Abstract
Cysteine is the most intrinsically nucleophilic amino acid in proteins, where its reactivity is tuned to perform diverse biochemical functions. The absence of a consensus sequence that defines functional cysteines in proteins has hindered their discovery and characterization. Here, we describe a proteomics method to quantitatively profile the intrinsic reactivity of cysteine residues en masse directly in native biological systems. Hyperreactivity was a rare feature among cysteines and found to specify a wide range of activities, including nucleophilic and reductive catalysis and sites of oxidative modification. Hyperreactive cysteines were identified in several proteins of uncharacterized function, including a residue conserved across eukaryotic phylogeny that we show is required for yeast viability and involved in iron-sulfur protein biogenesis. Finally, we demonstrate that quantitative reactivity profiling can also form the basis for screening and functional assignment of cysteines in computationally designed proteins, where it discriminated catalytically active from inactive cysteine hydrolase designs.
Large-scale scientific endeavors such as genome sequencing and structural genomics are providing a wealth of new information on the full complement of proteins present in eukaryotic and prokaryotic organisms. Many of these proteins, however, remain partly or completely unannotated with respect to their biochemical activities1. New methods are therefore needed to characterize protein function on a global scale. Much effort is currently devoted to the characterization of post-translational modification events because these covalent adducts can have profound and dynamic effects on protein activity2. Another oft-overlooked parameter that defines functional “hotspots” in the proteome is amino acid side-chain reactivity, which can vary by several orders of magnitude for a given residue depending on local protein microenvironment. Methods to measure side-chain reactivity en masse directly in complex biological systems have not yet been described, and as such, the reactive landscape of the proteome remains largely unexplored.
Among the protein-coding amino acids, cysteine is unique due to its intrinsically high nucleophilicity and sensitivity to oxidative modification. The pKa of the free cysteine thiol is between 8 and 9, meaning that only slight perturbations in the local protein microenvironment can result in ionized thiolate groups with enhanced reactivity at physiological pH3. Diverse families of enzymes utilize cysteine-dependent chemical transformations, including proteases, oxidoreductases, and acyltransferases4. In addition to its roles in catalysis, cysteine is subject to several forms of oxidative post-translational modification, including sulfenation (SOH), sulfination (SO2H), nitrosylation (SNO), disulfide formation, and glutathionylation, which endows it with the ability to serve as a regulatory switch on proteins that is responsive to cellular redox state5.
Functional cysteines, regardless of whether they are catalytic residues or sites of post-translational modification, do not conform to a canonical sequence motif, which complicates their systematic identification and characterization. pKa measurements can identify cysteine residues with heightened nucleophilicity (or “hyperreactive” cysteines6,7), but this requires purified protein and detailed kinetic and mutagenic experiments7,8 that cannot be performed on a proteome-wide scale. Additional methods have been introduced to computationally predict redox-active cysteines9, identify cysteines with specific modifications10-14, and qualitatively inventory electrophile-modified cysteines in proteomes15-18. Some of these studies have provided suggestive evidence that nucleophilic cysteines may possess a variety of important functions14-18, although the non-quantitative methods employed in each case precluded a robust and systematic evaluation of this potential relationship. We envisioned a different strategy to globally characterize cysteine functionality in proteomes based on quantitative reactivity profiling with isotopically labeled, small-molecule electrophiles.
A chemoproteomic method for quantifying cysteine reactivity
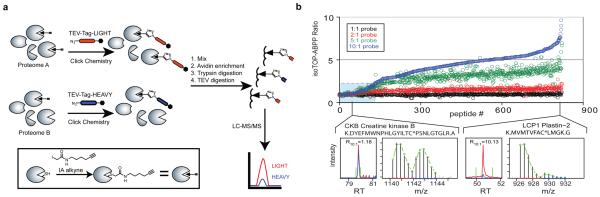
Our approach, termed isoTOP-ABPP (isotopic Tandem Orthogonal Proteolysis – Activity-Based Protein Profiling), has four features to enable quantitative analysis of native cysteine reactivity (Fig. 1a): [1] an electrophilic iodoacetamide (IA) probe to label cysteine residues in proteins that also has [2] an alkyne handle for “click chemistry” conjugation of probe-labeled proteins19 to [3] an azide-functionalized TEV-protease recognition peptide containing a biotin group for streptavidin-enrichment of probe-labeled proteins20 and [4] an isotopically-labeled valine for quantitative mass spectrometry (MS) measurements of IA-labeled peptides across multiple proteomes (Supplementary Fig. 1). Following tandem on-bead proteolytic digestions with trypsin and TEV protease15,20, probe-labeled peptides attached to isotopic tags are released and analyzed by liquid chromatography-high-resolution MS to identify IA-modified cysteines and quantify their extent of labeling based on MS2 and MS1 profiles, respectively. An isoTOP-ABPP ratio, R, is generated for each identified cysteine that reflects the difference in signal intensity between light and heavy tag-conjugated proteomes.
Figure 1. A quantitative approach to globally profile cysteine reactivity in proteomes.
a, isoTOP-ABPP involves proteome labeling, click chemistry-based incorporation of isotopically-labeled cleavable tags, and sequential on-bead protease digestions to afford probe-labeled peptides for MS analysis. The IA-probe is shown in the inset. b, Measured isoTOP-ABPP ratios for peptides from MCF7 cells labeled with four pairwise IA-probe concentrations (10:10 μM, 20:10 μM, 50:10 μM, 100:10 μM). The blue box highlights peptides with low isoTOP-ABPP ratios (R < 2.0). Chromatographs for CKB (low ratio) and LCP1 (high ratio) are shown, with elution profiles for heavy- and light-labeled peptides in blue and red, respectively, and green lines depicting peak-boundaries used for quantitation. Isotopic envelopes are shown for light- and heavy-labeled peptides with green lines representing predicted values. Additional chromatographs from isoTOP-ABPP experiments are in Supplementary Table 7.
We first verified the accuracy of isoTOP-ABPP by labeling varying amounts of a mouse liver proteome (1X, 2X, 4X) with the IA-probe followed by click chemistry conjugation with either the heavy- or light-variants of the azide-TEV-biotin tag. The observed signals for labeled cysteines closely matched the expected proteome ratios (R1:1 ≈ 1, R2:1 ≈ 2, or R4:1 ≈ 4, respectively; Supplementary Fig. 2). A representative MS/MS profile of an IA-labeled peptide from our proteomic experiments is provided in Supplementary Fig. 3.
In contrast to traditional cysteine-alkylating protocols for proteomics that use millimolar concentrations of IA to stoichiometrically modify all cysteines in denatured proteins21, we hypothesized that, by applying low (micromolar) concentrations of the IA-probe to native proteomes, differences in the extent of alkylation would reflect differences in cysteine reactivity, rather than abundance. This hypothesis predicts that the reactivity of cysteines can be measured on a proteome-wide scale in isoTOP-ABPP experiments that compare low versus high concentrations of IA-probe, where hyperreactive cysteines would be expected to label to completion at low probe concentrations (generating isoTOP-ABPP ratios with R[high]:[low] ≈ 1) and less reactive cysteines should show concentration-dependent increases in IA-probe labeling (generating isoTOP-ABPP ratios with R[high]:[low] >> 1) (Supplementary Fig. 4). We tested this idea by performing four parallel isoTOP-ABPP experiments with the soluble proteome of the human breast cancer cell line MCF7 using pair-wise IA-probe concentrations of 10:10 μM, 20:10 μM, 50:10 μM and 100:10 μM (light:heavy). More than 800 probe-labeled cysteines were identified on 522 proteins, the vast majority of which exhibited escalating isoTOP-ABPP ratios (Fig. 1b) expected for reactions that did not reach completion over the tested probe concentration range. In contrast, a small subset of cysteines (< 10%) showed nearly identical ratios at all probe concentrations tested (R1:1 ≈ R2:1 ≈ R5:1 ≈ R10:1 ≈ 1, Fig. 1b, shaded blue box). An expanded analysis of multiple human cancer line (Supplementary Fig. 5 and Supplementary Table 1) and mouse tissue (Supplementary Fig. 6 and Supplementary Table 2) proteomes treated with low (10 μM) and high (100 μM) IA-probe concentrations revealed consistent isoTOP-ABPP ratios for individual cysteine residues, indicating that the propensity of a cysteine to display high IA reactivity is an intrinsic property of the residue (and presumably its local protein environment), and not, in general, contingent on features specific to a particular cell or tissue. Additionally, isoTOP-ABPP ratios showed no correlation with either protein abundance or peptide ion intensity (Supplementary Fig. 7), indicating that they were independent of potential MS-based ionization sources for saturation. Finally, we confirmed that similar isoTOP-ABPP ratios were obtained for cysteines in reactions where time rather than the concentration of probe was varied (Supplementary Fig. 8 and Supplementary Table 3), confirming that low isoTOP-ABPP ratios reflect rapid reaction kinetics (hyperreactivity), rather than saturable binding interactions (see Supplementary Discussion).
Hyperreactivity is a predictor of cysteine functionality
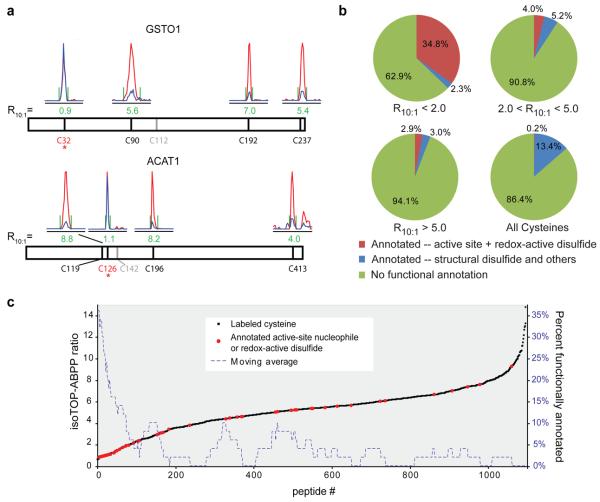
We next sought to assess the functional ramifications of the special subset of cysteines that showed hyperreactivity in isoTOP-ABPP experiments. We first noted that multiple sites of IA-probe labeling on the same protein often showed markedly different isoTOP-ABPP ratios. For example, the glutathione S-transferase GSTO1 was labeled on four cysteine residues, three of which exhibited high ratios (C90, C192, and C237 displayed ratios of R10:1 = 5.6, 7, and 5.4, respectively), while the fourth (C32) displayed a low ratio of R10:1 = 0.9 (Fig. 2a). Interestingly, C32 is the active-site nucleophile of GSTO122. Acetyl-CoA acetyltransferase-1 (ACAT1) was also labeled on four cysteines and three showed high ratios (C119, C196, and C413 displayed ratios of R10:1 = 8.8, 8.2, and 4, respectively), while the fourth, the active site nucleophile C12623, yielded a low ratio of R10:1 = 1.1 (Fig. 2a).
Figure 2. Hyperreactive cysteines are highly enriched in functional residues.
a, Chromatographs from an isoTOP-ABPP experiment using 100:10 μM IA-probe are shown for peptides from GSTO1 (top) and ACAT1 (bottom). The cysteine nucleophiles (asterisk) display low ratios (R10:1 ≈ 1), whereas other cysteines exhibit high ratios (R10:1 ≥ 4). b, Pie charts illustrating the percentage of functionally annotated cysteines for three isoTOP-ABPP ratio ranges, including an average derived from all cysteines in the UniProt database. c, Correlation of isoTOP-ABPP ratios with functional annotations from the UniProt database where active-site nucleophiles or redox-active disulfides are shown in red, and all other cysteines in black. A moving-average (window of 50) of functional residues is shown as a dashed-blue line, demonstrating a profound enrichment within R10:1 < 2.0. Data are from experiments in three human cancer cell lines (MCF7, MDA-MB-231 and Jurkat).
The aforementioned findings suggested that heightened IA-reactivity might be a good predictor of cysteine functionality in proteins. To more systematically examine this premise, we queried the UniProt database to retrieve functional annotation for the 1082 cysteine residues labeled by the IA-probe. This analysis revealed that the most hyperreactive cysteines were remarkably enriched in functional residues, with 35% of the cysteines with R10:1 < 2 being annotated as active-site nucleophiles or redox-active disulfides compared to 0.2% for all cysteine residues in the UniProt database (Fig. 2b, c; Supplementary Fig. 9; Supplementary Tables 4 and 5). Hyperreactive cysteines were also, as a group, more conserved across eukaryotic evolution (Supplementary Fig. 10). A broader survey of hyperreactive cysteines identified several that have been ascribed functional properties in the literature despite lacking annotation in UniProt (Supplementary Fig. 11). For example, a single hyperreactive cysteine C108 (R10:1 = 1.0) was identified in the uncharacterized protein D15Wsu75e. This protein and its orthologues are predicted to be cysteine proteases based on conservation of a prototypical Cys-His catalytic dyad24. Interestingly, C108 corresponds to the putative cysteine nucleophile of this catalytic motif and a recent crystal structure confirms the proximity of C108 to a conserved histidine (H38) (Supplementary Fig. 12). Thus, quantitative reactivity profiling supports structural predictions that D15Wsu75e is a functional cysteine protease.
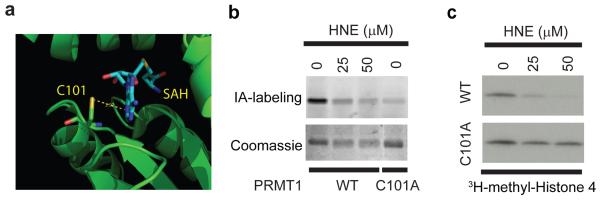
Hyperreactive cysteines also corresponded to sites for post-translational modification. For instance, C101 (R10:1 = 1.92) in the protein arginine methyltransferase PRMT1 has been identified as a site of modification by the endogenous oxidative product 4-hydroxy-2-nonenal (HNE)25. This cysteine, although nonessential for catalytic function, is an active site residue that makes direct contact with the S-adenosylmethionine cofactor26 (Fig. 3a). Interestingly, we found that HNE inhibited both the IA-labeling (Fig. 3b) and catalytic activity (Fig. 3c) of wild-type PRMT1. A C101A mutant of PRMT1 showed substantially reduced IA-labeling (Fig. 3b) and HNE sensitivity (Fig. 3c). These data indicate that PRMT1 may be regulated by oxidative stress pathways through selective HNE modification of its hyperreactive, active-site C101 residue. Additional hyperreactive cysteines represented sites for glutathionylation27 [CLIC1 (C24), CLIC3 (C25), and CLIC4 (C35); R10:1 = 2.02, 1.07, and 1.45, respectively] and nitrosylation28 (RTN3; C42, R10:1 = 0.78). These data, taken together, indicate that heightened reactivity is not only a feature of catalytic cysteines, but also “non-catalytic”, active-site cysteines, as well as those that undergo various forms of oxidative modification.
Figure 3. Functional characterization of the hyperreactive cysteines in PRMT1.
a, Crystal structure of rat PRMT126 (green, PDB: 1ORI) showing the hyperreactive cysteine C101 in contact with an S-adenosylhomocysteine (SAH) cofactor (cyan). b, Wild-type and C101A mutant of human PRMT1 were labeled with the IA-probe, followed by click-chemistry to incorporate a fluorescent rhodamine tag. In-gel fluorescence demonstrates robust labeling of the wild-type (WT) but not C101A mutant PRMT1 and IA-labeling of WT PRMT1 is inhibited by HNE. c, Catalytic activity of purified WT, but not C101A PRMT1 is inhibited by HNE as measured by monitoring transfer of 3H-methyl from 3H-S-adenosylmethionine (SAM) to a histone 4 substrate.
Functional significance of the hyperreactive cysteine in FAM96B
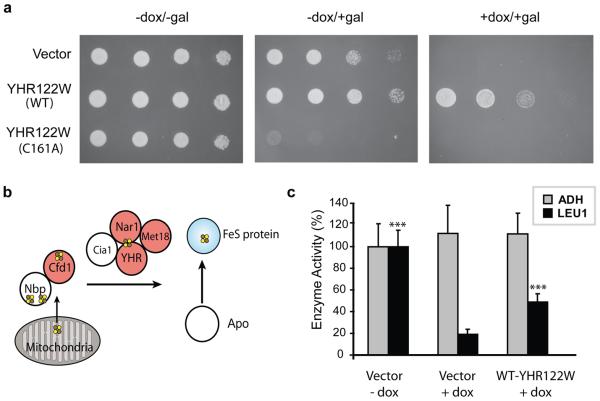
Intrigued by the diverse functional properties displayed by hyperreactive cysteines, we reasoned that critical activities might be inferred for such residues in hitherto uncharacterized proteins. A survey of the cysteines displaying low isoTOP-ABPP ratios uncovered the highly conserved C93 (R10:1 = 1.15) in the uncharacterized protein FAM96B (Supplementary Fig. 13). FAM96B has close orthologues in many organisms including the YHR122W protein from the budding yeast Saccharomyces cerevisiae, which displays 52% identity with human FAM96B, including conservation of C93 (the corresponding residue in YHR122W is C161). The gene encoding YHR122W is essential for yeast viability29, and we found that expression of WT-YHR122W, but not the C161A-YHR122W mutant could rescue a yeast strain in which the YHR122W gene was conditionally suppressed (Fig. 4a; Supplementary Fig. 14). These data confirm the importance of C93 for the in vivo function of YHR122W and, by extension, other members of the FAM96B family.
Figure 4. Functional characterization of YHR122W/FAM96B.
a, Expression of wild-type (WT) and C161A-YHR122W in a yeast strain with a doxycycline (dox)-repressable YHR122W gene demonstrated a dominant-negative phenotype upon induction of C161A-YHR122W expression (−dox/+gal, middle panel) and rescue of viability by expression of WT, but not C161A-YHR122W (+dox/+gal, right panel). b, The FeS cluster assembly pathway contains multiple proteins with hyperreactive cysteines (in red). YHR122W/FAM96B (YHR) is a putative member of this network based on protein-protein interaction studies (see http://www.yeastgenome.org/). This panel was adapted from reference 30. c, Doxycycline treatment of the YHR122W-repressable yeast strain significantly decreased the activity of the cytosolic FeS enzyme Leu141, and this activity is rescued by overexpression of WT-YHR122W. These treatments had no effect on the activity of the non-FeS enzyme alcohol dehydrogenase (ADH). ***, P < 0.001, t-test.
We also observed that expression of the C161A-YHR122W mutant caused a severe growth defect in non-suppressive media indicative of a dominant-negative phenotype (Fig. 4a, Supplementary Fig. 14). This result suggests that the YHR122W protein may engage in protein complexes that are sequestered by the C161A mutant, thereby disrupting the activity of the WT protein. Consistent with this premise, queries of the Saccharomyces genome databank (SGD) revealed that YHR122W has been found in several yeast-two hybrid studies to bind to proteins involved in iron-sulfur (FeS) cluster assembly, namely Nar1, Cia1, and Met1830 (Fig. 4b). We found that the activity of the FeS-client protein isopropylmalate isomerase (Leu1)31 was dramatically reduced in YHR122W-deleted yeast, and this reduction was substantially rescued by expression of the wild-type YHR122W protein (Fig. 4c). These data support a role for the YHR122W/FAM96B protein in FeS-protein biogenesis. We also note that reactive cysteines appear to be a common feature of proteins involved in FeS-protein biogenesis, including the human orthologues of Nar1, Met18, and Cfd1 (NARF, MMS19, and NUBP2, respectively) (R10:1 = 0.91, 2.2, and 2.9 respectively) (Supplementary Fig. 11) , where they may assist in the transfer of assembled FeS-clusters to client proteins32.
Predicting functional cysteines in designed proteins
The striking correlation between cysteine hyperreactivity and functionality observed in native proteomes inspired us to ask whether this relationship would extend to de novo designed proteins. We compared the IA-labeling of a dozen proteins that were computationally designed to act as cysteine hydrolases. These proteins originated from structurally distinct scaffolds and were all designed to contain cysteine-histidine dyads within an active site cavity (see Supplementary Methods for more details). Two of the designed proteins, ECH13 and ECH19, exhibited significant hydrolytic activity using a fluorogenic ester substrate, while the other ten designs were inactive (Fig. 5a; Supplementary Fig. 15a).
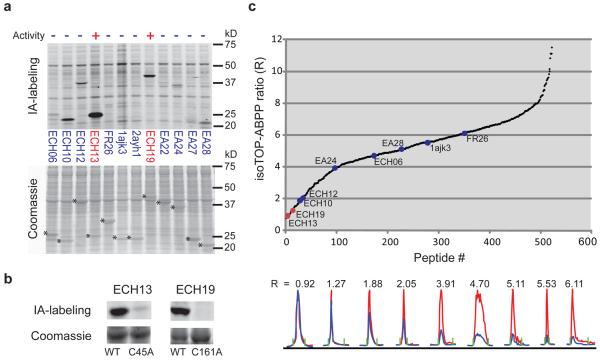
Figure 5. Quantitative reactivity profiling predicts functional cysteines in designed proteins.
a, In-gel fluorescence demonstrates robust IA labeling of two active cysteine hydrolases, ECH13 and ECH19 , relative to inactive designs (top panel). Hydrolysis activities of ECH13 and ECH19 measured as the ratio of velocities in the presence versus absence of purified enzymes were 71.64+/−6.94 and 104.15+/−10.78, respectively (see Supplementary Fig. 15a for substrate hydrolysis assay). Other designs showed no measurable hydrolysis activity over background (0.76 +/− 0.058 nmol/s). Asterisks designate Coomassie blue signals for protein designs (lower panel). b, IA-labeling is observed for ECH13 and ECH19, but not their active-site cysteine mutants C45A and C161A, respectively. c, Catalytic cysteines in ECH13 and ECH19 demonstrate low isoTOP-ABPP ratios (red) compared with other designs (blue). Chromatographs are shown for peptides from the nine designs identified in this experiment (bottom panel).
We first evaluated IA-labeling of protein designs using a clickable, fluorescent reporter tag and SDS-PAGE analysis, where similar amounts of each protein were tested in a homogeneous background proteome representing a mix of E. coli and human (MCF7 cell line) proteins. The two active protein designs ECH13 and ECH19 showed strong IA-labeling signals compared to inactive designs (Fig. 5a), and, in both cases, mutation of the active-site cysteine to alanine abolished labeling (Fig. 5b) and hydrolytic activity (data not shown). We next combined the proteomes containing all twelve protein designs, diluted them into a background human cell proteome, and analyzed the mixture by isoTOP-ABPP. Strikingly, both ECH13 and ECH19 showed isoTOP-ABPP ratios that were equivalent to the most hyperreactive cysteines in human and E. coli proteomes (R10:1 = 0.92 and 1.27, respectively), while the remaining inactive protein designs all showed higher ratios ranging from 1.88-6.11 (Fig. 5c; Supplementary Fig. 15b, c). These data thus reveal a strong correlation between cysteine hyperreactivity and hydrolytic activity across a diverse panel of protein designs and designate heightened cysteine nucleophilicity as a key feature of successful cysteine hydrolase designs.
Conclusions
Here, we have described a quantitative method to profile the intrinsic reactivity of cysteine residues in native proteomes. Measurement of the rate of alkylation by IA (or other carbon electrophiles) has been used by enzymologists to assess the nucleophilicity of cysteine residues in individual, purified proteins6. With isoTOP-ABPP, these studies can now be extended to quantitative, proteome-wide surveys of cysteine reactivity in complex biological systems. A key advantage of isoTOP-ABPP over more traditional proteomic methods that target cysteine-containing peptides14,18 is the use of an alkynylated IA-probe in place of more bulky biotinylated reagents, which have shown an impaired ability to label cysteines in native proteins14. Alkynylated IA-probes, due to their cell-permeability, also afford the opportunity to perform cysteine reactivity profiling in living systems. In pilot experiments, we have found that a large fraction of hyperreactive cysteines are labeled by the IA-probe in living cells (Supplementary Fig. 16). isoTOP-ABPP also distinguishes itself by selectively targeting probe-accessible cysteines in native proteins. In this way, structural cysteines engaged in disulfide bonds or buried within the body of a protein are avoided to provide preferential access to a specific fraction of cysteines that are profoundly enriched in functionality (i.e., the IA-probe labeled 1082 out of a total 8910 cysteines present on the 890 human proteins detected in this study). Projecting forward, it is possible that, by varying the nature of the electrophile, isoTOP-ABPP probes can be created that profile the reactivity of different subsets of cysteines, as well as other amino acids in proteomes, such as serine, threonine, tyrosine, and glutamate/aspartate, that have also been shown to react with small-molecule probes16,18,33-35.
We discovered that hyperreactivity can predict cysteine function in both native and designed proteins. That hyperreactivity was strongly correlated with catalytic activity in de novo designed cysteine hydrolases is interesting from the principles of both enzyme engineering and assay development, as it suggests that heightened cysteine nucleophilicity is a key feature of active catalysts and, accordingly, electrophile reactivity could serve as an effective primary screen for novel cysteine-dependent enzymes. We show that these screens can be performed directly in complex proteomes using either gel or MS (isoTOP-ABPP) detection platforms, thus offering a versatile and relatively high-throughput way to evaluate many protein designs in parallel. The isoTOP-ABPP platform has the additional advantage of reading out the relative cysteine reactivity of designs independent of their expression levels against a ‘background’ of native, hyperreactive cysteines for comparison. isoTOP-ABPP might also offer a complementary way to perform cysteine reactivity/accessibility experiments that monitor protein stability and ligand interactions36,37.
The relationship between cysteine reactivity and functionality extends beyond nucleophilic catalysis to include other enzymatic activities (oxidative/reductive), as well as sites of electrophilic and oxidative modification. Quantitative reactivity profiling thus distinguishes itself as a complementary and perhaps more inclusive strategy to survey cysteine function compared to previous computational9 and experimental11-14,17 methods that focus on specific cysteine-based activities or modification events. Considering further that hyperreactive cysteines corresponded to sites for glutathionylation27, nitrosylation28, and HNE-modification25, we speculate that cysteine nucleophilicity is a property that may have been selected for during evolution to offer points of protein control by oxidative stress pathways. Determining how the reactivity of cysteine residues is honed will require further investigation, but we anticipate that quantitative proteomic data, when integrated with the output of ongoing structural genomics programs, may eventually uncover unifying mechanistic principles that explain cysteine reactivity in proteins. In this regard, it is interesting to note that, while hyperreactive cysteines did not conform to any obvious consensus sequence motifs, many of these residues were found at the N-termini of α-helices (Supplementary Fig. 17). This finding is consistent with literature reports ascribing a role for α-helix dipoles in the stabilization of cysteine thiolate anions38.
Finally, it is important to stress that some functional cysteines may be inherently reactive, but inaccessible to our IA probe for steric reasons. Other cysteine-reactive electrophilic probes16,17 may prove more suitable for such cysteine residues. Also, hyperreactivity is not necessarily a defining feature for all functional cysteines. Some enzymes with catalytic cysteines may, for instance, exhibit reduced reactivity until binding their physiologic substrates or may rely more on substrate recognition than inherent catalytic power for function. This may be the case, for instance, with the E1-activating and E2-conjugating enzymes, which recognize a specific class of ubiquitinated substrates and possess active site cysteines that showed only moderate levels of electrophile reactivity (Supplementary Fig. 18). Other cysteines may display activities that are not dependent on their nucleophilicity. Our data do suggest, however, that those cysteines that are hyperreactive in proteomes likely perform important catalytic and/or regulatory functions for their parent proteins. The large number of newly discovered residues that fall into this category foretell a broad role for hyperreactive cysteines in mammalian biology.
METHODS SUMMARY
Probes and tags
The IA-probe and the light- and heavy-variants of the azide-TEV-biotin tags were synthesized as previously described20,39.
Sample preparation, mass spectrometry and data analysis
For concentration-dependent experiments, proteome samples in PBS were probe labeled with the desired probe concentration for 1 hour. Click chemistry was performed with either the light- or heavy-variants of the azide-TEV-biotin tags and the samples were mixed and subject to streptavidin enrichment and subsequent trypsin and TEV digestion. The resulting TEV digests were analyzed by Multidimensional Protein Identification Technology (MudPIT) on an LTQ-Orbitrap instrument. The resulting tandem MS data were searched using the SEQUEST algorithm40 using a concatenated target/decoy variant of the human, mouse and E. coli protein sequence databases. Quantification of light/heavy ratios (isoTOP-ABPP ratios, R) was performed using in-house software. Detailed information on sample preparation, mass spectrometry methods and data analysis is presented in the Supplementary Methods.
Complementation of S. cerevisiae YHR122W deletion mutant
cDNA encoding wild-type YHR122W was subcloned into the pESC_Leu vector (Stratagene). The YHR122W C161A mutant was generated using the Quickchange procedure (Stratagene). These constructs were introduced into a yeast Tet promoter Hughes (yTHC) strain harboring a conditional (doxycycline-dependent) disruption in the YHR122W gene (Open Biosystems). Growth of these transformed cell lines on +/− Gal/+/− Dox media were monitored for 3 days. These cell lines were also used to monitor Leu1 and ADH activity. Detailed information on the protocols used to subclone, transform, monitor the growth of the yeast strains and measure enzyme activity is available in the Supplementary Methods.
Methods
All compounds and reagents were purchased from Novabiochem, Sigma or Fisher, except where noted.
Preparation of mouse proteomes
Mouse tissues (heart and liver) were harvested and immediately flash frozen in liquid nitrogen. The tissues were then Dounce homogenized in 1X Phosphate Buffered Saline (PBS), pH 7.4. Centrifugation at 100,000 × g (45 min) provided soluble fractions (supernatant) and membrane fractions (pellet). Protein concentrations for each proteome were obtained using the Bio-Rad Dc Protein Assay and stored at −80 °C till use.
Preparation of human cancer cell line proteomes
MDA-MB-231 cells were grown in L15 media supplemented with 10% fetal bovine serum at 37 °C in a CO2-free incubator. Jurkat cells and MCF7 cells were grown in RPMI-1640 supplemented with 10% fetal bovine serum at 37 °C with 5% CO2. For in vitro labeling experiments, cells were grown to 100% confluency, washed three times with PBS and scraped in cold PBS. Cell pellets were isolated by centrifugation at 1400 × g for 3 min, and the cell pellets stored at −80 °C until further use. For in situ labeling of MDA-MB-231 and MCF7 cells, the cells were grown to 90% confluency, the media was removed and replaced with fresh media containing 10 μM IA-probe. The cells were incubated at 37 °C for 1 hour and harvested as detailed above. The harvested cell pellets were lysed by sonication and fractionated by centrifugation (100,000 × g, 45 min.) to yield soluble and membrane proteomes. The proteomes were diluted to 2 mg/ml and stored at −80 °C until use.
Protein labeling and click chemistry
Proteome samples were diluted to a 2 mg protein/mL solution in PBS. Each sample (2 × 0.5 mL aliquots) was treated with 10, 20, 50, or 100 μM of IA-probe using 5 μL of a 1, 2, 5, or 10 mM stock in DMSO. The labeling reactions were incubated at room temperature for 1 hour. Click chemistry was performed by the addition of 150 μM of either the Light-TEV-Tag or Heavy-TEV-Tag (15 μL of a 5 mM stock), 1 mM TCEP (fresh 50X stock in water), 100 μM ligand (17X stock in DMSO:t-Butanol 1:4) and 1 mM CuSO4 (50X stock in water). Samples were allowed to react at room temperature for 1 hour. After the click chemistry step, the light and heavy-labeled samples were mixed together and centrifuged (5900 × g, 4 min, 4 °C) to pellet the precipitated proteins. The pellets were washed twice in cold MeOH, after which the pellet was solubilized in PBS containing 1.2% SDS via sonication and heating (5 min, 80 °C).
For time course experiments, proteome samples were labeled with 100 μM of IA-probe (using 5 μL of a 10 mM stock in DMSO). After 6 minutes of probe labeling, an aliquot of the reaction was quenched by passaging the sample through a NAP-5 column (GE Healthcare) to remove excess, unreacted probe. After 60 minutes of probe labeling, the other sample was quenched as before and click chemistry was performed as described above.
Streptavidin enrichment of probe-labeled proteins
The SDS-solubilized, probe-labeled proteome samples were diluted with 5 mL of PBS for a final SDS concentration of 0.2%. The solutions were then incubated with 100 μL of streptavidin-agarose beads (Pierce) for 3 hours at room temperature. The beads were washed with 10 mL 0.2% SDS/PBS, 3 × 10 mL PBS and 3 × 10 mL H2O and the beads were pelleted by centrifugation (1300 × g, 2 min) between washes.
On-bead trypsin and TEV digestion
The washed beads from above were suspended in 500 μL of 6 M urea/PBS and 10 mM TCEP (from 20X stock in H2O) and placed in a 65 °C heat block for 15 minutes. 20 mM iodoacetamide (from 50X stock in H2O) was then added and allowed to react at 37 °C for 30 minutes. Following reduction and alkylation, the beads were pelleted by centrifugation (1300 × g, 2 min) and resuspended in 200 μL of 2 M urea/PBS, 1 mM CaCl2 (100X stock in H2O), and trypsin (2 μg). The digestion was allowed to proceed overnight at 37 °C. The digest was separated from the beads using a Micro Bio-Spin column and the beads were then washed with 3 × 500 μL PBS, 3 × 500 μL H2O, and 1 × 150 μL of TEV digest buffer. The washed beads were then resuspended in 150 μL of TEV digest buffer with AcTEV Protease (Invitrogen, 5 μL) for 12 hours at 29 °C. The eluted peptides were separated from the beads using a Micro Bio-Spin column and the beads washed with H2O (2 × 75 μL). Formic acid (15 μL) was added to the sample, which was stored at −20 °C until mass spectrometry analysis.
Liquid chromatography-mass spectrometry (LC-MS) analysis
LC-MS analysis was performed on an LTQ-Orbitrap mass spectrometer (ThermoFisher) coupled to an Agilent 1100 series HPLC. TEV digests were pressure loaded onto a 250 μm fused silica desalting column packed with 4 cm of Aqua C18 reverse phase resin (Phenomenex). The peptides were then eluted onto a biphasic column (100 μm fused silica with a 5 μm tip, packed with 10 cm C18 and 3 cm Partisphere strong cation exchange resin (SCX, Whatman) using a gradient 5-100% Buffer B in Buffer A (Buffer A: 95% water, 5% acetonitrile, 0.1% formic acid; Buffer B: 20% water, 80% acetonitrile, 0.1% formic acid). The peptides were then eluted from the SCX onto the C18 resin and into the mass spectrometer using four salt steps as previously described15,20. The flow rate through the column was set to ~0.25 μL/min and the spray voltage was set to 2.75 kV. One full MS scan (FTMS) (400-1800 MW) was followed by 18 data dependent scans (ITMS) of the nth most intense ions with dynamic exclusion disabled.
MS Data Analysis – peptide identification
The tandem MS data were searched using the SEQUEST algorithm40 using a concatenated target/decoy variant of the human and mouse IPI databases. A static modification of +57.02146 on Cys was specified to account for iodoacetamide alkylation and differential modifications of +464.28596 (Light probe modification) and +470.29977 (Heavy probe modification) were specified on cysteine to account for probe modifications with the either light or heavy variants of the IA-probe-TEV adduct. SEQUEST output files were filtered using DTASelect 2.042. Reported peptides were required to be fully tryptic and contain the desired probe modification and discriminant analyses were performed to achieve a peptide false-positive rate below 5%. The actual false positive rate was assessed at this stage according to established guidelines43 and found to be ~3.5%. Additional assessments of the false-positive rate were performed following the application of additional filters (described below) resulting in a final false-positive rate below 0.05%.
MS Data Analysis – quantification
Quantification of light/heavy ratios (isoTOP-ABPP ratios, R) was performed using in-house software written in the R programming language that utilizes routines from the open-source XCMS package44 for mass-spectrometry data analysis to read in raw chromatographic data in the mzXML format45. Each experiment consisted of two LC/LC-MS/MS runs: light:heavy 10 μM:10 μM, and light:heavy 100 μM:10 μM IA-probe concentration. Both runs were searched using SEQUEST and filtered with DTASelect as described above. Because the mass-spectrometer was configured for data-dependant fragmentation, peptides are not always identified in every run. As such, peptides were identified in either 1) only the 10 μM:10 μM run, 2) only the 100 μM:10 μM run, or 3) both runs. In the case of peptides that were sequenced in both runs, identification of the corresponding peaks was made by choosing peaks that co-elute with the peptide identification. In the case of probe-modified peptides that were sequenced in one, but not the other run, an algorithm was developed to identify the corresponding peak in the run without the SEQUEST identification. To accomplish this, the retention time of the “reference” peptide is used to position a retention time window (+/− 10 minutes) across the run lacking a peptide identification. Extracted ion chromatograms (+/− 10 ppm) of the target peptide m/z with both “light” and “heavy” modifications are generated within that window. The program then searches for candidate co-eluting pairs of light:heavy MS1 peaks, and for each candidate pair calculates the ratio of integrated peak area between the light and heavy peaks. Several filters are used to ensure that the correct peak-pair is identified: first, the extent of co-elution for each peak-pair is quantified using a Pearson correlation, an established method to gauge elution profile similarity46. Second, the predicted pattern of the isotopic envelope of the target peptide is generated and compared to the observed high-resolution MS1 spectrum. This comparison generates an ‘envelope correlation score’ (Env) which also enables confirmation of the monoisotopic mass and charge-state of each candidate peak. Peak-pairs that display poor co-elution scores, or that have the incorrect monoisotopic mass or charge, or whose isotopic envelopes are not well-correlated with the predicted envelope are eliminated from consideration. After application of these filters, in the rare case that multiple candidates still exist, then no peak is chosen and a ratio is not recorded. Usually, however, application of these filters results in a single candidate peak-pair and the ratio for this peak-pair is recorded for the peptide in the corresponding run. In this way, each experiment yields two ratios, one for the 10 μM:10 μM run and one for the 100 μM:10 μM run. Following application of these filters, the false-positive rate was re-assessed, and found to be less than 0.05% in all cases.
After ratios for unique peptide entries are calculated for each experiment, overlapping peptides with the same labeled cysteine (e.g., same local sequence around the labeled cysteines but different charge states, MudPIT segment numbers, or tryptic termini) are grouped together, and the median ratio from each group is reported as the final ratio (“R”). All of these values can be found in Supplementary Tables 1, 2 and 3 and representative chromatographs can be seen in Supplementary Table 7. Raw result files of peptide identification using SEQUEST can be found in Supplementary Table 9.
Functional annotation of labeled cysteines
For automated functional analyses, custom perl-scripts were developed to query the UniProtKB/Swiss-Prot protein knowledgebase release 57.4 (current as of 16-Jun-09). Sequence annotation in the (Features) section of the relevant UniProt entry was mined and any annotation corresponding to the labeled residue was collected. This functional annotation in its entirety can be found in Supplementary Tables 4 and 5.
Recombinant PRMT1 protein expression and purification
Full-length cDNA encoding human PRMT1 in pOTB7 was purchased from Open BioSystems and subcloned into pET-45b(+) (Novagen). BL21(DE3) E. coli containing this vector was grown in LB media containing 75 mg/L carbenicillin with shaking at 37 °C to an OD600 of 0.5. The cells were then induced with 1 mM IPTG and harvested 4 hours later by centrifugation. Cells were lysed by stirring for 20 mins at 4 °C in 50 mM Tris-HCl (pH 8.0) with 150 mM NaCl and supplemented with 1 mg/mL lysozyme and 1 mg/mL DNase I. The lysate was then sonicated and centrifuged at 10,000g for 10 min. Talon cobalt affinity resin (Clontech; 400 μL of slurry/g of cell paste) was added to the supernatant, and the mixture was rotated at 25 °C for 30 min. Beads were collected by centrifugation at 700g for 3 min, washed twice with Tris buffer, and applied to a 1 cm column. The column was washed twice with Tris buffer (10 mL/400 μL of resin slurry) and Tris buffer with 500 mM NaCl once. The bound protein was eluted by the addition of 100 mM imidazole (2 mL/400 μL of resin). Imidazole was removed by passage over a Sephadex G-25M column (GE Healthcare), and the eluate was concentrated using an Amicon centrifugal filter device (Millipore). Protein concentration was determined using the Bio-Rad DC Protein Assay kit. These conditions yielded PRMT1 at approximately 0.5 mg/L of culture. A C101A mutation was introduced into the pET-45b(+) construct described above using the Quikchange Site-Directed Mutagenesis Kit (Stratagene), and the resulting mutant protein was expressed identically and isolated with a similar yield.
In-gel fluorescence characterization of PRMT1
13 μg of recombinant PRMT1 (wild-type or C101A mutant) in 50 μL PBS buffer was pre-incubated with 0, 25 or 50 μM 4-hydroxy-2-nonenal (HNE, Calbiochem, 50 mM stock in ethanol) for 1 hour at room temperature and then was labeled with 100 nM of the IA-probe (5 μM stock in DMSO) and the reactions incubated for 1 hour at room temperature. Click chemistry was performed with 20 μM rhodamine-azide, 1 mM TCEP, 100 μM TBTA ligand and 1 mM CuSO4. The reaction was allowed to proceed at room temperature for 1 hour before quenching with 50 μL of 2X SDS-PAGE loading buffer (reducing). Quenched reactions were separated by SDS-PAGE (30 μL of sample/lane) and visualized in-gel using a Hitachi FMBio IIe flatbed laser-induced fluorescence scanner (MiraiBio, Alameda, CA).
PRMT1 in vitro methylation assays
500 ng of recombinant human PRMT1 (wild-type or C101A mutant) was pre-incubated with 4-hydroxy-2-nonenal (HNE, Calbiochem) for 30 minutes and methylation activity was monitored after addition of 1 mg of recombinant histone 4 (M2504S; NEB) and 3H-S-adenosylmethionine (SAM, 2 μCi) in methylation buffer (20 mM Tris [pH 8.0], 200mM NaCl, 0.4 mM EDTA). Reactions were incubated for 90 min at 30 °C and stopped with SDS sample buffer. SDS-PAGE gels were fixed with 10% acetic acid/10% methanol v/v, washed, and incubated with Amplify reagent (Amersham) before exposing at −80 °C.
Complementation of S. cerevisiae YHR122W deletion mutant
A cDNA encoding YHR122W was purchased as a full-length expressed sequence tag (Open Biosystems). The construct for subcloning into the yeast epitope tagging vector pESC-Leu (Stratagene) was generated by polymerase chain reaction (PCR) from the corresponding cDNA using the following primers:
Sense primer, 5′- GAAGCGGCCGCAATGTCTGAGTTTTTGAATGA-3′
Antisense primer, 5′- CCGACTAGTGCCTTACAAGTCACTAACATCTTAG-3′
The PCR product was digested with NotI-SpeI and subcloned into a NotI-SpeI digested pESC-Leu vector and sequenced. The YHR122W C161A mutant was generated using the Quickchange procedure (Stratagene). The mutant cDNA was sequenced and found to contain only the desired mutation.
Constructs containing wild-type and C161A mutant YHR122W were introduced into the yeast Tet promoter Hughes (yTHC) strain YSC1180-7428770 (Open Biosystems) using the reagents provided in the YeastmakerTM Yeast Transformation System 2 (Clontech). The yeast was grown in Synthetic Dextrose Minimal Medium (−Leu) and spot assays were performed in either Synthetic Dextrose Minimal Medium (−Leu) or Synthetic Galactose Minimal Medium (−Leu) + Agar plates +/− 50 μg/ml Doxycycline. The plates were cultured at 30 °C for three days.
Isopropylmalate isomerase (Leu1) assay
Yeast strains harboring either an empty vector or wild-type YHR122W (see section above) were cultured in Synthetic Dextrose Minimal Medium (−Leu) to OD600 of 1.0 and transferred into Synthetic Galactose Minimal Medium (−Leu) +/− 50 μg/ml Doxycycline for 12 hours. Yeast were lysed and Leu1 semi-purified by ammonium sulfate precipitation (40-70%). The activity assays were performed using DL-threo-3-isopropylmalic acid as the substrate and product formation was measured by monitoring absorbance at 235 nm for 10 minutes31.
Alcohol dehydrogenase (ADH) assay
Yeast cell lysates in 0.1M sodium pyrophosphate buffer (pH 9.2, 1.5 mL) were treated with 2 M Ethanol (0.5 mL) and 0.025 M NAD (1.0 mL) and ADH activity was measured by absorbance increase at 340 nm for 3 minutes47.
De novo designs of cysteine hydrolases and hydrolysis activity assays
We used the Rosetta computational enzyme design methodology48 to search a set of protein scaffolds for constellations of backbones capable of supporting an idealized transition state for ester hydrolysis derived from the geometries and mechanisms of natural cysteine hydrolases49. The idealized active site models feature a nucleophilic cysteine, a general base/acid histidine and at least one sidechain or backbone hydrogen bond donor as the oxyanion hole. The sequence of residues surrounding the putative active sites was optimized using the Rosetta design algorithm to maximize transition state stabilization50. A set of 12 designed proteins in 10 distinct scaffolds was chosen for experimental characterization. For each designed protein, synthetic genes were obtained and protein expression and purification was performed in E. coli as previously described50. Activity was measured with the substrate by following the initial (< 5% substrate conversion) increase in fluorescence due to the appearance of the product coumarin. A protein concentration of 20 μM and substrate concentration of 100 μM were used in 25 mM HEPES buffer, 150 mM NaCl, 1mM TCEP, pH 7.5. The background rate was measured under identical conditions but without the protein. Kunkel mutagenesis was used for creating point mutations in the active site residues. A detailed description of the design and characterization of the cysteine hydrolases will be presented elsewhere. Amino acid sequences of the 12 designs can be found in Supplementary Information.
In-gel fluorescence and isoTOP-ABPP characterization of designed proteins
For in-gel fluorescence studies, E.coli lysates overexpressing the designed proteins were diluted to 2 mg protein/mL in PBS. Each sample (25 μL) was mixed with 25μL of MCF7 human cell soluble proteome (2 mg/mL) and was labeled with 100 nM of the IA-probe (5 μM stock in DMSO) and the reactions incubated for 1 hour at room temperature. Click chemistry, SDS-PAGE separation and in-gel fluorescence visualization were performed as described in previous sections.
For isoTOP-ABPP studies, 10 uL of each of the E.coli lysates (2 mg protein/mL) overexpressing the designed constructs were mixed together and the total volume was brought to 1 mL by addition of 2 mg/mL of MCF7 soluble proteome. Time-dependent and concentration-dependent labeling with the IA-probe, click-chemistry, on-bead trypsin and TEV digestions, LC-MS runs, and MS data analysis were performed as described in previous sections.
Supplementary Material
Acknowledgements
We would like to thank Tamas Bartfai, Ian Wilson and members of the Cravatt Lab for comments and critical reading of the manuscript, Tianyang Ji for experimental assistance and Jasmine Gallaher for expression of designed proteins. This work was supported by the National Institutes of Health (CA087660, MH084512), a Pfizer Postdoctoral Fellowship (E.W.), a Koshland Graduate Fellowship in Enzyme Biochemistry (G.M.S.), a National Science Foundation predoctoral fellowship (D.A.B.) and the Skaggs Institute for Chemical Biology.
Footnotes
Supplementary Information includes Supplementary Discussion, Supplementary Methods, Supplementary Figs. 1-18, Supplementary Tables 1-9 and References.
Author Contributions B.F.C., E.W., and C.W. conceived the project and E.W., and C.W. performed mass spectrometry experiments and yeast growth/Leu1 assays. C.W., and G.M.S. performed computational data analyses. S.K, F.R. and D.B. performed computational design of cysteine hydrolases and measured activity using a fluorogenic assay. D.A.B. purified PRMT1 and M.B.D.D. and K.M performed PRMT1 activity assays. B.F.C., E.W., C.W., and G.M.S. analyzed data and wrote the manuscript.
The authors declare no competing financial interests.
References
- 1.Eisenberg D, Marcotte EM, Xenarios I, Yeates TO. Protein function in the post-genomic era. Nature. 2000;405:823–826. doi: 10.1038/35015694. [DOI] [PubMed] [Google Scholar]
- 2.Zhao Y, Jensen ON. Modification-specific proteomics: Strategies for characterization of post-translational modifications using enrichment techniques. Proteomics. 2009 doi: 10.1002/pmic.200900398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bulaj G, Kortemme T, Goldenberg DP. Ionization reactivity relationships for cysteine thiols in polypeptides. Biochemistry. 1998;37:8965–8972. doi: 10.1021/bi973101r. [DOI] [PubMed] [Google Scholar]
- 4.Giles NM, Giles GI, Jacob C. Multiple roles of cysteine in biocatalysis. Biochem. Biophys. Res. Commun. 2003;300:1–4. doi: 10.1016/s0006-291x(02)02770-5. [DOI] [PubMed] [Google Scholar]
- 5.Reddie KG, Carroll KS. Expanding the functional diversity of proteins through cysteine oxidation. Curr. Opin. Chem. Biol. 2008;12:746–754. doi: 10.1016/j.cbpa.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 6.Voss AA, Lango J, Ernst-Russell M, Morin D, Pessah IN. Identification of hyperreactive cysteines within ryanodine receptor type 1 by mass spectrometry. J Biol Chem. 2004;279:34514–20. doi: 10.1074/jbc.M404290200. [DOI] [PubMed] [Google Scholar]
- 7.Lewis CT, Seyer JM, Carlson GM. Cysteine 288: an essential hyperreactive thiol of cytosolic phosphoenolpyruvate carboxykinase (GTP) J Biol Chem. 1989;264:27–33. [PubMed] [Google Scholar]
- 8.Knowles JR. Intrinsic pKa-values of functional-groups in enzymes - improper deductions from pH-dependence of steady-state parameters. CRC Crit. Rev. Biochem. 1976;4:165–173. doi: 10.3109/10409237609105457. [DOI] [PubMed] [Google Scholar]
- 9.Fomenko DE, Xing W, Adair BM, Thomas DJ, Gladyshev VN. High-throughput identification of catalytic redox-active cysteine residues. Science. 2007;315:387–9. doi: 10.1126/science.1133114. [DOI] [PubMed] [Google Scholar]
- 10.Sethuraman M, et al. Isotope-coded affinity tag (ICAT) approach to redox proteomics: identification and quantitation of oxidant-sensitive cysteine thiols in complex protein mixtures. J Proteome Res. 2004;3:1228–33. doi: 10.1021/pr049887e. [DOI] [PubMed] [Google Scholar]
- 11.Baty JW, Hampton MB, Winterbourn CC. Proteomic detection of hydrogen peroxide-sensitive thiol proteins in Jurkat cells. Biochem J. 2005;389:785–95. doi: 10.1042/BJ20050337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salsbury FR, Jr., Knutson ST, Poole LB, Fetrow JS. Functional site profiling and electrostatic analysis of cysteines modifiable to cysteine sulfenic acid. Protein Sci. 2008;17:299–312. doi: 10.1110/ps.073096508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leonard SE, Reddie KG, Carroll KS. Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS Chem Biol. 2009;4:783–99. doi: 10.1021/cb900105q. [DOI] [PubMed] [Google Scholar]
- 14.Kim J-R, Yoon HW, Kwon K-S, Lee S-R, Rhee SG. Identification of proteins containing cysteine residues that are sensitive to oxidation by hydrogen peroxide at neutral pH. Anal. Biochem. 2000;283:214–221. doi: 10.1006/abio.2000.4623. [DOI] [PubMed] [Google Scholar]
- 15.Speers AE, Cravatt BF. A tandem orthogonal proteolysis strategy for high-content chemical proteomics. J Am Chem Soc. 2005;127:10018–9. doi: 10.1021/ja0532842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weerapana E, Simon GM, Cravatt BF. Disparate proteome reactivity profiles of carbon electrophiles. Nat. Chem. Biol. 2008;4:405–407. doi: 10.1038/nchembio.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dennehy MK, Richards KA, Wernke GR, Shyr Y, Liebler DC. Cytosolic and nuclear protein targets of thiol-reactive electrophiles. Chem Res Toxicol. 2006;19:20–9. doi: 10.1021/tx050312l. [DOI] [PubMed] [Google Scholar]
- 18.Shin N-Y, Liu Q, Stamer SL, Liebler DC. Protein targets of reactive electrophiles in human liver microsomes. Chem. Res. Toxicol. 2007;20:859–867. doi: 10.1021/tx700031r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Speers AE, Adam GC, Cravatt BF. Activity-based protein profiling in vivo using a copper(i)-catalyzed azide-alkyne [3 + 2] cycloaddition. J Am Chem Soc. 2003;125:4686–7. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 20.Weerapana E, Speers AE, Cravatt BF. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)--a general method for mapping sites of probe modification in proteomes. Nat Protoc. 2007;2:1414–25. doi: 10.1038/nprot.2007.194. [DOI] [PubMed] [Google Scholar]
- 21.Shiio Y, Aebersold R. Quantitative proteome analysis using isotope-coded affinity tags and mass spectrometry. Nat Protoc. 2006;1:139–45. doi: 10.1038/nprot.2006.22. [DOI] [PubMed] [Google Scholar]
- 22.Board PG, et al. Identification, Characterization, and Crystal Structure of the Omega Class Glutathione Transferases. J. Biol. Chem. 2000;275:24798–24806. doi: 10.1074/jbc.M001706200. [DOI] [PubMed] [Google Scholar]
- 23.Thompson S, et al. Mechanistic studies on beta-ketoacyl thiolase from Zoogloea ramigera: identification of the active-site nucleophile as Cys89, its mutation to Ser89, and kinetic and thermodynamic characterization of wild-type and mutant enzymes. Biochemistry. 1989;28:5735–5742. doi: 10.1021/bi00440a006. [DOI] [PubMed] [Google Scholar]
- 24.Iyer LM, Koonin EV. Aravind, L. Novel predicted peptidases with a potential role in the ubiquitin signaling pathway. Cell Cycle. 2004;3:1440–1450. doi: 10.4161/cc.3.11.1206. [DOI] [PubMed] [Google Scholar]
- 25.Codreanu SG, Zhang B, Sobecki SM, Billheimer DD, Liebler DC. Global analysis of protein damage by the lipid electrophile 4-hydroxy-2-nonenal. Mol Cell Proteomics. 2009;8:670–80. doi: 10.1074/mcp.M800070-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Cheng X. Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of Its binding to substrate peptides. Structure. 2003;11:509–520. doi: 10.1016/s0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrop SJ, et al. Crystal structure of a soluble form of the intracellular chloride ion channel CLIC1 (NCC27) at 1.4-A resolution. J Biol Chem. 2001;276:44993–5000. doi: 10.1074/jbc.M107804200. [DOI] [PubMed] [Google Scholar]
- 28.Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc. Natl Acad. Sci. USA. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 30.Lill R. Function and biogenesis of iron sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 31.Pierik AJ, Netz DJ, Lill R. Analysis of iron-sulfur protein maturation in eukaryotes. Nat Protoc. 2009;4:753–66. doi: 10.1038/nprot.2009.39. [DOI] [PubMed] [Google Scholar]
- 32.Netz DJA, Pierik AJ, Stumpfig M, Muhlenhoff U, Lill R. The Cfd1-Nbp35 complex acts as a scaffold for iron-sulfur protein assembly in the yeast cytosol. Nat. Chem. Biol. 2007;3:278–286. doi: 10.1038/nchembio872. [DOI] [PubMed] [Google Scholar]
- 33.Okerberg ES, et al. High-resolution functional proteomics by active-site peptide profiling. Proc. Natl Acad. Sci. USA. 2005;102:4996–5001. doi: 10.1073/pnas.0501205102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nazif T, Bogyo M. Global analysis of proteasomal substrate specificity using positional-scanning libraries of covalent inhibitors. Proc Natl Acad Sci U S A. 2001;98:2967–72. doi: 10.1073/pnas.061028898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen G, et al. Reactivity of functional groups on the protein surface: development of epoxide probes for protein labeling. J Am Chem Soc. 2003;125:8130–3. doi: 10.1021/ja034287m. [DOI] [PubMed] [Google Scholar]
- 36.Silverman JA, Harbury PB. Rapid mapping of protein structure, interactions, and ligand binding by misincorporation proton-alkyl exchange. J Biol Chem. 2002;277:30968–75. doi: 10.1074/jbc.M203172200. [DOI] [PubMed] [Google Scholar]
- 37.Isom DG, Vardy E, Oas TG, Hellinga HW. Picomole-scale characterization of protein stability and function by quantitative cysteine reactivity. Proc Natl Acad Sci U S A. 107:4908–13. doi: 10.1073/pnas.0910421107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kortemme T, Creighton TE. Ionisation of cysteine residues at the termini of model [alpha]-helical peptides. Relevance to unusual thiol pKa values in proteins of the thioredoxin family. J. Mol. Biol. 1995;253:799–812. doi: 10.1006/jmbi.1995.0592. [DOI] [PubMed] [Google Scholar]
- 39.Macpherson LJ, et al. Noxious compounds activate TRPA1 ion channels through covalent modification of cysteines. Nature. 2007;445:541–5. doi: 10.1038/nature05544. [DOI] [PubMed] [Google Scholar]
- 40.Eng JK, Mccormack AL, Yates JR. An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 41.Kispal G, Csere P, Prohl C, Lill R. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 1999;18:3981–9. doi: 10.1093/emboj/18.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Meth. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 44.Collins SR, et al. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol. Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 45.Pedrioli PGA, et al. A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotech. 2004;22:1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 46.Park SK, Venable JD, Xu T, Yates JR. A quantitative analysis software tool for mass spectrometry-based proteomics. Nat. Meth. 2008;5:319–322. doi: 10.1038/nmeth.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vallee BL, Hoch FL. Zinc, a Component of Yeast Alcohol Dehydrogenase. Proc Natl Acad Sci U S A. 1955;41:327–38. doi: 10.1073/pnas.41.6.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zanghellini A, et al. New algorithms and an in silico benchmark for computational enzyme design. Protein Sci. 2006;15:2785–94. doi: 10.1110/ps.062353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma S, Devi-Kesavan LS, Gao J. Molecular dynamics simulations of the catalytic pathway of a cysteine protease: a combined QM/MM study of human cathepsin K. J Am Chem Soc. 2007;129:13633–45. doi: 10.1021/ja074222+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang L, et al. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–91. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.