Abstract
Studies of gene flow between closely related taxa can provide insight into the genetic basis of speciation. To evaluate the importance of the X chromosome in reproductive isolation between subspecies of the European rabbit and to study the genomic scale over which islands of differentiation extend, we resequenced a total of 34 loci distributed along the X chromosome and chromosome 14. Previous studies based on few markers suggested that loci in centromeric regions were highly differentiated between rabbit subspecies, whereas loci in telomeric regions were less differentiated. Here, we confirmed this finding but also discovered remarkable variation in levels of differentiation among loci, with FST values from nearly 0 to 1. Analyses using isolation-with-migration models suggest that this range appears to be largely explained by differential levels of gene flow among loci. The X chromosome was significantly more differentiated than the autosomes. On chromosome 14, differentiation decayed very rapidly at increasing distances from the centromere, but on the X chromosome distinct islands of differentiation encompassing several megabases were observed both at the centromeric region and along the chromosome arms. These findings support the idea that the X chromosome plays an important role in reproductive isolation between rabbit subspecies. These results also demonstrate the mosaic nature of the genome at species boundaries.
Keywords: Gene flow, islands of differentiation, isolation-with-migration, reproductive isolation, speciation, X chromosome
Understanding the genetic basis of the origin of species is a central problem in evolution. Studies on the genetics of reproductive isolation have been performed using both laboratory crosses and naturally hybridizing species, and there are advantages to each approach. In the laboratory, it is possible to conduct repeatable experiments, controlling both the genetic and environmental backgrounds (e.g., Dobzhansky 1936; Ting et al. 1998; Barbash et al. 2003; Moehring et al. 2006; Masly and Presgraves 2007; Good et al. 2008; Mihola et al. 2009). As an alternative to laboratory crosses, the study of naturally occurring hybrids relies on the basic premise that different portions of the genome may vary in their permeability to foreign alleles (Endler 1977; Caisse and Antonovics 1978; Bengtsson 1979; Barton and Hewitt 1989; Harrison 1990; Wu 2001; Nosil et al. 2009; Via 2009). As a result, genes or genomic regions underlying reproductive isolation or adaptation can be identified as those showing reduced levels of introgression (e.g., Rieseberg et al. 1999; Machado et al. 2002; Payseur et al. 2004; Borge et al. 2005; Turner et al. 2005; Geraldes et al. 2006; Macholan et al. 2007; Teeter et al. 2008; Kulathinal et al. 2009; Nolte et al. 2009; Baxter et al. 2010), even in the absence of information about which phenotypes they control. This approach offers the advantages of more generations of recombination and the identification of hybrid incompatibilities that have been tested in a natural setting.
Both kinds of studies (laboratory crosses and natural populations) have revealed that loci contributing to reproductive isolation disproportionally accumulate on the X chromosome in male heterogametic systems (i.e., “the large X-effect”; Coyne and Orr 1989). Mapping studies show evidence for a large X-effect on both hybrid sterility and hybrid inviability (Dobzhansky 1936; Grula and Taylor 1982; Orr and Coyne 1989; True et al. 1996; Tao et al. 2003; Storchova et al. 2004; Good et al. 2008). For example, Masly and Presgraves (2007) described a much higher density of hybrid male sterility factors on the X chromosome when compared to the autosomes using introgression experiments in Drosophila. Moreover, the few genes underlying reproductive isolation that have been identified so far either map to the X chromosome or interact with genes on the X chromosome (e.g., Ting et al. 1998; Barbash et al. 2003; Presgraves et al. 2003). In addition, studies of hybrid zones have revealed reduced gene flow at X-linked loci relative to autosomal loci (Sperling and Spence 1991; Tucker et al. 1992; Munclinger et al. 2002; Besansky et al. 2003; Macholan et al. 2007; Geraldes et al. 2008a). Reduced introgression of the Z chromosome has also been observed across the hybrid zone of several birds (Borge et al. 2005; Carling and Brumfield 2008; Storchova et al. 2010) and Lepidopteran species (Putnam et al. 2007), supporting the more general phenomenon that sex chromosomes contribute strongly to the emergence of reproductive barriers.
Regions of restricted recombination have also been implicated in higher differentiation between species, and several distinct models suggest ways in which these regions may facilitate divergence in the presence of gene flow (Noor et al. 2001; Rieseberg 2001; Navarro and Barton 2003). These models posit that regions of reduced crossing-over may limit gene flow between incipient species by harboring a disproportionate share of linked genomic incompatibilities. Empirical support for these models is still fairly scarce but it derives both from mapping experiments (Noor et al. 2001; Brown et al. 2004) and from inferred levels of gene flow in nature (Rieseberg et al. 1999; Machado et al. 2002, 2007; Feder et al. 2003). Most examples involve chromosomal rearrangements, but recently, centromeric regions, which usually show low levels of recombination, have also been shown to be associated with lower rates of gene flow both in Anopheles mosquitoes (Stump et al. 2005; but see White et al. 2010) and in the European rabbit (Carneiro et al. 2009).
The European rabbit (Oryctolagus cuniculus) provides an opportunity to study patterns of introgression for different regions of the genome. Two subspecies occur in parapatry in the Iberian Peninsula (Fig. 1) and are thought to have diverged approximately 2 million years ago (Mya; Branco et al. 2000; Carneiro et al. 2009). They exhibit slight morphological differences in body size (Villafuerte 2002) and cranial measurements (Sharples et al. 1996) and crosses between them produce viable hybrids (N. Ferrand, unpubl. data). Experimental crosses in outdoor enclosures are currently underway to study hybrid fertility and viability in detail. Geraldes et al. (2008b) showed that O. c. algirus and O. c. cuniculus are characterized by contrasting patterns of differentiation at multiple loci. Although the majority of loci show low differentiation presumably due to high levels of gene flow, some loci, including the mtDNA (Branco et al. 2000), the Y chromosome (Geraldes et al. 2008b), and loci near centromeres of the X chromosome (Geraldes et al. 2006) and several autosomes (Carneiro et al. 2009), exhibit high differentiation.
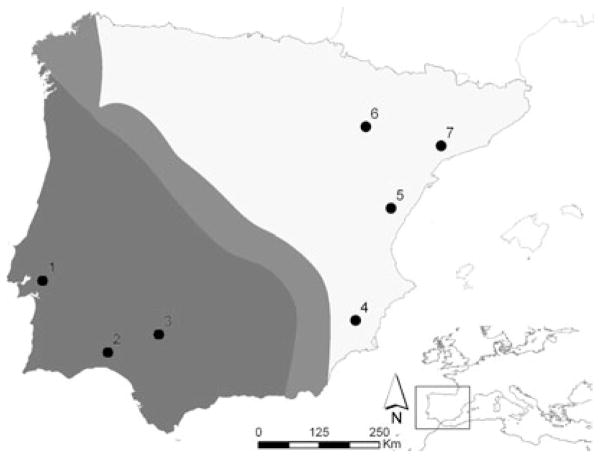
Figure 1.
Geographical location of the populations used in this study. Dark and light areas indicate the range of O. c. algirus and O. c. cuniculus, respectively. The putative hybrid zone is separating both ranges (adapted from Geraldes et al. (2008b)). Numbers in the figure correspond to populations as follows: 1- Pancas; 2- Huelva; 3- Sevilla; 4- Alicante; 5- Rosell; 6- Zaragoza; 7- Tarragona.
To evaluate the importance of the X chromosome in reproductive isolation between subspecies of the European rabbit and to study the genomic scale over which islands of differentiation extend, we resequenced a total of 34 loci distributed along the X chromosome and chromosome 14, and we reanalyzed published data from 10 autosomal loci for the same set of individuals. We were also interested in studying whether highly differentiated loci are restricted to centromeric regions, which are likely to experience reduced recombination.
Materials and Methods
INDIVIDUALS AND LOCI SAMPLED
A total of 22 male rabbits from seven localities across the Iberian Peninsula were used in this study (Fig. 1 and Table 1). Ten individuals were sampled from the southwestern part of the Iberian Peninsula, corresponding to the subspecies O. c. algirus, and 12 from the northeastern part of the Iberian Peninsula corresponding to the subspecies O. c. cuniculus. A single male Lepus granatensis was used as an outgroup. DNA was extracted using a standard phenol–chloroform DNA protocol (Sambrook and Russell 2001).
Table 1.
Individuals sampled and their geographic locations.
| Population | Population number1 | Sample size | Subspecies | Individual ID |
|---|---|---|---|---|
| Pancas | 1 | 3 | O. c. algirus | Pancas3, Pancas5, Pancas6 |
| Huelva | 2 | 4 | O. c. algirus | Huelva1, Huelva4, Huelva43, Huelva53 |
| Sevilla | 3 | 3 | O. c. algirus | Sevilla8, Sevilla12, Sevilla14 |
| Alicante | 4 | 4 | O. c. cuniculus | Alicante110, Alicante131, Alicante134, Alicante138 |
| Rosell | 5 | 1 | O. c. cuniculus | Rosell4 |
| Zaragoza | 6 | 2 | O. c. cuniculus | Zaragoza14, Zaragoza15 |
| Tarragona | 7 | 5 | O. c. cuniculus | Tarragona10, Tarragona102, Tarragona105, Tarragona107, Tarragona110 |
Population numbers are from Figure 1.
We obtained polymorphism data by resequencing 27 loci on the X chromosome and seven loci on chromosome 14. Loci were chosen at regular intervals along each chromosome using two different approaches. First, whenever possible we used loci that were mapped on the rabbit cytogenetic map (Chantry-Darmon et al. 2005) and were present in an earlier assembly of the rabbit genome sequence (2X coverage available in Ensembl; http://www.ensembl.org). Second, we chose loci from syntenic regions in human–rabbit comparisons. The recent release of the 7X assembly of the European rabbit genome sequence (https://www.broadinstitute.org) has allowed us to identify the physical position of most loci in the rabbit genome (Table 2). A few X-linked loci were not mapped in this new assembly and for these loci we assumed the same position relative to neighboring loci in the rabbit as in the human genome. In fact, with the exception of the region encompassing NKR to KLHL13, all loci on the rabbit X chromosome are in the same order as on the human X chromosome. Additionally, published DNA sequences for 10 autosomal loci from the same individuals (Carneiro et al. 2009) were combined with the loci obtained in this study (Table 2).
Table 2.
Genes and genomic locations of the loci used in this study.
| Gene | Chromosome | Chromosome location1 | Location in the gene2 | Reference |
|---|---|---|---|---|
| SHOX | X | Unknown | Intron 6 | This study |
| GLRA2 | X | 606,087–606,861 | Intron 9 | This study |
| PHKA2 | X | 4,723,497–4,725,600 | Introns 2/3 | Geraldes et al. (2006) |
| PDHA1 | X | 5,096,544–5,097,183 | Intron 8 | This study |
| POLA1 | X | 10,616,047–10,616,751 | Intron 10 | This study |
| IL1RAPL1 | X | 15,984,427–15,985,150 | Intron 11 | This study |
| TMEM47 | X | 20,817,981–20,818,776 | Intron 1 | This study |
| MAOA | X | 29,148,153–29,148,958 | Intron 12 | This study |
| TIMP1 | X | 32,657,446–32,658,226 | Intron 10 | This study |
| DGKK | X | Unknown | Intron 14 | This study |
| SMCX | X | 34,720,991–34,723,906 | Introns 2/3 | Geraldes et al. (2006) |
| ARHGEF9 | X | 42,303,886–42,304,638 | Intron 6 | This study |
| Centromere3 | X | Unknown | NA | NA |
| MSN | X | 44,317,226–44,320,307 | Introns 6/7/8 | Geraldes et al. (2006) |
| OPHN1 | X | 46,884,710–46,885,471 | Intron 18 | This study |
| OGT | X | 50,027,095–50,027,850 | Intron 15 | This study |
| NRK | X | 54,602,900–54,603,613 | Intron 15 | This study |
| AMOT | X | 61,383,648–61,384,060 | Intron 2 | This study |
| KLHL13 | X | 66,537,385–66,538,064 | Intron 7 | This study |
| PGK1 | X | 69,862,748–69,863,565 | Intron 6 | This study |
| CYLC1 | X | 74,678,953–74,679,740 | Intron 1 | This study |
| KLHL4 | X | 78,346,123–78,346,922 | Intron 13 | This study |
| PABPC5 | X | 83,299,091–83,299,832 | 3′ | This study |
| TNMD | X | 88,777,964–88,778,801 | Intron 6 | This study |
| DIAPH2 | X | 92,701,134–92,701,933 | Intron 2 | This study |
| GRIA3 | X | 97,949,748–97,950,466 | Intron 8 | This study |
| OCRL | X | 104,011,520–104,012,070 | Intron 22 | This study |
| GPC4 | X | 107,650,182–107,650,932 | Intron 8 | This study |
| HPRT1 | X | 108,777,703–108,779,229 | Intron 2 | Geraldes et al. (2006) |
| F9 | X | Unknown | Intron 1 | This study |
| FMR1 | X | Unknown | Intron 11 | This study |
| G6PD | X | Unknown | Intron 1 | This study |
| EXT1 | 3 | 133,930,356–133,931,580 | Intron 4 | Carneiro et al. (2009) |
| CYTC | 4 | 13,353,549–13,354,658 | Intron 1 | Carneiro et al. (2009) |
| LUM | 4 | 70,318,042–70,319,218 | Intron 1 | Carneiro et al. (2009) |
| UD14 | 7 | Unknown | Intron 4 | Carneiro et al. (2009) |
| ATP12A | 8 | 45,292,648–45,293,961 | Intron 2 | Carneiro et al. (2009) |
| PRL | 12 | 14,728,167–14,729,416 | 5′ | Carneiro et al. (2009) |
| T | 12 | 155,309,281–155,310,209 | 5′ | Carneiro et al. (2009) |
| MGST3 | 13 | 27,250,930–27,251,858 | Intron 1 | Carneiro et al. (2009) |
| SLC4A7 | 14 | 14,216,142–14,216,900 | Intron 16 | This study |
| Centromere3 | 14 | Unkown | NA | NA |
| STAG1 | 14 | 30,256,593–30,257,916 | Intron 6 | Carneiro et al. (2009) |
| GK5 | 14 | 35,607,955–35,608,670 | Intron 13 | This study |
| SIAH2 | 14 | 44,630,546–44,631,214 | 3′ | This study |
| KPNA4 | 14 | 54,925,301–54,926,023 | Intron 16 | This study |
| MYNN | 14 | 64,678,559–64,679,184 | Intron 6 | This study |
| NAALADL2 | 14 | 70,911,227–70,911,750 | Intron 10 | This study |
| GBE1 | 14 | 135,937,066–135,937,688 | Intron 17 | This study |
| TIAM1 | 14 | 161,363,063–161,364,253 | Intron 2 | Carneiro et al. (2009) |
Chromosome location was obtained at the Broad Institute website from the European rabbit 7X genome sequence assembly or from the rabbit physical map (Chantry-Darmon et al. 2005).
Location in the gene was obtained from the rabbit 2X genome sequence annotation available in Ensembl Build 48.1e.
Centromere location relative to other loci was obtained from the rabbit physical map (Chantry-Darmon et al. 2005).
Primers were designed from the European rabbit genome sequence (Table S1). Most loci have PCR primers located in flanking exons, but only intronic sequences were used in all the analyses below. DNA was PCR amplified in 25 μl reactions containing 100 ng of genomic DNA, 2 mM MgCl2, 0.5 μM of each primer, 200 μM DNTP, and 0.5 unit Taq polymerase. PCR cycling profile consisted of 35 cycles of 30 sec at 94°C, 30 sec at various annealing temperatures (Table S1), and 90 sec at 72°C, preceded by an initial denaturation step at 94°C for 2 min, and followed by a final extension of 5 min at 72°C. Sequencing was carried out using an ABI 3700 automated sequencer through the University of Arizona’s Genomic Analysis and Technology Core sequencing service. Sequences were edited, aligned, and assembled using phred/phrap/consed/polyphred (Nickerson et al. 1997; Ewing and Green 1998; Ewing et al. 1998; Gordon et al. 1998) together with auxiliary shell scripts and Perl programs kindly provided by August Woerner. Manual adjustments were further performed in some contigs using Bioedit (Hall 1999). Sequences have been deposited in GenBank under accession numbers HM027927-HM028639.
DATA ANALYSIS
Levels of variation and tests of neutrality
By using only males, we were able to directly recover haplotypes for X-linked loci. For autosomal loci, haplotypes were assigned by the computer program PHASE 2.1 (Stephens et al. 2001; Stephens and Donnelly 2003). We applied the algorithm five times for each locus to check for convergence across independent runs.
Most polymorphism and divergence analyses were performed using SITES (Hey and Wakeley 1997) or DnaSP 4.50.3 (Rozas et al. 2003). Insertion–deletion polymorphisms were excluded from all analyses. We estimated the neutral mutation parameter (4Neμ for autosomal loci and 3Neμ for X-linked loci), where Ne is the effective population size and μ is the mutation rate per site per generation, using two estimators: Watterson’s θw (1975), the proportion of segregating sites in a sample, and π (Nei 1987), the average number of pairwise differences between sequences in a sample. To investigate levels of linkage disequilibrium, we estimated the population recombination parameter (R = 4Nec for autosomal loci and R = 3Nec for X-linked loci, where c is the recombination rate per generation) between adjacent sites using γ (Hey and Wakeley 1997) and ρ (Hudson 2001). γ is a maximum-likelihood estimator developed using a coalescent model for a sample of four DNA sequences with recombination and ρ is adapted to a finite-sites model and is estimated by means of a composite likelihood method, as implemented in the LDhat 2.0 package (McVean et al. 2002).
We performed two tests of the neutral model based on the frequency spectrum of polymorphisms, Tajima’s D (Tajima 1989), and Fu and Li’s D* (Fu and Li 1993). To generate the null distribution of these statistics, we used coalescent simulations using DnaSP 4.50.3. We ran 104 coalescent simulations of the standard neutral model with constant population size, conditioned on the sample size and the observed estimates of θ, with the assumption of no recombination. In addition, ratios of polymorphism to divergence were compared with the expectations under a neutral model using a multilocus Hudson–Kreitman–Aguade (HKA) test (Hudson et al. 1987) as implemented in the HKA program (http://lifesci.rutgers.edu/heylab). We compared ratios of polymorphism for both subspecies together and separately to divergence between rabbit and Lepus. We also evaluated heterogeneity in the ratio of polymorphism to divergence between a pooled sample of X-linked versus autosomal loci.
Patterns of differentiation and gene flow
Several summary statistics were used to describe levels of genetic differentiation between the two rabbit subspecies. We estimated the fixation index (FST; Hudson et al. 1992), the net nucleotide divergence (Da; Nei 1987), and the average pairwise differences (Dxy; Nei 1987) between O. c. algirus and O. c. cuniculus. Divergence estimates of the outgroup were derived for each locus by estimating Dxy between all rabbit samples and L. granatensis. We also calculated fixed, shared, and exclusive polymorphisms between the two subspecies. To look at differentiation from a genealogical perspective, we reconstructed the evolutionary relationships among alleles by means of Median-joining networks (Bandelt et al. 1999) as implemented in the software NETWORK 4.2.0.1 (http://www.fluxus-technology.com).
To infer the presence of gene flow between O. c. algirus and O. c. cuniculus we used three approaches. First, we derived Nm values from FST estimates [FST = 1/(4Nm + 1) for autosomal loci, and FST = 1/(3Nm + 1) for X linked loci]. Second, we estimated the relative node depth (RND) statistic (Feder et al. 2005). This statistic was performed separately for each locus by dividing Dxy between O. c. algirus and O. c. cuniculus by Dxy between all rabbit samples and Lepus. Finally, we fit the dataset to an isolation-with-migration (IM) model using the programs IM and IMa to obtain coalescent-based estimates of gene flow (Nielsen and Wakeley 2001; Hey and Nielsen 2004, 2007). A recent simulation study demonstrated the robustness of this methodology in estimating demographic parameters, even when several of its underlying assumptions are moderately violated (Strasburg and Rieseberg 2010). We used IM and IMa to estimate the posterior density for population migration rates (2Nm), which is defined as the rate at which genes come into the population per generation. IMa was used to infer multilocus estimates of gene flow and IM was used to estimate the migration rate parameters for each locus individually. Using the same program, we conducted likelihood ratio tests (LRTs) comparing nested models of divergence with and without gene flow. In models without gene flow, migration parameters, m1 and/or m2, were forced to be zero. We were also interested in evaluating whether levels of gene flow between subspecies were asymmetrical. We compared the full IM model of divergence with a model in which migration parameters m1 and m2 were constrained to be identical. For all loci for which we estimated nonzero values of gene flow, we estimated the posterior distribution of the mean time of migration events. To convert these estimates to years, we estimated the neutral mutation rate for each locus as μ = D/2T, where D is the estimated Da (Nei 1987) between Oryctolagus and Lepus and T is 11.8 Mya, the published divergence time for the Oryctolagus-Lepus comparison (Matthee et al. 2004). We then calculated the geometric mean of the locus-specific mutation rates and we assumed a generation time of one year. The isolation-with-migration model implemented in IM and IMa assumes that there has been no recombination or gene conversion within locus. Therefore, the biggest region without four gametic types in each locus was obtained using the program IMgc (Woerner et al. 2007). We assessed convergence by inspecting the plots of parameter trend lines and by comparing the results across multiple runs with different priors and random seeds.
Results
LEVELS AND PATTERNS OF POLYMORPHISM
In the analyses reported below, we combined existing polymorphism data for the same set of individuals on multiple autosomes (Carneiro et al. 2009) with new resequencing data on 27 X-linked and seven autosomal loci for a total of 19,282 bp of X chromosome sequence and a total of 16,007 bp of autosomal sequence. Haplotype tables for each gene are depicted in Figure S1. As we used only males, haplotypes were recovered directly for X-linked loci with the exception of SHOX. In this gene, we detected heterozygote positions that are consistent with a pseudoautosomal location on the rabbit X chromosome as observed for several mammals (e.g., Helena Mangs and Morris 2007). Haplotypes at autosomal loci and at SHOX were inferred computationally.
Polymorphism, frequency-spectrum tests of neutrality, population recombination estimates and divergence are summarized in Table 3 and detailed in Tables S2 and S3. Levels of genetic variation were generally high. Averaging over all loci, levels of π (Nei 1987) and θ (Watterson 1975) were higher in O. c. algirus (π = 0.502%; θw = 0.589%) than in O. c. cuniculus (π = 0.458%; θw = 0.495%). Although this difference is not significant for π (Wilcoxon signed rank test, P = 0.26) and only marginally significant for θ (Wilcoxon signed rank test, P = 0.053), it is in agreement with previous inferences of a larger historical effective population size in O. c. algirus (Geraldes et al. 2008b; Carneiro et al. 2009). We also observed substantial heterogeneity in levels of genetic variation among loci. Values of π ranged from 0% to 1.377% in O. c. algirus and from 0.037% to 1.332% in O. c. cuniculus. Despite this heterogeneity a multilocus HKA test (Hudson et al. 1987) did not reject the null model when all sequences from the two subspecies were included (P = 0.98) or when each subspecies was considered separately (P = 0.56 in O. c. algirus; P = 0.22 in O. c. cuniculus). In line with levels of genetic variation, O. c. algirus also exhibited slightly but not significantly higher average values of the population recombination parameter than O. c. cuniculus for both γ (Hey and Wakeley 1997) and ρ (Hudson 2001; γ = 1.28% and ρ = 1.77% for O. c. algirus; γ = 0.93% and ρ = 1.28% for O. c. cuniculus, Wilcoxon signed rank test, P > 0.05 for both tests).
Table 3.
Summary of the analyses of polymorphism, frequency spectrum tests of neutrality, recombination and divergence in O. c. algirus (alg.) and O. c. cuniculus (cun.).
| Locus | Polymorphism | Frequency spectrum tests of neutrality | Recombination | Divergence | ||||
|---|---|---|---|---|---|---|---|---|
| Mean π (%)1 | Mean θ (%)2 | Mean DT3 | Mean DFL4 | Mean γ (%)5 | Mean ρ (%)6 | Mean Dxy (%)7 | ||
| X-linked* (19,282 bp) | All | 0.595 | 0.620 | −0.038 | −0.307 | 0.62 | 1.40 | 3.689 |
| alg | 0.420 | 0.471 | −0.577 | −0.473 | 1.42 | 1.56 | ||
| cun | 0.358 | 0.381 | −0.271 | −0.243 | 0.51 | 0.89 | ||
| Autosomal* (16,007 bp) | All | 0.691 | 0.939 | −0.974 | −0.952 | 1.46 | 2.42 | 4.093 |
| alg | 0.633 | 0.777 | −0.774 | −0.501 | 1.13 | 2.07 | ||
| cun | 0.617 | 0.677 | −0.390 | −0.359 | 1.34 | 1.83 | ||
| Overall* (35,289 bp) | All | 0.632 | 0.743 | −0.399 | −0.556 | 0.95 | 1.79 | 3.837 |
| alg | 0.502 | 0.589 | −0.641 | −0.484 | 1.28 | 1.77 | ||
| cun | 0.458 | 0.495 | −0.317 | −0.288 | 0.93 | 1.28 | ||
Unweighted average values across 27 X-linked loci, 17 autosomal loci, or 44 combined loci are given in each row.
The average number of pairwise differences in a sample (Nei 1987).
The proportion of segregating sites in a sample (Watterson 1975).
Fu and Li’s D* (Fu and Li 1993).
Maximum likelihood estimate of the population recombination parameter between adjacent sites (Hey and Wakeley 1997).
Composite likelihood method to estimate the population recombination parameter between adjacent sites, employing a finite sites mutation model (Hudson 2001).
Dxy is the average nucleotide divergence (Nei 1987) between all rabbit haplotypes and Lepus granatensis.
The distribution of allele frequencies as measured by Tajima’s D (Tajima 1989) and Fu and Li’s D* (Fu and Li 1993) was also highly variable among loci. Tajima’s D values ranged from −1.668 to 1.558 in O. c. algirus and from −1.777 to 2.139 in O. c. cuniculus. Fu and Li’s D* values ranged from −1.916 to 1.111 in O. c. algirus and from −2.053 to 1.416 in O. c. cuniculus. Regardless of this heterogeneity, the overall trend indicates a high proportion of rare polymorphisms in the dataset. First, in most loci showing a significant skew in the allelic frequency spectrum we observed negative values of one or both statistics. Second, using multilocus coalescent simulations, the observed mean values of Tajima’s D within subspecies were significantly negative for O. c. algirus (Tajima’s D = −0.641, P < 0.0001) and for O. c. cuniculus (Tajima’s D = −0.317, P = 0.018). This significant skew of the frequency distribution of polymorphisms toward rare variants, compared to the standard neutral expectations, is in agreement with previous studies (Carneiro et al. 2009) and suggests that both subspecies have most probably undergone a population size expansion in the recent past.
Next, we compared nucleotide polymorphism (θw) between X-linked and autosomal loci. Considering a standard neutral model with constant population size, random mating and no migration, and assuming a sex ratio of 1:1, we expect levels of X-linked variation to be 75% of autosomal levels. In our dataset, levels of X-linked variation were much lower than expected: 66% of autosomal levels for the global sample (θw X-linked = 0.620; θw autosomal = 0.939), 61% in O. c. algirus (θw X-linked = 0.471; θw autosomal = 0.777), and 56% in O. c. cuniculus (θw X-linked = 0.381; θw autosomal = 0.677). However, male-driven evolution will likely increase mutation rates on the autosomes relative to the X chromosome. In fact, the average divergence to Lepus was higher for the autosomes (Dxy = 4.093%) than for the X chromosome (Dxy = 3.689%). To take into account possible mutation rate differences among regions that could be responsible for the lower than expected X chromosome diversity, we divided our estimates of θw by the divergence to Lepus (Dxy, Nei 1987). Using our corrected estimates of nucleotide variation, levels of X-linked variation were 73% of autosomal levels in the global sample (θ w/Dxy X-linked = 0.168; θw/Dxy autosomal = 0.229), 67% in O. c. algirus (θw/Dxy X-linked = 0.128; θw/Dxy autosomal = 0.190) and 62% in O. c. cuniculus (θw/Dxy X-linked = 0.103; π autosomal = 0.165). Even though the values were still lower than expected, when we performed HKA tests comparing all X-linked loci to all autosomal loci, (taking into account differences in inheritance mode) we did not reject a neutral model (P > 0.5 for all three tests). It is noteworthy that the ratio of X/autosome diversity is slightly lower for each subspecies compared to the global sample. A similar pattern was seen for indel variation (data not shown). Distinct levels of gene flow between X-linked and autosomal loci could partly explain this discrepancy (see below).
MULTILOCUS PATTERNS OF DIFFERENTIATION AND GENE GENEALOGIES
Patterns of differentiation were highly variable among loci, with some loci showing little differentiation between subspecies and others showing strong differentiation (Table 4). Values of FST varied between 0.8% and 97.7% and Da values from 0% to 1.329%. This variation in differentiation can be visualized in the gene genealogies obtained for each of the 44 loci, where all three possible kinds of genealogies are represented (Fig. 2). Reciprocally monophyletic genealogies were observed in some loci (e.g., STAG1), whereas the great majority of loci displayed variable levels of shared variation, from paraphyletic (e.g., ARHGEF9) to polyphyletic genealogies (e.g., GK5).
Table 4.
Genetic differentiation and gene flow between O. c. algirus and O. c. cuniculus at 27 X-linked and 17 autosomal loci.
| Locus | Genetic differentiation | Gene flow | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chromosome | FST (%)4 | Da (%)5 | RND6 | Sxalg7 | Sxcun8 | Ss9 | Sf10 | Nm11 | 2Nm12,* | 2Nm13,* | Mean 2Nm | |
| SHOX | X | 90.7 | 0.939 | 0.14 | 5 | 2 | 0 | 5 | 0.03 | 0.00 | 0.00 | 0.00 |
| GLRA2 | X | 47.1 | 0.218 | 0.18 | 0 | 8 | 6 | 0 | 0.38 | 0.53 | 0.00 | 0.27 |
| PHKA22 | X | 2.7 | 0.008 | 0.11 | 45 | 35 | 60 | 0 | 12.22 | 10.22 | 0.28 | 5.25 |
| PDHA1 | X | 1.1 | 0.000 | 0.10 | 4 | 7 | 7 | 0 | 29.67 | 23.54 | 12.11 | 17.82 |
| POLA1 | X | 57.4 | 0.709 | 0.24 | 12 | 2 | 3 | 0 | 0.25 | 0.64 | 0.00 | 0.32 |
| IL1RAPL1 | X | 7.4 | 0.037 | 0.15 | 7 | 2 | 7 | 0 | 4.20 | 23.11 | 0.03 | 11.57 |
| TMEM47 | X | 42.6 | 0.184 | 0.17 | 5 | 5 | 2 | 0 | 0.45 | 0.00 | 0.36 | 0.19 |
| MAOA | X | 31.8 | 0.257 | 0.26 | 1 | 19 | 3 | 0 | 0.72 | 0.48 | 2.14 | 1.31 |
| TIMP1 | X | 6.5 | 0.036 | 0.09 | 6 | 6 | 8 | 0 | 4.81 | 0.18 | 12.18 | 6.18 |
| DGKK | X | 65.1 | 0.300 | 0.15 | 8 | 1 | 0 | 2 | 0.18 | 0.00 | 0.00 | 0.00 |
| SMCX2 | X | 68.0 | 0.531 | 0.45 | 35 | 12 | 9 | 0 | 0.16 | 0.00 | 0.40 | 0.20 |
| ARHGEF9 | X | 81.0 | 0.764 | 0.25 | 3 | 7 | 1 | 0 | 0.08 | 0.00 | 0.25 | 0.13 |
| Centromere | ||||||||||||
| MSN2 | X | 82.9 | 0.786 | 0.23 | 11 | 20 | 3 | 19 | 0.07 | 0.00 | 0.06 | 0.03 |
| OPHN1 | X | 14.3 | 0.093 | 0.21 | 9 | 5 | 6 | 0 | 1.99 | 0.00 | 0.69 | 0.35 |
| OGT | X | 73.7 | 0.267 | 0.11 | 1 | 5 | 0 | 2 | 0.12 | 0.00 | 0.00 | 0.00 |
| NRK | X | 53.5 | 0.539 | 0.35 | 8 | 7 | 1 | 1 | 0.29 | 0.00 | 0.00 | 0.00 |
| AMOT | X | 90.0 | 0.961 | 0.26 | 2 | 1 | 0 | 4 | 0.04 | 0.00 | 0.00 | 0.00 |
| KLHL13 | X | 79.1 | 0.836 | 0.46 | 10 | 2 | 0 | 5 | 0.09 | 0.00 | 0.00 | 0.00 |
| PGK1 | X | 46.1 | 0.565 | 0.28 | 18 | 8 | 3 | 0 | 0.39 | 1.08 | 0.00 | 0.54 |
| CYLC | X | 40.3 | 0.144 | 0.10 | 5 | 6 | 0 | 0 | 0.49 | 0.00 | 0.00 | 0.00 |
| KLHL4 | X | 20.1 | 0.185 | 0.19 | 8 | 9 | 8 | 0 | 1.33 | 0.56 | 0.30 | 0.43 |
| PABPC5 | X | 8.2 | 0.025 | 0.10 | 5 | 3 | 4 | 0 | 3.76 | 4.11 | 0.43 | 2.27 |
| TNMD | X | 7.1 | 0.000 | 0.48 | 2 | 2 | 9 | 0 | 5.06 | 4.52 | 5.41 | 4.97 |
| DIAPH2 | X | 18.4 | 0.133 | 0.22 | 2 | 6 | 9 | 0 | 1.48 | 0.04 | 3.52 | 1.78 |
| GRIA3 | X | 15.1 | 0.066 | 0.19 | 3 | 6 | 2 | 0 | 1.87 | 0.07 | 2.50 | 1.28 |
| OCRL | X | 29.6 | 0.342 | NA | 19 | 3 | 2 | 0 | 0.79 | 3.45 | 0.03 | 1.74 |
| GPC4 | X | 14.4 | 0.134 | 0.21 | 8 | 10 | 10 | 0 | 1.99 | 0.00 | 1.09 | 0.55 |
| HPRT12 | X | 2.2 | 0.027 | 0.28 | 9 | 15 | 38 | 0 | 14.99 | 3.47 | 0.95 | 2.21 |
| F9 | X | 80.0 | 0.570 | 0.20 | 2 | 4 | 0 | 4 | 0.08 | 0.00 | 0.00 | 0.00 |
| FMR1 | X | 90.1 | 1.197 | 0.51 | 4 | 2 | 0 | 8 | 0.04 | 0.00 | 0.00 | 0.00 |
| G6PD | X | 97.7 | 1.329 | 0.34 | 0 | 1 | 0 | 7 | 0.01 | 0.00 | 0.00 | 0.00 |
| EXT11 | 3 | 2.0 | 0.008 | 0.12 | 15 | 8 | 10 | 0 | 12.26 | 15.27 | 0.06 | 7.66 |
| CYTC1 | 4 | 6.4 | 0.052 | 0.14 | 19 | 10 | 14 | 0 | 3.65 | 6.48 | 0.41 | 3.45 |
| LUM1 | 4 | 3.7 | 0.034 | 0.24 | 13 | 16 | 30 | 0 | 6.49 | 0.07 | 12.56 | 6.31 |
| UD141 | 7 | 8.2 | 0.069 | NA | 16 | 17 | 19 | 0 | 2.82 | 5.12 | 0.07 | 2.59 |
| ATP12A1 | 8 | 26.9 | 0.278 | 0.25 | 23 | 19 | 18 | 0 | 0.68 | 0.00 | 0.48 | 0.25 |
| PRL1 | 12 | 1.9 | 0.012 | 0.15 | 22 | 27 | 12 | 0 | 12.90 | 2.93 | 4.51 | 3.72 |
| T1 | 12 | 8.0 | 0.119 | 0.19 | 16 | 18 | 22 | 0 | 2.86 | 4.46 | 0.00 | 2.23 |
| MGST31 | 13 | 42.7 | 0.605 | 0.31 | 19 | 20 | 9 | 0 | 0.34 | 0.00 | 0.00 | 0.00 |
| SLC4A7 | 14 | 20.6 | 0.094 | 0.12 | 7 | 8 | 3 | 0 | 0.97 | 2.12 | 0.18 | 1.15 |
| Centromere | ||||||||||||
| STAG11 | 14 | 72.9 | 0.967 | 0.37 | 18 | 16 | 1 | 9 | 0.09 | 0.00 | 0.00 | 0.00 |
| GK5 | 14 | 12.0 | 0.093 | 0.17 | 4 | 5 | 14 | 0 | 1.84 | 4.16 | 6.95 | 5.56 |
| SIAH2 | 14 | 3.7 | 0.027 | 0.14 | 19 | 8 | 8 | 0 | 6.46 | 4.93 | 0.00 | 2.47 |
| KPNA4 | 14 | 9.5 | 0.022 | 0.17 | 8 | 11 | 0 | 0 | 2.38 | 0.00 | 0.00 | 0.00 |
| MYNN | 14 | 14.0 | 0.083 | NA | 7 | 4 | 5 | 0 | 1.53 | 0.00 | 0.90 | 0.46 |
| NAALADL2 | 14 | 13.0 | 0.065 | 0.14 | 6 | 9 | 3 | 0 | 1.67 | 0.34 | 0.27 | 0.31 |
| GBE1 | 14 | 1.3 | 0.003 | 0.09 | 5 | 7 | 3 | 0 | 18.79 | 0.00 | 12.23 | 6.12 |
| TIAM11 | 14 | 0.8 | 0.006 | 0.20 | 21 | 8 | 21 | 0 | 33.00 | 16.45 | 1.15 | 8.80 |
| Mean X-linked3 | 44.7 | 0.401 | 0.23 | 5.8 | 5.1 | 3.4 | 1.4 | 2.24 | 2.31 | 1.52 | 1.91 | |
| Mean Autosomal | 14.6 | 0.149 | 0.19 | 14.0 | 12.4 | 11.3 | 0.5 | 6.40 | 3.67 | 2.34 | 3.00 | |
Data from Carneiro et al. (2009).
Data from Geraldes et al. (2006).
Loci from Geraldes et al. (2006) were excluded from mean values.
Calculated using the method proposed by Hudson et al. (1992).
Net nucleotide distance per base pair (Nei 1987) between O. c. algirus and O. c. cuniculus.
Dxy between O. c. algirus and O. c. cuniculus divided by Dxy between all rabbit samples and Lepus
No. of exclusive polymorphisms in O. c. algirus.
No. of exclusive polymorphisms in O. c. cuniculus.
No. of shared polymorphisms.
No. of fixed differences.
Calculated using the expression FST =1/(1+4Nm) for autosomal loci and FST =1/(1+3Nm) for X-linked loci.
ML estimates of population migration rate from O. c. cuniculus to O. c. algirus using the IM software.
ML estimates of population migration rate from O. c. algirus to O. c. cuniculus using the IM software.
Values of 0 correspond to the first bin of the parameter space surveyed for the migration parameters.
Figure 2.
Median-joining haplotype networks representing the phylogenetic relationships among all the alleles found in both subspecies of the European rabbit. Individuals with missing data were excluded. The size of the circles is proportional to the frequency of each haplotype. The population group of the individuals represented in each haplotype is denoted by black for O. c. algirus and white for O. c. cuniculus.
All measures of differentiation were consistent in revealing higher differentiation on the X chromosome compared to the autosomes (Table 4). Mean FST and net divergence (Da, Nei 1987) values were significantly higher for X-linked loci (FST = 44.7%, Da = 0.401%) than for autosomal loci (FST = 14.6%, Da = 0.149%; Mann–Whitney U-test, P < 0.01 for both tests). Likewise, the ratio of fixed differences to shared polymorphisms was significantly elevated for X-linked loci (0.39) compared to autosomal loci (0.05) (Fisher’s Exact Test, P < 0.001). Furthermore, nine of the 27 genealogies on the X chromosome were reciprocally monophyletic, and a few others showed a single mismatched haplotype in an otherwise well-sorted genealogy (Fig. 2). In contrast, just two of the 17 autosomal loci (STAG1 and MGST3) showed clear phylogenetic differentiation between subspecies, but only STAG1 was reciprocally monophyletic (9/18, 1/16; Fisher’s Exact Test, P < 0.035). Thus, although there were loci on the X chromosome and on the autosomes showing high levels of differentiation and loci showing low levels of differentiation, our results show that the X chromosome is more differentiated, on average, than the autosomes.
LEVELS AND PATTERNS OF GENE FLOW
Two possible factors, or some combination of both, could cause greater average differentiation on the X chromosome compared to the autosomes: (1) lower levels of gene flow on the X, and (2) faster rate of lineage sorting on the X. To test whether less gene flow occurred on the X chromosome compared to the autosomes we used three approaches. First, we derived Nm values per locus from FST estimates of differentiation with the assumption of migration–drift equilibrium. As expected based on FST estimates, mean Nm values were significantly lower for X-linked loci (Nm = 2.24) than for autosomal loci (Nm = 6.40; Mann–Whitney U-test, P = 0.003). However, the assumption of migration–drift equilibrium may not be met, and levels of gene flow on the autosomes derived from these estimates may still be overestimated by slower lineage sorting compared to the X chromosome.
Second, we estimated the relative node depth (RND) statistic (Feder et al. 2005; Table 4). We expect RND values per locus to be inversely proportional to the amount of gene flow after the split of the two subspecies. Mean RND values are higher but not significantly higher in X-linked loci (RND = 0.23) when compared to autosomal loci (RND = 0.19; Mann–Whitney U-test, P = 0.25). However, most of the highest RND values in our dataset are X-linked.
Third, we fitted an isolation-with-migration model (IM) model (Nielsen and Wakeley 2001; Hey and Nielsen 2004, 2007) to the 27 X-linked and to the 17 autosomal loci separately. This method assesses the relative roles of migration and isolation underlying the observed differentiation between populations, thus allowing a better distinction between ancestral polymorphism versus recent gene flow. LRTs (Hey and Nielsen 2007) revealed a significantly better fit to a model with gene flow suggesting that gene flow has occurred on the X chromosome, as well as on the autosomes (P < 0.001; Table S4). Furthermore, a model incorporating migration in both directions was also a significantly better fit than models in which one of the migration parameters was set to zero. In agreement with inferences of gene flow from FST, the IM analysis revealed lower levels of gene flow on the X chromosome compared to the autosomes. For X-linked loci, the estimated population migration rate (2Nm) was 0.38 (90% highest posterior density (HPD) interval: 0.16 to 0.77) per generation for O. c. algirus and 0.26 (90% HPD interval: 0.08 to 0.58) per generation for O. c. cuniculus. For autosomal loci, estimated 2Nm values were much higher: 1.69 (90% HPD interval: 0.60 to 3.18) and 0.83 (90% HPD interval: 0.27 to 1.88) in O. c. algirus and O. c. cuniculus, respectively. To test if loci for which we detected significant deviations from neutral expectations were influencing the IM simulations, we repeated the analyses excluding those loci showing significant values of Tajima’s D or Fu and Li’s D* when calculated for each subspecies separately. The resulting estimates were very similar (data not shown). To further explore the IM model, we estimated migration rates separately for each locus in a combined run of autosomal and X-linked loci (Table 4). Mean 2Nm values per locus were significantly lower for X-linked loci (2Nm = 1.92) than for autosomal loci (2Nm = 3.01; Mann–Whitney U-test, P = 0.047). Taken together, the greater differentiation of the X chromosome compared to the autosomes appears to be largely explained by differences in levels of gene flow.
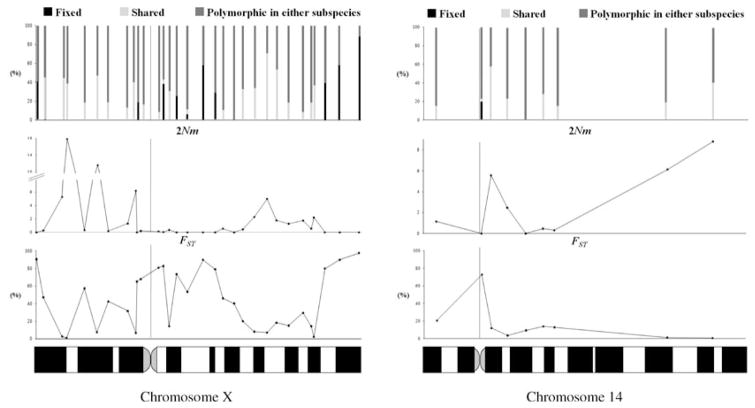
CHROMOSOMAL PATTERNS OF DIFFERENTIATION
A major goal of this study was to assess the genomic scale over which patterns of differentiation change along individual chromosomes. Figure 3 shows fixed, shared, and exclusive polymorphisms, average 2Nm values inferred using IM and FST values per locus, against position on chromosomes X and 14. This figure also includes four X-linked loci from a previous study sampled slightly differently (Geraldes et al. 2006; Table 2). On chromosome 14, a single locus located near the centromere (STAG1) showed high differentiation. In contrast, on the X chromosome multiple highly differentiated loci were detected along the entire chromosome, and some of these were found in clusters. For example, we observed one group of four and another group of three consecutive loci displaying reciprocally monophyletic genealogies with fixed differences between subspecies (OGT, NRK, AMOT, KLHL13 and F9, FMR1, G6PD; Figs. 2 and 3). We also observed two groups of four and one group of three consecutive loci with FST > 50% (DGKK, SMCX, ARHGEF9, MSN; OGT, NRK, AMOT, KLHL13 and F9, FMR1, G6PD). To test whether this pattern was spatially nonrandom, we generated 100,000 datasets randomizing the position of the loci with fixed and shared polymorphism, and found that clustering in one group of four and another group of three loci with fixed differences is unlikely by chance (P < 0.05). The observation of three groups of three or more consecutive loci with FST > 50% is also unlikely (P < 0.05). Moreover, our coalescent estimates of gene flow per locus revealed that several continuous regions on the X chromosome display estimates of 2Nm well below 1. These results suggest that some regions of the X chromosome represent continuous segments of high differentiation and reduced introgression encompassing several megabases.
Figure 3.
Fixed/shared/exclusive polymorphisms, average 2Nm values estimated using IM, and FST plotted against sequence position on chromosomes X and 14. Position of the centromere is indicated with a vertical line. Data on four X-linked loci from Geraldes et al. (2006) on a different set of individuals were also included.
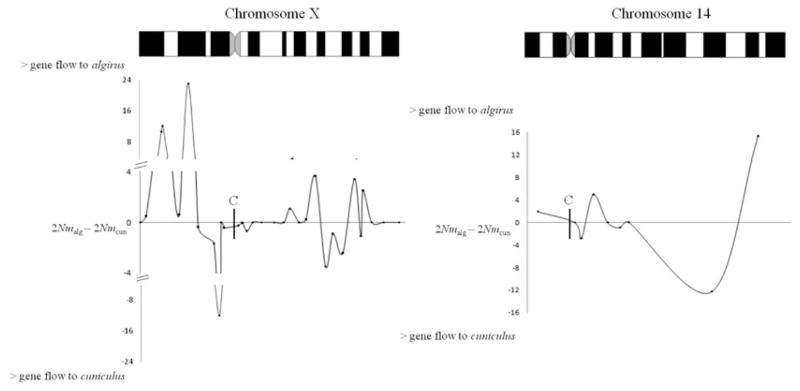
ASYMMETRIC LEVELS OF GENE FLOW AND DISTRIBUTION OF MIGRATION EVENTS THROUGH TIME
We were interested in testing whether our estimates of 2Nm suggest asymmetrical introgression. Although a combined IM run of 44 loci indicates higher gene flow from O. c. cuniculus to O. c. algirus (2Nm = 0.79; 90% HPD interval: 0.43 to 1.23) than in the opposite direction (2Nm = 0.51; 90% HPD interval: 0.26 to 0.84), when we formally tested whether a model with asymmetrical introgression is better than models in which gene flow is equal in both directions, we did not obtain significant results (LRT P > 0.5; Table S4). On the other hand, we highlight that when we look at each locus separately, migration estimates are usually highly asymmetric (Table 4). Interestingly, on the X chromosome consecutive loci tend to show an identical asymmetrical nature (Fig. 4). For example, in all loci close to the centromere we either inferred zero gene flow in both directions or higher gene flow from O. c. algirus to O. c. cuniculus.
Figure 4.
Plot of the difference between 2Nm values in O. c. algirus and 2Nm values in O. c. cuniculus per locus versus position on chromosomes X and 14. If the resulting value is positive higher gene flow was inferred from O. c. cuniculus to O. c. algirus than in the opposite direction. If the resulting value is negative higher gene flow was inferred from O. c. algirus to O. c. cuniculus than in the opposite direction. Position of the centromere (C) is indicated with a vertical line. Data on four X-linked loci from Geraldes et al. (2006) on a different set of individuals were also included.
Further insight can be gained into the history of gene flow between rabbit subspecies by looking at the timescale over which gene flow has occurred. We measured the distribution of the mean time of introgression events for each locus over the course of the IM simulations (Fig. S2). Posterior probability distributions for mean times of migration per locus varied from 0.279 Mya to 0.732 Mya (Fig. S2).
Discussion
This study describes DNA sequence diversity and divergence at multiple X-linked and autosomal loci in both subspecies of the European rabbit (O. c. cuniculus). Three main findings emerged: (1) levels of differentiation between subspecies were highly heterogeneous, and this variation is likely caused by differential levels of introgression among loci; (2) X-linked loci displayed higher levels of differentiation, on average, than autosomal loci; and (3) on chromosome 14, only the centromeric region displayed a high level of differentiation, while on the X chromosome several islands of differentiation extending for several megabases were observed, both near the centromere and on the chromosome arms. These findings support the idea that the X chromosome plays an important role in reproductive isolation in rabbits and shows that large islands of differentiation may exist between species that are experiencing substantial gene flow.
GENEALOGICAL PATTERNS AND THE SEMIPERMEABLE NATURE OF THE RABBIT HYBRID ZONE
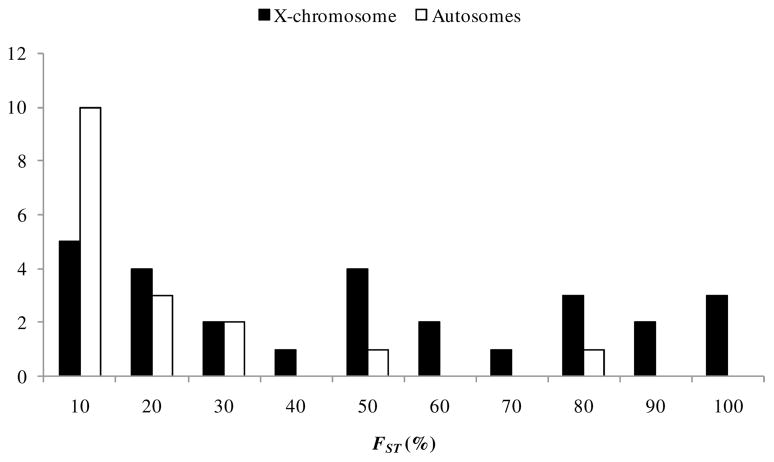
One of the more striking observations in this study is that genealogical patterns varied considerably among loci. Reciprocally monophyletic genealogies were observed at some loci, whereas the great majority displayed variable levels of shared variation. Notably, this genealogical discordance is reflected in a remarkable range of FST values (from 0.8% to 97.7%) and is observed both on the X chromosome, as well as on the autosomes (Fig. 5). There are two possible interpretations for the pattern of differentiation observed here. The first is that incomplete lineage sorting is pervasive across the genome because of the relatively recent origin of these taxa. Under this scenario, the few regions that attained reciprocal monophyly became differentiated. The second interpretation is that neutral or advantageous alleles crossed species boundaries and are homogenizing genetic variability across the genome, while certain regions are protected by selection from gene flow.
Figure 5.
Histogram of FST values between subspecies of rabbits (data from Table 4). Autosomal loci are shown in white, X-linked loci in black.
As this and other studies have shown (Geraldes et al. 2008b; Carneiro et al. 2009), the genomes of these two subspecies appear to be remarkably permeable with respect to gene flow. Our IMa runs for both autosomal and X-linked loci indicate that models incorporating gene flow are a significantly better fit to the data than models without gene flow. We inferred high levels of gene flow in both directions. In addition, locus-specific estimates of gene flow are strongly and negatively correlated with FST and Da values (2Nm × FST: r = −0.88, P < 0.001; 2Nm × Da: r = −0.797, P < 0.001). This strong inverse relationship between coalescent estimates of gene flow and measures of differentiation argues against the idea that most shared variation represents ancestral polymorphism. In addition, the genealogical patterns in Figure 2 suggest that differential lineage sorting alone cannot explain why levels of genetic variation between subspecies are highly variable among loci. For example, at several loci on the X chromosome and autosomes there are two divergent lineages that correspond to the two subspecies (e.g., SHOX, STAG1), whereas in others two highly divergent haplogroups are still present but no correspondence with the subspecies is observed (e.g., ARHGEF9, GK5). This second topology is particularly informative about introgression and suggests that these loci apparently attained reciprocal monophyly in the past but subsequently experienced introgression of well-differentiated haplotypes. In summary, although we cannot exclude the presence of ancestral variation, high gene exchange between rabbit subspecies seems likely. Large differences among loci in levels of genetic differentiation in the early stages of divergence, as documented here, have now been reported for many plant and animal taxa (e.g., Emelianov et al. 2004; Borge et al. 2005; Putnam et al. 2007; Minder and Widmer 2008; Kane et al. 2009; Maroja et al. 2009; see Nosil et al. 2009 and Noor and Bennett 2009 for recent reviews), suggesting that this pattern may be quite common in nature.
The evidence for introgressive hybridization between O. c. algirus and O. c. cuniculus raises the question of how long ago, and how many times, migration events have happened. On the one hand, full haplotype sharing for several loci (e.g., PDHA1, OCRL, LUM, TIAM1; Fig. 2 and Fig. S1) for which we inferred moderate-to-high estimates of the population recombination parameter suggests that some of the migration events happened very recently. These events are most likely associated with the postglacial secondary contact between the two subspecies that is thought to have occurred in the last few thousand years (Branco et al. 2002). On the other hand, our estimates of the mean times of migration events for each locus under an IM model ranged from 0.279 Mya to 0.732 Mya, suggesting that some gene flow is old and has probably occurred multiple times throughout the history of divergence of these two subspecies. In fact, Quaternary climatic oscillations were characterized by several interglacial periods that may have provided the opportunity for secondary contact and gene exchange on several occasions.
It is instructive to compare the genealogical patterns seen in rabbits to those seen in subspecies of the house mouse, the only other mammal for which such data exist in the context of hybridizing taxa. In a similar fashion, genealogical patterns between Mus musculus musculus and M. m. domesticus vary substantially among loci, with some genealogies being reciprocally monophyletic between subspecies, whereas others being paraphyletic or polyphyletic (Salcedo et al. 2007; Geraldes et al. 2008a). Despite this similarity, the two systems differ markedly in overall levels of differentiation. In mice, all X-linked loci and about half of autosomal loci were reciprocally monophyletic between subspecies. In rabbits, the two groups are genetically much less differentiated. In our dataset, only nine of the 27 X-linked loci and one of the 17 autosomal loci were reciprocally monophyletic. This is surprising because mouse subspecies diverged about 0.5 Mya whereas rabbit subspecies diverged about 2.0 Mya. The underlying cause for this substantial difference could be related to distinct levels of introgression in the two systems. A recent study in mice using multiple markers in an IM framework found low levels of gene flow from M. m domesticus to M. m. musculus (2Nm = 0.094) and no gene flow was detected in the opposite direction (Geraldes et al. 2008a). These estimates are substantially lower than levels of bidirectional gene flow inferred from our IM analysis in rabbits (2Nmalgirus = 0.79; 2Nmcuniculus = 0.51). Importantly, however, the higher levels of historical gene flow inferred for rabbits do not necessarily indicate that the rate of hybridization is currently higher; instead, it may be related to multiple opportunities for hybridization between rabbit subspecies during the last few thousand years together with a more ancient date for the current contact. In fact, Pool and Nielsen (2009) analyzed the length distribution of chromosome segments of migrant origin between mouse subspecies and argued that the rate of hybridization has increased in the recent past and has the potential to homogenize a portion of their genomes.
THE X CHROMOSOME AND SPECIATION
The X chromosome is known to play a disproportionately large role in the evolution of postzygotic reproductive isolation (Coyne and Orr 1989; Presgraves 2008). Consistent with this view, we observed a general trend for X-linked loci to display higher differentiation between rabbit subspecies than autosomal loci (Fig. 5). The IM analyses suggests that this higher differentiation is likely to be explained by lower levels of gene flow on the X chromosome (mean 2Nm = 0.32) compared to the autosomes (mean 2Nm = 1.26).
Although differentiation is on average higher and estimates of 2Nm lower on the X compared to the autosomes, these average values mask substantial variation observed among X-linked loci (Figs. 3 and 5). Several loci on the X chromosome showed little differentiation and 2Nm values much higher than 1 (Table 4). In fact, the highest 2Nm estimates in our dataset were X-linked (PDHA1, IL1RAPL). Differences in levels of differentiation among loci help to identify particular genomic regions contributing to reproductive isolation. On the X chromosome, we identified four regions of high differentiation (defined as having one or more loci with FST > 0.7): (1) the telomeric region of the short arm (SHOX), most likely corresponding to the pseudoautosomal region (PAR); (2) the centromeric region (DGKK-MSN); (3) a region in the middle of the long arm (OGT-KLHL13); and (4) the telomeric region of the long arm (F9-G6PD).
The fact that we observed high levels of differentiation at the PAR deserves comment. PARs are small homologous regions shared by the mammalian X and Y chromosomes within which obligatory recombination occurs in male meiosis, and they therefore play a key role in initiating XY pairing (e.g., Rappold 1993). Observations of reduced testis weight and dissociation of X and Y chromosomes during male meiosis in crosses between Mus domesticus and M. spretus led to the suggestion that a structural X–Y incompatibility due to divergence at the PAR could be responsible for male sterility between these species (Guenet et al. 1990; Matsuda et al. 1991; Hale et al. 1993). Our results also suggest that divergence at the PAR may play a role in reproductive isolation between rabbit subspecies. Furthermore, such a cytogenetic mechanism could also explain the reduced introgression observed for the Y-chromosome in rabbits (Geraldes et al. 2008b). Under this hypothesis, we expect X–Y asynapsis rates to be higher in male F1s resulting from crosses between subspecies compared to males resulting from crosses within subspecies.
ISLANDS OF DIFFERENTIATION BETWEEN SUBSPECIES
The chromosomal distribution of patterns of differentiation and gene flow observed in Figure 3 illustrate the mosaic nature of the rabbit genome with respect to gene flow between subspecies. Most interestingly, several regions resist the homogenizing influence of gene flow and retain strong divergence. Consistent with previous studies (Geraldes et al. 2006; Carneiro et al. 2009), we found an overrepresentation of highly differentiated loci situated close to the centromeres, where recombination is expected to be low, supporting recent speciation models (Noor et al. 2001; Rieseberg 2001; Navarro and Barton 2003). We note that this observation is also consistent with predictions coming from theories of centromere evolution (Henikoff et al. 2001), which posit that the rapidly evolving nature of centromeres may ultimately result in hybrid incompatibilities.
One of the main goals of our study was to interpret patterns of differentiation among loci taking into account their genome location, to begin to explore the genomic scale over which levels of differentiation and gene flow vary. We observed four regions with highly differentiated loci on the X chromosome, and each of these regions extends for several megabases. For example, the distance between OGT and KLHL13, located on the long arm of the X, is approximately 16 megabases. In contrast, on chromosome 14 only one island of differentiation was observed and was located near the centromere. Levels of differentiation were otherwise low throughout chromosome 14, suggesting that islands of differentiation on the autosomes might be much rarer and/or smaller than on the X.
It is important to note, however, that the current sampling of loci is not dense enough to identify all differentiated regions or to definitively confirm that the detected regions of high differentiation do not represent multiple independent regions separated by regions of low differentiation. We also note that the density of markers on the X is higher than on the autosomes, and this may contribute to some of the observed differences. It should be possible to address these issues by scanning the genome with a much higher density of markers.
Finally, we emphasize the asymmetrical estimates of gene flow in the same orientation for consecutive loci on the X chromosome (Fig. 4). This pattern is particularly striking near the centromere (Geraldes et al. 2006). Overall we infer more gene flow from O. c. cuniculus to O. c. algirus, but several loci close to the centromere either show no introgression or asymmetrical gene flow from O. c. algirus to O. c. cuniculus. This observation is in agreement with the asymmetrical nature of Dobzhansky–Muller incompatibilities under reciprocal introgression (Orr 1995; Gavrilets 1997). Importantly, the locus-specific estimates of gene flow obtained using IM provide testable predictions for studies of clinal patterns of variation in allele frequencies across the rabbit hybrid zone.
Supplementary Material
Figure S1. Polymorphisms for the 27 X-linked and 17 autosomal loci: (1) SHOX, (2) GLRA2, (3) PDHA1, (4) POLA1, (5) IL1RAPL1, (6) TMEM47, (7) MAOA, (8) TIMP1, (9) DGKK, (10) ARHGEF9, (11) OPHN1, (12) OGT, (13) NRK, (14) AMOT, (15) KLHL13, (16) PGK1, (17) CYLC1, (18) KLHL4, (19) PABPC5, (20) TNMD, (21) DIAPH2, (22) GRIA3, (23) OCRL, (24) GPC4, (25) F9, (26) FMR1, (27) G6PD, (28) EXT1, (29) CYTC, (30) LUM, (31) UD14, (32) ATP12A, (33) PRL, (34) T, (35) MGST3, (36) SLC4A7, (37) STAG1, (38) GK5), (39) SIAH2, (40) KPNA4, (41) MYNN, (42) NAALADL2, (43) GBE1, and (44) TIAM1.
Figure S2. Marginal posterior probability densities for the mean time of all inferred migration events per locus (A) from O. c. cuniculus to O. c. algirus and (B) from O. c. algirus to O. c. cuniculus.
Table S1. PCR primers and annealing temperatures.
Table S2. Analyses of polymorphism, frequency spectrum tests of neutrality, recombination and divergence in O. c. algirus (alg.) and O. c. cuniculus (cun.) for 27 X-linked loci.
Table S3. Analyses of polymorphism, frequency spectrum tests of neutrality, recombination and divergence in O. c. algirus (alg.) and O. c. cuniculus (cun.) for 17 autosomal loci.
Table S4. Tests of nested models with unidirectional gene flow, without gene flow and asymmetrical gene flow against the full isolation-with-migration model.
Acknowledgments
This work was partially supported by a Fundação para a Ciência e a Tec nologia Ph.D. grant to MC (SFRH/BD/23786/2005), by a Junta de Comunidades Castilla la Mancha grant to JABA supported by European Social Fund, by Research Projects to NF and RV (PTDC/BIA-BDE/72304/2006; CGL2009-11665; POII09-0099-2557), and by National Science Foundation and National Institutes of Health grants to MWN. We thank P. Tarroso for help with the map in Figure 1, members of the Nachman lab for valuable discussions, and M. Dean, A. Geraldes, F. Sequeira, Leonie Moyle, and two anonymous reviewers for valuable comments on the manuscript.
LITERATURE CITED
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Barbash DA, Siino DF, Tarone AM, Roote J. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc Natl Acad Sci USA. 2003;100:5302–5307. doi: 10.1073/pnas.0836927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH, Hewitt GM. Adaptation, speciation and hybrid zones. Nature. 1989;341:497–503. doi: 10.1038/341497a0. [DOI] [PubMed] [Google Scholar]
- Baxter SW, Nadeau NJ, Maroja LS, Wilkinson P, Counterman BA, Dawson A, Beltran M, Perez-Espona S, Chamberlain N, Ferguson L, et al. Genomic hotspots for adaptation: the population genetics of Müllerian mimicry in the heliconius melpomene clade. PLoS Genet. 2010;6:e1000794. doi: 10.1371/journal.pgen.1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson BO. Theoretical models of speciation. Zool Scripta. 1979;8:303–304. [Google Scholar]
- Besansky NJ, Krzywinski J, Lehmann T, Simard F, Kern M, Mukabayire O, Fontenille D, Toure Y, Sagnon NF. Semipermeable species boundaries between anopheles gambiae and anopheles arabiensis: evidence from multilocus DNA sequence variation. Proc Natl Acad Sci USA. 2003;100:10818–10823. doi: 10.1073/pnas.1434337100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borge T, Webster MT, Andersson G, Saetre GP. Contrasting patterns of polymorphism and divergence on the Z chromosome and autosomes in two Ficedula flycatcher species. Genetics. 2005;171:1861–1873. doi: 10.1534/genetics.105.045120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco M, Ferrand N, Monnerot M. Phylogeography of the European rabbit (Oryctolagus cuniculus) in the Iberian Peninsula inferred from RFLP analysis of the cytochrome b gene. Heredity. 2000;85 Pt 4:307–317. doi: 10.1046/j.1365-2540.2000.00756.x. [DOI] [PubMed] [Google Scholar]
- Branco M, Monnerot M, Ferrand N, Templeton AR. Postglacial dispersal of the European rabbit (Oryctolagus cuniculus) on the Iberian peninsula reconstructed from nested clade and mismatch analyses of mitochondrial DNA genetic variation. Evolution. 2002;56:792–803. doi: 10.1111/j.0014-3820.2002.tb01390.x. [DOI] [PubMed] [Google Scholar]
- Brown KM, Burk LM, Henagan LM, Noor MAF. A test of the chromosomal rearrangement model of speciation in Drosophila pseudoobscura. Evolution. 2004;58:1856–1860. doi: 10.1111/j.0014-3820.2004.tb00469.x. [DOI] [PubMed] [Google Scholar]
- Caisse M, Antonovics J. Evolution in closely adjacent plant populations. IX. Evolution of reproductive isolation in clinal populations. Heredity. 1978;40:371–384. [Google Scholar]
- Carling MD, Brumfield RT. Haldane’s rule in an avian system: using cline theory and divergence population genetics to test for differential introgression of mitochondrial, autosomal, and sex-linked loci across the passerina bunting hybrid zone. Evolution. 2008;62:2600–2615. doi: 10.1111/j.1558-5646.2008.00477.x. [DOI] [PubMed] [Google Scholar]
- Carneiro M, Ferrand N, Nachman MW. Recombination and speciation: loci near centromeres are more differentiated than loci near telomeres between subspecies of the European rabbit (oryctolagus cuniculus) Genetics. 2009;181:593–606. doi: 10.1534/genetics.108.096826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantry-Darmon C, Bertaud M, Urien C, Chadi-Taourit S, Perrocheau M, Rogel-Gaillard C, Hayes H. Expanded comparative mapping between man and rabbit and detection of a new conserved segment between HSA22 and OCU4. Cytogenet Genome Res. 2005;111:134–139. doi: 10.1159/000086382. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Two rules of speciation. In: Otte D, Endler J, editors. Speciation and its consequences. Sinauer Associates; Sunderland, MA: 1989. pp. 180–207 . [Google Scholar]
- Dobzhansky T. Studies on hybrid sterility. II. Localization of sterility factors in drosophila pseudoobscura hybrids. Genetics. 1936;21:113–135. doi: 10.1093/genetics/21.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelianov I, Marec F, Mallet J. Genomic evidence for divergence with gene flow in host races of the larch budmoth. Proc Biol Sci. 2004;271:97–105. doi: 10.1098/rspb.2003.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler JA. Geographic variation, speciation, and clines. Princeton Univ. Press; Princeton, NJ: 1977. [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Feder JL, Roethele JB, Filchak K, Niedbalski J, Romero-Severson J. Evidence for inversion polymorphism related to sympatric host race formation in the apple maggot fly, Rhagoletis pomonella. Genetics. 2003;163:939–953. doi: 10.1093/genetics/163.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JL, Xie XF, Rull J, Velez S, Forbes A, Leung B, Dambroski H, Filchak KE, Aluja M. Mayr, Dobzhansky, and Bush and the complexities of sympatric speciation in Rhagoletis. Proc Natl Acad Sci USA. 2005;102:6573–6580. doi: 10.1073/pnas.0502099102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilets S. Hybrid zones with Dobzhansky-type epistatic selection. Evolution. 1997;51:1027–1025. doi: 10.1111/j.1558-5646.1997.tb03949.x. [DOI] [PubMed] [Google Scholar]
- Geraldes A, Basset P, Gibson B, Smith KL, Harr B, Yu HT, Bulatova N, Ziv Y, Nachman MW. Inferring the history of speciation in house mice from autosomal, X-linked, Y-linked and mitochondrial genes. Mol Ecol. 2008a;17:5349–5363. doi: 10.1111/j.1365-294X.2008.04005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes A, Carneiro M, Delibes-Mateos M, Villafuerte R, Nachman MW, Ferrand N. Reduced introgression of the Y chromosome between subspecies of the European rabbit (Oryctolagus cuniculus) in the Iberian Peninsula. Mol Ecol. 2008b;17:4489–4499. doi: 10.1111/j.1365-294X.2008.03943.x. [DOI] [PubMed] [Google Scholar]
- Geraldes A, Ferrand N, Nachman MW. Contrasting patterns of introgression at X-linked loci across the hybrid zone between subspecies of the European rabbit (Oryctolagus cuniculus) Genetics. 2006;173:919–933. doi: 10.1534/genetics.105.054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good JM, Dean MD, Nachman MW. A complex genetic basis to X-linked hybrid male sterility between two species of house mice. Genetics. 2008;179:2213–2228. doi: 10.1534/genetics.107.085340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Grula JW, Taylor OR. Some characteristics of hybrids derived from the sulfur butterflies, Colias eurytheme and Colias philodice: phenotypic effects of the X-chromosome. Evolution. 1982;34:673–687. doi: 10.1111/j.1558-5646.1980.tb04006.x. [DOI] [PubMed] [Google Scholar]
- Guenet JL, Nagamine C, Simonchazottes D, Montagutelli X, Bonhomme F. Hst-3—an X-linked hybrid sterility gene. Genet Res. 1990;56:163–165. doi: 10.1017/s0016672300035254. [DOI] [PubMed] [Google Scholar]
- Hale DW, Washburn LL, Eicher EM. Meiotic abnormalities in hybrid mice of the C57Bl/6J X Mus-Spretus cross suggest a cytogenetic basis for Haldane rule of hybrid sterility. Cytogenet Cell Genet. 1993;63:221–234. doi: 10.1159/000133539. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user friendly biological sequence alignment editor and analyses program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Harrison RG. Hybrid zones: windows on evolutionary process. Oxf Surv Evol Biol. 1990;7:69–128. [Google Scholar]
- Helena Mangs A, Morris BJ. The human pseudoautosomal region (PAR): origin, function and future. Curr Genomics. 2007;8:129–136. doi: 10.2174/138920207780368141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- Hey J, Nielsen R. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics. 2004;167:747–760. doi: 10.1534/genetics.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Nielsen R. Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. Proc Natl Acad Sci USA. 2007;104:2785–2790. doi: 10.1073/pnas.0611164104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Wakeley J. A coalescent estimator of the population recombination rate. Genetics. 1997;145:833–846. doi: 10.1093/genetics/145.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Two-locus sampling distributions and their application. Genetics. 2001;159:1805–1817. doi: 10.1093/genetics/159.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane NC, King MG, Barker MS, Raduski A, Karrenberg S, Yatabe Y, Knapp SJ, Rieseberg LH. Comparative genomic and population genetic analyses indicate highly porous genomes and high levels of gene flow between divergent helianthus species. Evolution. 2009;63:2061–2075. doi: 10.1111/j.1558-5646.2009.00703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathinal RJ, Stevison LS, Noor MA. The genomics of speciation in Drosophila: diversity, divergence, and introgression estimated using low-coverage genome sequencing. PLoS Genet. 2009;5:e1000550. doi: 10.1371/journal.pgen.1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CA, Haselkorn TS, Noor MA. Evaluation of the genomic extent of effects of fixed inversion differences on intraspecific variation and interspecific gene flow in Drosophila pseudoobscura and D. persimilis. Genetics. 2007;175:1289–1306. doi: 10.1534/genetics.106.064758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CA, Kliman RM, Markert JA, Hey J. Inferring the history of speciation from multilocus DNA sequence data: the case of Drosophila pseudoobscura and close relatives. Mol Biol Evol. 2002;19:472–488. doi: 10.1093/oxfordjournals.molbev.a004103. [DOI] [PubMed] [Google Scholar]
- Macholan M, Munclinger P, Sugerkova M, Dufkova P, Bimova B, Bozikova E, Zima J, Pialek J. Genetic analysis of autosomal and X-linked markers across a mouse hybrid zone. Evolution. 2007;61:746–771. doi: 10.1111/j.1558-5646.2007.00065.x. [DOI] [PubMed] [Google Scholar]
- Maroja LS, Andres JA, Harrison RG. Genealogical discordance and patterns of introgression and selection across a cricket hybrid zone. Evolution. 2009;63:2999–3015. doi: 10.1111/j.1558-5646.2009.00767.x. [DOI] [PubMed] [Google Scholar]
- Masly JP, Presgraves DC. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biol. 2007;5:e243. doi: 10.1371/journal.pbio.0050243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Hirobe T, Chapman VM. Genetic-basis of x-y chromosome dissociation and male-sterility in interspecific hybrids. Proc Natl Acad Sci USA. 1991;88:4850–4854. doi: 10.1073/pnas.88.11.4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthee CA, van Vuuren BJ, Bell D, Robinson TJ. A molecular supermatrix of the rabbits and hares (Leporidae) allows for the identification of five intercontinental exchanges during the Miocene. Syst Biol. 2004;53:433–447. doi: 10.1080/10635150490445715. [DOI] [PubMed] [Google Scholar]
- McVean G, Awadalla P, Fearnhead P. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics. 2002;160:1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J. A mouse speciation gene encodes a meiotic histone h3 methyltransferase. Science. 2009;323:373–375. doi: 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- Minder AM, Widmer A. A population genomic analysis of species boundaries: neutral processes, adaptive divergence and introgression between two hybridizing plant species. Mol Ecol. 2008;17:1552–1563. doi: 10.1111/j.1365-294X.2008.03709.x. [DOI] [PubMed] [Google Scholar]
- Moehring AJ, Llopart A, Elwyn S, Coyne JA, Mackay TF. The genetic basis of postzygotic reproductive isolation between drosophila santomea and D. yakuba due to hybrid male sterility. Genetics. 2006;173:225–233. doi: 10.1534/genetics.105.052985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munclinger P, Bozikova E, Sugerkova M, Pialek J, Macholan M. Genetic variation in house mice (Mus, muridae, rodentia) from the Czech and Slovak republics. Folia Zoologica. 2002;51:81–92. [Google Scholar]
- Navarro A, Barton NH. Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evol Int J Org Evol. 2003;57:447–459. doi: 10.1111/j.0014-3820.2003.tb01537.x. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. Columbia Univ. Press; New York, NY: 1987. [Google Scholar]
- Nickerson DA, V, Tobe O, Taylor SL. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Wakeley J. Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics. 2001;158:885–896. doi: 10.1093/genetics/158.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte AW, Gompert Z, Buerkle CA. Variable patterns of introgression in two sculpin hybrid zones suggest that genomic isolation differs among populations. Mol Ecol. 2009;18:2615–2627. doi: 10.1111/j.1365-294X.2009.04208.x. [DOI] [PubMed] [Google Scholar]
- Noor MA, Bennett SM. Islands of speciation or mirages in the desert? Examining the role of restricted recombination in maintaining species. Heredity. 2009;103:439–44. doi: 10.1038/hdy.2009.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MA, Grams KL, Bertucci LA, Reiland J. Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci USA. 2001;98:12084–12088. doi: 10.1073/pnas.221274498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, Ortiz-Barrientos D. Divergent selection and heterogeneous genomic divergence. Mol Ecol. 2009;18:375–402. doi: 10.1111/j.1365-294X.2008.03946.x. [DOI] [PubMed] [Google Scholar]
- Orr HA. The population-genetics of speciation—the evolution of hybrid incompatibilities. Genetics. 1995;139:1805–1813. doi: 10.1093/genetics/139.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA, Coyne JA. The genetics of postzygotic isolation in the Drosophila virilis group. Genetics. 1989;121:527–537. doi: 10.1093/genetics/121.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur BA, Krenz JG, Nachman MW. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution. 2004;58:2064–2078. doi: 10.1111/j.0014-3820.2004.tb00490.x. [DOI] [PubMed] [Google Scholar]
- Pool JE, Nielsen R. Inference of historical changes in migration rate from the lengths of migrant tracts. Genetics. 2009;181:711–719. doi: 10.1534/genetics.108.098095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. Sex chromosomes and speciation in Drosophila. Trends Genet. 2008;24:336–343. doi: 10.1016/j.tig.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC, Balagopalan L, Abmayr SM, Orr HA. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature. 2003;423:715–719. doi: 10.1038/nature01679. [DOI] [PubMed] [Google Scholar]
- Putnam AS, Scriber JM, Andolfatto P. Discordant divergence times among Z-chromosome regions between two ecologically distinct swallowtail butterfly species. Evolution. 2007;61:912–927. doi: 10.1111/j.1558-5646.2007.00076.x. [DOI] [PubMed] [Google Scholar]
- Rappold GA. The pseudoautosomal regions of the human sex-chromosomes. Human Genet. 1993;92:315–324. doi: 10.1007/BF01247327. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH. Chromosomal rearrangements and speciation. Trends Ecol Evol. 2001;16:351–358. doi: 10.1016/s0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Whitton J, Gardner K. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics. 1999;152:713–727. doi: 10.1093/genetics/152.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Salcedo T, Geraldes A, Nachman MW. Nucleotide variation in wild and inbred mice. Genetics. 2007;177:2277–2291. doi: 10.1534/genetics.107.079988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. [Google Scholar]
- Sharples CM, Fa JE, Bell DJ. Geographic variation in size in the European rabbit Oryctolagus cuniculus (Lagomorpha: Leporidae) in western Europe and North Africa. Zool J Linn Soc. 1996;117:141–158. [Google Scholar]
- Sperling FAH, Spence JR. Structure of an asymmetric hybrid zone between 2 water strider species (hemiptera, gerridae, limnoporus) Evolution. 1991;45:1370–1383. doi: 10.1111/j.1558-5646.1991.tb02642.x. [DOI] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchova R, Gregorova S, Buckiova D, Kyselova V, Divina P, Forejt J. Genetic analysis of X-linked hybrid sterility in the house mouse. Mamm Genome. 2004;15:515–524. doi: 10.1007/s00335-004-2386-0. [DOI] [PubMed] [Google Scholar]
- Storchova R, Reif J, Nachman MW. Female heterogamety and speciation: reduced introgression of the Z chromosome between two species of nightingales. Evolution. 2010;64:456–471. doi: 10.1111/j.1558-5646.2009.00841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasburg JD, Rieseberg LH. How robust are “isolation with migration” analyses to violations of the IM model? A simulation study. Mol Biol Evol. 2010;27:297–310. doi: 10.1093/molbev/msp233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stump AD, Fitzpatrick MC, Lobo NF, Traore S, Sagnon N, Costantini C, Collins FH, Besansky NJ. Centromere-proximal differentiation and speciation in anopheles gambiae. Proc Natl Acad Sci USA. 2005;102:15930–15935. doi: 10.1073/pnas.0508161102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Zeng ZB, Li J, Hartl DL, Laurie CC. Genetic dissection of hybrid incompatibilities between drosophila simulans and D. mauritiana. II. Mapping hybrid male sterility loci on the third chromosome. Genetics. 2003;164:1399–1418. doi: 10.1093/genetics/164.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter KC, Payseur BA, Harris LW, Bakewell MA, Thibodeau LM, O’Brien JE, Krenz JG, Sans-Fuentes MA, Nachman MW, Tucker PK. Genome-wide patterns of gene flow across a house mouse hybrid zone. Genome Res. 2008;18:67–76. doi: 10.1101/gr.6757907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CT, Tsaur SC, Wu ML, Wu CI. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science. 1998;282:1501–1504. doi: 10.1126/science.282.5393.1501. [DOI] [PubMed] [Google Scholar]
- True JR, Weir BS, Laurie CC. A genome-wide survey of hybrid incompatibility factors by the introgression of marked segments of Drosophila mauritiana chromosomes into Drosophila simulans. Genetics. 1996;142:819–837. doi: 10.1093/genetics/142.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker PK, Sage RD, Warner J, Wilson AC, Eicher EM. Abrupt cline for sex-chromosomes in a hybrid zone between 2 species of mice. Evolution. 1992;46:1146–1163. doi: 10.1111/j.1558-5646.1992.tb00625.x. [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 2005;3:e285. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via S. Natural selection in action during speciation. Proc Natl Acad Sci USA. 2009;106:9939–9946. doi: 10.1073/pnas.0901397106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villafuerte R. Oryctolagus cuniculus. In: Palomo LJ, Gisbert P, editors. Atlas de los Mamíferos terrestres de España. Dirección General de Conservación de la Naturaleza-SECEM-SECEMU; Madrid, Spain: 2002. pp. 464–466 . [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- White BJ, Cheng C, Simard F, Costantini C, Besansky NJ. Genetic association of physically unlinked islands of genomic divergence in incipient species of anopheles gambiae. Mol Ecol. 2010;19:925–939. doi: 10.1111/j.1365-294X.2010.04531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woerner AE, Cox MP, Hammer MF. Recombination-filtered genomic datasets by information maximization. Bioinformatics. 2007;23:1851–1853. doi: 10.1093/bioinformatics/btm253. [DOI] [PubMed] [Google Scholar]
- Wu CI. The genic view of the process of speciation. J Evol Biol. 2001;14:851–865. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Polymorphisms for the 27 X-linked and 17 autosomal loci: (1) SHOX, (2) GLRA2, (3) PDHA1, (4) POLA1, (5) IL1RAPL1, (6) TMEM47, (7) MAOA, (8) TIMP1, (9) DGKK, (10) ARHGEF9, (11) OPHN1, (12) OGT, (13) NRK, (14) AMOT, (15) KLHL13, (16) PGK1, (17) CYLC1, (18) KLHL4, (19) PABPC5, (20) TNMD, (21) DIAPH2, (22) GRIA3, (23) OCRL, (24) GPC4, (25) F9, (26) FMR1, (27) G6PD, (28) EXT1, (29) CYTC, (30) LUM, (31) UD14, (32) ATP12A, (33) PRL, (34) T, (35) MGST3, (36) SLC4A7, (37) STAG1, (38) GK5), (39) SIAH2, (40) KPNA4, (41) MYNN, (42) NAALADL2, (43) GBE1, and (44) TIAM1.
Figure S2. Marginal posterior probability densities for the mean time of all inferred migration events per locus (A) from O. c. cuniculus to O. c. algirus and (B) from O. c. algirus to O. c. cuniculus.
Table S1. PCR primers and annealing temperatures.
Table S2. Analyses of polymorphism, frequency spectrum tests of neutrality, recombination and divergence in O. c. algirus (alg.) and O. c. cuniculus (cun.) for 27 X-linked loci.
Table S3. Analyses of polymorphism, frequency spectrum tests of neutrality, recombination and divergence in O. c. algirus (alg.) and O. c. cuniculus (cun.) for 17 autosomal loci.
Table S4. Tests of nested models with unidirectional gene flow, without gene flow and asymmetrical gene flow against the full isolation-with-migration model.