Abstract
Experimental evolution is relevant to ecology because it can connect physiology, and in particular metabolism, to questions in ecology. The investigation of the linkage between the environment and the evolution of metabolism is tractable because these experiments manipulate a very simple environment to produce predictable evolutionary outcomes. In doing so, microbial selection experiments can examine the causal elements of natural selection: how specific traits in varying environments will yield different fitnesses. Here, we review the methodology of microbial evolution experiments and address three issues that are relevant to ecologists: genotype-by-environment interactions, ecological diversification due to specialization, and negative frequency-dependent selection. First, we expect that genotype-by-environment interactions will be ubiquitous in biological systems. Second, while antagonistic pleiotropy is implicated in some cases of ecological specialization, other mechanisms also seem to be at work. Third, while negative frequency-dependent selection can maintain ecological diversity in laboratory systems, a mechanistic (biochemical) analysis of these systems suggests that negative frequency dependence may only apply within a narrow range of environments if resources are substitutable. Finally, we conclude that microbial experimental evolution needs to avail itself of molecular techniques that could enable a mechanistic understanding of ecological diversification in these simple systems.
Keywords: bacteria, ecological diversity, experimental evolution, genotype by environment, negative frequency-dependent selection, trade-offs
Introduction
A fundamental constraint of ecological patterns is organismal physiology, of which cellular metabolism is a key component. Many ecological and evolutionary phenomena are a direct consequence of variation in metabolism, including plant physiology, marine algal herbivory, the distribution and population genetics of marine invertebrates, flower phenology, the existence of geographic clines, the evolution of pathogenesis, local adaptation to abiotic stress, and range limits (Koehn et al. 1976, Place and Powers 1979, Miller and Hay 1996, Mitton 1997, Rausher et al. 1999, Day et al. 2001, Schmidt and Rand 2001, Canterbury 2002). Because changes in metabolism can alter the ecological patterns of a given species, one would like to know how the environment, a causal force of natural selection, can select changes in the metabolism of an organism. The relevance of changes in metabolism is less clear when higher levels of biological organization such as behavior, development, and structure, are the primary targets of selection.
Microbial selection experiments can investigate the linkage between the environment and the evolution of metabolism. In these experiments, a very simple environment can be manipulated to produce predictable evolutionary outcomes. Unlike other systems, selection experiments in microbes can incorporate both short-term ecological dynamics (changes in the abundance of genotypes) and longer-term evolutionary change (the rise of new genotypes by mutation) due to the short generation times and large population sizes of bacteria. Short-term experiments can examine how environmental complexity maintains diversity; in other words, what is the effect of defined variants in a specific environment? These experiments compete two variants over a short enough period of time so that no new mutations sweep through the populations (e.g., Dykhuizen and Davies 1980). Long-term experiments typically begin with no genetic variation; evolution comes from selection on de novo variation (e.g., Lenski et al. 1991). In natural populations, evolutionary change can happen on similar time scales as ecological dynamics and affect ecological phenomena (Endler 1986, Grant 1986). To ignore this change is to assume that species are static and do not evolve in response to the environment (Bohannan and Lenski 2000).
Most selection experiments can be divided into two categories: evolution-of-fitness experiments and evolution-of-trait experiments. Evolution-of-fitness experiments ask how fitness will change relative to an ancestor under a specific selective regime, without reference to traits or genetic differences (Travisano et al. 1995a). These experiments have yielded insights into the trajectory of fitness increases, as well as fitness tradeoffs (Travisano et al. 1995b, Travisano and Lenski 1996). One drawback of this approach is that it is difficult to determine the role that particular environmental parameters play in selecting various genetic changes, since the underlying genetic and physiological changes are not predicted a priori.
Evolution-of-trait experiments address the causes of natural selection: genetic variation and its relationship to the environment. These experiments examine which traits are selected in specific environments, and are highly amenable to hypothesis testing. Many experiments examining the evolution of traits studied selection on metabolic traits, owing to our understanding of the genetics of metabolism and quantitative theories of the interactions of metabolic enzymes (e.g., Dykhuizen and Dean 1994). These experiments can either be short term (Dykhuizen 1978, Dykhuizen and Davies 1980, Dykhuizen and Hartl 1980, Hartl and Dykhuizen 1985) or long term (Helling et al. 1987). Short-term evolution-of-trait experiments require assumptions about the evolutionary process and do not necessarily test what novel innovations might arise if the bacteria were allowed to evolve in that environment. Recent technological advances such as microarrays should allow one to determine which loci may change over time in a particular environment (Cho and Tiedje 2002), so more realistic experiments can be designed.
Here, we will review the methodology of evolution experiments using bacteria, and then address three issues on which a physiological approach to microbial experimental evolution can shed light: genotype by environment interactions, the evolution of ecological specialization, and negative frequency-dependent selection.
Methods
Batch culture
In batch culture, nongrowing cells are added to fresh growth medium. The growth medium is a chemically defined salt solution supplemented with a source of carbon and energy. After an initial lag period, where the cells are adapting physiologically to the new conditions, the cells begin to grow exponentially at the maximum growth rate until a resource becomes limiting. Once resources become limited, the growth rate slows until growth stops and the cells enter stationary phase. The cycle is then repeated by transferring a portion of the cells to fresh medium. This changing environment potentially selects for a decreased lag period, a higher maximum growth rate, higher growth when nutrients are limited, and survival during stationary phase; however, when the environment is replenished regularly, selection for higher maximum growth rate dominates (Vasi et al. 1994). Batch culture experiments are seasonal and r-selected (Dykhuizen 1990). The multiple physiological states that each cell experiences during every cycle of batch culture make this technique undesirable for examining cellular metabolism.
Chemostats
Chemostats are devices that enable microbial cultures to be maintained at a predetermined growth rate in a constant, homogeneous environment (Dykhuizen 1993). A peristaltic pump feeds fresh sterile medium into the growth chamber. An overflow siphon removes spent medium and cells to maintain a constant volume. Sterile air is forced into the bottom of the culture to maintain adequate oxygen and thorough mixing in the growth chamber. The growth medium is a chemically defined salt solution supplemented with a source of carbon and energy. Concentrations of the components of the fresh medium are such that only one is exhausted by the growing culture. The limiting nutrient is usually a sugar, such as glucose or lactose. When two carbon sources are used, the complexity of the environment has effectively increased. Resources in chemostats are limiting and constant, mutations that increase growth under limited resource availability are typically favored. The constancy of the resource also means that the bacteria will not enter multiple physiological states, which can confound an understanding of metabolic processes. Chemostat experiments are not seasonal and are K-selected (Dykhuizen 1990).
Genotype-by-Environment Interactions and Community Diversity
Genotype-by-environment (G × E) interactions have been typically thought of in terms of complex phenotypic characters, such as growth rate, seed set, or milk production. Generally, both the genetic differences, such as different breeds of animals or different ecotypes, and the environmental differences, such as different farms, soils, or elevations, are very complex. In experimental evolution, we can study single genetic changes in simple, well-defined environments, so that the basic properties of G × E interactions can be understood. However, the “character” used in these experiments is typically fitness. Unless the translation from phenotype to fitness is a monotonic function, extrapolation back to phenotype is impossible. This may not be as much of a problem as it first seems, since in the one case which has been examined, fitness was linearly proportional to metabolic flux (Dykhuizen and Dean 1990). This system involves the competition of various alleles of the lactose operon in sugar limited chemostats and was used by both Silva (1992) and Dean (1995) to investigate the molecular basis of G × E interactions.
Dean (1995), using data from Silva (1992), compared the fitnesses of seven different lactose operons when grown on five different β-galactoside sugars and found extensive G × E interactions. Typically, the interaction effect was as large or larger than the main genetic effect. To understand how this G × E effect arises, we need to attend to the details of the system. The lactose operon contains two genes important in the metabolism of lactose and the other four β-galactosides: (1) the lactose permease which brings the β-galactosides across the membrane and (2) the β-galactosidase which cleaves the β linkage to produce galactose and another compound. If different substrates change the rates of steps in a pathway, G × E interaction can be generated. G × E interaction can also be generated from the change of the kinetic properties of single enzymes. Some G × E interaction was due to variation in the lactose permease, but none in the β-galactosidase. Most of the G × E interaction is caused by changes in the distribution of the control of flow of metabolites through the steps of the pathway; i.e., the different β-galactosides tend to bottleneck at different steps in the pathway. Thus, G × E interaction is an emergent property of the metabolic system. G × E interaction could arise either because this is simply the nature of enzymes and pathways or because different lactose operons are adapted to different β-galactosides. We can rule out adaptation as, except for lactose, these β-galactosides are creations of chemists, not seen in nature (Dean 1995). Consequently, the G × E interaction cannot have evolved and must be an inherent part of this metabolic system. We postulate that G × E interactions frequently arise as a consequence of the properties of metabolic pathways, and that G × E interactions are expected for any set of metabolic genotypes in any set of environments. Further work needs to be done to test this statement.
G × E interactions are not only created by metabolism: evolution can increase G × E interactions if selection in one environment does not increase fitness evenly across a range of environments. In particular, if selection increases fitness in one environment while leaving it unchanged or decreasing it in other environments, G × E interactions (and potentially specialization) can increase. Bennett and Lenski (1993) tested this experimentally starting from a progenitor Escherichia coli that had been selected for 2000 generations in batch culture at 37°C. They then grew six replicate lines in each of three thermal environments (32°, 37°, and 42°C) for 2000 generations. For the lines evolved at 32°C, there was a significant improvement in fitness at 28°C and 32°C, but no significant improvement at any other temperature tested. One out of the six lines seems to have acquired a temperature sensitive mutation that decreased its fitness above 40°C. For the strains evolved at 42°C, there was a significant increase in fitness at 40°C and above, but no significant change below 40°C. For the strains evolved at 37°C, there was a small but significant increase in fitness at 37°C, but no change at any other temperature tested. Selection at a particular temperature improved fitness for a small range of temperatures around the selected temperature. This selection did not result in fitness decreases at other temperatures, so there are no trade-offs, yet the selection generated G × E interactions by restricting the extent of the response on the temperature gradient. Adaptation to a local environment can be expected to generate more G × E interaction than is inherent in the physiology.
All genetic differences across all environmental differences need not generate G × E interactions, as some genotypes are well buffered to genetic and environmental change. Another interesting experiment suggests that well-buffered systems may be the result of natural selection (Remold and Lenski 2001). The progenitor of this experiment was isolated from a population that had evolved for 10 000 generations in batch culture at 37°C, growing in a solution of glucose and inorganic salts. Genes were randomly knocked out from this founder, so that 26 different mutant strains were created which differed from the ancestor by a single genetic difference. The fitness of each of these 26 knockout mutations was compared to the ancestor at 28°C and 37°C for growth on glucose and on maltose, giving a total of four treatments. There is a small G × E effect for temperature on maltose, none on glucose, but a major G × E effect for the comparison of fitness between glucose and maltose. For all 26 knockout lines, the fitness on glucose is not significantly different from the ancestor at either temperature, while the fitness on maltose is significantly different from the ancestor at both temperatures for almost all the mutations. Four mutations are advantageous on maltose and the rest are detrimental. Only about a half-dozen genes are involved in the uptake and cleavage of maltose to produce two glucose molecules. While it was not checked, it is unlikely that any, let alone most, of the 26 mutations tested knocked out one of these half-dozen genes. Yet these mutations affect fitness on maltose, but not on glucose, even though maltose metabolism quickly enters the same pathway as glucose metabolism. We do not yet know why glucose metabolism is highly canalized, and maltose metabolism is not. This canalization on glucose is not due to the 10 000 generations growing on glucose, since, when the knockout mutations were transferred into the strain used to start the culture that evolved for the 10 000 generations, the resulting strains still show the canalized behavior (Elena and Lenski 2001). Presumably, the canalized behavior evolved earlier in response to the centrality of glucose metabolism and is not an inherent property of the metabolic system. If the canalized behavior is an inherent property, it is expected that all bacterial species would show it irrespective of how important glucose is in their resource base.
Based on the studies reported in this section, we propose that G × E interactions initially arise as a consequence of the properties of metabolic pathways and that selection in a particular environment will increase the G × E effect beyond what is inherent in the metabolic system. Thus, G × E interactions are likely for any set of metabolic genotypes in any set of environments. However, selection can also decrease G × E effects by selecting for canalized genotypes. Specialization and niche differentiation will be favored by both the metabolic system which initially creates fitness differences across environments and selection by increasing fitness in favored environments. If this view is correct, then to the degree metabolism drives ecological interactions, evolutionary biologists and ecologists need to explain generalization and canalization, rather than specialization.
The Evolution of Ecological Specialization
The extensive G × E interactions in bacteria show that, at least with respect to metabolism, organisms are likely to have maximal fitness in only parts of their niche. If, in adapting to this portion of the niche, the species loses fitness in other portions of its niche, it will become specialized. This pattern of a single mutation increasing fitness in the current habitat while lowering it in some other habitat, known as antagonistic pleiotropy (Rose 1991, Holt 1996), has been suggested as important in the specialization of many organisms and shown to be important in some. For example, when Hawthorne and Via (2001) examined two host races of the pea aphid Acyrthosiphon pisum pisum, they found several regions in the genome which both increased fitness on the native host, and decreased fitness on the alternate host. Antagonistic pleiotropy has also been implicated in limiting the host range of pathogenic bacteria. Numerous species of bacteria can enter a pathogenic niche only when they lose functions that would presumably be advantageous in other niches (Sokurenko et al. 1999). While these patterns have shown that antagonistic pleiotropy can promote specialization, it is not the only mechanism that can do so.
As a species begins to specialize on a subset of its niche, it may loose fitness in the unused portion of its niche due to the fixation of mutations that are neutral or nearly neutral in the current environment but detrimental in an alternate environment. The process, known as mutation accumulation (Rose 1991, Holt 1996), can occur by genetic drift, or by hitchhiking with an advantageous mutation (Maynard Smith and Haigh 1974). Bacteria that are highly specialized to their pathogenic or symbiotic lifestyles frequently have lost many physiological abilities that would be necessary to live without their host, due to a lack of effective selection to maintain those abilities (Moran and Wernegreen 2000, Ochman and Moran 2001). Finally, fitness increases in the current environment without a concordant loss of fitness in other environments can still lead to specialization (Fry 1996). Imagine that two competing species A and B have equal fitness in environments 1 and 2. If species A increases in fitness in environment 1, and species B does likewise in environment 2, then each species will begin to exclude the other species from its specialized environment even though neither has lost fitness in either environment. Hawthorne and Via (2001) found some regions of the pea aphid genome that enhance fitness on the preferred host plant, but do not affect fitness on the alternate host.
Experimental evolution studies have three distinct advantages in examining the ecology and evolution of specialization. First, the brief generation times, large population sizes, ease of replication, and ability to compete derived and ancestral populations enables direct testing of patterns of fitness change associated with specialization. Second, a detailed understanding of the changes in characters (as opposed to fitness) which cause specialization has been facilitated by the tremendous knowledge of genetics, biochemistry, and physiology in some bacterial species. Our understanding of metabolism has been particularly useful. Finally, microorganisms provide unparalleled opportunities to use genetic manipulations to directly test hypotheses about the functional basis of specialization, although these types of experiments are still rare.
The general prevalence of specialization as a direct consequence rather than a by-product of local adaptation (i.e., antagonistic pleiotropy vs. mutation accumulation or adaptation to specific habitats) is still unclear. Selection experiments in bacteria have found that antagonistic pleiotropy, while not universal, is clearly at work in some case. Adams and coworkers (Helling et al. 1987, Rosenzweig et al. 1994, Treves et al. 1998) provide strong evidence for antagonistic pleiotropy in specialization because they characterize the phenotypic changes involved. They found that after over 700 generations growing in glucose-limited chemostats, a stable polymorphism developed in many of their E. coli populations. This was surprising, as there did not appear to be multiple resources to partition in the chemostat in order to allow stable coexistence. Four separate clones were isolated that coexisted in this simple environment, and three were analyzed extensively. One clone specialized on glucose, one on acetate, and one on glycerol.
The first clone was very efficient at importing glucose, but secreted by-products of glycolysis (glycerol and acetate) into the medium (Rosenzweig et al. 1994). Secreting glycerol and acetate can increase the rate of glucose uptake, so this clone is a strong candidate for antagonistic pleiotropy leading to specialization on glucose. In order to conclude that antagonistic pleiotropy, rather than mutation accumulation, is the basis for this specialization, we would need to identify candidate mutations involved in this phenotype, then place these single mutations into the ancestral genetic background and determine if a single mutation has both phenotypic effects. One of the other clones has improved its ability to utilize glycerol, and another increased its uptake efficiency on acetate. Both retained their ability to use glucose, so these organisms may be a case of specialization due to fitness increases in a specialized environment. They seem to be unable to compete for glucose due to the dominant clones’ increased ability to use this sugar, and so have specialized on acetate and glycerol. In the case of the acetate specialist, a candidate mutation was identified, and competition experiments showed that acetate specialists without this mutation were unable to survive (Treves et al. 1998). These experiments are utterly amazing (D. Dykhuizen, personal observation), in that a stable community evolved from a single clone in the simplest environment we can imagine.
This simple experimental system still has a great deal to tell us about the evolution of ecological specialization. Using their knowledge of biochemistry, the original authors speculated about mechanisms promoting coexistence, and confirmed the mechanism for the case of the acetate specialist. Apart from understanding the functional basis of this metabolic polymorphism, this system could be used to explore the temporal dynamics of specialization. In particular, did the glucose specialist evolve first with the other specialists descending from that clone in response to the changed environment? If so, why did the acetate and glycerol specialists have the ancestral ability to utilize glucose? Could the enhanced ability to use these metabolic byproducts have already evolved before the glucose specialist, preadapting these strains to their specialized niches? Or is it possible that they all arose simultaneously? Repeating these experiments again with dense sampling of the dynamics could provide answers to these questions.
The evolution of specialization has also been studied extensively by Cooper and coworkers (Cooper and Lenski 2000, Cooper et al. 2001a, b). They examined 12 strains of E. coli which had grown on glucose at 37°C for 20 000 generations. All 12 strains evolved to be completely unable to use ribose as a carbon source. Cooper et al. (2001b) studied this loss of function, and demonstrated that the same mutation which eliminates the ability to use ribose increases fitness on glucose, providing strong evidence for antagonistic pleiotropy in the loss of this part of the ancestral niche. However, ribose might be an unusual case since the operon is constitutively expressed in many strains of E. coli B; a genetic loss of function might be the only way to “regulate” expression (Dykhuizen and Davies 1980).
An understanding of the general causes of specialization in these lines is still elusive, however. Cooper and Lenski (2000) showed that decreasing growth rates on a variety of alternative sugars roughly coincided with increase in fitness on glucose. In the same lines, decreases in maximum growth rates at extreme temperatures (20°, 40°, 41°, and 42°C) occurred around the time of increase in maximum growth rate at 37°C (Cooper et al. 2001a). While this pattern of increase in fitness in the evolved environment coupled with the decrease in function in other environments could be the result of antagonistic pleiotropy, mutation accumulation could create the same pattern due to hitchhiking. Hitchhiking occurs when a mutation that is neutral or detrimental is swept to fixation due to its linkage with an advantageous mutation. The rate of fixation of slightly deleterious mutations increases under hitchhiking (Birky and Walsh 1988). If the loss of function on alternative resources or at alternative temperatures were slightly deleterious, we could see a pattern of fitness change indistinguishable from the pattern generated by antagonistic pleiotropy. Remold and Lenski’s (2001) experiment argues that many mutations will have no effect on glucose utilization (which is well canalized), but should effect growth rate on other carbon sources (which are not as likely to be well canalized). The time to fixation of neutral mutations will decrease with hitchhiking, so it is possible that mutations that were neutral on glucose (but not neutral on other carbon sources) were fixed rapidly when the population underwent rapid adaptation to the glucose environment in the first 2000 generations. As the rate of evolution slowed, so did the time to fixation of neutral mutations. This lengthening of the time to fixation would result in a temporarily slower rate of fixation, in accord with the observed decrease in the rate of loss of function during later parts of the experiment. The relative importance of antagonistic pleiotropy versus mutation accumulation in specialization will be determined in part by the difference in canalization between phenotypes exposed to selection in the current niche, and those unused there.
Our knowledge of E. coli’s metabolism might lead to a more nuanced understanding of Cooper and coworkers’ experiments. For example, there were some carbon sources on which all lines initially decreased their growth, only to have some lines at least return to the level of the ancestor. Understanding why growth is decreased for the entire experiment on some resources, while rebounding in some lines on other resources will provide insights into the basis for long-term specialization. We also await a better understanding of the prevalence of different mechanisms of specialization. Antagonistic pleiotropy has been demonstrated by Cooper et al. (2001b) in the loss of ribose metabolism as well by Rosenzweig et al. (1994) in acetate and glycerol secretion. Other mechanisms have also been observed, such as the adaptation to a specific environment in the acetate specialist studied by Treves et al. (1998). Further tests, incorporating functional analyses in conjunction with patterns of fitness change will help us understand what the most prevalent mechanisms are. We will also be able to understand how those changes confer specific fitness consequences.
Negative Frequency Dependence
One frequently observed phenomenon in microbial experimental evolution is negative frequency dependence, wherein genotypes are favored when rare and selected against when common. These types of interactions have also been implicated in maintaining species diversity in ecological communities (Molofsky et al. 2002). In several different experimental systems, diversity has been maintained by negative frequency dependence (Paquin and Adams 1983, Rosenzweig et al. 1994, Turner et al. 1996, Elena and Lenski 1997, Rozen and Lenski 2000). Two mechanisms can explain coexistence: facilitation, where one type secretes a metabolic intermediate into the environment that can be used by another type (e.g., Rosenzweig et al. 1994, Turner et al. 1996) and demographic tradeoffs, where one type has a growth or survival advantage in a changing environment (e.g., high vs. low glucose concentrations; Turner et al. 1996). Recently, a third mechanism involving antagonism either due to a toxic secreted metabolite or elimination of a metabolite required for survival has been observed (Rozen and Lenski 2000).
The repeated observation of negative frequency-dependent interactions would suggest that such interactions are ubiquitous in maintaining diversity. A mechanistic model of negative frequency dependence in chemostats with two limiting sugars suggests that frequency dependence should only occur under a narrow range of environmental parameters when resources are interchangeable (Lunzer et al. 2002; Fig. 1). In this model, the two genotypes each prefer a different sugar, although each genotype can use the alternative carbon source. While coexistence is possible, it can only occur under a very limited subset of sugar concentrations. In more variable environments, the range of parameters that can maintain phenotypic variation might be exceeded (Lunzer et al. 2002). This suggests that demographic trade-offs may not maintain ecological diversity in many natural environments. The stability of cross-feeding under conditions of high environmental variability has yet to be empirically tested. An experiment that could address this would be to determine whether negative frequency dependence can evolve in a variable environment. In most microbial experimental evolution studies, the environment is a source of variation to be eliminated. By focusing on a mechanistic basis for coexistence, we are forced to confront the role of environmental heterogeneity in evolutionary processes.
Fig. 1.
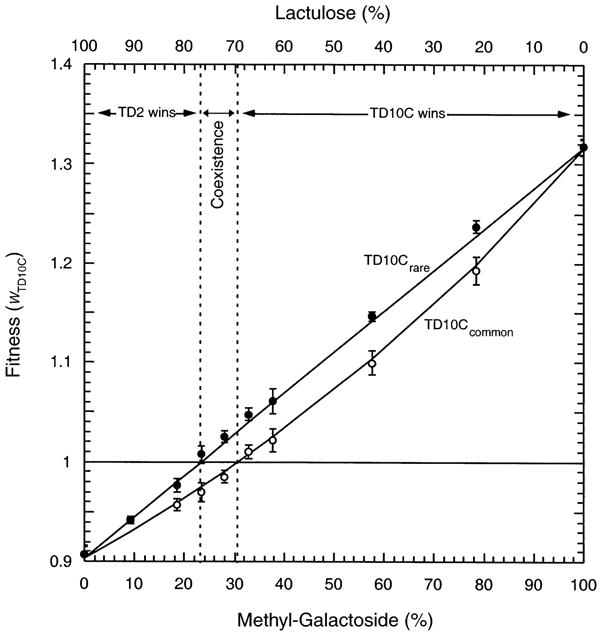
The observed fitness relations between strains TD10C and TD2 vary across the methyl-galactoside/lactulose resource axis and are also dependent on strain frequency. Strain TD2 is a derivative of the common laboratory strain of E. coli, and TD10C is isogenic to TD2 except for a region around the lactose operon. The operon was transduced in from a wild strain, is constitutive, and contains a permease which is 2.16 times more active that that in TD2 (Dykhuizen and Dean 1994). Fitnesses of TD10C were estimated using initial frequencies of 0–20% (rare; filled circles) or 80–100% (common; open circles) on various proportions of the sugars lactulose and methyl-galactoside. The total concentration of sugar entering the chemostat is held constant, and the percentage of methyl-galactoside refers to the percentage of the total sugar that is methyl-galactoside. Between 23% and 30.5% methyl-galactoside, the fitness of TD10C is > 1 when rare, and <1 when common. Here, TD10C and TD2 can coexist, maintained by frequency-dependent selection in a balanced polymorphism. The error bars designate 95% confidence intervals. When TD10C is common, fitness is not linear because the high frequency of TD10C alters the concentration of resource. This figure is redrawn from Fig. 9 of Lunzer et al. (2002).
Potential Problems with Selection Experiments
Is the artificial simplified nature of microbial experiments so unrealistic as to be misleading? We have suggested above that this might be true for the frequency dependence experiments. In this section, we discuss how some of these apparent weaknesses are either hidden strengths or opportunities to incorporate further environmental complexity. Most of the work presented here has taken great pains to eliminate environmental heterogeneity and phenotypic plasticity. While unrealistic, the advantage of simple systems is that the causal mechanisms of selection are more easily discovered. We think that a future productive avenue of research will be to add additional complexity, particularly temporal. Does the time scale on which the environment fluctuates affect niche diversification, and will those traits that evolve differ with changes in the periodicity of the environment?
A second complaint is that microbes are not relevant to most ecologists. Not only are they not multicellular, but they are asexual haploids. Setting aside the observation that the bulk of the planet’s biomass and biochemical diversity is microbial (Whitman et al. 1998), asexual haploid organisms can serve as excellent proxies for interspecies interactions. Since bacteria are haploid and easy to manipulate genetically, we can address the mechanistic bases of adaptation and diversification. For example, determining the role of pleiotropy in adaptive change is relatively easy compared to problems when using sexual diploids (e.g., Notley-McRobb et al. 2002). A third issue is the relative simplicity of the organism and system. Much of the evolution discussed in this paper results from one or few genetic changes. It is unclear to what extent these few changes are analogous to speciation in sexual diploids, although ecological divergence can happen rapidly and suddenly in multicellular eukaryotes (Thompson 1998, Hendry and Kinnison 1999).
Conclusions
While the concerns of experimental evolutionists may seem distant from the concerns of most ecologists, we believe that both groups are, in the end, interested in similar questions. The use of single-celled asexual prokaryotes growing in a constant, very simple, laboratory environment may be unappealing to some ecologists. However, these simplifications have enabled researchers to piece together a deep understanding of several issues of ecological importance. Experiments in these simple environments have shown that there may be simple rules for G × E interactions arising from metabolism. Likewise, these simple environments have provided us with the ability to understand the mechanistic basis of ecological specialization. On the other hand, metabolic analysis has shown that while negative frequency dependence is common in constant laboratory environments, it is unlikely to maintain diversity in variable natural environments. If microbial experimental evolution hopes to explain ecological diversity outside of the lab, it must investigate under what conditions laboratory environments provide insight into the natural world, and when they are misleading.
A variety of new molecular genetic techniques will broaden the field in several ways. First, a number of technologies will allow post-hoc exploration of evolved organisms to understand how traits have changed. For example, microarray technologies allow investigators to simultaneously monitor the expression of thousands of genes. Similarly, new techniques to locate point mutations in bacterial genomes (Sokurenko et al. 2001) will allow investigators to pinpoint the genetic basis of adaptation. The use of genetically altered microorganisms to test hypotheses about the evolution of traits will create a more convincing understanding of how traits have evolved. While we suspect that these techniques will never be employed by most ecologists, we believe that their use in understanding organisms will be appreciated by all.
Acknowledgments
This is contribution number 1101 in Ecology and Evolution, State University of New York at Stony Brook. The authors received partial funding for this work from NIH grants GM6073102 and GM6380001. D. Brisson and D. M. Stoebel received funding from NSF Graduate Research Fellowships. We thank J. M. Hoch, G. Hynes, S.-R. Liou, T. L. Morelli, and two anonymous reviewers for helpful comments on this document.
Literature Cited
- Bennett AF, Lenski RE. Evolutionary adaptation to temperature. II. Thermal niches of experimental lines of Escherichia coli. Evolution. 1993;47:1–12. doi: 10.1111/j.1558-5646.1993.tb01194.x. [DOI] [PubMed] [Google Scholar]
- Birky CW, Jr, Walsh JB. Effects of linkage on rates of molecular evolution. Proceedings of the National Academy of Sciences (USA) 1988;85:6414–6418. doi: 10.1073/pnas.85.17.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohannan BJM, Lenski RE. Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecology Letters. 2000;3:362–377. [Google Scholar]
- Canterbury G. Metabolic adaptation and climatic constraints on winter bird distribution. Ecology. 2002;83:946–957. [Google Scholar]
- Cho JC, Tiedje JM. Quantitative detection of microbial genes by using DNA microarrays. Applied and Environmental Microbiology. 2002;68:1425–1430. doi: 10.1128/AEM.68.3.1425-1430.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper VS, Bennett AF, Lenski RE. Evolution of thermal dependence of growth rate of Escherichia coli populations during 20 000 generations in a constant environment. Evolution. 2001a;55:889–896. doi: 10.1554/0014-3820(2001)055[0889:eotdog]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Cooper VS, Lenski RE. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature. 2000;407:736–739. doi: 10.1038/35037572. [DOI] [PubMed] [Google Scholar]
- Cooper VS, Schneider D, Blot M, Lenski RE. Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B. Journal of Bacteriology. 2001b;183:2834–2841. doi: 10.1128/JB.183.9.2834-2841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day WA, Fernandez RE, Maurelli AT. Pathoadaptive mutations that enhance virulence: genetic organization of the cadA regions of Shigella spp. Infection and Immunity. 2001;69:7471–7480. doi: 10.1128/IAI.69.12.7471-7480.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean AM. A molecular investigation of genotype by environment interactions. Genetics. 1995;139:19–35. doi: 10.1093/genetics/139.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen DE. Selection for tryptophan auxotrophs of Escherichia coli in glucose-limited chemostats as a test of the energy conservation hypothesis of evolution. Evolution. 1978;32:125–150. doi: 10.1111/j.1558-5646.1978.tb01103.x. [DOI] [PubMed] [Google Scholar]
- Dykhuizen DE. Experimental studies of natural selection in bacteria. Annual Review of Ecology and Systematics. 1990;21:373–398. [Google Scholar]
- Dykhuizen DE. Chemostats used for studying natural selection and adaptive evolution. Methods in Enzymology. 1993;224:613–631. doi: 10.1016/0076-6879(93)24046-w. [DOI] [PubMed] [Google Scholar]
- Dykhuizen DE, Davies M. An experimental model: bacterial specialists and generalists competing in chemostats. Ecology. 1980;61:1213–1227. [Google Scholar]
- Dykhuizen DE, Dean AM. Enzyme activity and fitness: evolution in solution. Trends in Ecology and Evolution. 1990;5:257–262. doi: 10.1016/0169-5347(90)90067-N. [DOI] [PubMed] [Google Scholar]
- Dykhuizen DE, Dean AM. Predicted fitness changes along an environmental gradient. Evolutionary Ecology. 1994;8:524–541. [Google Scholar]
- Dykhuizen DE, Hartl DL. Selective neutrality of 6PGD allozymes in E. coli and the effects of genetic background. Genetics. 1980;96:801–817. doi: 10.1093/genetics/96.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena SF, Lenski RE. Test of synergistic interactions among deleterious mutations in bacteria. Nature. 1997;390:395–398. doi: 10.1038/37108. [DOI] [PubMed] [Google Scholar]
- Elena SF, Lenski RE. Epistasis between new mutations and genetic background and a test of genetic canalization. Evolution. 2001;55:1746–1752. doi: 10.1111/j.0014-3820.2001.tb00824.x. [DOI] [PubMed] [Google Scholar]
- Endler JA. Natural selection in the wild. Princeton University Press; Princeton, New Jersey, USA: 1986. [Google Scholar]
- Fry JD. The evolution of host specialization: are trade-offs overrated? American Naturalist. 1996;148:S84–S107. [Google Scholar]
- Grant PR. Ecology and evolution of Darwin’s Finches. Princeton University Press; Princeton, New Jersey, USA: 1986. [Google Scholar]
- Hartl DL, Dykhuizen DE. The neutral theory and the molecular basis of preadaptation. In: Ohta T, Aoki K, editors. Population genetics and molecular evolution. Springer-Verlag; Berlin, Germany: 1985. pp. 107–124 . [Google Scholar]
- Hawthorne DJ, Via S. Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature. 2001;412:904–907. doi: 10.1038/35091062. [DOI] [PubMed] [Google Scholar]
- Helling RB, Vargas CN, Adams J. Evolution of Escherichia coli during growth in a constant environment. Genetics. 1987;116:349–358. doi: 10.1093/genetics/116.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry AP, Kinnison MT. The pace of modern life: measuring rates of contemporary microevolution. Evolution. 1999;53:1637–1653. doi: 10.1111/j.1558-5646.1999.tb04550.x. [DOI] [PubMed] [Google Scholar]
- Holt RD. Demographic constraints in evolution: towards unifying the evolutionary theories of senescence and niche conservatism. Evolutionary Ecology. 1996;10:1–11. [Google Scholar]
- Koehn RK, Milkman R, Mitton JB. Population genetics of marine pelecypods. 4. Selection, migration and genetic differentiation in the blue mussel Mytilus edulis. Evolution. 1976;30:2–32. doi: 10.1111/j.1558-5646.1976.tb00878.x. [DOI] [PubMed] [Google Scholar]
- Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2000 generations. American Naturalist. 1991;138:1315–1341. [Google Scholar]
- Lunzer M, Natarajan A, Dykhuizen DE, Dean AM. Enzyme kinetics, substitutable resources and competition: from biochemistry to frequency-dependent selection in lac. Genetics. 2002;162:485–499. doi: 10.1093/genetics/162.1.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J, Haigh J. The hitch-hiking effect of a favourable gene. Genetical Research. 1974;23:23–35. [PubMed] [Google Scholar]
- Miller MW, Hay ME. Coral–seaweed–grazer–nutrient interactions on temperate reefs. Ecological Monographs. 1996;66:323–344. [Google Scholar]
- Mitton JB. Selection in natural populations. Oxford University Press; New York, New York, USA: 1997. [Google Scholar]
- Molofsky J, Bever JD, Antonovics J, Newman TJ. Negative frequency dependence and the importance of spatial scale. Ecology. 2002;83:21–27. [Google Scholar]
- Moran NA, Wernegreen JJ. Lifestyle evolution in symbiotic bacteria: insights from genomics. Trends in Ecology and Evolution. 2000;15:321–326. doi: 10.1016/s0169-5347(00)01902-9. [DOI] [PubMed] [Google Scholar]
- Notley-McRobb L, King T, Ferenci T. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. Journal of Bacteriology. 2002;184:806–811. doi: 10.1128/JB.184.3.806-811.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Moran NA. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science. 2001;292:1096–1098. doi: 10.1126/science.1058543. [DOI] [PubMed] [Google Scholar]
- Paquin C, Adams J. Frequency of fixation of adaptive mutations is higher in evolving diploid than haploid yeast populations. Nature. 1983;302:495–500. doi: 10.1038/302495a0. [DOI] [PubMed] [Google Scholar]
- Place AR, Powers DA. Genetic variation and relative catalytic efficiencies: lactate dehydrogenase B allozymes of Fundulus heteroclitus. Proceedings of the National Academy of Sciences (USA) 1979;76:2354–2358. doi: 10.1073/pnas.76.5.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausher MD, Miller RE, Tiffin P. Patterns of evolutionary rate variation among genes of the anthocyanin biosynthetic pathway. Molecular Biology and Evolution. 1999;16:266–274. doi: 10.1093/oxfordjournals.molbev.a026108. [DOI] [PubMed] [Google Scholar]
- Remold SK, Lenski RE. Contribution of individual random mutations to genotype-by-environment interactions in Escherichia coli. Proceedings of the National Academy of Sciences (USA) 2001;98:11 388–11 393. doi: 10.1073/pnas.201140198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MR. Evolutionary biology of aging. Oxford University Press; New York, New York, USA: 1991. [Google Scholar]
- Rosenzweig RF, Sharp RR, Treves DS, Adams J. Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli. Genetics. 1994;137:903–917. doi: 10.1093/genetics/137.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen DE, Lenski RE. Long-term experimental evolution in Escherichia coli. VIII. Dynamics of a balanced polymorphism. American Naturalist. 2000;155:24–35. doi: 10.1086/303299. [DOI] [PubMed] [Google Scholar]
- Schmidt PS, Rand DM. Adaptive maintenance of genetic polymorphism in an intertidal barnacle: habitat-and life-stage-specific survivorship of Mpi genotypes. Evolution. 2001;55:1336–1344. doi: 10.1111/j.0014-3820.2001.tb00656.x. [DOI] [PubMed] [Google Scholar]
- Silva PJN. Thesis. State University of New York; Stony Brook, Stony Brook, New York, USA: 1992. Natural selection of the lac operon of Escherichia coli. [Google Scholar]
- Sokurenko EV, Hasty DL, Dykhuizen DE. Pathoadaptive mutations: gene loss and variation in bacterial pathogens. Trends in Microbiology. 1999;7:191–195. doi: 10.1016/s0966-842x(99)01493-6. [DOI] [PubMed] [Google Scholar]
- Sokurenko EV, Tchesnokova V, Yeung AT, Oleykowski CA, Trintchina E, Hughes KT, Rashid RA, Brint JM, Moseley SL, Lory S. Detection of simple mutations and polymorphisms in large genomic regions. Nucleic Acids Research. 2001;29:E111. doi: 10.1093/nar/29.22.e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JN. Rapid evolution as an ecological process. Trends in Ecology and Evolution. 1998;13:3329–3332. doi: 10.1016/s0169-5347(98)01378-0. [DOI] [PubMed] [Google Scholar]
- Travisano M, Lenski RE. Long-term experimental evolution in Escherichia coli. IV. Targets of selection and the specificity of adaptation. Genetics. 1996;143:15–26. doi: 10.1093/genetics/143.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travisano M, Mongold JA, Bennett AF, Lenski RE. Experimental tests of the roles of adaptation, chance, and history in evolution. Science. 1995a;267:87–90. doi: 10.1126/science.7809610. [DOI] [PubMed] [Google Scholar]
- Travisano M, Vasi F, Lenski RE. Long-term experimental evolution in Escherichia coli. III. Variation among replicate populations in correlated responses to novel environments. Evolution. 1995b;49:189–200. doi: 10.1111/j.1558-5646.1995.tb05970.x. [DOI] [PubMed] [Google Scholar]
- Treves DS, Manning S, Adams J. Repeated evolution of an acetate-crossfeeding polymorphism in long-term populations of Escherichia coli. Molecular Biology and Evolution. 1998;15:789–797. doi: 10.1093/oxfordjournals.molbev.a025984. [DOI] [PubMed] [Google Scholar]
- Turner PE, Souza V, Lenski RE. Tests of ecological mechanisms promoting the stable coexistence of two bacterial genotypes. Ecology. 1996;77:2119–2129. [Google Scholar]
- Vasi F, Travisano M, Lenski RE. Long-term experimental evolution in Escherichia coli. II. Changes in life-history traits during adaptation to a seasonal environment. American Naturalist. 1994;144:432–456. [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proceedings of the National Academy of Sciences (USA) 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]


