Abstract
Recently, we identified proteins that co-purify with the human spliceosome using mass spectrometry. One of the identified proteins, CDC5L, corresponds to the human homologue of the Schizosaccharomyces pombe CDC5+ gene product. Here we show that CDC5L is part of a larger multiprotein complex in HeLa nuclear extract that incorporates into the spliceosome in an ATP-dependent step. We also show that this complex is required for the second catalytic step of pre-mRNA splicing. Immunodepletion of the CDC5L complex from HeLa nuclear extract inhibits the formation of pre-mRNA splicing products in vitro but does not prevent spliceosome assembly. The first catalytic step of pre-mRNA splicing is less affected by immunodepleting the complex. The purified CDC5L complex in HeLa nuclear extract restores pre-mRNA splicing activity when added to extracts that have been immunodepleted using anti-CDC5L antibodies. Using mass spectrometry and database searches, the major protein components of the CDC5L complex have been identified. This work reports a first purification and characterization of a functional, human non-snRNA spliceosome subunit containing CDC5L and at least five additional protein factors.
Keywords: CDC5L/mass spectrometry/pre-mRNA splicing/spliceosome
Introduction
Nuclear pre-mRNA splicing is the process by which the introns (intervening sequences) in the primary transcript are specifically removed and the coding sequences joined to form mature mRNA, which is subsequently transported into the cytoplasm for protein synthesis. Splicing takes place in the nucleus, via a two-step transesterification mechanism, and this is catalysed by a large RNA–protein complex termed the spliceosome. The major subunits of spliceosomes are the U1, U2, U5 and U4/U6 small nuclear ribonucleoprotein particles (snRNPs), each of which contains the corresponding snRNA and a set of snRNP proteins (reviewed by Krämer, 1996; Will and Lürhmann, 1997). In addition, other non-snRNP protein splicing factors are also required for spliceosome formation and splicing (Will and Lürhmann, 1997; Staley and Guthrie, 1998). Spliceosome assembly occurs in a pathway that involves the sequential binding of snRNP and protein splicing factors to conserved intron sequences to form the active complex. This is a multistep process and separate ATP-dependent complexes designated A, B and C have been identified in vitro (reviewed by Reed and Palandjian, 1997). The C complex contains the active spliceosome while the A and B complexes contain assembly intermediates. The detailed roles played by many spliceosomal proteins in the pre-mRNA splicing mechanism are not yet fully understood.
The protein composition of mammalian spliceosomes has been studied using complexes purified from HeLa nuclear extracts by a combination of gel filtration and affinity chromatography (Reed, 1990; Bennett et al., 1992). More recently in our laboratories, a large-scale analysis of spliceosome-associated proteins has been carried out using mass spectrometry and database searches (Neubauer et al., 1998). One of the spliceosomal proteins identified by mass spectrometry, spot number 61, is the human homologue of the CDC5+ gene product in Schizosaccharomyces pombe.
The human spliceosomal protein in spot number 61 (called CDC5L gene product; Groenen et al., 1998) had previously been isolated in a yeast two-hybrid screen of a HeLa cDNA library using cytoplasmic domain amino acids of the human thrombin receptor as bait and identified as the human homologue of the fission yeast CDC5 gene product (Bernstein and Coughlin, 1997). Although the function of the CDC5L protein was not determined, the authors reported that it is rapidly translocated from the cytoplasm into the cell nucleus upon serum stimulation of CV-1 cells (Bernstein and Coughlin, 1997). It has been suggested that CDC5L may have a role in transcription because of sequence similarities in certain domains with the proto-oncogenic transcription factor c-Myb (Ohi et al., 1994; Bernstein and Coughlin, 1997). The Arabadopsis thaliana homologue (AtCDC5), when overexpressed in S.pombe, is able to complement the growth-defective phenotype of an S.pombe CDC5+ temperature-sensitive mutant. AtCDC5 was also reported to have a sequence-specific DNA-binding activity (Hirayama and Shinozaki, 1996). A CDC5L homologue was identified in Xenopus embryos as a mitotically phosphorylated protein (Stukenberg et al., 1997). A genetic analysis of S.pombe for cell division cycle mutants indicated that the CDC5+ gene encodes an essential protein and that the gene’s function may be necessary in the G2 phase of the cell cycle (Ohi et al., 1994). Recently, it has been suggested that overexpression of CDC5L in mammalian cells shortened the G2 phase of the cell cycle. A dominant-negative mutant of the protein lacking the C-terminal activation domain slowed G2 progression and delayed entry into mitosis (Bernstein and Coughlin, 1998). The CDC5L protein is highly conserved in eukaryotes (Ohi et al., 1998) and it has been shown that in yeast it is part of a multiprotein complex that is important for pre-mRNA splicing (Burns et al., 1999; McDonald et al., 1999; Tsai et al., 1999). It has been shown that the CDC5L protein co-purifies with the spliceosome using HeLa nuclear extracts and that the protein co-localizes with other splicing factors in mammalian cells (Neubauer et al., 1998; Burns et al., 1999). Recently, it has been suggested that CDC5L is in a complex with protein phosphatase 1 (PP1) and that the interaction between the proteins in the complex is mediated by the nuclear regulator of PP1, called NIPP1 (Boudrez et al., 2000).
In order to characterize the role of human CDC5L in the pre-mRNA splicing mechanism, we have studied CDC5L and analysed its interaction with other protein factors in HeLa nuclear extract. We show that the catalytic steps of splicing are inhibited (with the second catalytic step being more sensitive) upon immunodepletion of CDC5L from HeLa nuclear extracts, whereas spliceosome assembly still occurs. Splicing capability can be restored to immunodepleted extracts by addition of a purified multiprotein complex containing CDC5L. This complex contains previously known spliceosomal proteins and several novel proteins that are highly conserved across species. Using two different purification strategies and mass spectrometry analysis we show that the CDC5L complex is made up of a core of six proteins that are stably associated throughout the various purification steps.
Results
ATP-dependent association of CDC5L with the spliceosome complex
CDC5L contains two RNA-binding motifs and binds adeno-pre-mRNA strongly in vitro (data not shown). We studied its association with adeno-pre-mRNA under splicing conditions, to verify its association with the spliceosomal complex in HeLa nuclear extract. Anti-CDC5L antibodies were used to immunoprecipitate the protein from a splicing reaction and the presence of co-immunoprecipitated adeno-mRNA determined by denaturing PAGE (Figure 1A). The data show that pre-mRNA, as well as splicing intermediates and products, are all co-immunoprecipitated by anti-CDC5L antibodies (lanes 7 and 8). The specificity of this immunoprecipitation reaction is demonstrated by the fact that in the presence of an anti-CDC5L specific peptide, immunoprecipitation of adeno-pre-mRNA and splicing intermediates and products was blocked (lanes 5 and 6) or reduced to background levels (lane 4).
Fig. 1. Specific immunoprecipitation of the spliceosome by anti-CDC5L. (A) pre-mRNA splicing reactions/spliceosomes were immunoprecipitated using 5–10 µg of anti-CDC5L antibody in the presence and absence of the antibody specific peptide (10-fold molar excess) and the immunoprecipitated RNA revealed by autoradiography. Lane 1 contained the pre-mRNA transcript used in the splicing reactions. Lanes 2 and 3 show the input splicing reaction. Lane 4 contains a mock immunoprecipitation using pre-immune IgG. Lanes 5 and 6 are duplicate immunoprecipitation reactions performed with anti-CDC5L antibodies in the presence of the antibody specific peptide. Lanes 7 and 8 are duplicate spliceosome immunoprecipitations using anti-CDC5L antibody. The symbols on the right of the figure represent the pre-mRNA band and the different splicing intermediates and products. All the other bands in the figure have been produced by RNA partial degradation. (B) Splicing reactions were prepared as above, but in the presence or absence of ATP and immunoprecipitation reactions performed using anti-CDC5L antibody. The amount of radioactivity immunoprecipitated was measured as relative fluorescence units (RFU) using a PhosphorImager (Molecular Dynamics) and the results presented on a bar chart. The bars marked 1, 3 and 4 represent the amount of radioactivity immunoprecipitated when the splicing reaction is carried out in the presence of ATP. However, in the case of bar 3, the immunoprecipitation was performed using anti-CDC5L antibody pre-blocked with specific peptide, whereas bar 4 represents a mock immunoprecipitation of the spliceosome using pre-immune IgG. Bar 2 shows the amount of radioactivity measured from the immunoprecipitation reaction using anti-CDC5L antibody in the absence of ATP, whereas bar 1 represents a similar immunoprecipitation reaction of a pre-mRNA splicing assay performed in the presence of ATP.
Since spliceosome assembly is ATP dependent (reviewed by Krämer, 1995; Reed and Palandjian, 1997), the specific association of CDC5L with the spliceosome complex was tested further using in vitro splicing reactions performed either with or without ATP (Figure 1B). The data show that there is enhanced co-immunoprecipitation of adeno-RNA with CDC5L when the splicing reaction is performed in the presence of ATP (bar 1) compared with when ATP was absent from the splicing reaction (bar 2). In the presence of an anti-CDC5L specific peptide, the amount of RNA co-immunoprecipitated with the protein is greatly reduced, even though the splicing reaction in this case contains ATP (bar 3). Similar results were obtained from three independent experiments, indicating an increase of ≥10-fold in the co-immunoprecipitation of adeno-RNA by anti-CDC5L antibodies in the presence of ATP. In summary, our data indicate that CDC5L associates specifically with adeno-pre-mRNA in HeLa nuclear extract under splicing conditions.
Immunodepletion of CDC5L from HeLa nuclear extract inhibits pre-mRNA splicing
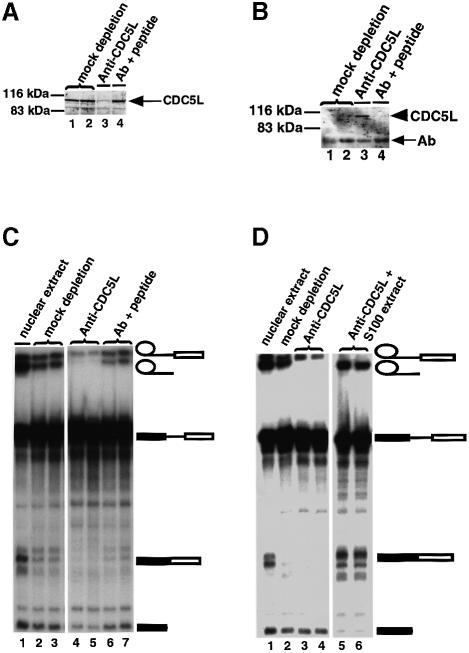
We next examined whether the presence of CDC5L in HeLa nuclear extract is important for splicing activity by immunodepletion with anti-CDC5L antibodies (Figure 2). Analysis of the depleted HeLa extract and parallel control extracts showed that CDC5L in the nuclear extracts is effectively removed by this procedure (Figure 2A, lane 3). Analysis of the beads from the immunodepletion experiments revealed the immunoprecipitated CDC5L (Figure 2B, lane 3). When the anti-CDC5L antibody was pre-blocked with the cognate CDC5L peptide, immunoprecipitation of CDC5L was blocked (Figure 2A and B, lanes 4). Nuclear extract supernatants from both the immunodepleted and the control (i.e. peptide pre-blocked antibody) samples were used in pre-mRNA splicing assays (Figure 2C). In all the splicing experiments we observed that both catalytic steps of pre-mRNA splicing were reduced when we used HeLa nuclear extracts depleted of CDC5L. However, we consistently observed that the formation of splicing products was inhibited (Figure 2C, lanes 4 and 5) whereas the level of splicing intermediates was less reduced compared with the mock depleted extracts (Figure 2C, compare lanes 2 and 3 with 4 and 5). A control extract that was incubated with antibody blocked with cognate CDC5L peptide was still able to splice adeno-pre-mRNA (Figure 2C, lanes 6 and 7). This indicates that the inhibition of pre-mRNA splicing results from either removal of CDC5L itself from the anti-CDC5L immunodepleted nuclear extracts or removal of one or more CDC5L-associated factors.
Fig. 2. Immunodepletion of CDC5L in HeLa nuclear extract inhibits pre-mRNA splicing. (A) Supernatants from immunodepletion experiments were probed by western blotting using anti-CDC5L antibody and the protein revealed using ECL (Amersham). Lanes 1 and 2 are duplicates containing supernatants of mock immunodepletions using pre-immune IgG. Lane 3 contains the supernatant from anti-CDC5L antibody immunodepleted nuclear extract and lane 4 is similar to lane 3 except that the antibody was pre-incubated with specific peptide before use in the immunodepletion experiment. The arrow indicates the CDC5L bands. (B) Proteins from the beads corresponding to the immunodepletions in (A) were separated by SDS–PAGE, transferred to nitrocelluose and probed with anti-CDC5L antibody as above. Lane numbers are identical to (A) except that the samples in each lane contain protein immunoprecipitated onto beads by bound antibody. The arrowhead indicates the CDC5L band while the arrow (Ab) shows bands corresponding to antibody heavy chain polypeptides. CDC5L is only detected in lane 3. (C) The supernatants in (A) were used in pre-mRNA splicing reactions, and the splicing intermediates and products separated on a 10% polyacrylamide–8 M urea denaturing gel. The symbols on the right of the figure represent the pre-mRNA and the different splicing intermediates and products. Bands not marked by symbols correspond to partial degradation products of the pre-mRNA. Lane 1 contains a pre-mRNA splicing control using untreated nuclear extract. Lanes 2 and 3 are duplicates from mock depletion experiments. The lanes marked 4 and 5 contained duplicates of splicing reactions using nuclear extracts depleted of CDC5L, while lanes 6 and 7 contain splicing reactions using nuclear extract treated with anti-CDC5L antibody pre-blocked with the antibody specific peptide. (D) S100 extract restores splicing activity to nuclear extract depleted with anti-CDC5L antibodies. Lane 1 shows a splicing reaction with untreated nuclear extract. Lane 2 contains a mock depletion using sheep pre-immune IgG. Lanes 3 and 4 show duplicate pre-mRNA splicing reactions using nuclear extracts immunodepleted with anti-CDC5L antibodies, and lanes 5 and 6 represent duplicate splicing reactions using nuclear extracts immunodepleted with anti-CDC5L antibodies to which have been added ∼16–20 µg of S100 cytoplasmic extract.
We next attempted to restore splicing activity to the immunodepleted HeLa nuclear extract by adding recombinant, Escherichia coli expressed CDC5L protein. However, when purified recombinant CDC5L protein alone was added back to the immunodepleted nuclear extract, pre-mRNA splicing activity was not restored (data not shown). We cannot exclude the possibility that recombinant CDC5L protein expressed in E.coli is inactive for splicing, although we observed that it has strong RNA binding activity in vitro (data not shown). However, we consider it more likely that the failure of recombinant CDC5L alone to restore pre-mRNA splicing activity to immunodepleted nuclear extracts may be because it acts as part of a larger complex that is co-depleted by the anti-CDC5L antibody. Indeed, it has been shown recently that the CDC5L homologue in S.pombe is part of a 40S multiprotein complex (McDonald et al., 1999).
When a HeLa S100 cytoplasmic extract, which lacks splicing activity, was added to the extracts immunodepleted using anti-CDC5L antibodies, pre-mRNA splicing activity was efficiently restored (Figure 2D, lanes 5 and 6) and blotting experiments confirmed that this S100 extract contains CDC5L (data not shown). Nuclear extracts immunodepleted using anti-CDC5L antibodies that had no S100 cytoplasmic extract added showed inhibition of formation of pre-mRNA splicing products (Figure 2D, lanes 3 and 4). These data demonstrate that the nuclear extract immunodepleted using anti-CDC5L antibodies fails to splice due to the absence of one or more components that are present in S100 extract.
Effect of the immunodepletion of CDC5L in nuclear extract on spliceosome assembly
We analysed whether immunodepletion of CDC5L from nuclear extract inhibits the formation of splicing products because it prevents spliceosome assembly. Splicing complexes formed in control and CDC5L immunodepleted extracts (Figure 3) were analysed by non-denaturing gel electrophoresis. This showed that spliceosome assembly was not blocked in HeLa nuclear extracts depleted with the anti-CDC5L antibody (Figure 3, lane 4). This is consistent with the observation that the first catalytic step of pre-mRNA splicing can still occur, albeit at lower efficiency, in the immunodepleted nuclear extract.
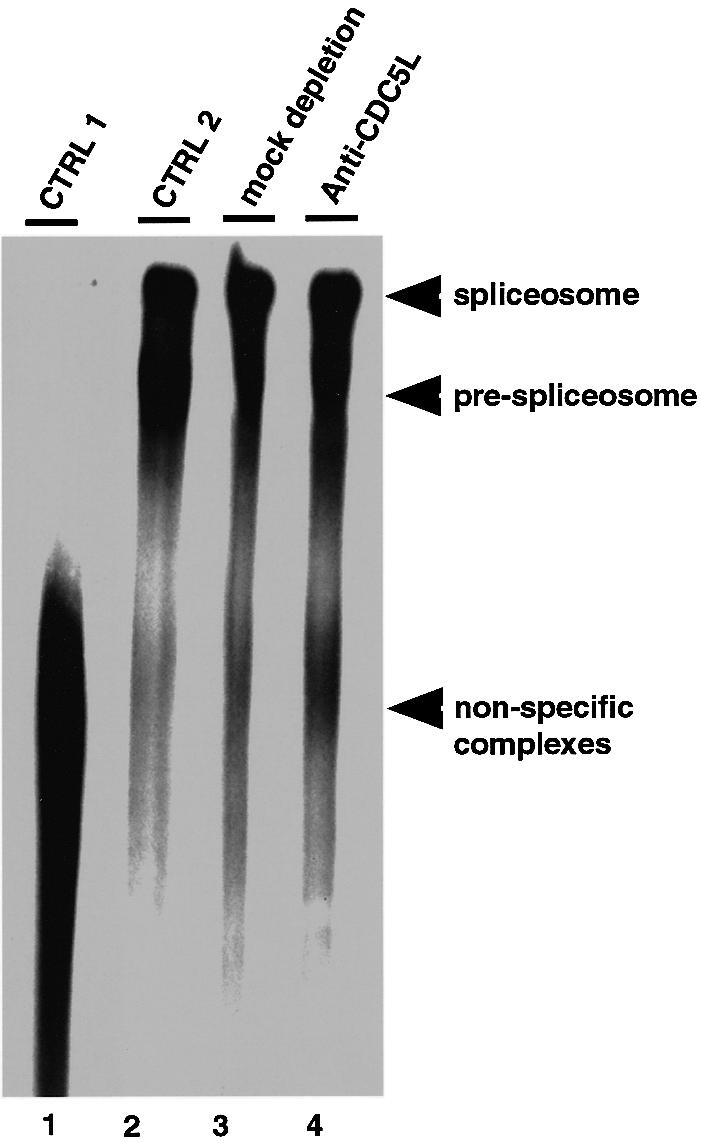
Fig. 3. Polyacrylamide/agarose composite native gel of spliceosome complexes detected by autoradiography. Lane 1, marked CTRL 1, represents a control containing a splicing reaction incubated on ice for 1 h. Lane 2 contained a normal splicing reaction (incubated for 1 h at 30°C) using untreated nuclear extract. Lane 3 contained a splicing assay using nuclear extract mock depleted with pre-immune IgG, while lane 4 contained a splicing reaction using CDC5L immunodepleted nuclear extract. Arrowheads indicate the different complexes separated on the gel.
Purification of CDC5L-associated proteins in HeLa nuclear extract
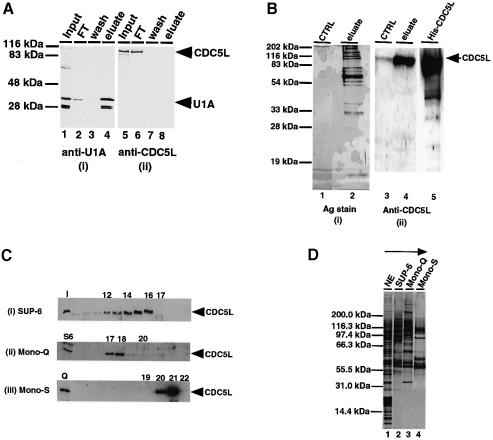
To examine whether CDC5L is part of a complex that contains other splicing factors, we first tested whether it is stably associated with snRNPs. Therefore, snRNP proteins were purified by affinity chromatography on an anti-2,2,7-trimethylguanosine cap column as described (Bach et al., 1990). Purified snRNP proteins were separated by SDS–PAGE, transferred onto a nitrocellulose membrane and probed with both anti-CDC5L antibodies and with polyclonal anti-U1A rabbit antiserum as a positive control (Figure 4A). As expected, the U1A protein, which is part of the U1 snRNP complex, was detected in the snRNP protein eluate from the column [Figure 4A(i), lane 4]. In contrast, CDC5L is absent from the snRNP eluate fraction (Figure 4A, lane 8) and quantitatively present in the flow-through fraction (Figure 4A, lane 6). These data indicate that the protein is not a stable snRNP component.
Fig. 4. Purification of a multiprotein complex containing CDC5L. (A) Probing of purified snRNPs for the presence of CDC5L. Total snRNPs in HeLa nuclear extract were purified as described in Materials and methods. About 15–20 µg of purified snRNPs fractions were separated on a 12% SDS–PAGE gel and the fractions probed with anti-U1A and anti-CDC5L antibodies. Lanes 1 and 5 contain the input nuclear extract. Lanes 2 and 6 contain the column flow-through fractions, while lanes 3 and 7 have the column wash fractions. Lanes 4 and 8 contain the column eluate obtained using m3G-cap dinucleotide. The fractions in panel (i) were probed with anti-U1A antibody and the fractions in panel (ii) probed with anti-CDC5L antibody. Arrowheads on the right of the figure indicate the bands representing U1A and CDC5L. (B) Purification of the CDC5L complex by immunoaffinity chromatography. The complex was purified by passing 80–100 mg of HeLa nuclear extract through a column containing covalently coupled anti-CDC5L antibody. The column was eluted using glycine (see Materials and methods) and the eluate separated on a 12% SDS–PAGE gel and the gel either silver stained (i) or western blotted and probed with anti-CDC5L antibody (ii). Lanes 1 and 3 contained the control samples, i.e. eluate from the column pre-blocked with antibody specific peptide. Lanes 2 and 4 contained the eluate from the antibody column and lane 5 contained purified, E.coli expressed, His6-tagged CDC5L. (C) Identification of chromatographic fractions containing CDC5L in the tandem purification procedure. Aliquots of fractions from each purification step were separated on a 4–12% pre-cast SDS–PAGE gel (Novex) and the presence of CDC5L protein confirmed by probing western blots with anti-CDC5L antibody. The numbers at the top of each panel represent the fraction numbers and the arrowheads on the right of each panel indicate the position of CDC5L in the gel. Panel (i) contained fractions from the gel filtration column (Superose-6). The lane marked I contained the input nuclear extract. Panel (ii) shows fractions from the Mono-Q column and panel (iii) contains fractions from the Mono-S column. The lanes marked S6 and Q represent pools of Superose-6 and Mono-Q fractions containing CDC5L that were used as the input for the purification procedure. (D) Tandem chromatographic purification of the CDC5L-associated complex in HeLa nuclear extract. Aliquots of pooled CDC5L-containing fractions from each purification step were separated on a 4–12% pre-cast SDS–PAGE gel (Novex) and the protein bands revealed by silver staining. Lane 1 contained the input nuclear extract. Lanes 2–4 contained pools of the Superose 6, Mono-Q and Mono-S column fractions containing CDC5L, respectively. The arrow at the top of the figure indicates the purification steps of the CDC5L complex from HeLa nuclear extract. The first step involves gel filtration (Superose-6 or SUP-6) and the last step is an ion exchange purification of the complex on a Mono-S column.
We next decided to purify the CDC5L complex from HeLa nuclear extract and test its ability to restore splicing activity to extracts immunodepleted using the anti-CDC5L antibody. The complex was purified either by affinity chromatography using anti-CDC5L antibodies (Figure 4B) or by tandem chromatographic steps involving gel filtration and ion exchange chromatographic procedures (Figure 4C). The fractions collected were tested for the presence of CDC5L by transferring the proteins onto a nitrocellulose membrane and probing with anti-CDC5L antibodies [Figure 4B(ii) and C]. As judged by silver staining [Figure 4B(i) and D], we estimate that ∼25–30 proteins from HeLa nuclear extract co-purify with CDC5L under these conditions.
Purified CDC5L complex restores splicing activity to immunodepleted HeLa nuclear extracts
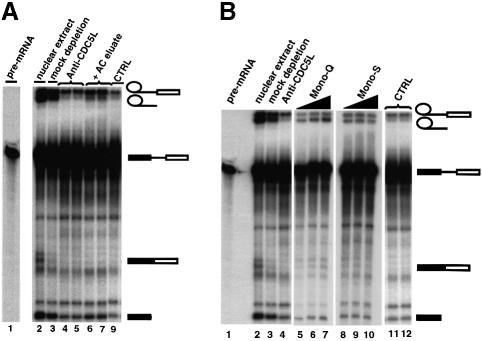
We tested whether purified CDC5L isolated from HeLa cells as a multiprotein complex would restore pre-mRNA splicing activity to extracts immunodepleted with anti-CDC5L antibodies. Chromatographic fractions containing the peak levels of CDC5L protein were added back to the immunodepleted extracts (Figure 5). Addition of fractions eluted from the affinity purified CDC5L preparation showed a small but reproducible increase in the formation of pre-mRNA splicing product (Figure 5A, lanes 6 and 7, and data not shown). Using different elution conditions (i.e. anti-CDC5L cognate peptide or 100 mM glycine–HCl pH 2.5) made no significant difference to the overall efficiency of restoration of splicing activity that was observed.
Fig. 5. Purified CDC5L-associated complex restores splicing activity to nuclear extract depleted of the protein. The symbols on the right of the figure represent the pre-mRNA and the different splicing intermediates and products. Bands not marked by symbols correspond to partial degradation products of the pre-mRNA. (A) About 0.2 µg of eluate from the anti-CDC5L affinity column was added back to the immunodepleted extract before the extracts were used in pre-mRNA splicing reactions. The pre-mRNA splicing intermediates and products were separated on a 10% polyacrylamide–8 M urea denaturing gel. Lane 1 contained ∼15% of the input pre-mRNA. Lanes 2 and 3 contained untreated and mock-depleted nuclear extracts, respectively. Lanes 4 and 5 contain duplicate splicing reactions using nuclear extract immunodepleted using anti-CDC5L antibodies. Lanes 6 and 7 have duplicate splicing reactions using the same nuclear extract as above except that these lanes also have the CDC5L-containing eluate from the affinity column. Lane 9, marked CTRL, contains a control splicing reaction using immunodepleted nuclear extract as above except that here the eluate from the antibody specific peptide pre-blocked column (see Figure 4B, lane 1) was used in the add back. (B) Fractions from the Mono-Q and Mono-S columns were used to restore splicing activity to CDC5L immunodepleted nuclear extract. Lane 1 contains ∼15% of the input pre-mRNA used in the splicing reaction. Lanes 2 and 3 have untreated and mock depleted nuclear extracts, respectively. Lane 4 contains a splicing reaction using nuclear extract immunodepleted using anti-CDC5L antibodies. The same nuclear extract was used for the splicing reactions in lanes 5–12. In the add back experiments, the reactions in lanes 5–7 also had 0.5, 1 and 2 µl, respectively, of the pooled Mono-Q fractions containing CDC5L and lanes 8–10 had similar amounts of the pooled CDC5L-containing Mono-S fractions, respectively. Lane 11 is a control add back experiment containing a pool of Mono-Q fractions 16 and 19 that lack CDC5L, while lane 12 contains a pool of Mono-S column fractions 19 and 22 that is deficient in CDC5L (see Figure 4C above).
When peak fractions from the Mono-Q and Mono-S columns containing the CDC5L protein were added back to extracts immunodepleted with anti-CDC5L antibodies, pre-mRNA splicing activity was also increased (Figure 5B, lanes 5–10). The efficiency of the restoration of splicing activity to CDC5L immunodepleted nuclear extracts was comparable between the Mono-Q and Mono-S fractions containing the peak levels of CDC5L. However, the affinity chromatography column fractions were less efficient in restoring splicing activity to immunodepleted nuclear extracts. This may be due to some loss of activity during the purification/elution of bound proteins or dissociation of factors from the complex. Elution of bound proteins using anti-CDC5L specific peptides proved to be an inefficient recovery method, whereas elution from the affinity column using glycine–HCl pH 2.5 was found to be very efficient, but these harsh elution conditions may cause the denaturation and inactivation of some essential components in the CDC5L complex. Taken together, these data indicate that a protein complex containing CDC5L can increase pre-mRNA splicing activity in nuclear extracts that have been immunodepleted using anti-CDC5L antibodies.
Identification of the CDC5L complex-associated proteins by mass spectrometry
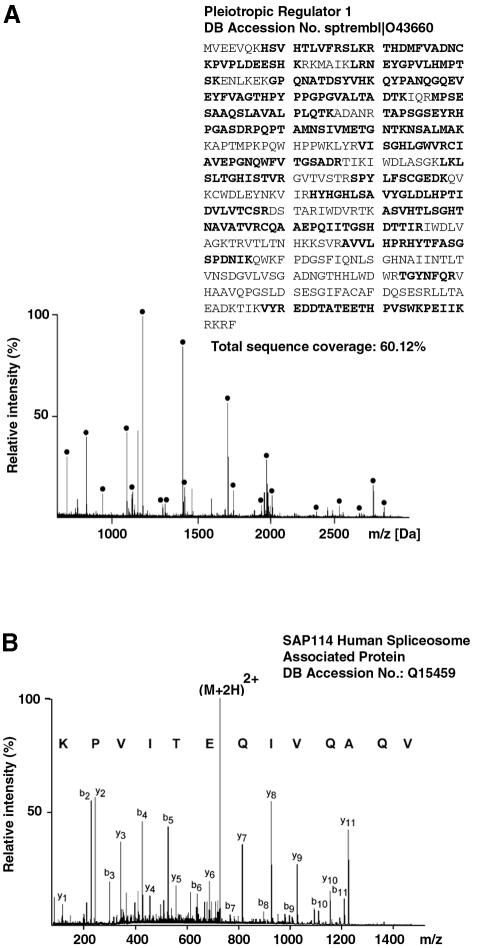
The proteins found to co-purify with CDC5L during either ion exchange chromatography or immunoaffinity purification were analysed by a two-tier mass spectrometric approach in conjunction with sequence database searching as described in Materials and methods. The proteins identified are listed in Tables I–III. An example of protein identification by matrix-assisted laser desorption/ionization (MALDI) peptide mass mapping is given in Figure 6A. Signals in the spectrum correspond to tryptic peptides generated from a protein by in-gel trypsin digestion. The measured peptide masses are used to search a sequence database for proteins that would give rise to a similar tryptic peptide mass map. Data derived from this spectrum unambiguously identified the human pleiotropic regulator 1 (PRL1) (sptrembl: O43660) that we had previously identified as a spliceosomal protein (Neubauer et al., 1998). Signals marked with bullets correspond to tryptic peptides of this protein and all matching peptides cover a total of 60% of the protein sequence.
Table I. CDC5L-associated proteins known to be associated with the spliceosome.
| Protein grouping | SwissProt accession No. | Mol. wt. (kDa) | Orthologues (yeast) | Comments |
|---|---|---|---|---|
| snRNP associated proteins | ||||
| SAP155 | O75533 | 146.5 | ScSAP155 | |
| SAP145 | Q13435 | 88.9 | CUS1 | |
| SAP114 | Q15439 | 88.9 | PRP21 | |
| SAP49 | Q15427 | 44.4 | HSH49 | |
| SMD1 | P13641 | 13.3 | YGR074W | |
| SMD2 | P43331 | 13.9 | YLR275W | |
| SMD3 | P43330 | 13.5 | YLR147C | |
| U2A | P09661 | 28.4 | ||
| U1A | P09012 | 31.3 | MUD1 | |
| Other previously described spliceosomal proteins | ||||
| CDC5L* | Q99974 | 92.5 | Cef1 | high sequence conservation between species |
| ASF/SF2 | Q07955 | 27.7 | ||
| SC35 | Q01130 | 25.5 | ||
| PRL1* | O43660 | 57.5 | PRP46 | 70% homology to yeast protein |
| hnRNP-G | P38159 | 42.4 | Hrp1p | |
| PSF | P23246 | 76.1 | ||
| SPF27* |
O75934 |
26.2 |
NUF1 |
|
| SRM160 | O60585 | 93.6 | contains SR repeat domain | |
*Core protein in the CDC5L-associated protein complex.
Table III. Ribosomal and other proteins that co-purify with the CDC5L complexa.
| Other proteins | Accession No. (SwissProt) | Mol. wt by sequence (kDa) | Homologues (yeast) |
|---|---|---|---|
| Ribosomal proteins | |||
| L12 | P30050 | 17.8 | RPL12A |
| L13 | P26373 | 24.2 | RPL13A |
| S16 | P17008 | 16.4 | RPS16A |
| S25 | P25111 | 13.8 | RPS25A |
| Possible contaminants | |||
| COPD-like protein | P48444 | 57.7 | RET2 or YFR051C |
| TALIN | Q9UHH8 | 271.8 | |
aIt is possible that these proteins are contaminants in the preparation.
Fig. 6. Mass-spectrometric identification of CDC5L-associated proteins. (A) MALDI peptide mass mapping. Peptide masses derived from MALDI mass spectra were used to query an amino acid sequence database. Proteins are identified by correlating the measured peptide masses to a theoretical tryptic digest of all proteins present in the database. In the example shown, human pleiotropic regulator 1 (PRL1) was unambiguously identified in the search and signals corresponding to tryptic peptides of this protein are marked with bullets (60% sequence coverage). (B) Peptide sequencing by nanoelectrospray tandem mass spectrometry. Peptides selected for sequencing from the unseparated peptide mixture undergo collisional dissociation within the collision cell of a tandem mass spectrometer to produce sequence-specific fragment ions. Fragment ion spectra contain signals corresponding to C-terminal (yn-type) as well as N-terminal (bn-type) fragment ions. This information can be assembled into a peptide sequence tag and searched in amino acid as well as nucleotide sequence databases. The information contained in the spectrum shown unambiguously identified the sequence AQVIQETIVPK from the SAP114 human spliceosome-associated protein. Note that the sequence in the spectrum was derived from the C-terminal fragment ion series and hence is printed in the C- to N-terminal direction. Fragment ions are labelled according to Biemann (1990).
Proteins that were either of too low abundance or not present in the NRDB, and could therefore not be identified by MALDI peptide mass mapping, were partially sequenced by nanoelectrospray tandem mass spectrometry on a quadrupole time-of-flight mass spectrometer. An example of a typical result of such an experiment is shown in Figure 6B. A doubly charged peptide (M+2H)2+ was selected from the unseparated peptide mixture and fragmented inside the instrument to yield a series of singly charged fragment ions containing either the N-terminus (b-type) or the C-terminus (y-type) of the peptide (nomenclature according to Biemann, 1990). From the exact mass difference between ions of the same series, a partial amino acid sequence was derived and, together with the fragment ion mass information, assembled into a peptide sequence tag (Mann and Wilm, 1994). The database query retrieved the peptide VQAQVIQETIVPK and comparison of that sequence with the tandem mass spectrum identified the SAP114 human spliceosome-associated protein (swissnew Q15459) with certainty. Several novel putative spliceosomal proteins were identified in the same way by searching peptide sequence tags in the dbEST, NRDB or human genome sequence databases. Whether or not all of these are bona fide components of the spliceosome requires further studies.
During our analysis of the CDC5L-containing complex we identified 17 proteins (Table I) that have previously been shown to be associated with the spliceosome or are essential for pre-mRNA splicing (Lamm and Lamond, 1993; Neubauer et al., 1998; Venables et al., 2000). Thirteen novel putative spliceosomal proteins or proteins that have not previously been shown to have a role in pre-mRNA splicing were also identified (Table II). We have called some of the newly identified proteins that have not been previously named CCAPs (CDC5L complex-associated proteins (CCAP7). One of these proteins (CCAP7) contained the RS repeat sequence motifs characteristic of members of the SR family of pre-mRNA splicing factors (reviewed in Fu, 1995). The other novel proteins all contain previously described domain sequence motifs and/or putative enzyme activities (Table II) that are suggestive of their involvement in a multiprotein/nucleic acid complex and pre-mRNA biogenesis/processing. In addition, a further group of proteins, including four ribosomal proteins, was also identified in the CDC5L complex preparation (Table III). These may be contaminants and have not been investigated further at this stage. Our identification of the proteins in the CDC5L complex by mass spectrometry revealed five proteins that were consistently found to be associated with CDC5L, irrespective of the purification method (Tables I and II, marked with an asterisk). We call these proteins ‘core’ members of the complex. On the other hand, some of the other proteins that co-purified with CDC5L appear not to be stably associated with this complex under all the different purification conditions tested. For example, U1A co-purifies with the complex under our affinity purification conditions and we have observed that this protein will interact directly with CDC5L in vitro (data not shown). U1A co-purifies with CDC5L under gel filtration chromatography conditions, but separates during our ion exchange chromatography experiments (data not shown). Because the ion exchange chromatographic fractions that contain CDC5L but lack some of the ‘non-core’ proteins will restore splicing activity to extracts immunodepleted using anti-CDC5L antibodies, this implies that it is mainly removal of the core proteins during immunodepletion of CDC5L from HeLa nuclear extracts that is responsible for the observed inhibitory effects on pre-mRNA splicing. For example, we have observed that the core protein PRL1 is completely depleted from HeLa nuclear extract upon immunodepletion of CDC5L (data not shown). So far, we have been unable to test the efficiency of removal of the other core proteins on depletion of CDC5L in HeLa nuclear extract because we do not yet have the appropriate reagents for detecting these proteins biochemically. Like U1A, the other snRNP-associated proteins identified as co-purifying with CDC5L eluted at different peaks from that of CDC5L during the ion exchange chromatographic procedures (data not shown).
Table II. Novel CDC5L complex proteins and other proteins that although present in the complex have not previously been shown to be associated with the spliceosome.
| Protein name | SwissProt/DDBJ/EMBL/GenBank accession No. | Mol. wt (kDa) | Orthologues (yeast) | Comments/motifs |
|---|---|---|---|---|
| TOP IIα | Q9UP44 | 175.1 | TOP2p | TOP I activates SR proteins by phosphorylation and interacts with PSF/p54nrb. |
| PP2c (WIP1) | O15297 | 67.4 | PTC1 | Induced in response to ionizing radiation (Fiscella et al., 1997). |
| DDP kinase | Q13327 | 470.4 | TOR1p | |
| PP1-α | P08129 | 38.2 | GLC7 | Very high sequence conservation (93% homology between yeast and human). |
| NF45 | Q12905 | 44.9 | ||
| CCAP1* (HS7C) | P11142 | 71.1 | Ssa2p | 75% homology to Drosophila protein (TrEMBL Q9VG73). |
| CCAP2 (HSPC 148) | AAF29112 (DDBJ/EMBL/GenBank) | 26.6 | SPBC337.06c | Putative RNA helicase with DExD motif; has ATP/GTP-binding site motif A (P-loop). |
| CCAP3* | Q9UMS4 | 55.6 | PRP19-like | Contains several WD40a sequence motifs. |
| CCAP4 | Q15394 | 49 | Has leucine zipper pattern DNA binding motif; ATP/GTP-binding site (P-loop) motif A. | |
| CCAP5 | Q9Y4G5 | 239.9 | Contains several putative PP1-bindingb domain sequence motifs; ATP/GTP binding site motif (P-loop); leucine zipper pattern. | |
| CCAP6* | Q92616 | 266.3 | Has RNP-1 and DNA binding motifs (leucine zipper pattern); putative histidine acid phosphatase. | |
| CCAP7 | Q9UPN6 | 140.8 | Has SR repeat motif domain; ATP/GTP binding site (P-loop) and RNP-1 sequence motifs. | |
| CCAP8 | O60289 | 145.4 | Contains RNA binding and two WWc domain motifs. |
*Core protein in the CDC5L-associated protein complex.
aReviewed by Smith et al. (1999).
cEspanel and Sudol (1999) and references therein.
Put together, our data indicate that there exists a CDC5L ‘core’ complex in HeLa nuclear extract containing at least six proteins that interact with each other and with other splicing factors and enzymes (including protein phosphatases, kinases and helicases) with an essential role in the second catalytic step of pre-mRNA splicing.
Discussion
We had previously shown using mass spectrometry that the protein CDC5L co-purifies with human spliceosomes isolated from HeLa nuclear extracts (Neubauer et al., 1998). In this study we have shown that CDC5L, which is the human homologue of the S.pombe CDC5+ gene product, is one of the core members of a large multiprotein complex that is essential for the formation of pre-mRNA splicing products but not needed for spliceosome assembly. We have purified the complex from HeLa nuclear extract and shown that the purified complex will restore splicing activity to nuclear extracts depleted using anti-CDC5L antibodies. We have identified using mass spectrometry most of the components of the CDC5L complex isolated from HeLa nuclear extract. These data show that the complex contains known spliceosome-associated proteins as well as several novel proteins that have not previously been described as components of the pre-mRNA splicing machinery.
The CDC5+ gene product (CDC5L in humans) was first identified in a genetic screen in S.pombe for cell division cycle mutants. These experiments indicated that the protein in yeast may be involved in the regulation of the cell division cycle (Ohi et al., 1994). A role in the cell division cycle has also been suggested for this protein in humans because overexpression of the protein in mammalian cells leads to a shortening of the G2 phase of the cycle, while a dominant-negative mutant of the protein lacking the C-terminal activation domain affected the G2 phase by delaying entry into mitosis (Bernstein and Coughlin, 1998). Similar observations have been made in other species, i.e. A.thaliana and Drosophila melanogaster (Ohi et al., 1998 and references therein). It has also been suggested that CDC5L is involved in mitogen-activated signalling (Bernstein and Coughlin, 1997). When CV1 or COS-7 cells were transfected with a plasmid encoding a tagged CDC5L protein and the cells grown in serum-free medium, CDC5L was found exclusively in the cytoplasm. However, upon serum stimulation of the transfected cells, it was found to translocate to the nucleus. We have shown in this study that the CDC5L protein in HeLa nuclear extract is part of a functional multiprotein complex that is needed for the second catalytic step of pre-mRNA splicing. Several recent studies have also implicated CDC5L in splicing in both yeast and mammalian cells (Burns et al., 1999; McDonald et al., 1999). This does not exclude the possibility that CDC5L protein also plays an additional direct role in regulating cell cycle progression. An alternative possibility is that the cell cycle defects observed on mutating CDC5 (S.pombe)/CDC5L are an indirect consequence of changes in the splicing machinery, and hence expression of other cell cycle regulatory factors, mediated by CDC5L.
Removal of CDC5L from HeLa nuclear extract by immunodepletion inhibits pre-mRNA splicing in vitro. We were unable to restore splicing activity to the depleted extract by adding back E.coli expressed CDC5L alone. The purified recombinant protein shows strong specific RNA binding activity in vitro (data not shown). Nonetheless, we cannot exclude the fact that its failure to restore splicing may be due to intrinsic inactivity of the recombinant factor, e.g. because of either incorrect folding or lack of post-translational modification. A more likely explanation is that the CDC5L protein functions as a member of a complex containing other factors and one or more of these additional components are required for activity. Indeed, it has been shown recently that the yeast homologue of CDC5L co-purifies in a 40S multiprotein complex (McDonald et al., 1999) and our inability to restore splicing activity to immunodepleted extracts by adding the recombinant CDC5L protein alone is consistent with this result. In support of this model, we have identified a large protein complex containing CDC5L in HeLa nuclear extracts, and have substantially purified it and characterized its protein composition using mass spectrometry. This study identifies a core of at least six protein factors that are stable components of the CDC5L complex.
Our data on the immunodepletion of CDC5L suggest that this complex has an essential function in the second catalytic step of splicing. The presence of the splicing factor PSF and the PP1 α isoform in the CDC5L complex is consistent with this finding. The data presented here show a first biochemical association of the PP1 α isoform with the pre-mRNA splicing machinery or an active complex containing pre-mRNA splicing factors. The presence of this enzyme in the complex is consistent with the link between the effect of immunodepletion of the CDC5L complex in nuclear extracts and the effect of phosphatase inhibitors on pre-mRNA splicing (Mermoud et al., 1992; Tazi et al., 1992; Misteli and Spector, 1996). It has been suggested that PSF is needed for the second catalytic step of pre-mRNA splicing (Gozani et al., 1994). Interestingly, PSF has been shown to interact with PP1 (Hirano et al., 1996); thus, PSF may target PP1 to the CDC5L complex, where it dephosphorylates one or more factors that stimulate the second catalytic step of pre-mRNA splicing. Previous studies using protein phosphatase inhibitors have indicated that protein phosphatase activities (PP1 and PP2A) are required for the catalytic steps of pre-mRNA splicing but not spliceosome assembly (Mermoud et al., 1992; Tazi et al., 1992). This is similar to the effect of depleting CDC5L from nuclear extract and raises the interesting possibility that the effect of PP1 inhibitors on splicing may be mediated, at least in part, by the CDC5L complex.
Recently, it has been suggested that CDC5L interacts with the PP1 regulatory protein NIPP1 and that this interaction only occurs when CDC5L is phosphorylated (Boudrez et al., 2000). We have recently shown that NIPP1 co-localizes with pre-mRNA splicing factors in HeLa cell nuclei. Immunodepletion of NIPP1 from nuclear extracts or addition of a ‘dominant-negative’ mutant lacking a functional PP1 binding site greatly reduces pre-mRNA splicing activity in vitro (Trinkle-Mulcahy et al., 1999). However, we did not identify NIPP1 here in our analysis of CDC5L interacting proteins. It is possible that both PSF and NIPP1 may be involved in regulation of the CDC5L complex pre-mRNA splicing mechanism, e.g. by modulating the dephosphorylation/phosphorylation of components of the spliceosome complex or perhaps of CDC5L itself. NIPP1 and PSF could either modulate the phosphorylation of different phosphoprotein substrates in the spliceosome complex or, alternatively, the two PP1-binding proteins may cooperate in the dephosphorylation of a single substrate. More work still needs to be done to understand how these PP1 targeting proteins may regulate the dephosphorylation/phosphorylation of proteins in the pre-mRNA splicing pathway and which components of the pathway are substrates for these events. Another protein phosphatase identified in our complex is a protein phosphatase 2c (PP2C) isoform WIP1. This PP2C isoform is not sensitive to okadaic concentrations that will inhibit PP1 or protein phosphatase 2a (PP2A) in vitro. WIP1 is induced in mammalian cells in response to ionizing radiation in a p53-dependent manner (Fiscella et al., 1997), but has not previously been shown to be involved in pre-mRNA splicing. PP2C γ, a closely related protein, associates with the spliceosome and is needed during the early stages of spliceosome assembly (Murray et al., 1999). It is possible that the two isoforms of PP2C may be involved in different steps of the pre-mRNA splicing pathway or may be required to dephosphorylate specific proteins in the pathway. The protein substrates for these dephosphorylation/phosphorylation events in the pre-mRNA splicing cycle are still to be determined.
The possible involvement of the CDC5L complex in the regulation of phosphorylation activity is also consistent with the identification of the protein kinase DNA-PK in the complex. DNA-PK is composed of a large catalytic subunit of ∼470 kDa and the protein has been shown to interact with the antigen response element-binding proteins NF90 and NF45. DNA-PK acts as a serine/threonine protein kinase and the absence of this enzyme in the cell confers sensitivity to ionizing radiation and defects in DNA double-strand break repair (Ting et al., 1998 and references therein). NF90 and NF45 exist as a heterodimer that binds to DNA and with high avidity to double-stranded RNA (Satoh et al., 1999). The cellular functions of DNA-PK, NF90 and NF45 are unclear, although it has recently been suggested that the enzyme may have a role in the modulation of chromatin structure and function (Galande and Kohwi-Shigematsu, 2000). The presence of NF45 in our CDC5L protein complex and its ability to bind double-stranded RNA as well as DNA suggest the possible involvement of this complex in tethering the spliceosome to the DNA template during transcription. CDC5L also contains putative DNA-binding motifs and it is possible that the protein may be involved in DNA–protein interactions during coupled transcription/pre-mRNA splicing. Consistent with this idea, we identified several proteins with putative leucine zipper DNA-binding motifs as well as ATP/GTP-binding site motif sequences. The presence of topoisomerase IIα (TOP IIα) in the complex is indicative of a possible dual role for the CDC5L complex in the cell nucleus. It has been suggested that TOP IIα may be involved in transcription because of its enhanced expression during mitosis (Boland et al., 2000).
The human CDC5L complex contains only three clear homologues of the proteins described for the yeast CDC5p complex (McDonald et al., 1999), i.e. PRL1, CDC5L and Sm D3. We have so far not found homologues of the other proteins identified as part of the yeast CDC5p complex in our analysis of the complex in HeLa nuclear extract. This may be due to species differences, but it is also possible that not all of the proteins in the yeast (S.pombe) and/or human complex have been identified.
Our identification of proteins that co-purify with CDC5L also contained several proteins of the SF3a and SF3b complex. The SF3 complex is associated with U2 snRNP and has been shown to contain two sub-complexes, SF3a and SF3b (Krämer and Utans, 1991; Brossi et al., 1993). SF3a contains three polypeptides (Bennett and Reed, 1993; Krämer et al., 1994), while SF3b is made up of four characterized polypeptide subunits (Champion-Arnaud and Reed, 1994; Gozani et al., 1996; Wang et al., 1998; Das et al., 1999). The homologues of these subunits have been identified in yeast and Xenopus (Habara et al., 1998 and references therein). In budding yeast, SF3a has been shown to contain the proteins encoded by the genes prp9, prp11 and prp21 (Wiest et al., 1996). The presence of these proteins in the purified CDC5L complex suggests an interaction between this complex and U2 snRNP. Consistent with this observation, U2 snRNA co-purified with the CDC5 complex in S.pombe (McDonald et al., 1999). However, we show here that CDC5L is absent from purified snRNP preparations affinity purified from HeLa nuclear extracts. Also, purification of the complex by multiple ion exchange chromatographic steps led to loss of the snRNP-associated proteins (Sm proteins, U1A, U2A and the SF3 complex proteins), although the purified complex could still restore splicing activity to extracts immunodepleted using anti-CDC5L antibodies. These data indicate that the CDC5L complex, at least in HeLa cells, is not stably associated with snRNPs and the observed interactions may be only transient.
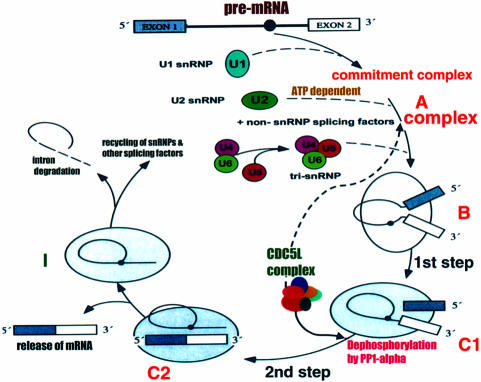
As mentioned in Results, our chromatographic analysis of the CDC5L-associated protein complex revealed five proteins that remained associated with CDC5L during all the purification steps. We propose that these proteins constitute a ‘core’ complex containing at least six proteins. This core complex in turn interacts with other splicing factors, including snRNPs, SR proteins and regulatory enzymes such as protein phosphatases and kinases, to modulate the second catalytic step of pre-mRNA splicing. This hypothesis is supported by the fact that immunodepletion of CDC5L did not lead to depletion of snRNP-associated proteins (data not shown). On the other hand, immunodepletion of CDC5L resulted in depletion of the core protein PRL1 (data not shown). We are currently preparing reagents in order to study the role of each of these core proteins in the CDC5L complex in order to gain more insight into how this complex functions in the process of pre-mRNA splicing. The diagram shown in Figure 7 describes the possible role of the CDC5L complex in the pre-mRNA splicing pathway based on the data currently available.
Fig. 7. Schematic representation of the pre-mRNA splicing cycle showing the steps at which the CDC5L complex joins the cycle and facilitates catalysis. The diagram is similar to that produced in Lamm and Lamond (1993) with some modifications. The letters A, B, C1, C2 and I indicate the different steps in the cycle and the complexes formed. The continuous arrow originating from the CDC5L-associated complex in the figure shows the stage in the cycle at which the complex is required for splicing progression, whereas the broken arrow shows the step at which CDC5L joins the cycle. Although our data suggest that the CDC5L complex is needed for progression through the second catalytic step of pre-mRNA splicing, we observed (by immunoprecipitations of pre-spliceosomal/spliceosomal complexes with anti-CDC5L at different time points; data not shown) that the protein associates with the pre-mRNA splicing machinery quite early on in the pathway in an ATP-dependent manner, perhaps concomitant with U2 snRNP binding. The reason for this early association of the CDC5L protein complex with pre-mRNA is still unclear. Dotted lines between steps in the figure indicate that several events may be involved.
Materials and methods
cDNA cloning and sequencing
The CDC5L cDNA was cloned from a HeLa cDNA library (Clontech) by PCR. Primers were designed for the N- and C-termini of the proteins using previously deposited sequences for these cDNAs in the DDBJ/EMBL/GenBank database (accession No. U86753). The CDC5L PCR primers contained SalI sites added to their 5′-ends. The PCR products were purified on a Qiagen PCR purification column according to the manufacturer’s instructions. Purified PCR products were digested with the appropriate enzymes and cloned using standard methods into the compatible sites of the vectors pGEX-4T1 (Pharmacia) and pET-30a (Novagen). The plasmid DNA was sequenced on an Applied Biosytems 377 automated DNA sequencer using the Taq dye terminator cycle sequencing method according to the manufacturer’s instructions. pGEX-4T1 and pET30a were used for expression of recombinant protein in E.coli.
Expression of recombinant proteins in E.coli
cDNAs cloned into the pGEX-4T1 vector were used to transform E.coli BL21(DE3). Overnight cultures were grown from single colonies, then diluted 1:10 in fresh LB medium with ampicillin (100 µg/ml) and grown at ∼30°C to an OD600 of 0.7–1.0 before induction with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Three hours post-induction, cells were pelleted and resuspended in 10 ml of phosphate-buffered saline (PBS), 0.5% Triton X-100 containing protease inhibitor cocktail (Boehringer). Cell lysis was achieved by sonication. The cell debris was removed by centrifugation at 10 000 g for 10 min. Pre-swollen glutathione–Sepharose beads pre-equilibrated in PBS were added to the supernatant (1 ml/l of culture). The beads were incubated with the crude protein extract for 2 h at 4°C with rocking. Beads were collected and washed three times in PBS, 0.5% Triton X-100 followed by three washes in PBS. Proteins were eluted from the beads by incubating in 25 mM glutathione in 50 mM Tris–HCl pH 8.0. The proteins were dialysed into a buffer containing 20 mM HEPES pH 8.0, 20% glycerol, 5 mM MgCl2, 100 mM KCl, 0.2 mM EDTA, 1 mM dithiothreitol (DTT) and stored at –80°C.
pET-30a cDNA clones were treated as above for expression except that 1 mM IPTG was used for induction and the cells were grown at 37°C prior to induction at 30°C. The expressed protein was bound to Ni-NTA–agarose beads (Qiagen) and the beads washed with buffer containing 20 mM imidazole, 1 mM phenylmethylsulfonyl fluoride (PMSF), 50 mM NaH2PO4, 300 mM NaCl pH 8.0. Recombinant protein was eluted from beads using the same buffer as above except that the concentration of imidazole was increased to 250 mM.
Antibody production and affinity purification
Synthetic peptides were designed from the protein sequence of CDC5L and synthesized by Immune Systems. Peptides used for antibody production were: EMSP1, 238-DFRKLRQQDLD-248; EMSP2, 281-QTSGVSEFTKKRSK-294; EMSP3, 315-QASEIARQTAEE-326. The synthesized peptides were conjugated to bovine serum albumin or keyhole limpet haemocyanin with glutaraldehyde using standard methods (Harlow and Lane, 1988). Recombinant protein or a mixture of the conjugated peptides was used to inoculate sheep (SAPU) for the production of polyclonal antisera. The antiserum was affinity purified against the peptides coupled by 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) to diaminodipropylamine immobilized on the azalactone-activated support 3M Emphaze Biosupport Medium AB1 (Pierce), according to the manufacturer’s recommendations. Affinity purified antibodies were dialysed against PBS containing 0.02% sodium azide and stored at –80°C.
Splicing assays
Nuclear extracts used in the splicing assays were obtained commercially from the Computer Cell Culture Centre (Mons, Belgium). Splicing assays were performed using uniformly labelled, capped pre-mRNAs incubated with nuclear extract as described (Lamond et al., 1987). For splicing reactions containing the protein phosphatase inhibitor okadaic acid (Sigma), assays were performed as described previously (Mermoud et al., 1992). The adeno-pre-mRNA was transcribed from Sau3AI-digested plasmid pBSAd1 (Konarska and Sharp, 1987). The splicing reactions were loaded onto a 10% polyacrylamide–8 M urea denaturing gel and run in 1× TBE in order to separate the splicing products. When samples were to be used for the analysis of splicing complexes, reactions were terminated by addition of heparin to a final concentration of 5 mg/ml. The reactions were loaded onto polyacrylamide/agarose composite gels (Konarska and Sharp, 1986) and run for ∼5 h at 25 mA.
Immunoprecipitation and immunodepletion
Immunoprecipitation of the spliceosomal proteins from nuclear extract was carried out using affinity purified peptide antibodies. Aliquots of 100 µl of nuclear extract (4–5 mg/ml) were pre-cleared for 1 h at 4°C on 25 µl of settled protein G–Sepharose (Pharmacia) or protein G–agarose beads (Boehringer) that had been pre-incubated with ∼10 µg of sheep pre-immune IgG. The pre-cleared nuclear extract was diluted 10 times (except in immunodepletion experiments) with PBS buffer containing 0.5% Triton X-100 before adding to protein G–Sepharose or protein G–agarose beads (25 µl) that had been pre-incubated with ∼60 pmol of antibody for 1 h at 4°C. Immunoprecipitations were carried out at 4°C for 2–16 h. The immunoprecipitates were washed with 1 ml of PBS containing 0.5% Triton X-100 three times at 4°C. Protein G beads carrying the immune complexes were collected after each wash by centrifugation at ∼1500 g for 1 min. For immunodepletion experiments, a higher amount of antibody (0.66 nmol) was used during the immunoprecipitation step.
Chromatographic purification of the CDC5L complex in HeLa nuclear extract
The CDC5L-containing complex was purified from ∼10 mg of nuclear extract. Briefly, 2 ml of HeLa cells nuclear extract (5 mg/ml) were fractionated on a Superose-6 HR 10/30 column equilibrated with 150 mM KCl in TM10 buffer (50 mM Tris–HCl pH 7.9, 12.5 mM MgCl2, 1 mM EDTA, 10% glycerol, 1 mM DTT, 1 mM sodium metabisulfite, 1 mM PMSF, 0.015% NP-40) at a flow rate 0.1 ml/min (10 runs at 0.2 ml of extract/run). Four fractions from each run containing the CDC5 complex were pooled, diluted three times with TM10 buffer and loaded onto a Mono-Q HR5/5 column equilibrated with 50 mM KCl in TM10 buffer. After washing with the same buffer, the column was eluted with a linear gradient of KCl in TM10 buffer (50 mM–0.35 M, 20 column vols, 1 ml/min flow rate, 0.5 ml fraction size). The CDC5L complex activity was eluted at 188 mM KCl; 2–3 fractions contained most of this activity. These fractions were pooled, desalted on a Hi-Trap G25 column (3 × 5 ml columns, 2 ml/min flow rate) that had previously been equilibrated with 25 mM KCl in TM10 buffer. The desalted CDC5L complex was then loaded onto a Mono-S PC 1.6/5 column equilibrated with the same buffer as above at 200 µl/min flow rate. The column was eluted with a linear gradient of KCl in TM10 buffer (25 mM–1 M, using 45 column vols of elution buffer at a flow rate of 100 µl/min and collecting 100 µl fractions). The CDC5L-containing complex was eluted as a single peak at 0.32 M KCl.
For affinity chromatography, the antibodies were covalently coupled to Affi-Gel Hz (Bio-Rad) according to the manufacturer’s instructions. The coupled anti-CDC5L antibodies/beads were then used to purify the CDC5-associated proteins in HeLa nuclear extract using the same conditions as in the immunoprecipitation experiments above. Purified proteins were eluted with 100 mM glycine pH 2.5 or antibody specific peptide. Fractions to be used in add back experiments were dialysed in splicing buffer (100 mM KCl, 5 mM MgCl2, 1 mM DTT, 20–25% glycerol) or PBS and stored at –80°C.
SDS–PAGE and western blotting
SDS–PAGE gel analysis was performed as described previously (Laemmli, 1970).
For immunoblotting, the washed immunoprecipitates were resuspended in 50 µl of 2× SDS–PAGE loading buffer and heated at 95°C for 5 min. Protein samples (usually 10–25 µl) were loaded onto an 8/12% SDS–PAGE gel or a 4–12% gradient gel (Novex). The separated proteins were transferred onto Hybond-C Extra membrane (Amersham) by electroblotting for antibody probing or stained with silver or colloidal Coomassie blue (Novex) according to the manufacturer’s instructions. The membranes carrying the transferred proteins were blocked with 5% non-fat milk powder in PBS, 0.3% Tween-20. The membranes were incubated with primary antibody for 1–16 h at 4°C or room temperature, washed with blocking buffer and incubated with the appropriate secondary antibody. The primary antibodies were used at the following dilutions: anti-CDC5L, 1:1000; anti-U1A polyclonal serum 856, 1:2000 (Kambach and Mattaj, 1992). Protein bands were detected by developing blots with the ECL kit (Amersham) according to the manufacturer’s instructions.
Purification of U snRNPs
U snRNPs were purified as described by Bach et al. (1990). Aliquots of 20 ml of nuclear extract (5 mg/ml) were diluted with 50 ml of buffer containing 20 mM HEPES, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA before purification on an anti-3mG antibody (Oncogene Science) column.
Protein identification by mass spectrometry and database searching
Gel-separated proteins were reduced, alkylated and digested in-gel as described previously (Shevchenko et al., 1996) and subsequently identified by a two-tier mass spectrometric approach. In the first round, small aliquots (1–2%) of the generated peptide mixtures were analysed by MALDI mass spectrometry on a Bruker REFLEX III (Bruker Daltonik, Bremen, Germany) to yield a peptide mass map (Vorm et al., 1994). The list of mono-isotopic peptide masses obtained was used to query a non-redundant protein sequence database (NRDB, >500 000 entries) maintained and updated regularly at the European Bioinformatics Institute (EBI, Hinxton, UK). Proteins were identified by correlating the measured peptide masses with theoretical digests of all proteins present in the database (Henzel et al., 1993; James et al., 1993; Mann et al., 1993; Yates et al., 1993).
Proteins that could not be identified by a list of peptide masses alone were subjected to nanoelectrospray tandem mass spectrometry (Wilm et al., 1996). Protein digests were desalted and concentrated using micro-columns packed with ∼100 nl of POROS R2 perfusion chromatography material (Perseptive Biosystems, Framingham, MA). Peptides were eluted with 60% MeOH, 5% HCOOH directly into nanospray capillaries (Protana, Odense, Denmark). Peptide sequencing by tandem mass spectrometry was performed using a nanoelectrospray ion source (Protana, Odense, Denmark) coupled to a quadrupole time-of-flight mass spectrometer (Shevchenko et al., 1997) (QSTAR; Sciex, Toronto, Canada). The sequence and mass information contained in the tandem mass spectra were assembled into peptide sequence tags (Mann and Wilm, 1994) and used for queries in the NRDB, dbEST (expressed sequence tags database, NCBI) and human genome (NCBI) databases. Proteins were identified by comparing the retrieved sequences with the mass spectrometric data. All database searches were performed using the program Pepsea (Protana, Odense, Denmark).
Acknowledgments
Acknowledgements
We thank Dr Iain Mattaj for generously providing us with anti-U1 antibody. We acknowledge the assistance provided by Mr J.Morrison in subcloning and sequencing of the CDC5L cDNA and Ursula Ryder in purification of snRNPs from HeLa nuclear extract. We also thank the members of the Lamond and Mann laboratories for helpful advice and encouragement during these studies. J.C.B.M.Z. is a Wellcome Trust Senior Research Fellow. A.I.L. is a Principal Research Fellow of the Wellcome Trust.
References
- Bach M., Krol,A. and Luhrmann,R. (1990) Structure-probing of U1 snRNPs gradually depleted of the U1-specific proteins A, C and 70k. Evidence that A interacts differentially with developmentally regulated mouse U1 snRNA variants. Nucleic Acids Res., 18, 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M. and Reed,R. (1993) Correspondence between a mammalian spliceosome component and an essential yeast splicing factor. Science, 262, 105–108. [DOI] [PubMed] [Google Scholar]
- Bennett M., Michaud,S., Kingston,J. and Reed,R. (1992) Protein components specifically associated with prespliceosome and spliceosome complexes. Genes Dev., 6, 1986–2000. [DOI] [PubMed] [Google Scholar]
- Bernstein H.S. and Coughlin,S.R. (1997) A putative human transcription factor implicated in mitogen-activated signaling. J. Biol. Chem., 272, 5833–5837. [DOI] [PubMed] [Google Scholar]
- Bernstein H.S. and Coughlin,S.R. (1998) A mammalian homolog of fission yeast CDC5 regulates G2 progression and mitotic entry. J. Biol. Chem., 273, 4666–4671. [DOI] [PubMed] [Google Scholar]
- Biemann K. (1990) Nomenclature for peptide fragment ions. Methods Enzymol., 193, 886–887. [DOI] [PubMed] [Google Scholar]
- Boland M.P., Fitzgerald,K.A. and O’Neill,L.A. (2000) Topoisomerase II is required for mitoxantrone to signal NFκB activation in HL60 cells. J. Biol. Chem., 275, 25231–25238. [DOI] [PubMed] [Google Scholar]
- Boudrez A. et al. (2000) NIPP1-mediated interaction of protein phosphatase-1 with CDC5L, a regulator of pre-mRNA splicing and mitotic entry. J. Biol. Chem., 275, 25411–25417. [DOI] [PubMed] [Google Scholar]
- Brossi R., Groning,K., Behrens,S.E., Luhrmann,R. and Krämer,A. (1993) Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science, 262, 102–105. [DOI] [PubMed] [Google Scholar]
- Burns C.G., Ohi,R., Krainer,A.R. and Gould,K.L. (1999) Evidence that Myb-related CDC5 proteins are required for pre-mRNA splicing. Proc. Natl Acad. Sci. USA, 96, 13789–13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion-Arnaud P. and Reed,R. (1994) The prespliceosome components SAP 49 and SAP 145 interact in a complex implicated in tethering U2 snRNP to the branch site. Genes Dev., 8, 1974–1983. [DOI] [PubMed] [Google Scholar]
- Das B.K., Xia,L., Palandjian,L., Gozani,O., Chyung Y. and Reed R. (1999) Characterisation of a protein complex containing spliceosomal proteins SAPs 49, 130, 145 and 155. Mol. Cell. Biol., 19, 6796–6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff M.P., Johnson,D.F., Moorhead,G., Cohen,P.T., Cohen,P. and Barford,D. (1997) Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J., 16, 1876–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espanel X. and Sudol,M. (1999) A single point mutation in a group I WW domain shifts its specificity to that of group II WW domains. J. Biol. Chem., 274, 17284–17289. [DOI] [PubMed] [Google Scholar]
- Fiscella M., Zhang,H., Fan,S., Sakaguchi,K., Shen,S., Mercer,W., Vande Woude,G., O’Connor,P.M. and Appella,E. (1997) Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc. Natl Acad. Sci. USA, 94, 6048–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.-D. (1995) The superfamily of arginine/serine-rich splicing factors. RNA, 1, 663–680. [PMC free article] [PubMed] [Google Scholar]
- Galande S. and Kohwi-Shigematsu,T. (2000) Caught in the act: binding of Ku and PARP to MARs reveals novel aspects of their functional interaction. Crit. Rev. Eukaryot. Gene Expr., 10, 63–72. [PubMed] [Google Scholar]
- Gozani O., Patton,J.G. and Reed,R. (1994) A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J., 13, 3356–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O., Feld,R. and Reed,R. (1996) Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev., 10, 233–243. [DOI] [PubMed] [Google Scholar]
- Groenen P.M., Vanderlinden,G., Devriendt,K., Fryns,J.-P. and Van de Ven,W.J. (1998) Rearrangement of the human CDC5L gene by a t(6;19)p21;q13.1 in a patient with multicystic renal dysplasia. Genomics, 49, 218–229. [DOI] [PubMed] [Google Scholar]
- Habara Y., Urushiyama,S., Tani,T. and Ohshima,Y. (1998) The fission yeast prp10+ gene involved in pre-mRNA splicing encodes a homologue of highly conserved splicing factor, SAP155. Nucleic Acids Res., 26, 5662–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Henzel W.J., Billeci,T.M., Stults,J.T., Wong,S.C., Grimley,C. and Watanabe,C. (1993) Identifying proteins from two-dimensional gels by molecular mass searching of peptide fragments in protein sequence databases. Proc. Natl Acad. Sci. USA, 90, 5011–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K., Erdodi,F., Patton,J.G. and Hartshorne,D.J. (1996) Interaction of protein phosphatase type 1 with a splicing factor. FEBS Lett., 389, 191–194. [DOI] [PubMed] [Google Scholar]
- Hirayama T. and Shinozaki,K. (1996) A cdc5+ homolog of a higher plant, Arabidopsis thaliana. Proc. Natl Acad. Sci. USA, 93, 13371–13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Quadroni,M., Carafoli,E. and Gonnet,G. (1993) Protein identification by mass profile fingerprinting. Biochem. Biophys. Res. Commun., 195, 58–64. [DOI] [PubMed] [Google Scholar]
- Kambach C. and Mattaj,I.W. (1992) Intracellular distribution of the U1A protein depends on active transport and nuclear binding to U1 snRNA. J. Cell Biol., 118, 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M.M. and Sharp,P.A. (1986) Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell, 46, 845–855. [DOI] [PubMed] [Google Scholar]
- Konarska M.M. and Sharp,P.A. (1987) Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell, 49, 763–774. [DOI] [PubMed] [Google Scholar]
- Krämer A. (1995) The biochemistry of pre-mRNA splicing. In Lamond,A.I. (ed.), Pre-mRNA Processing. Springer-Verlag, Heidelberg, Germany, pp. 39–64. [Google Scholar]
- Krämer A. (1996) The structure and function of proteins in mammalian pre-mRNA splicing. Annu. Rev. Biochem., 65, 367–409. [DOI] [PubMed] [Google Scholar]
- Krämer A. and Utans,U. (1991) Three protein factors (SF1, SF3 and U2AF) function in pre-mRNA splicing complex formation in addition to snRNPs. EMBO J., 10, 1503–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A., Legrain,P., Mulhauser,F., Groning,K., Brosi,R. and Bilbe,G. (1994) Splicing factor SF3a60 is the mammalian homologue of PRP9 of S. cerevisiae: the conserved zinc finger-like motif is functionally exchangeable in vivo. Nucleic Acids Res., 22, 5223–5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lamm G.M. and Lamond,A.I. (1993) Non-snRNP protein splicing factors. Biochim. Biophys. Acta, 1173, 247–265. [DOI] [PubMed] [Google Scholar]
- Lamond A.I., Konarska,M.M. and Sharp,P.A. (1987) A mutational analysis of spliceosome assembly: evidence for splice site collaboration during spliceosome formation. Genes Dev., 1, 532–543. [DOI] [PubMed] [Google Scholar]
- Mann M. and Wilm,M. (1994) Error tolerant identification of peptides in sequence databases by peptide sequence tags. Anal. Chem., 66, 4390–4399. [DOI] [PubMed] [Google Scholar]
- Mann M., Hojrup,P. and Roepstorff,P. (1993) Use of mass spectrometric molecular weight information to identify proteins in sequence databases. Biol. Mass Spectrom., 22, 338–345. [DOI] [PubMed] [Google Scholar]
- McDonald W.H., Ohi,R., Smelkova,N., Frendewey,D. and Gould,K.L. (1999) Myb-related fission yeast cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol. Cell. Biol., 19, 5352–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermoud J.E., Cohen,P. and Lamond,A.I. (1992) Ser/Thr-specific protein phosphatases are required for both catalytic steps of pre-mRNA splicing. Nucleic Acids Res., 20, 5263–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. and Spector,D.L. (1996) Serine/threonine phosphatase 1 modulates the subnuclear distribution of pre-mRNA splicing factors. Mol. Biol. Cell, 7, 1559–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M.V., Kobayashi,R. and Krainer,A.R. (1999) The type 2C Ser/Thr phosphatase PP2Cγ is a pre-mRNA splicing factor. Genes Dev., 13, 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer G., King,A., Rappsilber,J., Calvio,C., Watson,M., Ajuh,P., Sleeman,J., Lamond,A. and Mann,M. (1998) Mass spectrometry and EST-database searching allows characterisation of the multi-protein spliceosome complex. Nature Genet., 20, 46–50. [DOI] [PubMed] [Google Scholar]
- Ohi R., McCollum,D., Hirani,B., Den Haese,G.J., Zhang,X., Burke,J.D., Turner,K. and Gould,K.L. (1994) The Schizosacharomyces pombe cdc5+ gene encodes an essential protein with homology to c-Myb. EMBO J., 13, 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi R., Feoktistova,A., McCann,S., Valantine,V., Look,A.T., Lipsick,J.S. and Gould,K.L. (1998) Myb-related Schizosacharomyces pombe cdc5p is structurally and functionally conserved in eukaryotes. Mol. Cell. Biol., 18, 4097–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. (1990) Protein composition of mammalian spliceosomes assembled in vitro. Proc. Natl Acad. Sci. USA, 87, 8031–8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. and Palandjian,L. (1997) Spliceosome assembly. In Krainer,A.R. (ed.), Eukaryotic mRNA Processing. Oxford University Press, New York, NY, pp. 130–173. [Google Scholar]
- Satoh M., Shaheen,V.M., Kao,P.N., Okano,T., Shaw,M., Yoshida,H., Richards,H.B. and Reeves,W.H. (1999) Autoantibodies define a family of proteins with conserved double-stranded RNA-binding domains as well as DNA binding activity. J. Biol. Chem., 274, 34598–34604. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm,M., Vorm,O. and Mann,M. (1996) Mass spectro metric sequencing of proteins in silver-stained polyacrylamide gels. Anal. Chem., 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Chernushevich,I., Ens,W., Standing,K.G., Thomson,B., Wilm,M. and Mann,M. (1997) Rapid ‘de novo’ peptide sequencing by a combination of nanoelectrospray, isotopic labelling and a quadrupole/time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom., 11, 1015–1024. [DOI] [PubMed] [Google Scholar]
- Smith T.F., Gaitatzes,C., Saxena,K. and Neer,E.J. (1999) The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci., 24, 181–185. [DOI] [PubMed] [Google Scholar]
- Staley J.P. and Guthrie,C. (1998) Mechanical devices of the spliceosome: motors, clocks, springs and things. Cell, 92, 315–326. [DOI] [PubMed] [Google Scholar]
- Stukenberg P.T., Lustig,K.D., McGarry,T.J., King,R.W., Kuang,J. and Kirschner,M.W. (1997) Systematic identification of mitotic phosphoproteins. Curr. Biol., 7, 338–348. [DOI] [PubMed] [Google Scholar]
- Tazi J., Daugeron,M.C., Cathala,G., Brunel,C. and Jeanteur,P. (1992) Adenosine phosphorothioates (ATP α S and ATP τ S) differentially affect the two steps of mammalian pre-mRNA splicing. J. Biol. Chem., 267, 4322–4326. [PubMed] [Google Scholar]
- Ting N.S., Kao,P.N., Chan,D.W., Lintott,L.G. and Lees-Miller,S.P. (1998) DNA-dependent protein kinase interacts with antigen receptor response element binding proteins NF90 and NF45. J. Biol. Chem., 273, 2136–2145. [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., Ajuh,P., Prescott,A., Claverie-Martin,F., Cohen,S., Lamond,A.I. and Cohen,P. (1999) Nuclear organisation of NIPP1, a regulatory subunit of protein phosphatase 1 that associates with pre-mRNA splicing factors. J. Cell Sci., 112, 157–168. [DOI] [PubMed] [Google Scholar]
- Tsai W.Y. et al. (1999) Cef1p is a component of the Prp19p-associated complex and essential for pre-mRNA splicing. J. Biol. Chem., 274, 9455–9462. [DOI] [PubMed] [Google Scholar]
- Venables J.P., Elliott,D.J., Makarova,O.V., Makarov,E.M., Cooke,H.J. and Eperon,I.C. (2000) RBMY a probable human spermatogenesis factor and other hnRNP-G proteins interact with Tra2β and affect splicing. Hum. Mol. Genet., 9, 685–694. [DOI] [PubMed] [Google Scholar]
- Vorm O., Roepstorff,P. and Mann,M. (1994) Improved resolution and very high sensitivity in MALDI TOF of matrix surfaces made by fast evaporation. Anal. Chem., 66, 3281–3287. [Google Scholar]
- Wang C., Chua,K., Seghezzi,W., Lees,E., Gozani,O. and Reed,R. (1998) Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev., 12, 1409–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiest D.K., O’Day,C.L. and Abelson,J. (1996) In vitro studies of the Prp9⋅Prp11⋅Prp21 complex indicate a pathway for U2 small nuclear ribonucleoprotein activation. J. Biol. Chem., 271, 33268–33276. [DOI] [PubMed] [Google Scholar]
- Will C.L. and Lürhmann,R. (1997) snRNP structure and function. In Krainer,A.R. (ed.), Eukaryotic mRNA Processing. Oxford University Press, New York, NY, pp. 130–173. [Google Scholar]
- Wilm M., Shevchenko,A., Houthaeve,T., Breit,S., Schweigerer,L., Fotsis,T. and Mann,M. (1996) Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature, 379, 466–469. [DOI] [PubMed] [Google Scholar]
- Yates J.R., Speicher,S., Griffin,P.R. and Hunkapiller,T. (1993) Peptide mass maps: a highly informative approach to protein identification. Anal. Biochem., 214, 397–408. [DOI] [PubMed] [Google Scholar]