C264E2 and L271E2 and another: C257E2, I267E2, F268E2 and P269E2) on the top of the TM helical bundle appear to minimize the exposure to water and contribute to receptor stabilization. P269E2 also forms an inter-loop hydrophobic cluster with the TM residues L1903.26, L1933.29 and L2534.62, suggesting that the C-terminal of E2 in CB1(E2disulfide) is closely packed to TMH. Finally, the aromatic network at the receptor extracellular region in CB1(E2disulfide), which is formed by the E2 aromatic residues F268E2 and H270E2, is highly extensive (see below).
Considering that the square of each buried hydrophobic surface area is estimated to contribute to the free energy of protein folding (24 cal/mol),54 the E2 hydrophobic residues folded into the receptor core significantly contribute to receptor stabilization. Similarly, considering that multiple salt bridges compensate for the loss of entropy by a single salt bridge,55 the multiple salt bridges centered at D1762.63/K1923.28 in CB1(E2dithiol) and by D184E1/K1923.28 and E258E2/R1863.22 in CB1(E2disulfide) appear to be important for receptor stabilization. In support, recent mutation studies suggest that D1762.63 and D184E1 of the CB1 receptor contribute to receptor structure and function through charge interaction.56,57
CD spectroscopy of the E2 peptide of the CB1 receptor
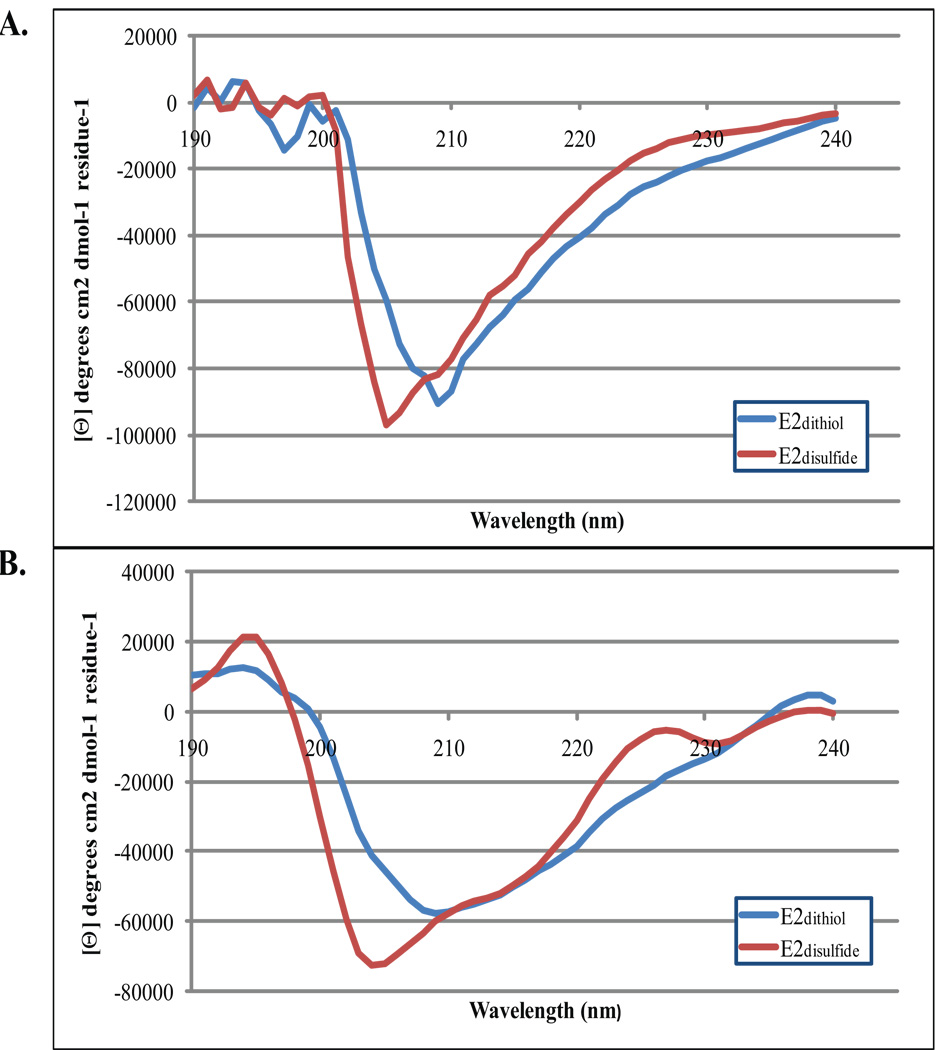
It is well known that a-helical secondary structure gives rise to a positive CD-band (positive ellipticity) at 195–196 nm and two negative bands around 209 and 222 nm. The CD-spectrum of E2dithiol displayed a positive band at 195 nm and a distinct negative one at 209 nm (Fig. 3A). Furthermore, a very weak band can be seen around 220 nm in this CD-spectrum. Results from deconvolution of the E2dithiol CD-spectrum are listed in Table SIII and a calculated spectrum is displayed in Fig. 3B. Results from analysis of E2dithiol using CONTIN L suggest that E2dithiol has an α-helical content around 80 %. Because the normalized root mean square deviation (NMRSD) is very high (0.3) for the deconvolution of the CD-spectrum for E2dithiol, the calculated secondary structure is probably not going to be in complete agreement with the actual one.53 However, both the experimental and calculated spectra clearly indicate the presence of some α-helical structure in E2dithiol. Deconvolution of the E2disulfide spectrum suggests α-helical content close to 60 % (Table IV). It should be noted that CONTIN L also predicts the presence of a turn (43 %) in this peptide. Again the NMRSD-value is very high so no conclusions can be made about the ‘correct’ amount of α-helix in E2disulfide. The experimental spectrum (Fig. 3A) features two of the bands indicative of α-helix (196 and 209 nm). The 222 nm band cannot be seen in the experimental or calculated spectra (Figs. 3A and 3B). However, because the E2disulfide spectrum has some of the bands typical of α-helix, the presence of α-helix cannot be ruled out in this case. In addition, the presence of some α-helix is supported, but not proven, by deconvolution of the E2dithiol CD spectrum.
FIGURE 3.
Circular dichroism spectroscopy of peptide fragments of E2dithiol and E2disulfide A. Far UV CD spectra of the E2dithiol (in blue) and E2disulfide (in red). CD-bands with mean residue ellipticities ([Θ]-values) that are positive at 195 nm and negative at 209 nm are visible in both spectra indicative of α-helix formation by both E2dithiol and E2disulfide. The 222 nm band typical of α-helicies is barely visible in these spectra. The strong negative band at 205 nm in the E2disulfide spectrum remains to be explained. B. Deconvolution of the CD-spectra further indicates that α-helical secondary structure elements contribute to the CD-spectrum of E2dithiol. All three bands typical of α-helical secondary structure are visible, including the band at 222 nm. Deconvolution of the CD-spectrum for E2disulfide did not yield any additional information about the secondary structure although α-helix formation is likely also in this peptide as indicated by the positive and negative [Θ]-values at 195 nm and 209 nm respectively. Output from CONTIN L show that α-helix contributes to 80 % of the E2dithiol secondary structure.
Aromatic stacking
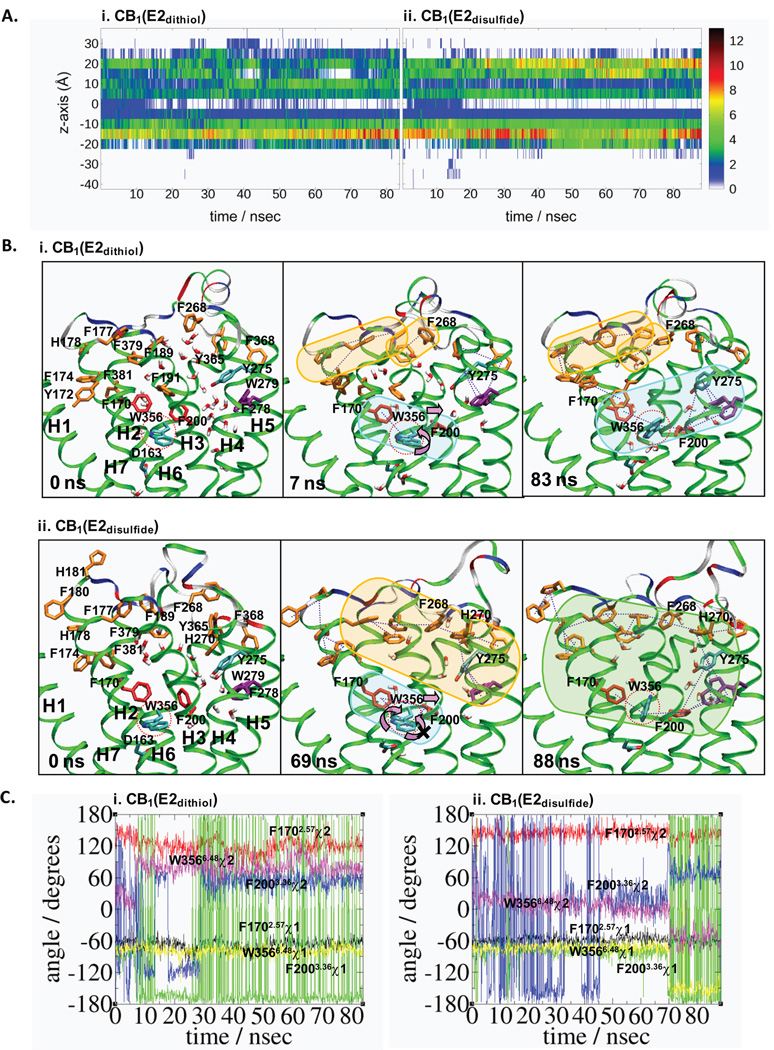
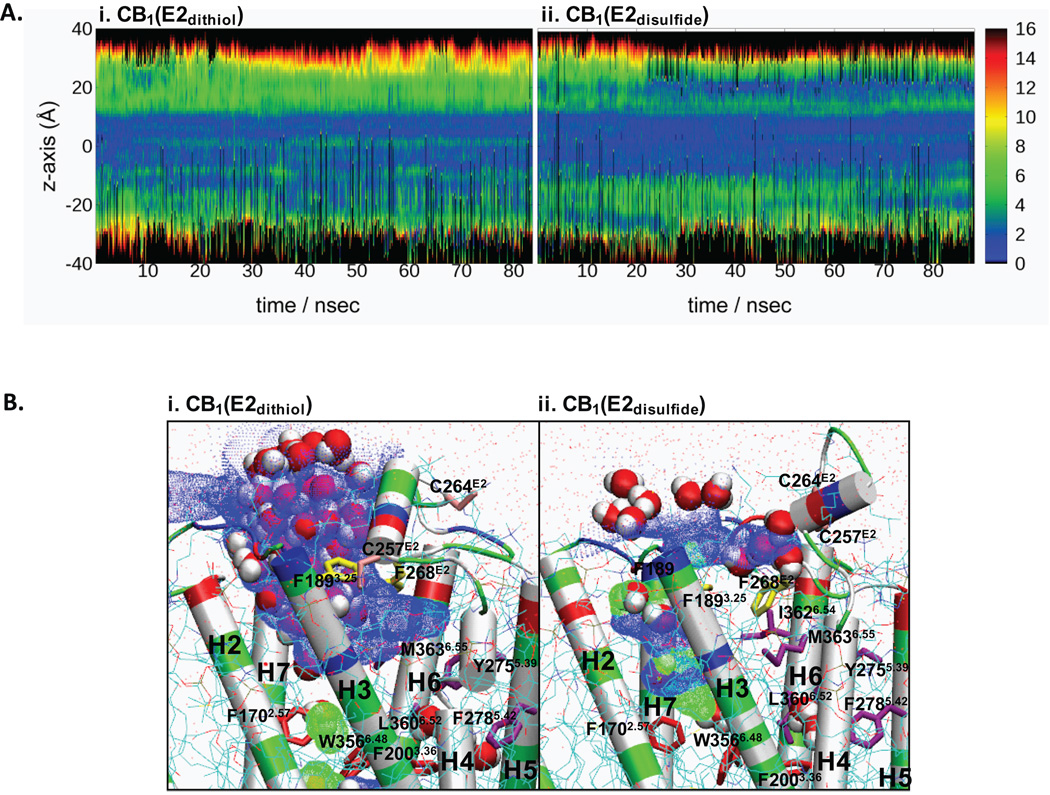
Due to the importance of aromatic stacking for stabilizing the receptor bundle structure in IMPs,58 we examined the pattern of aromatic stacking in both CB1(E2dithiol) and CB1(E2disulfide). As shown in Fig. 4A, a significant amount of receptor aromatic stacking occurs in the membrane region between −20 and +30 Å along the Z-axis. An extensive aromatic stacking network exists in the region between +5 and +15 Å, centering at W3566.48: F1702.57/W3566.48/F2003.36/F2785.42(W2795.43)/Y2755.39 in both CB1(E2dithiol) and CB1(E2disulfide).
FIGURE 4.
Aromatic stacking interactions in CB1(E2dithiol) and CB1(E2disulfide). A. Concentration of aromatic stacking during the 83 ns MD simulation of CB1(E2dithiol) (i) and the 88 ns simulation of CB1(E2disulfide) (ii). Aromatic stacking was defined as any two aromatic residues (F, Y, W, or H) for which the centroid to centroid distance was < 8.0 Å. Z-axis position of each stack was determined by the z coordinate of one participating residue. The concentration of aromatic stacking is plotted in ranges of 5 Å with the concentration indicated by the following colors in decreasing order: black, red, orange, yellow, green, blue and white (no aromatic stacking). B. The aromatic stacking (by dotted lines) networks in CB1(E2dithiol) (i) and CB1(E2disulfide) (ii). The snapshot at 6 ns of the simulation of CB1(E2dithiol), when the W3566.48 indole ring flipping occurs (i.e., the χ2 angle changes from ~ 0 degree to ~ +70 degrees), and the snapshot at 26 ns of the simulation of CB1(E2disulfide), when the W3566.48 indole ring flipping occurs (i.e., the χ1 angle changes from ~ −70 degree to ~ −150 degrees and the χ2 angle changes from ~ 0 degree to ~ −70 degrees), are shown in addition to the snapshots at the beginning and at the end of the simulations. The toggle switch W3566.48 is circled (in red dot). Only the side chains of these aromatic residues, without H atoms, are shown. The water molecules located within 12 Å of W3566.48 are also shown. The aromatic stacking networks are indicated by the circles: the network around F268E2 and H270E2, orange; the network around W3566.48, blue; and the network combining these two networks through H270E2/Y2755.39 aromatic stacking, in green. Color coding for the protein (in cartoon) is the same as in Fig. 2. C. The χ1 and χ2 angles of F1702.57, F2003.36 and W3566.48 during the duration of 83 ns MD simulation for CB1(E2dithiol) (i) and during the duration of 88 ns MD simulation for CB1(E2disulfide) (ii). Color coding: the χ1 angle of F1702.57, black; the χ2 angle of F1702.57, red; the χ1 angle of F2003.36, green; the χ2 angle of F2003.36, blue; the χ1 angle of W3566.48, yellow; and the χ2 angle of W3566.48, magenta.
The number of aromatic stacking interactions in the extracellular top layer around +20 Å is maintained low in CB1(E2dithiol), yet gradually increased in CB1(E2disulfide) as the simulation continues (Fig. 4A). A detailed examination reveals that in this region the aromatic stacking network by F268E2/H270E2/Y2755.39 is well maintained only in CB1(E2disulfide) but is unavailable in CB1(E2dithiol) due to the location of H270E2: H270E2 in CB1(E2dithiol), exposed to water, is not able to form aromatic stacking to Y2755.39, while H270E2 in CB1(E2disulfide), positioned toward the helical inner core region, forms aromatic stacking to Y2755.39 and tightly connects the aromatic stacking network centered at F268E2 in the extracellular region to the aromatic stacking network centered at W3566.48 in the helical inner core region (Fig. 4B). Overall, the aromatic stacking at the extracellular region is better developed in CB1(E2disulfide) than in CB1(E2dithiol) due to distinct E2 structures.
Toggle switch W3566.48
W3566.48 of the CB1 receptor has been proposed as a toggle switch for receptor activation,62 similar to W2866.48 of β2AR.63 The X-ray structure of rhodopsin14 reveals that W2656.48 is stabilized by aromatic stacking to F2616.44 and Y2686.51, highly homologous residues in many GPCRs.64 The CB1 receptor, however, lacks the corresponding aromatic residues at the 6.44 and 6.51 positions and, consequently, is expected to have unique interaction patterns for W3566.48.
As shown in Fig. 4B, F1702.57 and F2003.36 of the CB1 receptor form aromatic stacking to W3566.48, suggesting their role in stabilizing W3566.48. To gain insight into how F1702.57 and F2003.36 stabilize W3566.48 in CB1(E2dithiol) and CB1(E2disulfide), the χ1 and χ2 angles of F1702.57, F2003.36 and W3566.48 are analyzed (Fig. 4C). Closely surrounded by the neighboring residues, the aromatic ring of F1702.57 appears to be rather fixed, with the χ1 and χ2 angles of F1702.57 showing little change throughout the simulations of both CB1(E2dithiol) and CB1(E2disulfide). Of interest, it appears that the preferred value for the χ1 angle of F2003.36 is ~ +180 degrees, common to both CB1(E2dithiol) and CB1(E2disulfide). In CB1(E2dithiol), as the χ1 angle of F2003.36 changes to ~ +180 degrees early in the simulation, the χ1 angle of W3566.48 maintains at ~ −70 degrees. In CB1(E2disulfide), however, as the χ1 angle of F2003.36 changes to ~ +180 degrees late in the simulation, the χ1 angle of W3566.48 changes from ~ −70 degrees to ~ −150 degrees. A close examination reveals that F2003.36 in CB1(E2dithiol), flexible without any steric conflict with W3566.48, achieves its preferred χ1 angle without any modification in the χ1 angle of W3566.48, while F2003.36 in CB1(E2disulfide), restricted in motion by W3566.48, achieves its preferred χ1 angle with a significant modification in the χ1 angle of W3566.48. As a result, the χ1 angle of W3566.48 in CB1(E2dithiol) becomes ~ −70 degrees, indicative of the receptor in its inactive state63, while the χ1 angle of W3566.48 in CB1(E2disulfide) becomes ~ −150 degrees, indicative of the receptor in its active-like state63. Overall, differences in the χ1 angle of W3566.48 shown in CB1(E2dithiol) and CB1(E2disulfide) indicate distinct E2 structures play a role in modulating the receptor conformational change required for its activation.65
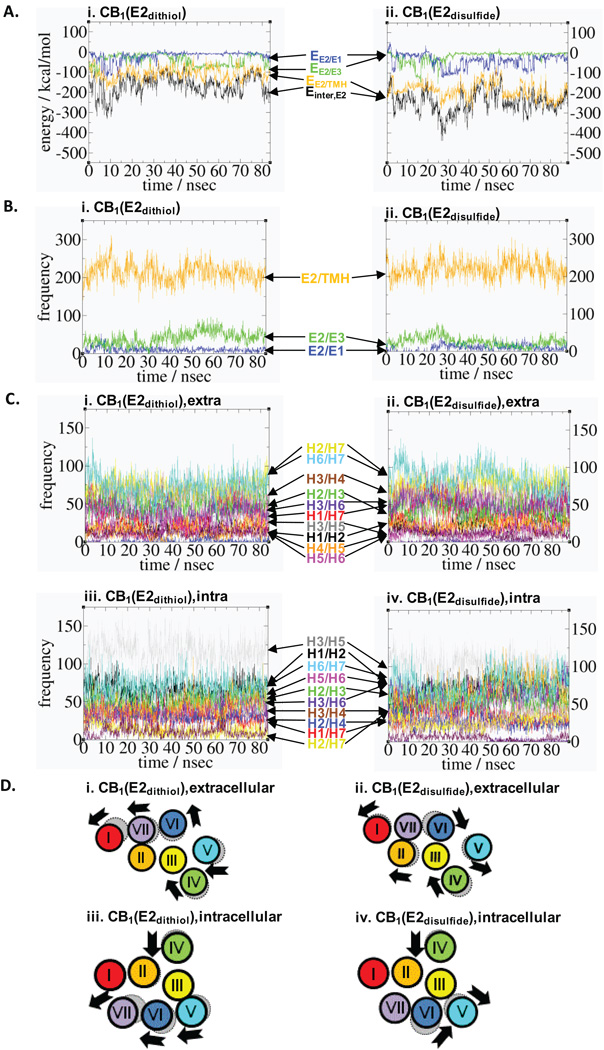
Interaction energy analysis
To examine how distinct E2 structures exhibit different degrees of receptor stabilization, the inter-molecular interaction energy between E2 and the rest of the receptor (Einter,E2) were estimated (Fig. 5A and Table SII). Einter,E2 is −147.0 kcal/mol for CB1(E2dithiol) (i.e., −104.6 kcal/mol by TMH; −13.5 kcal/mol by E1; and −28.9 kcal/mol by E3), compared with −252.9 kcal/mol for CB1(E2disulfide) (i.e., −226.0 kcal/mol by TMH; −22.5 kcal/mol by E1; and −4.1 kcal/mol by E3), indicating that the receptor is more stabilized by E2 in CB1(E2disulfide) than E2 in CB1(E2dithiol). E2 in CB1(E2disulfide), with one helical turn, interacts more closely with TMH than E2 in CB1(E2dithiol), with two helical turns, possibly due to its flexibility that allows to be closely packed to TMH. In support, only in CB1(E2disulfide) but not in CB1(E2dithiol), the C-terminal residue P269E2 forms a tight hydrophobic cluster with some residues from H3 and H4 as described above. E2 interacts favorably with E3 in CB1(E2dithiol) due to the salt bridge by D266E2/K370E3, which is absent in CB1(E2disulfide), and E2 interacts favorably with E1 in CB1(E2disulfide) due to the salt bridge by K183E1/D266E2, which is absent in CB1(E2dithiol). Overall, distinct E2 structures interact differently with the TM helical bundle as well as E1/E3.
FIGURE 5.
Modifications of the helical topology caused by the inter-molecular interactions with distinct E2 structures of the CB1 receptor. A. Non-bonding interaction energies (in kcal/mol), which is a summation of the electrostatic and the van der Waals components, between E2 and the other parts of the receptor (Einter,E2) (in black), including E1 (EE2/E1) (in blue), E3 (EE2/E3) (in green) and TMH (EE2/TMH) (in orange), are displayed for CB1(E2dithiol) (i) and for CB1(E2disulfide) (ii). B. The contact numbers between E2 and E1 (in blue), E3 (in green) or TMH (in orange) in CB1(E2dithiol) (i) and CB1(E2disulfide) (ii). A criterion of 3.5 Å was used between non-bonded atoms. C. The contact numbers between TMH in the extracellular half of CB1(E2dithiol) (i) and CB1(E2disulfide) (ii) and in the intracellular half of CB1(E2dithiol) (iii) and CB1(E2disulfide) (iv). A criterion of 3.5 Å was used between non-bonded atoms. Color coding: H1/H2, black; H1/H7, red; H2/H3, green; H2/H4, blue; H2/H7, yellow; H3/H4, brown; H3/H5, grey; H3/H6, violet; H4/H5, magenta; H5/H6, orange; and H6/H7, turquoise. D. The extracellular TM helical bundle structures of CB1(E2dithiol) (i) and CB1(E2disulfide) (ii) and the intracellular helical bundle structures of CB1(E2dithiol) (iii) and CB1(E2disulfide) (iv) predicted by their contact numbers are shown in comparison with the CB1 receptor model32 whose TM helical bundle structure was same both in CB1(E2dithiol) and CB1(E2disulfide) at the beginning of the respective simulations. The TM helical boundaries of the CB1 receptor are defined same as in Fig. 3. Color coding for the TMH (in ribbon): H1, red; H2, orange; H3, yellow; H4, green; H5, cyan; H6, blue; and H7, purple.
Contact number analysis
To examine how distinct E2 structures modify the TM helical topology, the contact numbers were counted (Figs. 5B and 5C and Table SII). These contact numbers include not only those between E2 and the rest of the receptor but also the inter-helical contacts. The E2/TMH contact number is higher in CB1(E2disulfide) than in CB1(E2dithiol), which is also reflected in the E2/TMH interaction energies. The E2/E3 contact number is higher in CB1(E2dithiol) than in CB1(E2disulfide), which is also reflected in their E2/E3 interaction energies. Of interest, the E2/E1 contact number is almost identical in both CB1(E2dithiol) and CB1(E2disulfide), but the E2/E1 interaction energy is lower in CB1(E2disulfide) than in CB1(E2dithiol), due to the salt bridge by K183E1/D266E2 in CB1(E2disulfide).
Because most of TMH are not exactly perpendicular to the membrane, the TM helical topology at the extracellular region is different from that at the intracellular region. Thus, the inter-helical contact numbers are analyzed by considering two separate parts: the extacellular half and the intracellular half. As shown in Fig. 5C and Table SII, the high inter-helical contact numbers shown for H2/H7 on the extracellular side and for H1/H2 and H3/H5 on the intracellular side are in agreement with the notion that the TM helical bundle of the CB1 receptor is primarily stabilized by H1/H2, H2/H7 and H3/H5.32
To examine the role of distinct E2 structures in TM helical rearrangement, the predicted TM helical structures in CB1(E2dithiol) and CB1(E2disulfide) suggested by the contact number analysis are compared with TMH in the CB1 receptor model32 (Fig. 5D) the template structure before the long simulation. For CB1(E2dithiol), H4/H5 move in toward the core and H1/H6/H7 move out of the core on the extracellular side, while H7 moves out of the core and H4/H5/H6 move toward the core on the intracellular side (Figs. 5Di & 5Diii). For CB1(E2disulfide), H4/H6 moves in toward the core and H1/H2/H5 move out of the core on the extracellular side, while H4/H6 moves into the core and H5 moves out of the core on the intracellular side (Figs. 5Dii & 5Div). Overall, the contact number analysis reveals that distinct E2 structures, with different degrees of molecular interaction with the rest of the receptor, induce unique alterations in the TM helical topology.
Core crevice analysis
Since the distinct E2 structures interact differently with the receptor bundle and uniquely alter the TM helical topology, we performed a core crevice analysis to examine key differences in the binding pockets in CB1(E2dithiol) and CB1(E2disulfide). As shown in Fig. 6A, common to both CB1(E2dithiol) and CB1(E2disulfide), the core crevice is almost completely closed at the inner core region between −10 Å and +10 Å along the Z-axis. In contrast, in the extracellular region between +10 Å and +20 Å, the water molecules are able to enter into the ligand binding core crevice. A close examination reveals that in CB1(E2dithiol), E2, positioned near the extracellular ends of H3/H4/H5, occupies only half of the extracellular pore, leaving the extracellular pore formed by H2/H3/H7 accessible to water. The binding core crevice in CB1(E2dithiol) with the predicted crevice diameters of > 6 Å appears to be fully developed to include the region deep in the binding pocket (Fig. 6Bi). Similarly, E2 in CB1(E2disulfide), positioned over H3/H4, leaves the extracellular pore region formed by H2/H3/H7 relatively open to water access. However, the predicted crevice diameters of ≤ 6 Å in CB1(E2disulfide) appear to restrict ligand access to the binding pocket (Fig. 6Bii). Differences in core crevice of CB1(E2dithiol) and CB1(E2disulfide) demonstrate the topologically distinct rearrangement of TMH initiated by distinct E2 structures, suggesting the role of E2 not only in the availability but also the dimensions of the ligand binding pore.
FIGURE 6.
Core crevice analysis of CB1(E2dithiol) and CB1(E2disulfide). A. Pore diameter along the Z axis during the 83 ns MD simulation of CB1(E2dithiol) (i) and the 88 ns simulation of CB1(E2disulfide) (ii). Colors indicate the diameter of the pore in angstroms (red: 14 Å; orange: 12 Å; yellow; 9 Å etc.). Pore diameter was determined using HOLE with sampling done every 0.5 Å along the z-axis; a midpoint between H3 and H7 was used to help define the pore (c-point). B. The solvent accessible pore (in blue dots for low radius surface and in green dots for mid radius surface) created by using HOLE at the extracellular core region in E2dithiol (i) at the end of 83 ns of MD simulation and in E2disulfide (ii) at the end of 88 ns of MD simulation. Color coding for residues (in stick): the residues forming the aromatic cluster at the top of the extracellular pore region, yellow; F1893.25 and F268E2, located at the entrance of the pocket and known as crucial for ligand recognition,56,61; the residues forming the water-accessible binding surface in the CB2 receptor,30 purple; and F1702.57, F2003.36 and W3566.48, red. Colored according to the atom type, water molecules inside and outside the receptor crevice are represented in space filling and line, respectively, while the lipid molecules are represented in lines. Color coding for the protein (in cartoon): hydrophobic, white; hydrophilic, green; positively charged, blue; and negatively charged, red.
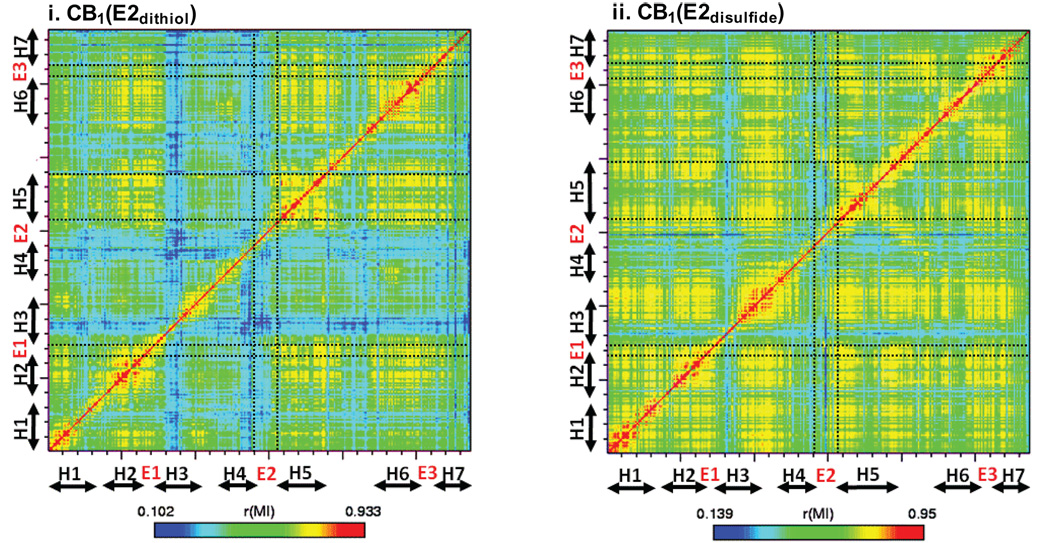
Cross-correlation analysis
To gain some insights into the role of distinct E2 structures in unique modifications in the TM helical topology, we also performed cross-correlation analysis for CB1(E2dithiol) and CB1(E2disulfide). The pattern of correlation of CB1(E2dithiol) quite differs from that of CB1(E2disulfide) (Fig. 7): only limited TMH, including H2, H5 and H6, are correlated in CB1(E2dithiol), while most of TMH are correlated in CB1(E2disulfide). Common to both CB1(E2dithiol) and CB1(E2disulfide), the N-terminal region of E2, containing the helical segment, is not involved in coupling. In contrast, the C-terminal region of E2 in CB1(E2dithiol) is coupled to H5, while the same region in CB1(E2disulfide) is coupled not only to most of TM helices but also to E1 and E3. Of interest, the segment spanning from the E2 C-terminal region to the N-terminal region of I3 in CB1(E2disulfide) appears to be closely coupled together, possibly due to H5 fluctuation initiated by E2 fluctuation (Fig. 2Ai), as indicated by the moderate E2/H5 coupling (Fig. 7ii). Overall, these results strongly suggest that distinct E2 structures are uniquely coupled to the TM helical bundle to modify TM helical topology.
FIGURE 7.
Cross correlation of CB1(E2dithiol) and CB1(E2disulfide). X- and y-axes are the receptor H1 through H7; red indicates highly correlated movement (1.0) and blue indicates less correlated (0.0). The E2/H5 correlation is indicated by a dotted circle. Cross correlation was performed with g_correlation 52 to create cross-correlation matrices from ~ 12,000 coordinates recorded every 2 ps from the last 24 ns MD simulations of CB1(E2dithiol) and CB1(E2disulfide), respectively.
DISCUSSION
The presence of a helical segment within E2 both in CB1(E2dithiol) (i.e., C257E2-V263E2) and in CB1(E2disulfide) (i.e., C257E2-L260E2), as seen in the X-ray structures of βARs15,16 and as predicted by several popular secondary prediction programs (Table I), is strongly suggested by the CD spectra of two E2 peptides (Fig. 3). It appears that the E2 helical segment in an amphipathic alignment, located in the membrane-water interfacial region, plays a role in stabilizing receptor structure. Furthermore, it appears that the degree of the helical content determines the flexibility of E2, which is important for its interaction with TMH (see below).
It has been reported that GPCR E2 plays an important role in receptor activation,65 primarily by coupling to the TM helical domain.13,66,67 The present study strongly suggests that E2 of the CB1 receptor is able to rearrange TMH through E2/TMH coupling. Examining the degree of E2 interaction with TMH in CB1(E2dithiol) and CB1(E2disulfide), we demonstrate that distinct E2 structures of the CB1 receptor uniquely modify the TM helical topology. The strong coupling between E2 and TMH observed in CB1(E2disulfide) (Fig. 7ii) appears to be attributed to the flexible nature of E2, which allows the C-terminal region of E2 to be inserted into the extracellular H3/H5 region for close interactions with the extracellular end residues of TMH: 1) the hydrophobic packing by P269E2 to the TM hydrophobic residues L1903.26, L1933.29 and L2534.62; and/or 2) the aromatic stacking centering at F268E2/H270E2/Y2755.39. As a result, H5 moves out from H3 and at the same time H6 moves in toward H3 at the extracellular region (Fig. 5Dii). These helical movements are evidenced by TMH/E3 fluctuation caused by E2 fluctuation (Fig. 2Aii). As demonstrated in a recent NMR study61, the outward movement of H5 and the inward movement of H6 at the extracellular region appear to be necessary for an efficient coupling of E2 to TMH. Conversely, in CB1(E2dithiol), E2 with the much reduced flexibility is less inserted into the extracellular H3/H5 region. As a result, H5 moves in toward H3 and at the same time H6 moves away from H3 at the extracellular region (Fig. 5Di). Thus, it appears that the E2 C-terminal residues determine the E2 conformation, which is important for its coupling to TMH and thereby for receptor activation. The importance of the E2 C-terminal region for the CB1 receptor activation has been suggested by a recent study by Ahn et al.61 where the Ala mutations of the C-terminal residues of E2 in the CB1 receptor significantly reduced the agonist binding but retained the inverse agonist binding. It appears that the C-terminal residues of E2 not only provide the binding contact sites exclusively for agonist binding but also initiate the conformational change in TMH required for receptor activation. It should be noted that although the coupling between E2 and TMH expects to be weak in the absence of the ligand, E2 conformational rearrangement upon ligand binding, as demonstrated in several experimental studies of other GPCRs,18,19,68 and the subsequent coupling to TMH, especially the segments of H5, are presumed to induce a ligand-specific conformational change in the CB1 receptor.
Considering that the CB1 receptor binding pocket consists of the TM helical core residues at the extracellular region mainly from H3/H5/H6/H7,69,70 the outward movement of H6 from the core and the inward movement of H5 in CB1(E2dithiol) and the outward movement of H5 and the inward movement of H6 in CB1(E2disulfide) (Fig. 5D) would uniquely modify the ligand binding pocket located in the extracellular core region of the receptor (Fig. 6B). It is known for the CB1 receptor that F1893.25, W2554.64, F268E2 and Y2755.39 are crucial for agonist recognition.56,60,61 Since the aromatic stacking networks by F1893.25/F268E2 connected to Y2755.39 are only available for CB1(E2disulfide) (Fig. 4B), CB1(E2disulfide) appears to be more suitable than CB1(E2dithiol) in maintaining the agonist binding pocket geometry. Similarly, it has been suggested that antagonist binding is more sensitive than agonist binding to the cleavage of the E2 disulfide bond of the CB1 receptor.21 Thus, CB1(E2disulfide) appears to be more suitable than CB1(E2dithiol) in maintaining the antagonist binding pocket geometry. Thus, it is conceivable that the outward movement of H5 and the inward movement of H6 at the extracellular region shown in CB1(E2disulfide) is beneficial to ligand binding, while the outward movement of H6 and the inward movement of H5 at the extracellular region shown in CB1(E2dithiol) is detrimental to ligand binding. Considering that I3, which connects H5 and H6 and forms one of the main interfaces to the coupled G-protein, is crucial for transferring the molecular signal from ligand binding at the extracellular face during receptor activation,71 the inward movement of H5 and H6 in CB1(E2dithiol) and the outward movement of H5 and the inward movement of H6 in CB1(E2disulfide) at the intracellular region would lead to distinct I3 conformations as E2 conformation-specific molecular signals.
In the present study, CB1(E2disulfide) appears to be more biologically relevant than CB1(E2dithiol). One of the main reasons for the preference of CB1(E2disulfide) over CB1(E2dithiol) is that CB1(E2disulfide) contains an extensive aromatic stacking network covering the region between +5 Å and +20 Å, while CB1(E2dithiol) lacks such aromatic stacking (Fig. 4B). It appears that the aromatic residues in this region not only contribute to stabilizing the ligand binding pocket through aromatic stacking but also serve as the initial ligand contact site at the extracellular top surface of the receptor. It appears that the aromatic stacking network centered at F1893.25 and F268E2 in the extracellular region at the entrance of the ligand binding pocket plays a role as a gate keeper, selectively allowing the right type of ligand to enter into the binding pocket region (Fig. 6B). Thus, upon ligand entry the aromatic cluster is rearranged in such a way that the entrance of the binding pocket is opened for ligand access. It is interesting to note that CB1(E2dithiol) represents a gate-open form as the aromatic stacking by F1893.25/F268E2 is absent, while CB1(E2disulfide) represents a gate-closed form as the aromatic stacking by F1893.25/F268E2 is present. Similarly, a recent study of β2AR suggests that the E2/H7 hydrophobic junction of the ligand-free receptor is partially broken upon ligand entry, while the hydrophobic cleft formed by H2/H3/H7 serves as a specific ligand-entry site.72 It appears that the binding core crevice shown in the present receptor models (Fig. 6) are strongly supported by experimental findings30,60,61,73 of key residues shaping the ligand binding pocket: for the extracellular part, F268E2 and F1893.25; for the middle part, I3626.54 and M3636.55; and for the inner core part, Y2755.39, L3596.51 and L3606.52.
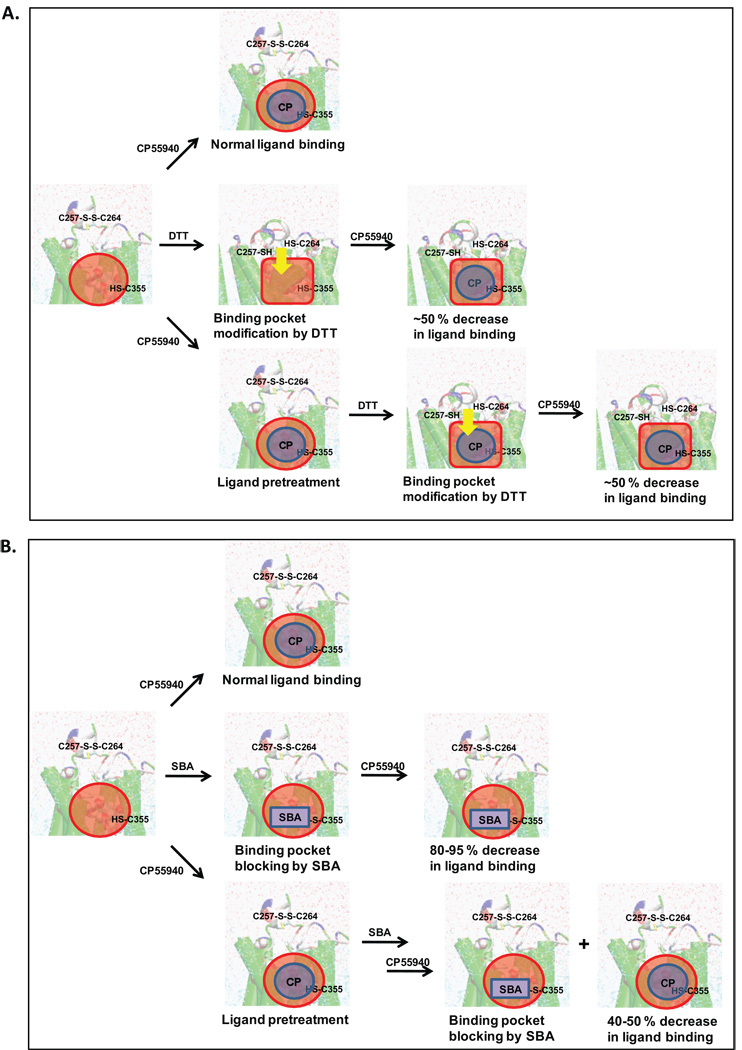
Another key reason for the preference of CB1(E2disulfide) over CB1(E2dithiol) is that CB1(E2disulfide) contains the disulfide bond in E2, while CB1(E2dithiol) does not. An early ligand binding study by Martin and his colleagues31 showed that dithiothreitol (DTT), a disulfide reducing agent, decreased the binding affinity of CP55940 by ~ 50 % regardless of the pretreatment of CP55940 to protect the binding site, indicating that a disulfide bond that affects CP55940 binding “exists” and such disulfide bond is not located at the CP55940 binding pocket. In CB1(E2disulfide), the E2 disulfide bond is located in the extracellular top region (Fig. 6Bii) off the CP55940 binding pocket and exposed to solvent, indicating that the disulfide bond is easily accessible to DTT for reduction. In contrast, in CB1(E2dithiol) the only residues that are somewhat close to form a disulfide bond are C3556.47 and C3867.42. However, the formation of the disulfide bond by these Cys residues appears to contradict the experimental results suggesting that the disulfide bond is not located at the CP55940 binding pocket,31 for C3556.47 is known to be a binding pocket residue.74 From the above-mentioned ligand binding study,31 it was also shown that sulfhydryl blocking agents (SBAs) decreased the binding affinity of CP55940 by > 90 % but significantly reduced the decrease in CP55940 binding (< 50 %) by the pretreatment of CP55940. These results suggest that a reactive free thiol of the receptor exists near or at the CP55940 ligand binding site. The Cys residues located near the CP55940 binding site include C3556.47 and C3867.42 at the helical core. Among these Cys residues, C3556.47 would be a better candidate than C3867.42 in providing the reactive free thiol to SBAs for the following reasons: 1) C3556.47 is known to form part of the CP55940 binding site;74 and 2) it is known that the sulfhydryl blocking of C3867.42 did not affect CP55940 binding.21
As shown in Fig. 8, the experimental results reported by Martin and his colleagues31 can be explained as follows: the observed ~ 50 % decrease in CP55940 binding affinity, regardless of the pretreatment of CP55940, by DTT (i.e., the reduction of the disulfide bond) is less likely to be due to the direct modification of the binding pocket but rather indirect modification as the E2 disulfide bond is reduced. Such indirect modification of the binding pocket is indicated by the results of the present study demonstrating that distinct E2 structures are able to modify the TM helical topology through the E2/H5 coupling (Fig. 7). As discussed before, CB1(E2disulfide), which is equivalent to the receptor before DTT reduction, is more suitable than CB1(E2dithiol), which is equivalent to the receptor after DTT reduction, in maintaining the agonist binding pocket geometry. It should be noted that the protection of the ligand binding pocket by the pretreatment of the ligand does not prevent the ligand binding pocket from the modification caused by the reduction of the E2 Cys disulfide bond (Fig. 8A), indicating that the impact of E2 structural change on the receptor TM helical bundle is rather significant. In contrast, the finding that the decrease in CP55940 binding by SBAs was significantly reduced by the pretreatment of CP55940 is due to the protection of the binding pocket by CP55940 that prevents the modification of the binding pocket by SBAs, leading to a significant reduction of the decrease in CP55940 binding (Fig. 8B).
FIGURE 8.
The role of E2 disulfide bond of the CB1 receptor in modification of ligand binding. A. The modification of the binding of CP55940 (as indicated by the blue circle) to the CB1 receptor by dithiothreitol (DTT). The observed 50 % decrease in CP55940 binding affinity,31 regardless of the pretreatment of CP55940, by DTT is due to indirect modification of the binding pocket, schematically shown from the red circle to the red square shape in the TM core induced (as indicated by the yellow arrow) by E2 conformational change as its disulfide bond is reduced to free thiols. B. The modification of the binding of CP55940 to the CB1 receptor by sulfhydryl blocking agents (SBAs) (as indicated by the blue rectangle). Pretreatment of CP55940 significantly attenuates the effects of SBAs, which reduce CP55940 binding affinity;31 this can be interpreted as the protection of the binding pocket by CP55940 without or with little modification of the binding pocket by SBAs. Explanation is based upon the suggestions of the present studies that E2disulfide is the biologically relevant form and that a conformational change caused by the reduction of E2 disulfide bond can modify the ligand binding pocket.
It is very intriguing to see that CB1(E2disulfide) is converted to an active-like state, while CB1(E2dithiol) maintains the inactive state, as judged from the the χ1 angle of W3566.48.63 The flipping of the indole ring of W3566.48 (i.e., the χ1 angle = ~ −150 degrees) occurs at the late stage of the simulation and is maintained for the rest of the simulation, indicating the resulting structure is somewhat stable in spite of the continued fluctuation. It should be noted, however, that the salt bridge between R2143.50 and D3386.30, which has been proposed as the ionic lock75 for the corresponding residues R1313.50 and E2686.30 in β2AR that retains the receptor in the inactive state, is maintained in CB1(E2disulfide), suggesting that CB1(E2disulfide) resembles the receptor in its inactive state but possibly at the early intermediate stage of the active state. Comparison of CB1(E2dithiol) with CB1(E2disulfide) suggests that the W3566.48 ring flipping in CB1(E2disulfide) occurs due to the steric conflict between W3566.48 and F2003.36 caused by the inward movement of H6 toward H3 at the extracellular region (Figure 5Dii). Thus, it is possible that one of the key steps at the early stage of receptor activation involves the initial ligand binding to the E2 C-terminal (aromatic) residues61 and the subsequent rearrangement of TMH, H5 and H6 in particular, through E2 coupling, leading to the turning on of the toggle switch W3566.48 (i.e., the ring flipping).62 It should be noted that W3566.48 and F2003.36 are known to be important for ligand binding and receptor function.60,61,73,76 The proposed receptor activation by the direct coupling between E2 and the TM helical bundle13,66,67 is applicable to the CB1 receptor, as demonstrated by the present correlation analysis (Fig. 7). Further study of the TM helical bundle of the CB1 receptor may provide insight into the molecular mechanism of the receptor activation initiated by W3566.48, the residue proposed as a toggle switch in the CB1 receptor.62
CONCLUSION
In the present study, two alternative forms, E2dithiol and E2disulfide, are considered as distinct E2 structures of the CB1 receptor. It is demonstrated that the CB1 receptor can have an α-helical segment within E2, as predicted by several secondary structure prediction programs and similar to βARs,15,16 which remains stable in the membrane-water interfacial region. It is also demonstrated that different TM helical topologies, including the binding pocket crevice at the extracellular region and the receptor interface to the coupled G-protein at the intracellular region, are induced by distinct E2 structures through the direct coupling between E2 and TMH. The present results suggest the critical role of the CB1 E2 in stabilizing the receptor, regulating ligand binding through modification of the binding pocket, and ultimately modulating receptor activation, as shown in other GPCRs.5–7,13,65,77 Further studies on the role of E2 of the CB1 receptor are warranted; particularly comparisons of the ligand-bound form with the present ligand-free form.
Supplementary Material
ACKNOWLEDGEMENTS
This work was partially supported by the NIDA K01-DA020663 & by the NCSA under MCB080037N and utilized the Teragrid BigRed IBM e1350 (Indiana University) and LoneStar Dell PowerEdge 1955 (The University of Texas at Austin). The authors thank Drs. L. Pedersen and L. Perera for many helpful discussions. The authors also thank Dr. S. Shaikh for assisting of the g_correlation analysis.
REFERENCES
- 1.Matsuda LA, Lolait SJ, Brownstein MJ, Young AC, Bonner TI. Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature. 1990;346:561–564. doi: 10.1038/346561a0. [DOI] [PubMed] [Google Scholar]
- 2.Gether U. Uncovering molecular mechanisms involved in activation of G protein-coupled receptors. Endocr Rev. 2000;21:90–113. doi: 10.1210/edrv.21.1.0390. [DOI] [PubMed] [Google Scholar]
- 3.Elofsson A, von Heijne G. Membrane protein structure: prediction versus reality. Annu Rev Biochem. 2007;76:125–140. doi: 10.1146/annurev.biochem.76.052705.163539. [DOI] [PubMed] [Google Scholar]
- 4.Bowie JU. Solving the membrane protein folding problem. Nature. 2005;438:581–589. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]
- 5.Marti T. Refolding of bacteriorhodopsin from expressed polypeptide fragments. J Biol Chem. 1998;273:9312–9322. doi: 10.1074/jbc.273.15.9312. [DOI] [PubMed] [Google Scholar]
- 6.de Planque MR, Kruijtzer JA, Liskamp RM, Marsh D, Greathouse DV, Koeppe RE, 2nd, de Kruijff B, Killian JA. Different membrane anchoring positions of tryptophan and lysine in synthetic transmembrane alpha-helical peptides. J Biol Chem. 1999;274:20839–20846. doi: 10.1074/jbc.274.30.20839. [DOI] [PubMed] [Google Scholar]
- 7.Klco JM, Wiegand CB, Narzinski K, Baranski TJ. Essential role for the second extracellular loop in C5a receptor activation. Nat Struct Mol Biol. 2005;12:320–326. doi: 10.1038/nsmb913. [DOI] [PubMed] [Google Scholar]
- 8.Ulmschneider MB, Tieleman DP, Sansom MS. The role of extra-membranous inter-helical loops in helix-helix interactions. Protein Eng Des Sel. 2005;18:563–570. doi: 10.1093/protein/gzi059. [DOI] [PubMed] [Google Scholar]
- 9.Ji TH, Grossmann M, Ji I. G protein-coupled receptors I Diversity of receptor-ligand interactions. J Biol Chem. 1998;273:17299–17302. doi: 10.1074/jbc.273.28.17299. [DOI] [PubMed] [Google Scholar]
- 10.Shi L, Javitch JA. The second extracellular loop of the dopamine D2 receptor lines the binding-site crevice. Proc Natl Acad Sci USA. 2004;101:440–445. doi: 10.1073/pnas.2237265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scarselli M, Li B, Kim SK, Wess J. Multiple residues in the second extracellular loop are critical for M3 muscarinic acetylcholine receptor activation. J Biol Chem. 2007;282:7385–7396. doi: 10.1074/jbc.M610394200. [DOI] [PubMed] [Google Scholar]
- 12.Avlani VA, Gregory KJ, Morton CJ, Parker MW, Sexton PM, Christopoulos A. Critical role for the second extracellular loop in the binding of both orthosteric and allosteric G protein-coupled receptor ligands. J Biol Chem. 2007;282:25677–25686. doi: 10.1074/jbc.M702311200. [DOI] [PubMed] [Google Scholar]
- 13.Ahuja S, Hornak V, Yan EC, Syrett N, Goncalves JA, Hirshfeld A, Ziliox M, Sakmar TP, Sheves M, Reeves PJ, Smith SO, Eilers M. Helix movement is coupled to displacement of the second extracellular loop in rhodopsin activation. Nat Struct Mol Biol. 2009;16:168–175. doi: 10.1038/nsmb.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 Å crystal structure. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 15.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AG, Tate CG, Schertler GF. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banères JL, Mesnier D, Martin A, Joubert L, Dumuis A, Bockaert J. Molecular characterization of a purified 5-HT4 receptor: a structural basis for drug efficacy. J Biol Chem. 2005;280:20253–20260. doi: 10.1074/jbc.M412009200. [DOI] [PubMed] [Google Scholar]
- 19.Khasawneh FT, Huang JS, Turek JW, Le Breton GC. Differential mapping of the amino acids mediating agonist and antagonist coordination with the human thromboxane A2 receptor protein. J Biol Chem. 2006;281:26951–26965. doi: 10.1074/jbc.M507469200. [DOI] [PubMed] [Google Scholar]
- 20.Shire D, Calandra B, Delpech M, Dumont X, Kaghad M, Le Fur G, Caput D, Ferrara P. Structural features of the central cannabinoid CB1 receptor involved in the binding of the specific CB1 antagonist SR 141716A. J Biol Chem. 1996;271:6941–6946. doi: 10.1074/jbc.271.12.6941. [DOI] [PubMed] [Google Scholar]
- 21.Fay JF, Dunham TD, Farrens DL. Cysteine residues in the human cannabinoid receptor: only C257 and C264 are required for a functional receptor, and steric bulk at C386 impairs antagonist SR141716A binding. Biochemistry. 2005;44:8757–8769. doi: 10.1021/bi0472651. [DOI] [PubMed] [Google Scholar]
- 22.de Bakker PI, DePristo MA, Burke DF, Blundell TL. Ab initio construction of polypeptide fragments: Accuracy of loop decoy discrimination by an all-atom statistical potential and the AMBER force field with the Generalized Born solvation model. Proteins. 2003;51:21–40. doi: 10.1002/prot.10235. [DOI] [PubMed] [Google Scholar]
- 23.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 24.Cole C, Barber JD, Barton J. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raghava GPS. Protein secondary structure prediction using nearest neighbor and neural network approach. CASP4. 2000:75–76. [Google Scholar]
- 26.Karnik SS, Sakmar TP, Chen HB, Khorana HG. Cysteine residues 110 and 187 are essential for the formation of correct structure in bovine rhodopsin. Proc Natl Acad Sci USA. 1988;85:8459–8463. doi: 10.1073/pnas.85.22.8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davidson FF, Loewen PC, Khorana HG. Structure and function in rhodopsin: replacement by alanine of cysteine residues 110 and 187, components of a conserved disulfide bond in rhodopsin, affects the light-activated metarhodopsin II state. Proc Natl Acad Sci USA. 1994;91:4029–4033. doi: 10.1073/pnas.91.9.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hwa J, Klein-Seetharaman J, Khorana HG. Structure and function in rhodopsin: Mass spectrometric identification of the abnormal intradiscal disulfide bond in misfolded retinitis pigmentosa mutants. Proc Natl Acad Sci USA. 2001;98:4872–4876. doi: 10.1073/pnas.061632798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karnik SS, Gogonea C, Patil S, Saad Y, Takezako T. Activation of G-protein-coupled receptors: a common molecular mechanism. Trends Endocrinol Metab. 2003;14:431–437. doi: 10.1016/j.tem.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 30.Nebane NM, Hurst DP, Carrasquer CA, Qiao Z, Reggio PH, Song ZH. Residues accessible in the binding-site crevice of transmembrane helix 6 of the CB2 cannabinoid receptor. Biochemistry. 2008;47:13811–13821. doi: 10.1021/bi8007802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu R, Hubbard JR, Martin BR, Kalimi MY. Roles of sulfhydryl and disulfide groups in the binding of CP-55,940 to rat brain cannabinoid receptor. Mol Cell Biochem. 1993;121:119–126. doi: 10.1007/BF00925970. [DOI] [PubMed] [Google Scholar]
- 32.Shim J-Y. The transmembrane helical domain of the cannabinoid CB1 receptor. Biophys J. 2009;96:3251–3262. doi: 10.1016/j.bpj.2008.12.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ballesteros JA, Weinstein H. Integrated methods for modeling G-protein coupled receptors. In: PM Conn, SC Sealfon., editors. Methods in Neuroscience. San Francisco: Academic Press; 1995. pp. 366–428. [Google Scholar]
- 34.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J, Im W, Brooks CL., 3rd Balancing solvation and intramolecular interactions: toward a consistent generalized Born force field. J Am Chem Soc. 2006;128:3728–3736. doi: 10.1021/ja057216r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buck M, Bouguet-Bonnet S, Pastor RW, MacKerell AD., Jr Importance of the CMAP correction to the CHARMM22 protein force field: dynamics of hen lysozyme. Biophys J. 2006;90:L36–L38. doi: 10.1529/biophysj.105.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A Program for Macromolecular Energy Minimization, and Dynamics Calculations. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 38.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck J, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher IWE, Roux B, Schlenkrich M, Smith J, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 39.Feller S, MacKerell AD., Jr An Improved Empirical Potential Energy Function for Molecular Simulations of Phospholipids. J Phys Chem B. 2000;104:7510–7515. [Google Scholar]
- 40.Saam J, Tajkhorshid E, Hayashi S, Schulten K. Molecular dynamics investigation of primary photoinduced events in the activation of rhodopsin. Biophys J. 2002;83:3097–3112. doi: 10.1016/S0006-3495(02)75314-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: The Langevin piston method. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- 42.Hoover WG. Canonical dynamics: Equilibrium phase-space distributions. Phys Rev A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 43.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle-mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 44.Tuckerman M, Berne BJ. Reversible multiple time scale molecular dynamics. J Chem Phys. 1992;97:1990–2001. [Google Scholar]
- 45.Fiser A, Do RKG, Sali A. Modeling of loops in protein structures. Prot Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darve E, Pohorille A. Calculating free energies using average force. J Chem Phys. 2001;115:9169–9183. [Google Scholar]
- 47.Hénin J, Chipot C. Overcoming free energy barriers using unconstrained molecular dynamics simulations. J Chem Phys. 2004;121:2904–2914. doi: 10.1063/1.1773132. [DOI] [PubMed] [Google Scholar]
- 48.Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 49.McGuffin LJ, Jones DT. Improvement of the GenTHREADER method for genomic fold recognition. Bioinformatics. 2003;19:874–881. doi: 10.1093/bioinformatics/btg097. [DOI] [PubMed] [Google Scholar]
- 50.Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- 51.Lange OF, Grubmüller H. Generalized Correlation for Biomolecular Dynamics. Proteins. 2006;62:1053–1061. doi: 10.1002/prot.20784. [DOI] [PubMed] [Google Scholar]
- 52.Smart OS, Goodfellow JM, Wallace BA. The Pore Dimensions of Gramicidin A. Biophys J. 1993;65:2455–2460. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitmore L, Wallace BA. DICHROWEB an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chothia C. Structural invariants in protein folding. Nature. 1975;254:304–308. doi: 10.1038/254304a0. [DOI] [PubMed] [Google Scholar]
- 55.Fersht A. Structure and Mechanism in Protein Science. New York: W. H. Freeman and Company; 1999. pp. 508–539. [Google Scholar]
- 56.Murphy JW, Kendall DA. Integrity of extracellular loop 1 of the human cannabinoid receptor 1 is critical for high-affinity binding of the ligand CP 55,940 but not SR 141716A. Biochem Pharmacol. 2003;65:1623–1631. doi: 10.1016/s0006-2952(03)00155-2. [DOI] [PubMed] [Google Scholar]
- 57.Kapur A, Samaniego P, Thakur GA, Makriyannis A, Abood ME. Mapping the structural requirements in the CB1 cannabinoid receptor transmembrane helix II for signal transduction. J Pharmacol Exp Ther. 2008;325:341–348. doi: 10.1124/jpet.107.133256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson RM, Hecht K, Deber CM. Aromatic and cation-pi interactions enhance helix-helix association in a membrane environment. Biochemistry. 2007;46:9208–9214. doi: 10.1021/bi7008773. [DOI] [PubMed] [Google Scholar]
- 59.Song ZH, Feng W. Absence of a conserved proline and presence of a conserved tyrosine in the CB2 cannabinoid receptor are crucial for its function. FEBS Lett. 2002;531:290–294. doi: 10.1016/s0014-5793(02)03537-8. [DOI] [PubMed] [Google Scholar]
- 60.McAllister SD, Rizvi G, Anavi-Goffer S, Hurst DP, Barnett-Norris J, Lynch DL, Reggio PH, Abood ME. An aromatic microdomain at the cannabinoid CB(1) receptor constitutes an agonist/inverse agonist binding region. J Med Chem. 2003;46:5139–5152. doi: 10.1021/jm0302647. [DOI] [PubMed] [Google Scholar]
- 61.Ahn KH, Bertalovitz AC, Mierke DF, Kendall DA. Dual Role of the Second Extracellular Loop of the Cannabinoid Receptor One: Ligand Binding and Receptor Localization. Mol Pharmacol. 2009;76:833–842. doi: 10.1124/mol.109.057356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singh R, Hurst DP, Barnett-Norris J, Lynch DL, Reggio PH, Guarnieri F. Activation of the cannabinoid CB1 receptor may involve a W6 48/F3 36 rotamer toggle switch. J Pept Res. 2002;60:357–370. doi: 10.1034/j.1399-3011.2002.21065.x. [DOI] [PubMed] [Google Scholar]
- 63.Shi L, Liapakis G, Xu R, Guarnieri F, Ballesteros JA, Javitch JA. Beta2 adrenergic receptor activation Modulation of the proline kink in transmembrane 6 by a rotamer toggle switch. J Biol Chem. 2002;277:40989–40996. doi: 10.1074/jbc.M206801200. [DOI] [PubMed] [Google Scholar]
- 64.Baldwin JM, Schertler GF, Unger VM. An alpha-carbon template for the transmembrane helices in the rhodopsin family of G-protein-coupled receptors. J Mol Biol. 1997;272:144–164. doi: 10.1006/jmbi.1997.1240. [DOI] [PubMed] [Google Scholar]
- 65.Massotte D, Kieffer BL. The second extracellular loop: a damper for G protein-coupled receptors? Nat. Struct Mol Biol. 2005;12:287–288. doi: 10.1038/nsmb0405-287. [DOI] [PubMed] [Google Scholar]
- 66.Goodwin JA, Hulme EC, Langmead CJ, Tehan BG. Roof and floor of the muscarinic binding pocket: variations in the binding modes of orthosteric ligands. Mol Pharmacol. 2007;72:1484–1496. doi: 10.1124/mol.107.038265. [DOI] [PubMed] [Google Scholar]
- 67.Jäger D, Schmalenbach C, Prilla S, Schrobang J, Kebig A, Sennwitz M, Heller E, Tränkle C, Holzgrabe U, Höltje HD, Mohr K. Allosteric small molecules unveil a role of an extracellular E2/transmembrane helix 7 junction for G protein-coupled receptor activation. J Biol Chem. 2007;282:34968–34976. doi: 10.1074/jbc.M705563200. [DOI] [PubMed] [Google Scholar]
- 68.Ruan KH, Cervantes V, Wu J. Ligand-specific conformation determines agonist activation and antagonist blockade in purified human thromboxane A2 receptor. Biochemistry. 2009;48:3157–3165. doi: 10.1021/bi801443g. [DOI] [PubMed] [Google Scholar]
- 69.Reggio PH. Ligand-ligand and ligand-receptor approaches to modeling the cannabinoid CB1 and CB2 receptors: achievements and challenges. Curr Med Chem. 1999;6:665–683. [PubMed] [Google Scholar]
- 70.Kapur A, Hurst DP, Fleischer D, Whitnell R, Thakur GA, Makriyannis A, Reggio PH, Abood ME. Mutation studies of Ser7.39 and Ser2.60 in the human CB1 cannabinoid receptor: evidence for a serine-induced bend in CB1 transmembrane helix 7. Mol Pharmacol. 2007;71:1512–1524. doi: 10.1124/mol.107.034645. [DOI] [PubMed] [Google Scholar]
- 71.Howlett AC, Padgett LW, Shim J-Y. Cannabinoid agonist-selective regulation of G-protein coupling. In: PH Reggio., editor. The Cannabinoid Receptors. New York: Humana Press; 2008. pp. 173–193. [Google Scholar]
- 72.Wang T, Duan Y. Ligand entry and exit pathways in the beta2-adrenergic receptor. J Mol Biol. 2009;392:1102–1115. doi: 10.1016/j.jmb.2009.07.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McAllister SD, Tao Q, Barnett-Norris J, Buehner K, Hurst DP, Guarnieri F, Reggio PH, Nowell Harmon KW, Cabral GA, Abood ME. A critical role for a tyrosine residue in the cannabinoid receptors for ligand recognition. Biochem Pharmacol. 2002;63:2121–2136. doi: 10.1016/s0006-2952(02)01031-6. [DOI] [PubMed] [Google Scholar]
- 74.Picone RP, Khanolkar AD, Xu W, Ayotte LA, Thakur GA, Hurst DP, Abood ME, Reggio PH, Fournier DJ, Makriyannis A. (−)-7'-Isothiocyanato-11-hydroxy-1',1'-dimethylheptylhexahydrocannabinol (AM841), a high-affinity electrophilic ligand, interacts covalently with a cysteine in helix six and activates the CB1 cannabinoid receptor. Mol Pharmacol. 2005;68:1623–1635. doi: 10.1124/mol.105.014407. [DOI] [PubMed] [Google Scholar]
- 75.Ballesteros JA, Jensen AD, Liapakis G, Rasmussen SG, Shi L, Gether U, Javitch JA. Activation of the beta 2-adrenergic receptor involves disruption of an ionic lock between the cytoplasmic ends of transmembrane segments 3 and 6. J Biol Chem. 2001;276:29171–29177. doi: 10.1074/jbc.M103747200. [DOI] [PubMed] [Google Scholar]
- 76.Shen CP, Xiao JC, Armstrong H, Hagmann W, Fong TM. F200A substitution in the third transmembrane helix of human cannabinoid CB1 receptor converts AM2233 from receptor agonist to inverse agonist. Eur J Pharmacol. 2006;531:41–46. doi: 10.1016/j.ejphar.2005.12.026. [DOI] [PubMed] [Google Scholar]
- 77.Tastan O, Klein-Seetharaman J, Meirovitch H. The effect of loops on the structural organization of alpha-helical membrane proteins. Biophys J. 2009;96:2299–2312. doi: 10.1016/j.bpj.2008.12.3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.