Abstract
S. cerevisiae telomerase binds and preferentially elongates short telomeres, events that require the checkpoint kinase Tel1. We show that the Mre11 complex bound preferentially to short telomeres, which can explain the preferential binding of Tel1 to these ends. Compared to wild type length telomeres, short telomeres generated by incomplete replication had low levels of the telomerase inhibitory protein Rif2. Moreover, in the absence of Rif2, Tel1 bound equally well to short and wild type length telomeres, arguing that low Rif2 content marks short telomeres for preferential elongation. Using congenic strains, a double strand break bound ≥140 times as much Mec1 in the first cell cycle after breakage as did a short telomere in the same time frame. Replication protein A binding was also much lower at short telomeres. The absence of Mec1 at short telomeres can explain why they do not trigger a checkpoint-mediated cell cycle arrest.
Keywords: telomere, telomerase, yeast, checkpoint, double strand breaks, RPA, Cdc13, γH2AX
In the 1930’s when Herman Muller was unable to generate terminally deleted chromosomes in irradiated flies, he reasoned that telomeres, a name he coined for the natural ends of chromosomes, must be essential to maintain stable chromosomes1. Soon after, Barbara McClintock, working with corn, found that broken chromosomes fuse with other breaks while telomeres do not fuse either with each other or with double strand breaks (DSBs). Together these data suggested that telomeres are important to distinguish natural chromosome ends from DSBs. The cell’s ability to distinguish telomeres and DSBs is particularly remarkable given that many proteins involved in sensing and repairing DNA damage also affect telomeres2. Molecular studies show that telomeres in most organisms consist of repeated DNA in which the strand that comprises the 3′ end of the chromosome is G-rich and extended to form a 3′ single-strand tail. In Saccharomyces cerevisiae, the ~300 bp C1–3A/TG1–3 duplex region and the TG1–3 single strand tail are bound respectively by two sequence specific DNA binding proteins, Rap1 and Cdc133.
Another key function of telomeres is to compensate for the incomplete replication of chromosome ends1. Due to the biochemical properties of DNA polymerases, a short gap is left at the 5′ ends of newly replicated strands when the most distal RNA primer is removed. In most eukaryotes, this end replication problem is solved by a telomere dedicated reverse transcriptase called telomerase. However, unlike conventional semi-conservative DNA replication, telomerase does not act on each telomere in every cell cycle. Rather in yeasts and mammals, telomerase preferentially elongates the shortest telomeres in the cell4–6. In yeast, the frequency and extent of elongation, as well as telomerase processivity, are all greater at telomeres shorter than 125 bps5,7.
The chromatin structure of short telomeres has been investigated to determine if it differs from that of wild-type (WT) telomeres in a manner that might explain why telomerase preferentially lengthens the former. Indeed, two telomerase subunits, Est2 and Est1, bind preferentially to short telomeres, as does the Tel1 kinase8–10. Moreover, at a truncated left telomere of chromosome VII, preferential binding of Est2 and Est1 is dependent on Tel1, the yeast equivalent of the ATM checkpoint kinase, and Tel1 binding requires the carboxyl end of Xrs28, a subunit of the heterotrimeric Mre11 complex. Tel1 is also required for processive telomerase action at short (≤125 bps) telomeres7.
Here we show that the MRX complex, comprised of Mre11, Rad50, and Xrs2, bound preferentially to short telomeres, a result that can explain how Tel1 and hence telomerase are targeted to short telomeres. However, this result raises the question of how MRX recognizes short telomeres. Rap1 and its associated proteins, Rif1 and Rif2, two negative regulators of telomerase, are brought to telomeres in a mutually exclusive manner by protein-protein interactions with the carboxyl terminus of Rap13. As Rap1 binding sites are distributed at ~18 bp intervals throughout the yeast telomere11, by definition telomeres lose Rap1 binding sites as they shorten. Thus, differences in Rif1 and/or Rif2 occupancy are an appealing explanation for how cells distinguish short from WT length telomeres.
Here we report that Rif2 (but not Rif1) was less abundant at two natural telomeres shortened from their ends by incomplete replication. In addition, Tel1 no longer bound preferentially to short telomeres in rif2Δ (but not rif1Δ) cells. Thus, Rif1 and Rif2 act by different mechanisms to inhibit telomerase, and the two proteins appear to be distributed differently along the length of the telomere. Likewise, when a DSB is introduced adjacent to a 162 bp tract of telomeric DNA, Rif2 inhibits telomere addition to the break while Rif1 has a much more modest effect, again suggesting that the two proteins act by different mechanisms12. Moreover, while the major checkpoint kinase Mec1 bound robustly to an induced DSB, Mec1 binding was ≥140 times lower at a short telomere in a tel1Δ strain where Mec1 is required for telomerase activity13–14. Replication protein A (RPA), a sequence non-specific, single strand DNA binding protein with essential roles in DNA replication and repair, showed a pattern similar to that of Mec1: high binding to DSBs and low binding to telomeres. Taken together, our data not only provide a molecular explanation for how short telomeres are targeted for preferential elongation, they also suggest a mechanism for how cells distinguish telomeres from DSBs.
Results
Mre11 complex binds preferentially to short VII-L telomeres
In previous work, we found that preferential binding of telomerase to a short chromosome VII-L telomere lacking subtelomeric repeats requires the Tel1 kinase, which itself binds preferentially to this telomere8. Since Tel1 binding to DSBs15–16 and short telomeres8 requires the carboxyl end of Xrs2, we asked if the MRX complex also binds preferentially to a short telomere. Each of the three MRX subunits, Mre11, Rad50, and Xrs2, was tagged at its carboxyl terminus with 13 MYC epitopes and expressed from its own promoter at its endogenous chromosomal locus.
To examine MRX binding to short versus WT length telomeres, we used a strain with an inducible short telomere4 (Fig. 1a). In the experimental version of this strain (Fig.1a), the left telomere on chromosome VII contains two recognition sites for the site-specific FLP recombinase (FRT sites). FLP is expressed under the control of a galactose inducible promoter. FLP-mediated recombination between the two FRT sites excises a sub-telomeric fragment that reduces the size of the VII-L telomere to only ~100 bps, compared to ~300 bps of telomeric repeats on all other telomeres (Fig. 1a; Supplementary Fig. 1). As a control, we used an otherwise isogenic strain that also has two FRT sites in the sub-telomeric region of the VII-L chromosome (Fig. 1a), but in which FLP action does not affect the length of the VII-L telomere4 (Supplementary Fig. 1, left).
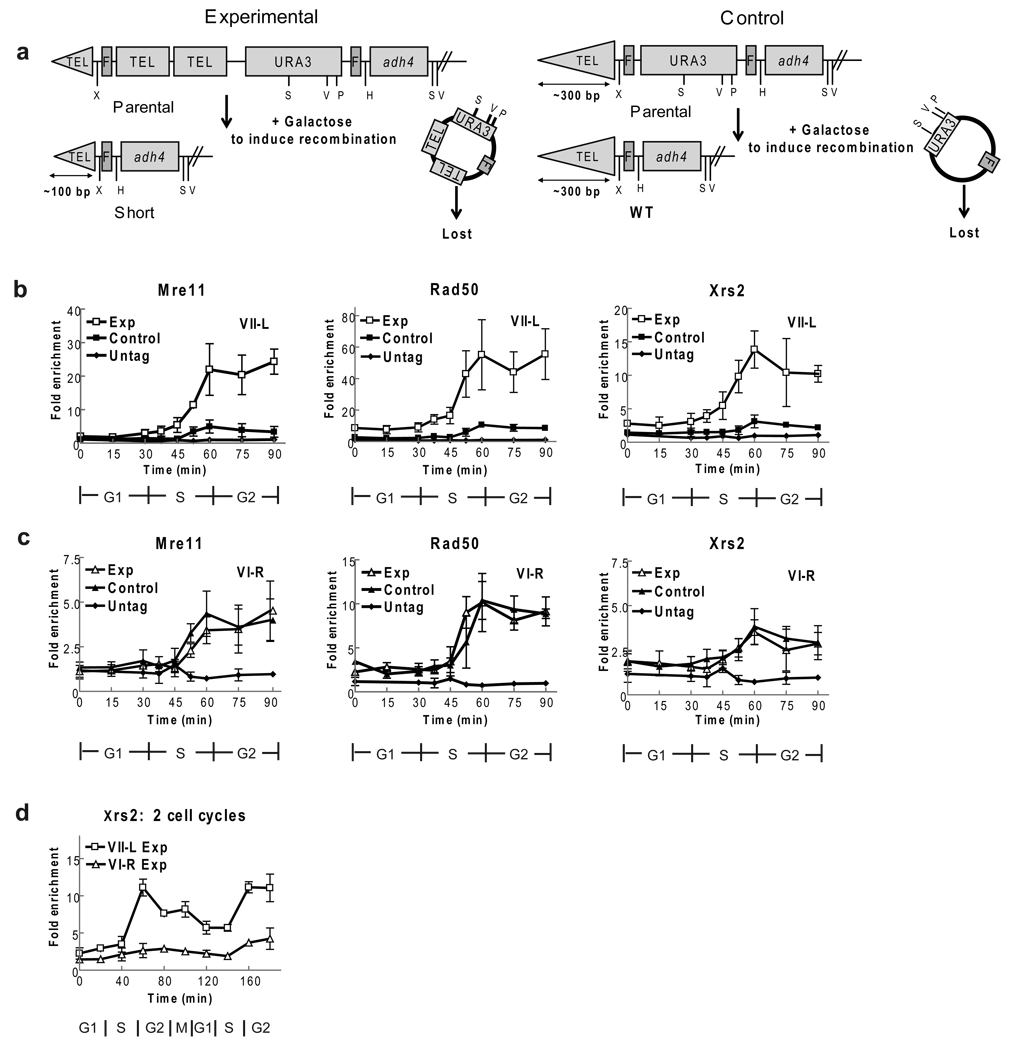
Figure 1. MRX binds preferentially to short telomeres.
(a) Schematic short telomere assay: structures of VII-L end before (parental) and after FLP recombination in experimental (left) and control (right) strains. Recombination generates VII-L telomere with ~100 bp telomeric DNA in experimental strain (short) or ~300 bps (WT) telomere in control strain. Restriction enzyme sites: XhoI (X), StuI (S), EcoRV (V), PstI (P), HindIII (H). (b) Experimental and control strains expressing Mre11-MYC, Rad50-MYC, or Xrs2-MYC were arrested, FLP was induced, and cells were removed from both alpha factor and galactose (0 min), and proceeded through synchronous cell cycle. Filled and open squares indicate, respectively, average fold enrichment of tagged protein ± one standard deviation to short (open) or WT (closed) VII-L telomere compared to binding to non-telomeric ARO1 locus in same sample. Fold enrichment scale is not the same on three panels. (c) Using same samples as in panel b, binding of indicated protein to WT length VI-R telomere was determined in experimental (open triangles) and control (closed triangles) strains. Scale for fold enrichment is different from other panels. Binding of each protein to VI-R telomeres in two strains was not significantly different at any time point (p-values all ≥ 0.12). (d) Xrs2 binds preferentially to short telomeres for at least two cell cycles. Experimental strain expressing Xrs2-MYC was synchronized. Samples were taken over two cell cycles and processed for ChIP. Average levels Xrs2-MYC binding to short VII-L (open squares) and WT length VI-R (open triangles) telomeres are shown.
The experimental and control strains, which expressed the same epitope tagged protein, were arrested in parallel in late G1 phase by incubation with the yeast pheromone alpha factor. Galactose was added to the G1 arrested cells to induce FLP, and the extent of recombination was assessed by Southern analysis (Supplementary Fig. 1). After recombination, cells were released from G1 arrest and followed through the subsequent synchronous cell cycle8. Samples were taken at regular intervals and processed for chromatin immuno-precipitation (ChIP) to determine the association of the epitope tagged protein with the VII-L telomeres and fluorescent activated cell sorting (FACS) to assess cell cycle position. As an additional control, we examined association of epitope tagged proteins with the chromosome VI-R telomere, which is of WT length in both the experimental and control strains.
The profiles of telomere binding were very similar for each of the MRX subunits (Fig. 1b,c). At the WT length VII-L telomere in the control strain, binding was low throughout the cell cycle, although there was significant telomere binding for each subunit at the 60 min time point, coincident with the time of telomere replication (Fig. 1b, filled squares; see Figure legends for p-values). Similarly low but significant binding of each MRX subunit was detected at the WT VI-R telomere in both the experimental (open triangles) and control (closed triangles) strains (Fig. 1c). However, each MRX subunit showed robust binding to the short VII-L telomere (Fig. 1b, open squares). This binding was significant even in S phase but increased dramatically as cells neared the end of the cell cycle. Binding of each MRX subunit was four to six times higher to the short VII-L telomere than to the WT length VII-L telomere in the control strain or to the VI-R telomeres in either the control or experimental strain. Xrs2-MYC binding remains high for at least two cell cycles after telomere shortening (Fig. 1d). Since the carboxyl end of Xrs2 is required for Tel1 telomere binding8, the preferential binding of Xrs2 to the short VII-L telomere can explain the preferential binding of Tel1 to these ends.
Mec1 does not bind short telomeres even in tel1Δ cells
Telomeres can be maintained by telomerase in tel1Δ cells as long as MEC1 is present17. To determine if Mec1, like Tel1, binds preferentially to short telomeres, we introduced three HA epitopes into an internal region of the protein (Mec1-HA18). As with the MRX experiment (Fig. 1), cells were arrested with alpha factor, and Mec1 association with telomeres was determined throughout the cell cycle in both the control and experimental strains (Fig. 2).
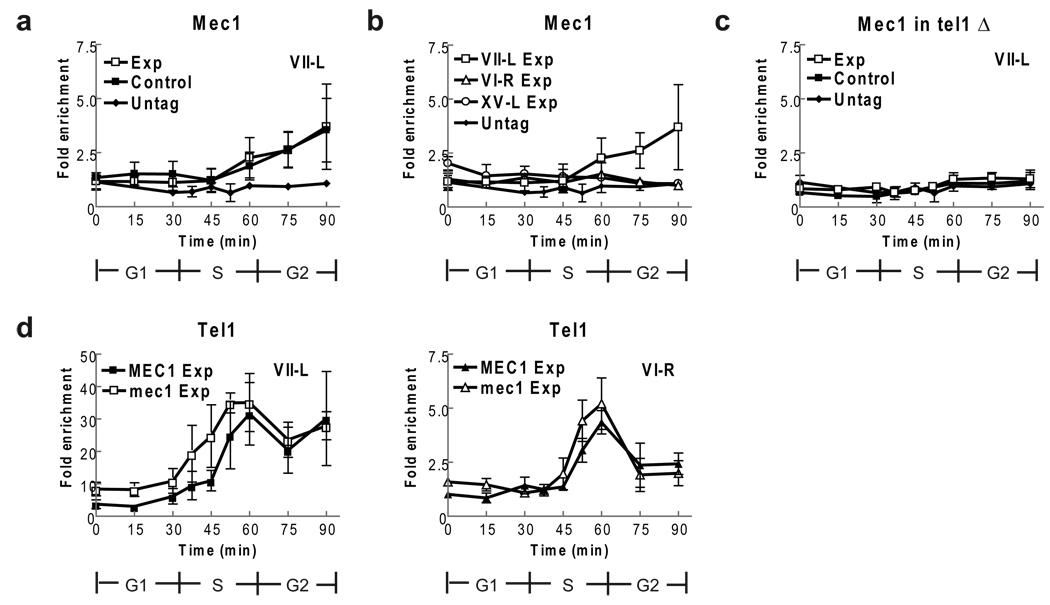
Figure 2. Mec1-HA does not bind preferentially to short telomeres, even in tel1Δ cells.
The control and experimental strains expressing Mec1-HA (a–c) or Tel1-HA (d) were synchronized and processed as described (Fig. 1). (a) Mec1-HA binding to the VII-L telomeres occurred at low levels at 60, 75 and 90 mins. The timing and level of Mec1-HA binding to the short VII telomere (open squares) and the WT length VII-L telomere (closed squares) were indistinguishable at all time points (p-values from 0.19 to 0.96). (b) Mec1-HA binding to the WT length VI-R and XV-L telomeres was indistinguishable from binding to the short VII-L telomere (taken from panel a and shown for comparison) except at 75 and 90 min (p= 0.01 and 0.03). Only data from the experimental strain are shown. (c) The experiment in panel a was carried out in tel1Δ versions of the control and experimental strains expressing Mec1-HA. Binding to the short and WT length VII-L telomeres was indistinguishable from binding to the non-telomeric ARO1 locus in the same samples (to which the values are normalized). (d) Experiments to determine if Tel1-HA binding is affected by absence of Mec1 were done in sml1Δ derivative of the WT and mec1Δ strains53. Tel1-HA binding to the short VII-L telomere or to the WT VI-R telomeres (panel d) was indistinguishable in sml1Δ (MEC1) and mec1Δ sml1Δ (mec1) cells (p-values from 30 to 90 min were 0.08 to 0.84).
Mec1-HA binding to the short (open squares) and WT (closed squares) length VII-L telomeres was indistinguishable (Fig. 2a). Binding to both VII-L telomeres was at background levels early in the cell cycle and through much of S phase. As cells completed S phase, Mec1-HA binding increased modestly until at the end of the cell cycle, it was four-fold higher than at the non-telomeric ARO1 control sequence. Thus, although Mec1-HA binds to both the control and experimental VII-L telomeres, it does not bind preferentially to short telomeres.
We also examined Mec1 binding to two WT length telomeres, VI-R (open triangles) and XV-L (open circles) (Fig. 2b). Since the pattern and extent of Mec1-HA binding to the two telomeres was indistinguishable in the two strains, only the data for the experimental strain are shown. This binding was indistinguishable from Mec1-HA binding to the non-telomeric ARO1 locus to which the values are normalized. Although Mec1-HA binding to the short VII-L telomere was modestly higher than to the VI-R or XV-L telomeres, this difference was significant only at the 75 min (p= 0.01) and 90 min (p= 0.03) time points. Thus, significant Mec1-HA binding was detected only at telomeres that were acted upon by the FLP recombinase, and even this binding was significant only at two time points.
We also examined Mec1-HA binding to the short and WT length VII-L telomeres in tel1Δ cells where Mec1 is essential for telomere maintenance (Fig. 2c). Mec1-HA binding was not detectable at either the control or experimental VII-L telomere in tel1Δ cells (Fig. 2c). These data suggest that the low Mec1-HA binding to the FLP-generated short and WT length VII-L telomeres was a Tel1-mediated response and likely due to FLP-mediated DSB breakage at individual FRT sites in the absence of synapsis19. Finally, in the experimental strain, Tel1 binding to the short VII-L or WT VI-R telomere was not significantly higher in the absence of Mec1 (Fig. 2d).
Mec1 binding is much higher at DSBs than at short telomeres
To serve as a positive control for detecting Mec1 at telomeres (Fig. 2a), we determined levels of Mec1 binding at an induced DSB, which is known to bind Mec120–23. For these experiments, we used strains that were isogenic to the short telomere strain except they contained a galactose-inducible HO endonuclease instead of a galactose-inducible FLP recombinase and a recognition site for the HO endonuclease ~13 kb from the VII-L telomere and no chromosomal FLP sites24. Two different DSB strains were used (Fig. 3a): TG80-HO contained 80 bp of TG1–3 telomeric DNA adjacent to the HO site, while N80-HO had 80 bp of unrelated DNA adjacent to the HO site24.
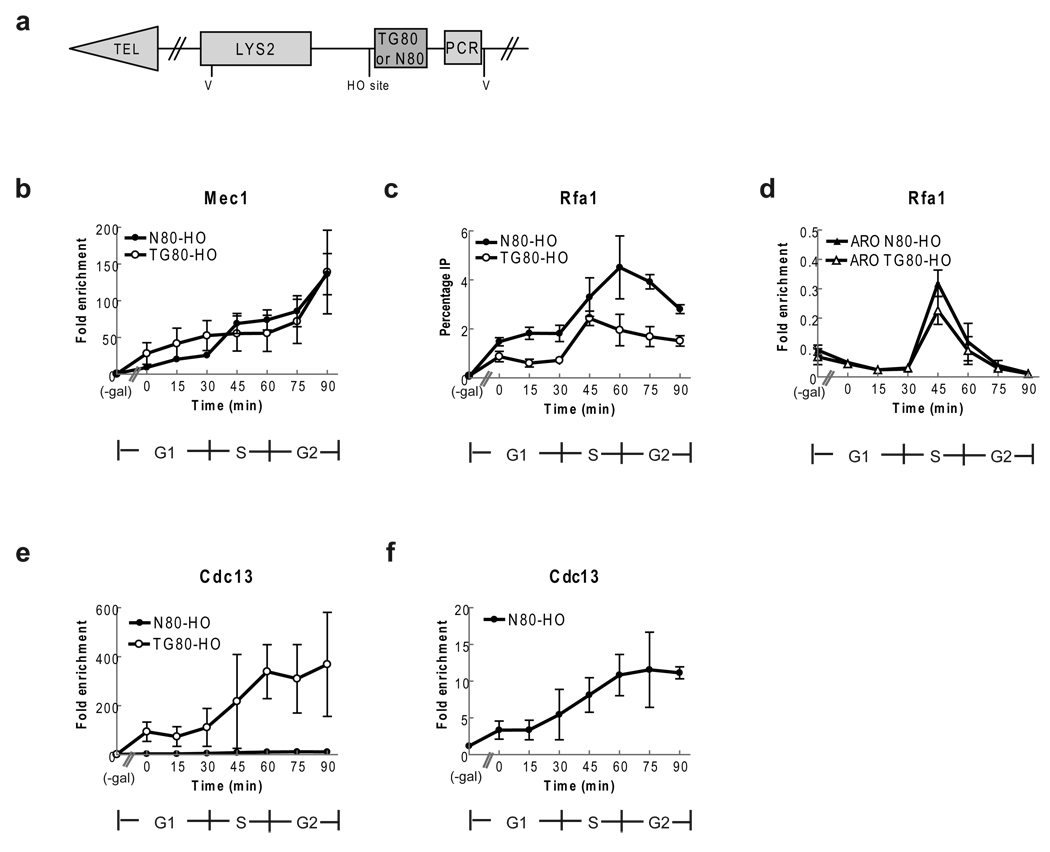
Figure 3. Mec1 and Rfa1 bind DSBs, even when the break is adjacent to telomeric DNA.
(a) Schematic of chromosome VII-L end in DSB strains24. TG80-HO contains 80 bp TG1–3 on centromere side of HO site; N80-HO contains 80 bp lambda DNA. V (EcoRV) sites (b) Mec1-HA binding to DSBs. For b, c, cells were synchronized and processed as described in (Fig. 1) except an extra sample was taken from G1 arrested cells before addition of galactose. Mec1 binding was determined at N80-HO (closed circles) and TG80-HO (open circles) before and after HO induction. Results are average fold enrichment ± one standard deviation over binding to control site ARO1. (c) Rfa1-MYC binding to DSBs. Rfa1-MYC binding was determined at N80-HO (closed circles) and TG80-HO (open circles) before and after induction of HO. Results are average percent of DNA in input. Rfa1-MYC binding to N80-HO was higher than at TG80-HO break (p-values 0.00037 to 0.017), in all post-galactose samples except the 45 min time point (p= 0.093). (d) Samples used in panel c were examined for Rfa1-MYC binding to internal ARO locus in both TG80 (open triangles) and N80 (closed triangles) strains. The level and timing of Rfa1-MYC binding was equivalent in the two strains. (e) Cdc13-MYC binding to TG80 (open circles) and N80 (closed circles) in first cell cycle after breakage. (f) The same samples in panel e for the N80 strain are shown with expanded scale.
Cells expressed the same Mec1-HA construct and were synchronized in the same manner as in the short telomere experiments with HO expression induced in G1 arrested cells and turned off before cells were allowed to progress through the cell cycle (Fig. 2; Supplementary Fig. 2). As expected, Mec1-HA did not associate with the HO site in either strain in the absence of HO expression (Fig. 3b, (-gal) time point). However, in both strains, Mec1-HA associated strongly and to a similar extent and duration with the HO induced breaks: binding was 20-fold over background in late G1 phase, increased to 80-fold at late S/G2 phase, and peaked at 140-fold by the end of the cell cycle. Thus, Mec1-HA binding was similarly high whether the DSB was adjacent to telomeric or non-telomeric DNA, and binding was much higher than binding to short telomeres (compare Fig. 2a to Fig. 3b).
We also examined Cdc13 binding to the induced TG80 and N80 DSBs (Fig. 3). Cdc13 binding was not detectable before HO cleavage in either strains (-gal). In the TG80 break, Cdc13 binding was detectable even in G1 phase (0 min time point) and increased as cells progressed through the cell cycle, peaking at ~400 fold compared to binding to the ARO control sequence late in the cell cycle (Fig. 3e, open circles). This level of Cdc13 binding is very similar to the level of Cdc13 binding detected at both short and wild type length telomeres in late S/G2 phase in a congenic strain that has been synchronized and analyzed in the same manner8. In contrast, Cdc13 binding to the N80 DSB was much lower, peaking at about 12 fold higher than the control (Fig. 3e,f closed circles; 3f scale is expanded compared to e). Using asynchronous cultures, Cdc13 binding to the TG80 DSB12,25 and to a non-telomeric, non-repairable DSB has been reported previously26.
RPA and γH2AX do not bind preferentially to short telomeres
RPA, a heterotrimeric complex that binds in a sequence non-specific manner to single strand DNA, is essential for DNA replication, repair, and recombination27–28. Because RPA is recruited to DSBs29–31 and is suggested to have a role in telomerase recruitment32, we examined its association with DSBs in the induced HO break system and with telomeres in the short telomere assay using synchronized cells and the same epitope tagged version of its largest subunit, Rfa1-MYC, in both experiments.
The signal in our telomere ChIPs is usually normalized to the amount of non-telomeric ARO1 DNA in the same immuno-precipitate (IP). However, Rfa1-MYC should bind to every nuclear DNA sequence during its time of replication, including ARO1. As ARO1 and telomeres replicate at different times in the S phase, the data for Rfa1-MYC binding are presented as percentage IP in both the induced DSB (Fig. 3) and short telomere experiments (Fig. 4).
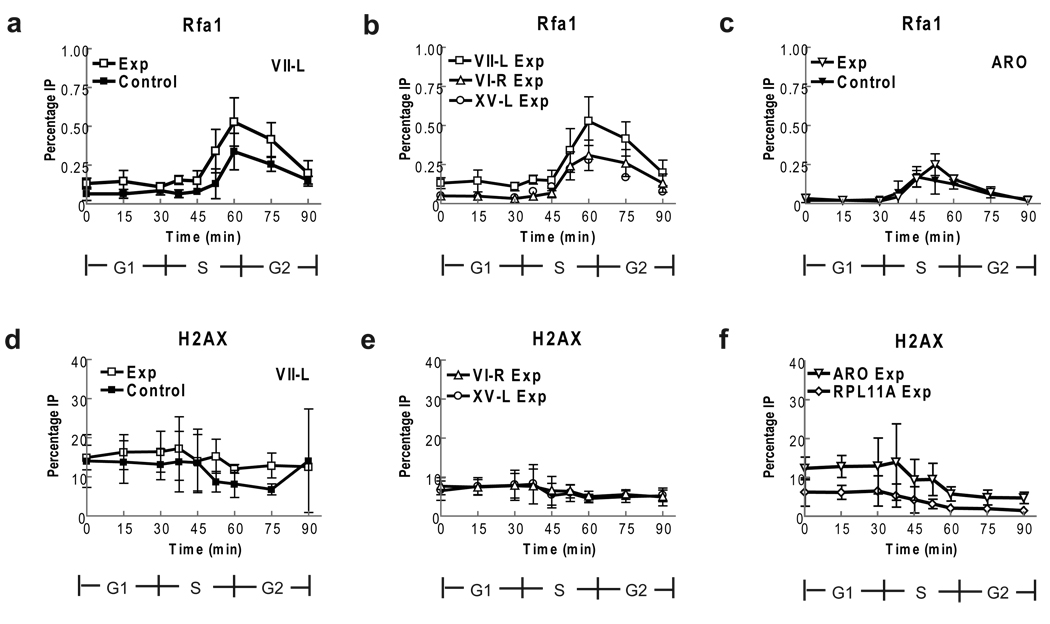
Figure 4. Rfa1 binding and H2A phosphorylation are similar at short and WT telomeres.
Rfa1-MYC cells were treated as in (Fig. 1) or immuno-precipitated with anti-γ H2Aserum (Panels d–f). Data are average percent immuno-precipitated DNA ± one standard deviation. (a) Rfa1-MYC binding to short and WT length VII-L telomeres. Average enrichment at short VII-L was modestly higher than for WT VII-L but difference was only significant at 37.5 min (p= 0.02; p-values for other time points were 0.08 to 0.42). Rfa1-MYC binding occurred at time of telomere replication (60–75 min). (b) Rfa1-MYC binding to short VII-L telomere (open squares; data are from panel a), WT length VI-R (open triangles) and XV-L (open circles) telomeres in experimental strain was indistinguishable except at early points (p-values for 45–90 min 0.03 to 0.45). Data from control strain are also indistinguishable (p-values 0.17 to 0.98) from binding in experimental strain. (c) Rfa1-MYC binding to ARO1 locus is same in experimental (open triangles) and control (closed triangles) strains (p-values 0.19 to 0.95). (d) H2A phosphorylation was similar at short (open squares) and WT length (closed squares) VII-L telomeres except at 75 min (p= 0.04). (e) γ-H2AX levels at VI-R and XV-L telomeres are constant throughout cell cycle. Binding to telomeres is shown only for the experimental strain but values for both telomeres were indistinguishable in the control versus experimental strains (p= 0.08 to 0.98). (f) γH2A phosphorylation at RPL11A (diamonds) and ARO1 (triangles) in experimental strain.
In both the N80-HO (closed circles) and TG80-HO strains (open circles), Rfa1-MYC binding was not detected at the HO recognition site before HO expression (Fig. 3c). However, Rfa1-MYC was DSB associated throughout the cell cycle with low binding in G1 phase that increased as cells progressed through the cell cycle. Although robust Rfa1-MYC binding was seen at both DSBs, at most time points, Rfa1-MYC binding was about two times higher at the N80-HO break than at the TG80-HO break, which is adjacent to telomeric sequence. In contrast, Rfa1-MYC binding to the internal ARO locus was low throughout the cell cycle except at the 45 min time point when binding was similarly high in the two DSB strains but much lower than to the DSB in the same cells (Fig. 3d).
Using the induced telomere assay, we examined the association of Rfa1-MYC with three telomeres, VII-L (Fig. 4a), VI-R (Fig. 4b, open triangles) and XV-L (Fig. 4b, open circles) in both the experimental and control strains. Because the data for the VI-R and XV-L telomeres were identical in the two strains (data not shown), only the data from the experimental strain are shown for these telomeres. In both strains, Rfa1-MYC binding to the three WT length and the short VII-L telomeres occurred during a discrete interval in late S phase, peaking at 60 min, consistent with the expected time of telomere replication (Fig. 4a,b). Rfa1-MYC binding to the ARO1 locus also occurred during a limited but earlier period in S phase (Fig. 4c), and its timing and extent of binding was similar to what was seen in the DSB strains (Fig. 3d). The level of Rfa1-MYC binding to the three WT length telomeres and to the ARO1 locus was similar, ranging from 0.25% (ARO1) to 0.37% (telomere VI-R). The level of binding to the short VII-L telomere was modestly higher late in the cell cycle compared to WT VII-L, but this difference was not significant. Moreover, Rfa1-MYC binding to telomeres was ~5–9 times lower at telomeres than at DSBs (compare Fig. 4a to 3c; maximal binding to telomeres was 0.5% while binding to the N80-HO and the TG80-HO breaks was, respectively, 4.5% and 2.3%). Thus, preferential binding of RPA to short telomeres is unlikely to mark them for preferential lengthening by telomerase as suggested by earlier work32.
An early response to DNA damage is the replacement of canonical H2A by an H2A variant called H2AX, which is then phosphorylated (referred to as γ-H2AX)33. Since the sole version of H2A in yeast is analogous to the H2AX of other eukaryotes, yeast H2A is phosphorylated rather than replaced upon DNA damage34. Because telomeres have high γ-H2AX levels35–36, we asked if this modification marks short telomeres for telomerase elongation. Using the inducible short telomere assay, we determined levels of γ-H2AX at the short and control VII-L telomeres, at two native WT length telomeres (VI-R and XV-L), and at two non-telomeric loci (ARO1 and RPL11A) in both the control and experimental strains (Fig. 4d – f). As with the Rfa1-MYC data (Fig. 4a – c), γ-H2AX values are presented as percent target sequence in the immuno-precipitate (Percentage IP).
For all loci, the level of γ-H2AX was fairly constant from late G1 phase through the end of the cell cycle, with a very modest decline after S phase (Fig. 4d – f). The level of γ-H2AX at the VII-L telomere was not affected by telomere length (Fig. 4d: open squares, short telomere; closed squares, WT length VII-L telomere; the only significant difference is at 75 min; p= 0.04). However, the levels of γ-H2AX at both VII-L telomeres were about twice as high as at either telomere VI-R or XV-L suggesting that FLP action increases γ-H2AX levels (Fig. 4e) (as well as Mec1-HA binding; Fig. 2b). Nonetheless, H2A phosphorylation does not mark short telomeres for preferential elongation by telomerase.
Loss of Rif2, not Rif1, occurs as telomeres shorten
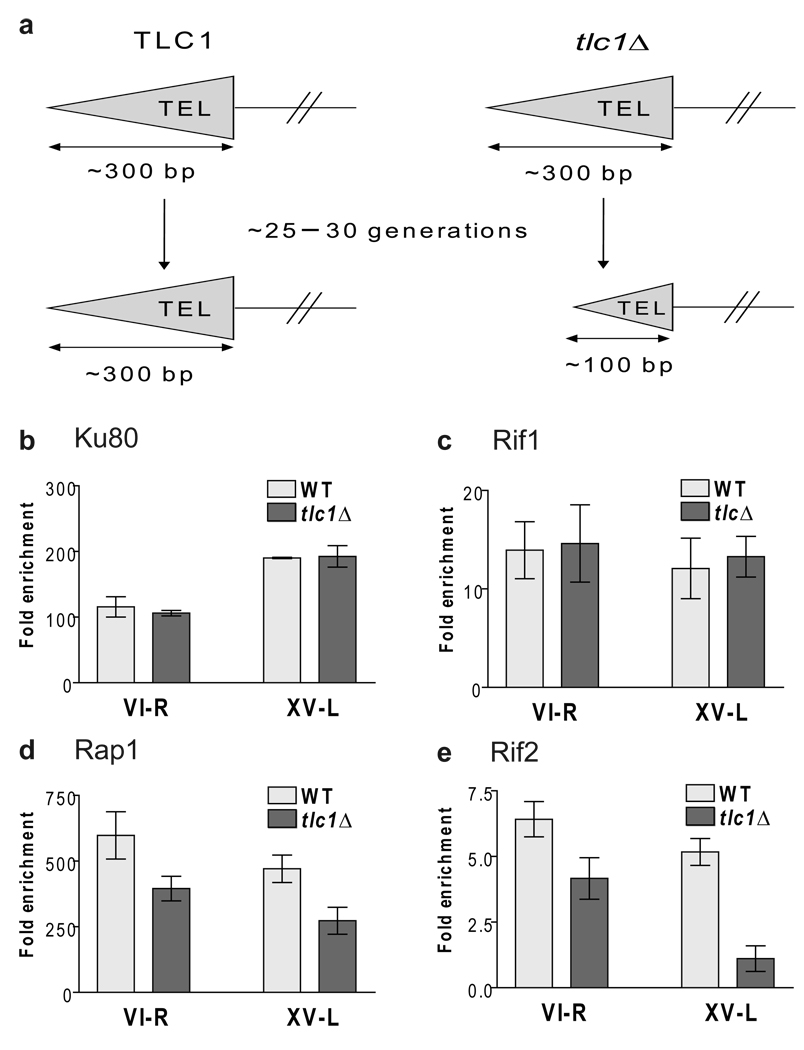
When telomeres are shortened by internal deletion as they are in the inducible short telomere system (Fig. 1a), Rif2 content is lower at short than at WT telomeres, while Rif1 levels are similar at both8. We wished to test the model that depletion of Rif2 is the signal that marks short telomeres for preferential telomerase elongation. However, first it was important to determine if telomeres shortened from their ends by incomplete replication, the normal mechanism of telomere shortening, show the same chromatin composition as telomeres shortened by FLP-mediated internal deletion. We constructed diploids that were homozygous for either Rif1-MYC, Rif2-MYC, or Yku80-MYC and heterozygous for a deletion of TLC1, the telomerase RNA gene (Fig. 5a). Diploids were sporulated, tetrads were dissected and individual tlc1Δ or TLC1 spore clones were grown for a total of ~25–30 generations until the tlc1Δ telomeres had shortened to ~150 bps (Supplementary Fig. 3). The same samples were examined by ChIP to assess levels of Rap1 (using a polyclonal anti-Rap1 serum) or MYC-tagged proteins at both the VI-R and XV-L telomeres (Fig. 5b – e).
Figure 5. Rif2 (and Rap1) but not Rif1 or Yku80 occupancy are reduced at short telomeres.
(a) Schematic of assay. (b–e) Rif2 but not Rif1 content is lower at short versus WT length telomeres. Samples from three (Yku80 and Rap1) or five (Rif1 and Rif2) independent TLC1 or tlc1Δ spore clones were subjected to ChIP after ~25–30 generations of spore outgrowth. Samples expressing MYC-tagged proteins (panels b–e) were immuno-precipitated with an anti-Myc antibody. Samples from an untagged strain were immuno-precipitated with anti-Rap1 antibody (panel d). Purified DNA was analyzed by quantitative PCR to determine the level of protein binding to VI-R and XV-L telomeres. Data are expressed as the average fold enrichment of telomeric sequence over the non-telomeric ARO1 sequence in the same immuno-precipitate. Error bars indicate one standard deviation from average. The differences between binding levels of Yku80 (p= 0.354, VI-R; 0.840, XV-L) and Rif1 (p= 0.731, VI-R; 0.492, XV-L) at WT and short telomeres were not significant. Binding levels of Rap1 (p= 0.026, VI-R; 0.009, XV-L) and Rif2 (p= 1.2×10−3, VI-R; 1.3×10−6, XV-L) were significantly different at WT and short tlc1Δ telomeres.
At both the VI-R and XV-L telomeres, Yku80-MYC (Fig. 5b) and Rif1-MYC (Fig. 5c) levels were indistinguishable at WT length and shortened telomeres (see Fig. 5 legends for p-values). However, Rap1 binding at short telomeres was reduced (66%, telomere VI-R; 58%, XV-L) (Fig. 5d). Rif2 levels were also lower at the short VI-R (65% of WT levels) and XV-L (21% of WT) telomeres (Fig. 5e).
Preferential Tel1 binding to short telomeres requires Rif2
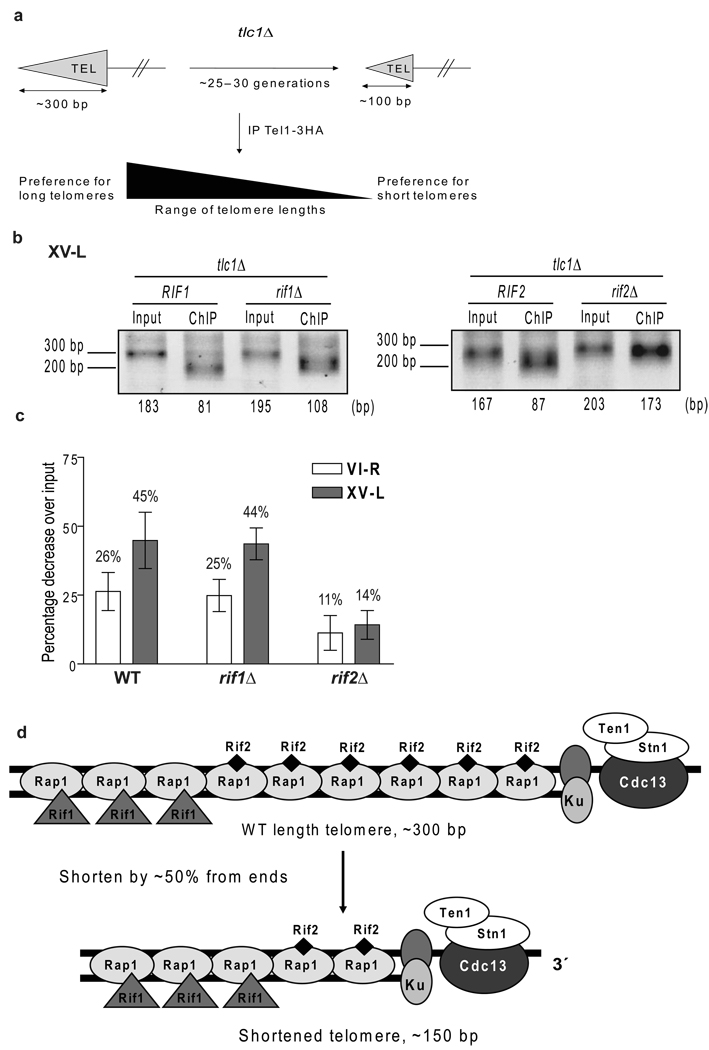
If Rif2 depletion is the signal that marks short telomeres for elongation, the ability of Tel1 to distinguish between short and WT length telomeres might be compromised in rif2Δ but not rif1Δ cells (Fig. 6). To test this possibility, we constructed diploid strains that were heterozygous for a deletion of both TLC1 and for either RIF1 or RIF2. Both haploid parents also expressed Tel1-HA. Diploids were sporulated, tetrads were dissected, and individual tlc1Δ spore clones that were either WT, rif1Δ, or rif2Δ were grown for a total of ~25–30 generations to an average telomere length of ~150 bps (as in Supplementary Fig. 4). After immuno-precipitating the samples with anti-HA to precipitate Tel1-HA associated DNA, both immuno-precipitate and input DNA were subjected to telomere PCR and then gel electrophoresis to determine telomere sizes (see Fig. 6b for representative gels). We examined the lengths of both telomeres VI-R and XV-L in the anti-Tel1 immuno-precipitates. Because the sizes of individual telomeres change in a stochastic manner37, we compared telomere lengths pre and post-immuno-precipitation from a given spore clone. The data are presented as the average percent decrease in length of the Tel1 immuno-precipitate sample compared to that of the input DNA for the individual spore clones examined (Fig. 6c).
Figure 6. Preferential binding of Tel1 to short telomeres is lost in rif2Δ cells.
(a) Schematic of assay. (b–c) After ~30 cell generations, 3–9 independent spores from tlc1Δ RIF1 RIF2, tlc1Δ rif1Δ, and tlc1Δ rif2Δ, all expressing Tel1-HA, were subjected to ChIP with anti-HA antibody. Average length of telomeric DNA in each sample was determined (representative gels for telomere XV-L, panel b). Data are expressed as percent decrease in the average telomere length of the immuno-precipitate compared to length of input. Error bars, one standard deviation from mean. Percent differences correspond to the following average absolute differences in telomere length in input versus Tel1 immuno-precipitated DNA: WT: 38 (VI-R); 76 (XV-L) bps; rif1Δ: 33 bps (VI-R); 78 (XV-L); rif2Δ: 18 (VI-R); 26 (XV-L) bps. (d) Model for Rif1 and Rif2 distribution along telomere. WT length telomere (top) of ~300 bp C1–3A/TG1–3 duplex DNA contains ~17 Rap1 binding sites (not all are shown), and a short single strand TG1–3 tail. Short and WT telomeres contain equal number of heterodimeric Ku complexes and Cdc13-Stn1-Ten1 complexes. For simplicity, we show one Ku and one Cdc13 complex per telomere. Rif1 and Rif2 come to the telomere by interaction with Rap1. As telomeres shorten, they lose Rif2 before Rif1, suggesting that Rif1 is positioned centromere proximal to Rif2. Thus, 300 bp WT and 150 bp short telomere would have ~the same amount of Ku, Cdc13, and Rif1. The shorter telomere has ~50% of Rap1 and Rif2 levels as at WT telomere.
As expected, in WT cells, Tel1 bound preferentially to short telomeres as the average length of telomeres was shorter in the Tel1 immuno-precipitate than in the input sample (26% shorter for telomere VI-R; 45% shorter for telomere XV-L) (Fig. 6c, VI-R, white bar; XV-L, solid bar). Very similar results were obtained in rif1Δ cells where the DNA in the anti-Tel1 immuno-precipitate was 25% (telomere VI-R) and 44% (telomere XV-L) shorter than in the input samples (Fig. 6c, VI-R, white bar; XV-L, solid bar). When the same experiment was done with rif2Δ cells (Fig. 6c, VI-R, white bar; XV-L, solid bar), telomeres in the Tel1-HA immuno-precipitate were still shorter than in the input DNA, but the effect was greatly attenuated (11% shorter for telomere VI-R; 14% shorter for telomere XV-L). Indeed, in rif2Δ cells, the difference in length between the input and immuno-precipitated DNA samples was not significant for either telomere (VI-R, p= 0.377; XV-L, p= 0.218). The average percent difference in length for rif2Δ cells was significantly different from both WT (VI-R, p= 0.002; XV-L, <10−4) and rif1Δ (VI-R, p= 0.008; XV-L, p= 0.0003) at both telomeres while the differences between rif1Δ and WT cells were not significant (VI-R, p= 0.699; XV-L, p= 0.814). These data support a model in which reduced Rif2 content is a signal that marks short telomeres for preferential Tel1 binding and telomerase elongation.
Discussion
A short telomere generated by an internal deletion (Fig. 1a) is lengthened at a faster rate than a WT length telomere for multiple cell cycles after shortening4. Here we show that Mre11, Rad50, and Xrs2 each bound preferentially to these short telomeres (Fig. 1b). In contrast, RPA (Fig. 4a) and γ-H2AX (Fig. 4d) associated equally well with short and WT length telomeres while Mec1 binding was very low at all telomeres (Fig. 2a – c). These results differ from those from another lab that showed robust Mec1 binding and low Tel1 binding to bulk telomeres in late S/G2 phase38–39. In a tel1Δ strain where Mec1 is essential for telomerase action13–14, no Mec1 telomere binding was detected (Fig. 2c). As a positive control, we monitored Mec1 binding to a DSB in an identical experimental situation and found that it was ≥140-fold higher than to a short telomere in the first cell cycle after telomere shortening or break induction (compare Fig. 2c and 3b). Since levels of Cdc13 binding to the TG80 DSB (Fig. 3e) were similar to levels of Cdc13 at short and WT length telomeres8, the ability to detect Mec1 at DSBs but not at telomeres was not due to the DSB being a better substrate for ChIP. In contrast to Mec1, Tel1 binding to short telomeres is extremely robust8. Mec1 must either bind very transiently and/or to only a small subset of telomeres or act by phosphorylating its targets when they are not telomere associated. The fact that telomeres are much shorter in tel1Δ compared to wild type cells indicates that Mec1 is less efficient than Tel1 at promoting telomerase-mediated telomere lengthening, behavior that can be explained by its extremely low telomere binding.
Est2 and Est1 do not bind preferentially to the inducible short VII-L telomere in the absence of Tel1 or in an xrs2-664 mutant that lacks the portion of the protein that interacts with Tel1 at DSBs8. Moreover, short telomeres are not processively lengthened in tel1Δ cells7. Therefore, the preferential binding of the MRX complex to short telomeres (Fig. 1b) is sufficient to explain how Tel1 and hence Est2 and Est1 act preferentially at short telomeres. MRX is not brought to short telomeres by differential Mec1, RPA, or γ-H2aX levels, as these proteins were either absent (Mec1) or equally abundant at short and WT length telomeres (Fig. 2a, Fig. 4a – d).
Since Rif2 was distributed differently on short versus WT length telomeres (Fig. 5; see also Fig. 6d), we asked if Rif2 is important to direct Tel1 to short telomeres by determining the lengths of telomeres in anti-Tel1 immuno-precipitates from WT, rif1Δ and rif2Δ cells (Fig. 6). Telomeres in the anti-Tel1 immuno-precipitates were ~25% (telomere VI-R) or ~45% (telomere XV-L) shorter than bulk telomeres in both WT and rif1Δ cells. Since Tel1 still bound preferentially to short telomeres in rif1Δ cells, Rif1 must inhibit telomerase at a step downstream of Tel1 binding. In contrast, in rif2Δ cells, the sizes of both the VI-R and XV-L telomeres were not significantly different in the anti-Tel1 immuno-precipitate compared to input DNA (Fig. 6c). Thus, in the absence of Rif2, Tel1 was unable to distinguish short from WT length telomeres. These data suggest that differential distribution of Rif2 on short versus WT length telomeres is required to direct MRX, Tel1, and telomerase to short telomeres. In vitro studies reveal that Rif2 (but not Rif1) interacts with the carboxyl terminus of Xrs2, and this interaction can prevent Xrs2 from interacting with Tel112. Thus, as telomeres shorten and lose Rif2, MRX should be more effective at recruiting Tel1.
Rif1 and Rif2 must inhibit telomerase by different mechanisms as Rif1 levels were the same at WT (~300 bps) and short (~150 bps) telomeres. Rif1 has 14 S/TQ sites, which are recognition sites for ATM kinases, while Rif2 has none. Rif1 is phosphorylated on at least one of these sites in vivo40. An appealing model is that telomere-associated Rif1 is phosphorylated by Tel1, and this phosphorylation reduces its inhibition of telomerase. In addition, the loss of Rif2 but not Rif1 as telomeres shorten from ~300 to ~150 bps (Fig. 5) suggests that the two proteins are distributed differently along the yeast telomere with Rif1 positioned centromere proximal to Rif2 (Fig. 6d). Earlier studies are consistent with the idea that Rif1 and Rif2 act by different mechanisms and also suggest that Rif2 is more potent than Rif1 at inhibiting telomerase12,41.
The data in this paper are relevant to an understanding of how cells distinguish telomeres from DSBs. The early events in DSB processing and telomerase-mediated lengthening are remarkably similar. At both, MRX binds and recruits Tel1, and at both, DNA resection occurs via the collaborative and partially overlapping actions of Sae2, Exo1, and Sgs142–44. However, because DSBs occur throughout the genome, the resection-generated 3′ single strand tails at these ends are not sequence specific whereas the ~50–100 base long resection-generated telomeric 3′ tails are exclusively TG1–3 DNA. We confirmed that DSBs are RPA-associated, even when the break was next to an internal tract of telomeric DNA (Fig. 3c). In contrast, at telomeres, single strand tails are Cdc13-associated45. Although RPA was detected at telomeres late in the S phase, this binding was not higher at the short versus control VII-L telomere (Fig. 4a). The most likely explanation for this telomeric RPA is that it occurs during semi-conservative replication of telomeric DNA. Although we cannot exclude the possibility that there is some RPA binding to the resection-generated TG1–3 tails, there was ~8-times more RPA at DSBs than at telomeres (Fig. 3c). Likewise, Mec1 binding was essentially undetectable at short telomeres while it bound robustly to DSBs. The fact that RPA recruits Mec1 to resected DSBs46–47 yet Mec1 was not at telomeres provides further evidence that the low level of RPA seen at telomeres was associated with conventional forked replication intermediates rather than bound to resection-generated TG1–3 tails.
Mec1 binding was similarly high at the TG80-HO and N80-HO DSBs (Fig. 3b), a different result from that in earlier studies that found much lower Mec1 binding when a break is adjacent to an internal tract of telomeric DNA21,25. However, these earlier studies used asynchronous cells and examined Mec1 binding at 1 to 6 hrs after inducing the DSB. In our study, the DSB was introduced in G1 arrested cells, and the nuclease turned off and cells released into the cell cycle only after most of the cells had an HO-induced DSB (Supplementary Fig. 2). Thus, in our experiments, RPA and Mec1 binding to the DSB were examined during the first cell cycle after break formation, before checkpoint-mediated arrest (Fig. 3b). Since the cell cycle arrest is not as lengthy when a DSB is adjacent to telomeric DNA48, the difference between the two experiments is likely explained by the different protocols used to detect Mec1 binding. Although there was high RPA association with the TG80-HO break, the level of RPA association was decreased by about 50% at this break than at N80-HO (Fig. 3c), perhaps because Cdc13 competes with RPA for binding to the TG1–3 tails generated by resection of the TG80-HO break (Fig. 3) or inhibits resection of the DSB (or both).
In mammals49–50 and yeasts, the processes that occur at DSBs and replicating telomeres are similar. Although a single short yeast telomere can trigger at least one step in the DNA damage signaling cascade, Rad53 phosphorylation51, it does not elicit a checkpoint mediated cell cycle arrest as, by several criteria, the cell cycle following its induction is of normal length8. In contrast, a single DSB elicits a strong checkpoint response52. Our data suggest that the lack of cell cycle arrest in response to a short telomere is due to the fact that resected telomeres are coated by Cdc13, not RPA, and hence do not recruit Mec1, whose presence is necessary to trigger a full checkpoint response.
Methods
(Detailed methods are available in Supplementary Information).
Yeast strains are listed in Supplementary Table 1
The inducible short telomere experiments were carried out in bar1Δ∷KAN154 derivatives of the W303 strain Lev2204. Tel1 and Mec1 were internally tagged with three HA epitopes as in18. Mre11, Rad50, Xrs2, and Rfa1 were tagged at their C-termini with thirteen MYC epitopes. Cdc13 was tagged at its C-terminus with nine MYC epitopes45. TEL1 was deleted in the Mec1-HA strain and replaced with HIS3. The DSB experiments were performed in derivatives of W303 strains analogous to those in ref.24 (YAB285 and YAB1083) that contained bar1Δ∷NAT (and sml1Δ∷HIS3 in the Mec1-HA strains). These strains contain a galactose-inducible HO gene at the leu2 locus on chromosome III and a modified chromosome VII-L in which an HO endonuclease recognition site was between the ADH4 and MNT2 loci, as described24. TG80-HO strains contain 80 bp of TG1–3 repeats on the centromere-proximal side of the HO recognition site; in N80-HO strains, this 80 bp sequence is replaced with 80 bp of lambda DNA.
Experiments to determine protein composition of telomeres shortened from their ends used spore clones obtained from sporulation of tlc1Δ∷LEU2/TLC1 diploids that were homozygous for MYC tagged RIF1, RIF2, or KU80, with tagging done as described previously8. tlc1Δ∷LEU2 and TLC1 spores clones carrying the appropriate tagged genes were grown for 25–30 generations in log phase growth before determining telomere length by Southern blot analysis and protein content by ChIP. To determine the effects of RIF genes on Tel1 binding to telomeres of different lengths, diploids homozygous for Tel1-HA and heterozygous for tlc1Δ, rif1Δ and rif2Δ were sporulated and spore clones were grown for a total of ~25–30 generations and processed for ChIP.
Synchrony methods
Galactose induction and cell synchronization in both the induced short telomere and induced DSB strains were done essentially as described8. Samples were taken at least every 15 min and processed for flow cytometry, Southern blot analysis to determine percent recombination or percent DSB formation, and ChIP.
Telomere PCR
Samples were processed for telomere PCR using minor modifications of the methods described in10. The primers used in this study are listed in Supplementary Table 2 The PCR products were resolved on a 3% (w/v) MetaPhor (Lonza) agarose gel using 1 Kb Plus DNA Ladder as a marker (Invitrogen). The lengths of the telomere PCR products were determined by the AlphaImager 3400 Molecular Weight Analysis program. For all experiments, an aliquot of the telomere PCR products were gel-purified, cloned and sequenced to verify that they contained telomeric repeats. Of the 100 sequenced clones, 90% contained telomeric DNA while 10% had no insert.
Chromatin immuno-precipitation (ChIP)
All ChIPs were performed as described8,55–56. Anti-sera were anti-MYC (Clontech monoclonal Ab #631206), anti-HA (Santa Cruz, monoclonal Ab #SC7392X), anti-H2A phosphoS129 (Abcam polyclonal Ab #ab15083) or an affinity purified polyclonal anti-Rap1 serum57. The amount of DNA in ChIP and input samples was quantitated using real-time PCR (BioRad iCycler). In most cases, percentage IP was normalized to the amount of the non-telomeric ARO1 sequence in the immuno-precipitate and input samples. However, for the Rfa1-Myc and the γH2AX experiments, results are presented as percent immuno-precipitate (amount of target sequence in IP/amount in input sample).
Statistical Testing
For all ChIP experiments, samples from each time point were amplified in duplicate or triplicate to obtain an average value for each sample. Each synchrony was repeated at least three times. For determining protein content at telomeres shortened from their ends, ChIPs were carried out on three or more independent spore clones for each genotype. Data are presented as the mean plus or minus one standard deviation, and a two-tailed Student’s t-test was used to determine statistical significance. A p-value ≤ 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank D. Shore and A. Bianchi (University of Geneva) for strains and advice on the DSB assay, K. Runge for advice on telomere PCR, T. Petes (Duke University) for strains, M. Jayaram for discussions about FLP-induced DSBs, and C. Webb and Y. Wu for helpful comments on the manuscript. This work was supported by grants from the National Institutes of Health grant GM43265 (VAZ), postdoctoral fellowships from the NIH (MS), Deutsche Forschungsgemeinschaft (KP) and NJCCR (KP) and predoctoral fellowships from NJCCR (JP, JSM) and the NIH (JSM). JSM did the experiments in Fig. 5 and 6, JAP did the experiments in Fig. 3, AC and MS did experiments in Fig. 1, 2, and 4, and KP helped with analysis of H2A phosphorylation. All authors participated in design and interpretation of experiments and in the preparation of the manuscript.
REFERENCES
- 1.Gall JG. In: Telomeres. Blackburn EH, Greider CW, editors. Cold Spring Harbor Laboratory Press; 1995. pp. 1–10. [Google Scholar]
- 2.Sabourin M, Zakian VA. ATM-like kinases and regulation of telomerase: lessons from yeast and mammals. Trends Cell Biol. 2008;18:337–346. doi: 10.1016/j.tcb.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vega L, Mateyak M, Zakian V. Getting to the end: telomerase access in yeast and humans. Nat Rev Mol Cell Biol. 2003;4:948–959. doi: 10.1038/nrm1256. [DOI] [PubMed] [Google Scholar]
- 4.Marcand S, Brevet V, Gilson E. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 1999;18:3509–3519. doi: 10.1093/emboj/18.12.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 6.Hemann MT, Strong MA, Hao LY, Greider CW. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- 7.Chang M, Arneric M, Lingner J. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 2007;21:2485–2494. doi: 10.1101/gad.1588807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabourin M, Tuzon C, VA Z. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol Cell. 2007;27:550–561. doi: 10.1016/j.molcel.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bianchi A, Shore D. Increased association of telomerase with short telomeres in yeast. Genes Dev. 2007;21:1726–1730. doi: 10.1101/gad.438907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hector R, et al. Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol Cell. 2007;27:851–858. doi: 10.1016/j.molcel.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Gilson E, Roberge M, Giraldo R, Rhodes D, Gasser SM. Distortion of the DNA double helix by RAP1 at silencers and multiple telomeric binding sites. J. Mol. Biol. 1993;231:293–310. doi: 10.1006/jmbi.1993.1283. [DOI] [PubMed] [Google Scholar]
- 12.Hirano Y, Fukunaga K, Sugimoto K. Rif1 and rif2 inhibit localization of tel1 to DNA ends. Mol Cell. 2009;33:312–322. doi: 10.1016/j.molcel.2008.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritchie KB, Mallory JC, Petes TD. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Cell. Biol. 1999;19:6065–6075. doi: 10.1128/mcb.19.9.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arneric M, Lingner J. Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO Rep. 2007;8:1080–1085. doi: 10.1038/sj.embor.7401082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakada D, Hirano Y, Tanaka Y, Sugimoto K. Role of the C terminus of Mec1 checkpoint kinase in its localization to sites of DNA damage. Mol Biol Cell. 2005;16:5227–5235. doi: 10.1091/mbc.E05-05-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakada D, Matsumoto K, Sugimoto K. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 2003;17:1957–1962. doi: 10.1101/gad.1099003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craven RJ, Greenwell PW, Dominska M, Petes TD. Regulation of genome stability by TEL1 and MEC1, yeast homologs of the mammalian ATM and ATR genes. Genetics. 2002;161:493–507. doi: 10.1093/genetics/161.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mallory JC, Petes TD. Protein kinase activity of tel1p and mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proceedings of the National Academy of Science. 2000;97:13749–13754. doi: 10.1073/pnas.250475697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voziyanov Y, Lee J, Whang I, Jayaram M. Analyses of the first chemical step in Flp site-specific recombination: Synapsis may not be a pre-requisite for strand cleavage. Journal of Molecular Biology. 1996;256:720–735. doi: 10.1006/jmbi.1996.0120. [DOI] [PubMed] [Google Scholar]
- 20.Dubrana K, van Attikum H, Hediger F, Gasser SM. The processing of double-strand breaks and binding of single-strand-binding proteins RPA and Rad51 modulate the formation of ATR-kinase foci in yeast. J Cell Sci. 2007;120:4209–4220. doi: 10.1242/jcs.018366. [DOI] [PubMed] [Google Scholar]
- 21.Hirano Y, Sugimoto K. Cdc13 telomere capping decreases Mec1 association but does not affect Tel1 association with DNA ends. Mol Biol Cell. 2007;18:2026–2036. doi: 10.1091/mbc.E06-12-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondo T, Wakayama T, Naiki T, Matsumoto K, Sugimoto K. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science. 2001;294:867–870. doi: 10.1126/science.1063827. [DOI] [PubMed] [Google Scholar]
- 23.Rouse J, Jackson SP. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol Cell. 2002;9:857–869. doi: 10.1016/s1097-2765(02)00507-5. [DOI] [PubMed] [Google Scholar]
- 24.Bianchi A, Negrini S, Shore D. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol. Cell. 2004;16:139–146. doi: 10.1016/j.molcel.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 25.Negrini S, Ribaud V, Bianchi A, Shore D. DNA breaks are masked by multiple Rap1 binding in yeast: implications for telomere capping and telomerase regulation. Genes Dev. 2007;21:292–302. doi: 10.1101/gad.400907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 2009;23:912–927. doi: 10.1101/gad.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 28.Iftode C, Daniely Y, Borowiec JA. Replication protein A (RPA): the eukaryotic SSB. Crit Rev Biochem Mol Biol. 1999;34:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- 29.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 30.Ira G, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Haber JE. Role of Saccharomyces single-stranded DNA-binding protein RPA in the strand invasion step of double-strand break repair. PLoS Biol. 2004;2:E21. doi: 10.1371/journal.pbio.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schramke V, et al. RPA regulates telomerase action by providing Est1p access to chromosome ends. Nat. Genet. 2004;36:46–54. doi: 10.1038/ng1284. [DOI] [PubMed] [Google Scholar]
- 33.Foster ER, Downs JA. Histone H2A phosphorylation in DNA double-strand break repair. FEBS J. 2005;272:3231–3240. doi: 10.1111/j.1742-4658.2005.04741.x. [DOI] [PubMed] [Google Scholar]
- 34.Downs JA, Lowndes NF, Jackson SP. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408:1001–1004. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- 35.Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J Cell Biol. 2007;178:209–218. doi: 10.1083/jcb.200612031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szilard R, et al. Systematic identification of fragile sites via genome-wide location analysis of gamma-H2AX. Nat Struct Mol Biol. 2010;17:299–305. doi: 10.1038/nsmb.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shampay J, Blackburn EH. Generation of telomere-length heterogeneity in Saccharomyces cerevisiae. Proceedings of the National Academy of Science. 1988;85:534–538. doi: 10.1073/pnas.85.2.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takata H, Kanoh Y, Gunge N, Shirahige K, Matsuura A. Reciprocal association of the budding yeast ATM-related proteins Tel1 and Mec1 with telomeres in vivo. Mol Cell. 2004;14:515–522. doi: 10.1016/s1097-2765(04)00262-x. [DOI] [PubMed] [Google Scholar]
- 39.Takata H, Tanaka Y, Matsuura A. Late S phase-specific recruitment of Mre11 complex triggers hierarchical assembly of telomere replication proteins in Saccharomyces cerevisiae. Mol Cell. 2005;17:573–583. doi: 10.1016/j.molcel.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 40.Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci U S A. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levy DL, Blackburn EH. Counting of Rif1p and Rif2p on Saccharomyces cerevisiae telomeres regulates telomere length. Mol Cell Biol. 2004;24:10857–10867. doi: 10.1128/MCB.24.24.10857-10867.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonetti D, Martina M, Clerici M, Lucchini G, Longhese MP. Multiple pathways regulate 3' overhang generation at S. cerevisiae telomeres. Mol Cell. 2009;35:70–81. doi: 10.1016/j.molcel.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 43.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taggart AKP, Teng S-C, Zakian VA. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- 46.Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- 47.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 48.Michelson RJ, Rosenstein S, Weinert T. A telomeric repeat sequence adjacent to a DNA double-stranded break produces an anticheckpoint. Genes Dev. 2005;19:2546–2559. doi: 10.1101/gad.1293805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 50.Verdun RE, Crabbe L, Haggblom C, Karlseder J. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol Cell. 2005;20:551–561. doi: 10.1016/j.molcel.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 51.Viscardi V, Bonetti D, Cartagena-Lirola H, Lucchini G, Longhese MP. MRX-dependent DNA damage response to short telomeres. Mol Biol Cell. 2007;18:3047–3058. doi: 10.1091/mbc.E07-03-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sandell LL, Zakian VA. Loss of a yeast telomere: arrest, recovery and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 53.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 54.Lorenz MC, et al. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene. 1995;158:113–117. doi: 10.1016/0378-1119(95)00144-u. [DOI] [PubMed] [Google Scholar]
- 55.Goudsouzian L, Tuzon C, Zakian VA. S. cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol Cell. 2006;24:603–610. doi: 10.1016/j.molcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 56.Fisher T, Taggart A, Zakian V. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat Struct Mol Biol. 2004;11:1198–1205. doi: 10.1038/nsmb854. [DOI] [PubMed] [Google Scholar]
- 57.Conrad MN, Wright JH, Wolf AJ, Zakian VA. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell. 1990;63:739–750. doi: 10.1016/0092-8674(90)90140-a. [DOI] [PubMed] [Google Scholar]
- 58.Longtine M, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 59.Sabourin M, Tuzon C, Fisher T, Zakian V. A flexible protein linker improves the function of epitope-tagged proteins in Saccharomyces cerevisiae. Yeast. 2007;24:39–45. doi: 10.1002/yea.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.