Abstract
The highly conserved and ubiquitously expressed 14-3-3 proteins regulate differentiation, cell cycle progression and apoptosis by binding intracellular phosphoproteins involved in signal transduction. By screening in vitro translated cDNA pools for the ability to bind 14-3-3, we identified a novel transcriptional co-activator, TAZ (transcriptional co-activator with PDZ-binding motif) as a 14-3-3-binding molecule. TAZ shares homology with Yes-associated protein (YAP), contains a WW domain and functions as a transcriptional co-activator by binding to the PPXY motif present on transcription factors. 14-3-3 binding requires TAZ phosphorylation on a single serine residue, resulting in the inhibition of TAZ transcriptional co-activation through 14-3-3-mediated nuclear export. The C-terminus of TAZ contains a highly conserved PDZ-binding motif that localizes TAZ into discrete nuclear foci and is essential for TAZ-stimulated gene transcription. TAZ uses this same motif to bind the PDZ domain-containing protein NHERF-2, a molecule that tethers plasma membrane ion channels and receptors to cytoskeletal actin. TAZ may link events at the plasma membrane and cytoskeleton to nuclear transcription in a manner that can be regulated by 14-3-3.
Keywords: 14-3-3/co-activator/PEBP2/PY motif/Runx
Introduction
Regulating the subcellular localization of kinases, phosphatases and transcription factors is critical for the organization of cell signaling events. This regulation often involves the formation of multimolecular signaling complexes at particular spatial positions within the cell. In many cases, these protein–protein assemblies are formed through the binding of modular domains on one molecule to short sequence motifs on another. For example, membrane-associated molecules use PDZ domains to bind the C-termini of cytosolic effector proteins and form signal transduction complexes at the plasma membrane (Fanning and Anderson, 1999), while 14-3-3 proteins bind phosphorylated protein ligands to localize them in the cytosol. PDZ domains, originally identified in the post-synaptic density protein PSD95, the Drosophila tumor suppressor protein Dlg1 and the tight junction protein ZO-1, have now been identified in >600 proteins in the non-redundant database, in organisms ranging from bacteria to man (Schultz et al., 2000). Although the vast majority of PDZ domain-containing proteins are membrane associated, a few have been identified that either reside in, or transit to, the nucleus in particular cell types (Poulat et al., 1997; Thomas et al., 1999).
In contrast to PDZ domain proteins, the majority of 14-3-3 proteins and their bound ligands reside within the cell cytoplasm. 14-3-3s are an ancient, ubiquitously expressed, highly conserved family of ∼30 kDa proteins, comprising seven highly homologous members in mammals (denoted β, γ, ε, η, σ, τ and ζ), and 2–12 members in plants, yeast and fungi. By forming homo- and heterodimeric cup-shaped structures, 14-3-3 proteins bind to a large number of signaling molecules to regulate growth, apoptosis and cell cycle progression (Aitken, 1996; Fu et al., 2000). Though the molecular details are still being uncovered, in many cases 14-3-3 proteins function by targeting their bound ligands for cytoplasmic sequestration (Peng et al., 1997; Dalal et al., 1999).
Most proteins bind to 14-3-3 after they have been phosphorylated on serine or threonine residues, although a few proteins have been shown to bind to 14-3-3 via non-phospho motifs (Muslin et al., 1996; Yaffe et al., 1997; Fu et al., 2000). We previously identified two distinct phosphoserine/threonine motifs with consensus sequences, R-(S/Ar)-(+/Ar)-pS-(LEAM)-P and R-X-(Ar/S)-(+/Ar)-pS-(LEAM)-P, respectively, that are present in the majority of 14-3-3-binding proteins and are recognized by all 14-3-3 isoforms (Yaffe et al., 1997; Rittinger et al., 1999). (Amino acids in the motifs are denoted in single-letter amino acid code. Ar denotes the aromatic residues, F, Y, W and H, and pS denotes phosphoserine or phosphothreonine.) Each monomeric subunit of the 14-3-3 dimer possesses a single phosphoserine/threonine-binding pocket lying along opposite edges of a large central channel. Thus each 14-3-3 dimer can accommodate up to two distinct phosphoserine/threonine-containing motifs on either the same or different molecules (Yaffe et al., 1997). Induced conformational changes within the ligand that occur upon 14-3-3 binding are likely to play an important functional role in subcellular localization through exposing or masking nuclear import or export sequences within the bound ligands (Rittinger et al., 1999).
In the present study, we used a small pool screening approach to identify novel 14-3-3-binding proteins. One of the molecules that we identified functions as a transcriptional regulator in mammalian cells. Intriguingly, the transcriptional co-activation function of this protein is critically dependent upon the C-terminal residues, which constitute a PDZ domain-binding motif, and is responsible for localizing the protein into both punctate nuclear foci and plasma membrane-associated complexes. 14-3-3 binding was found to inhibit both the nuclear localization and transcriptional co-activation function. This molecule, which we have named TAZ, appears to be a newly identified evolutionarily conserved paralog of Yes-associated protein (YAP). Competition between PDZ domain-mediated membrane and nuclear targeting along with 14-3-3 binding/cytoplasmic sequestration provides a mechanism for the spatial control of TAZ function.
Results
Identification of TAZ as a 14-3-3-binding protein
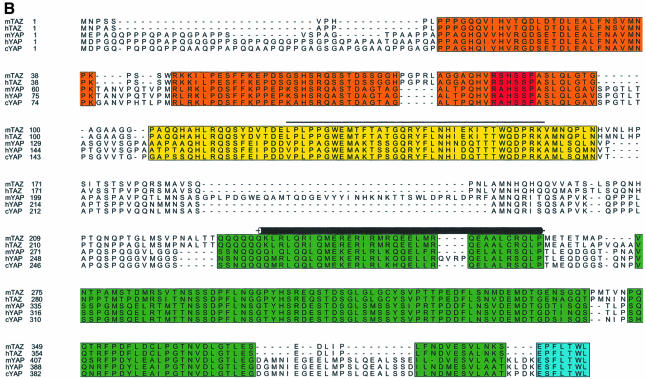
To search for novel 14-3-3-binding proteins, we performed small pool cDNA screening (Lustig et al., 1997). A total of 680 cDNA pools were translated in vitro using rabbit reticulocyte lysates, and the translated proteins (>20 000 total) were assayed for their ability to bind to 14-3-3. Using this approach, five distinct 14-3-3-binding proteins were identified. One of those, a 45 kDa protein in pool #636 (Figure 1A), is described in detail here. The plasmid encoding this protein contained a 1446 bp cDNA with a 1095 bp open reading frame (ORF). A search of the expressed sequence tag (EST) database identified two clones, one from mouse (DDBJ/EMBL/GenBank accession No. AI317016) and one from human (DDBJ/EMBL/GenBank accession No. AI2452255), with homology to the novel cDNA. Examination of these ESTs suggested that our isolated cDNA was partial, coding for the C-terminal 365 amino acids of a 400 amino acid human protein. The mouse EST contained a 1885 bp cDNA with a complete 1185 bp ORF. The initiating ATG codon was within a classic Kozak sequence and was preceded by an in-frame stop codon. The human EST clone contained the 5′ sequence nearly identical to that of the mouse EST, along with a partial ORF that overlapped our isolated cDNA with 100% nucleotide identity. To confirm that these clones encoded the full-length cDNA, 5′ RACE was performed, and the recovered cDNAs revealed no upstream methionines. The protein was named TAZ (transcriptional co-activator with PDZ-binding motif, see below). Human and mouse TAZ show 91% amino acid identity, with the remaining 9% consisting primarily of conservative substitutions (Figure 1B).
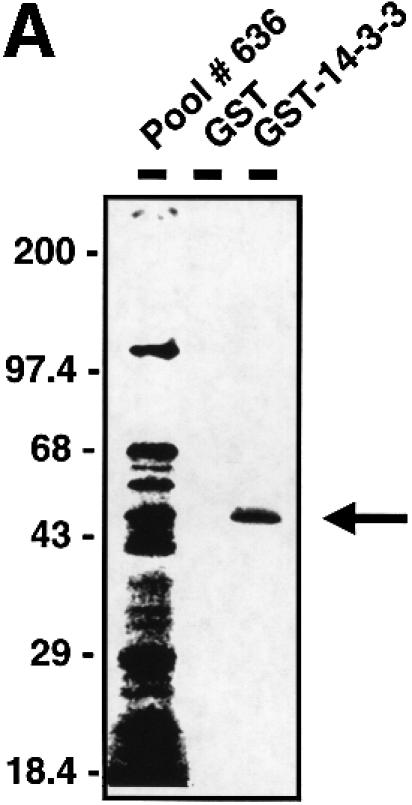
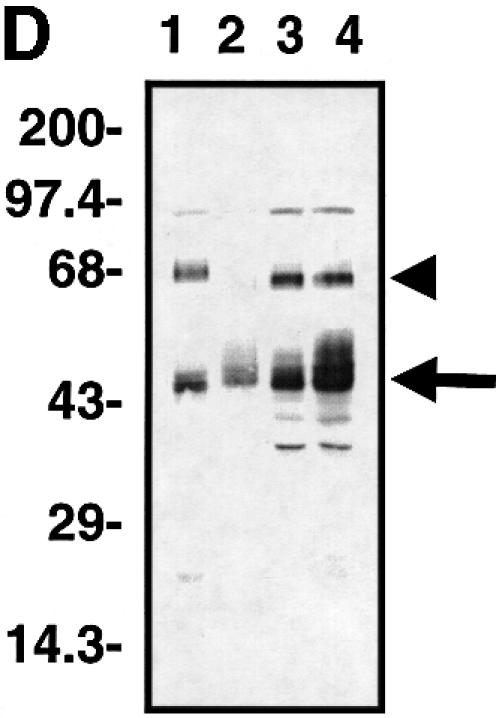
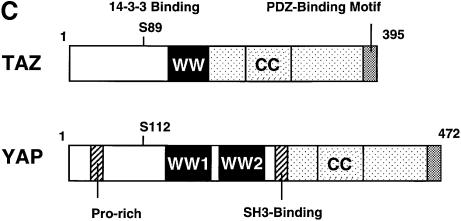
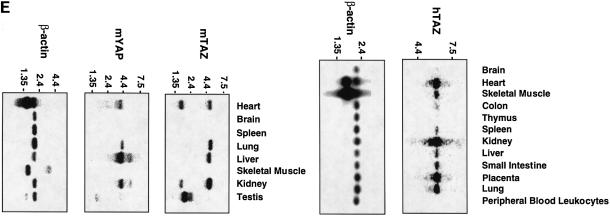
Fig. 1. Characterization of TAZ. (A) Isolation of TAZ as a 14-3-3-binding protein. 35S-labeled proteins from cDNA pool #636 were incubated with GST or GST–14-3-3-beads and bound proteins analyzed by SDS–PAGE/autoradiography. The arrow indicates a partial fragment of human TAZ. The mobility of molecular mass standards in kDa is indicated. (B) Sequence alignment of mouse and human TAZ with mouse, human and chicken YAP. The region surrounding the WW domain (indicated by a line) is boxed in yellow, the transcriptional activation domain in green, and the N-terminal region of homology in orange. The 14-3-3-binding site is indicated in red, and the conserved PDZ-binding motif is boxed in blue. The coiled-coil domain is shown as a cylinder. (C) The domain structure of mouse TAZ and mouse YAP65. CC, coiled-coil domain; SH3-Binding, SH3-binding motif; Pro-rich, proline-rich sequence; S89 of TAZ and S112 of YAP are the 14-3-3-binding site. Transactivation domains are dotted. (D) Immunoblot of endogenous and overexpressed TAZ: MDCK (lane 1); NIH-3T3 (lane 2); 293T (lane 3); and 293T cells transfected with pEF-TAZ (lane 4). The arrow indicates TAZ, and the arrowhead indicates YAP. The mobility of molecular mass standards is indicated. No immunostaining was observed using the pre-immune serum (data not shown). (E) Expression of TAZ determined by northern blot. A mouse (left panels) and human tissue blot (right panels) are probed as indicated. The positions of RNA size markers in kilobases are shown.
A BLAST search revealed significant homology between TAZ and YAP (Sudol, 1994, 1995) with 45% amino acid identity between human TAZ and human YAP in a pattern that is widely distributed across both sequences (Figure 1B). Similar levels of sequence homology were observed between mouse and human TAZ, and the mouse and chicken isoforms of YAP. In addition, comparison of the domain architecture in TAZ and YAP revealed a striking degree of conservation (Figure 1B and C). Both TAZ and YAP contain a central WW domain, followed by a highly conserved C-terminal sequence, which contains a region predicted to form a two-stranded coiled-coil (Lupas et al., 1991). Both human and mouse TAZ, along with human and chicken YAP, contain a single WW domain, whereas mouse YAP contains two WW domains. TAZ and YAP also contain a highly conserved sequence block that lies N-terminal to the WW domain, with limited homology to the Caenorhabditis elegans nuclear receptor NHR-6 (DDBJ/EMBL/GenBank accession No. AF083224). TAZ also contains a PDZ-binding motif at its C-terminus, and a potential 14-3-3-binding motif within the conserved N-terminal sequence block (Figure 1B).
There are several differences between TAZ and YAP that are likely to be related to distinct functions performed by each protein. For example, YAP contains an SH3-binding motif with the sequence PVKQPPPLAPQSP as well as a proline-rich region at its N-terminus, both of which are absent in TAZ (Figure 1B and C). Furthermore, YAP was identified originally by virtue of its ability to bind to the SH3 domain of Yes, while TAZ, which lacks both the SH3-binding motif and proline-rich N-terminus of YAP, is unable to bind the Yes SH3 domain in vitro (data not shown).
Rabbit polyclonal antibodies were generated against a peptide corresponding to the C-terminal 15 amino acids of TAZ. These antibodies recognized the identical series of closely spaced immunoreactive protein bands migrating at ∼50 kDa in lysates from MDCK, NIH-3T3 and 293T cells (Figure 1D, lanes 1–3). The endogenous immunoreactive protein co-migrates exactly with the overexpressed protein encoded by our cDNA (Figure 1D, lane 4). An immunoreactive band corresponding to endogenous TAZ was also detected in lysates from COS7, HepG2, CHO and HeLa cells, but not in lysates from Jurkat cells (data not shown). The pattern of closely spaced immunoreactive bands suggests that TAZ undergoes post-translational modifications such as phosphorylation. Both antibodies also recognized a 65 kDa protein, which was recognized by an anti-YAP antibody and migrates with the same mobility as that of exogenously expressed YAP (data not shown). Because of the cross-reactivity observed with these antibodies, most additional experiments were performed using epitope-tagged TAZ.
The tissue-specific expression of TAZ was investigated by northern blotting (Figure 1E). All human tissues, except thymus and peripheral blood leukocytes, express a 6.0 kb TAZ transcript, with the highest levels of expression in kidney, followed by the heart, placenta and lung. Similar results were seen in the mouse, where a 5.5 kb transcript was observed with the highest levels in kidney, lung, liver and heart, and a 2.2 kb transcript in testis. In addition, a minor 2.0 kb band was also observed in some mouse tissues. In contrast, the mouse blot contained a 4 kb YAP transcript in most tissues, with the highest expression in liver, followed by kidney, heart and lung.
The TAZ gene was mapped to chromosome 3q24 using the radiation hybrid panel method (Walter et al., 1994). To date, no chromosomal alterations associated with any known human disease have been identified at this site.
14-3-3 binding requires TAZ phosphorylation on Ser89
We examined whether phosphorylation is required for the interaction of TAZ with 14-3-3. Our finding that TAZ prepared by in vitro translation binds to 14-3-3 suggested either that TAZ phosphorylation was not required or that TAZ is phosphorylated by an endogenous kinase present within the reticulocyte lysate. To test this, we used a broad-spectrum Ser/Thr protein kinase inhibitor, K252a. The presence of K252a during the in vitro translation had no effect on the total amount of radiolabeled TAZ protein produced; however, K252a resulted in a marked dose-dependent inhibition of TAZ binding to 14-3-3 (Figure 2A). Thus, phosphorylation by Ser/Thr kinase(s) within the reticulocyte lysate is required for TAZ to interact with 14-3-3. To investigate whether TAZ binding occurs through the central binding groove of the 14-3-3 dimer, we investigated the binding of TAZ to two 14-3-3 mutant constructs that disrupt the phosphopeptide-binding pocket, E180K (Chang and Rubin, 1997; Rittinger et al., 1999) and K49E (Zhang et al., 1997). As seen in Figure 2B, both of the 14-3-3 mutations eliminated the binding of TAZ to 14-3-3.
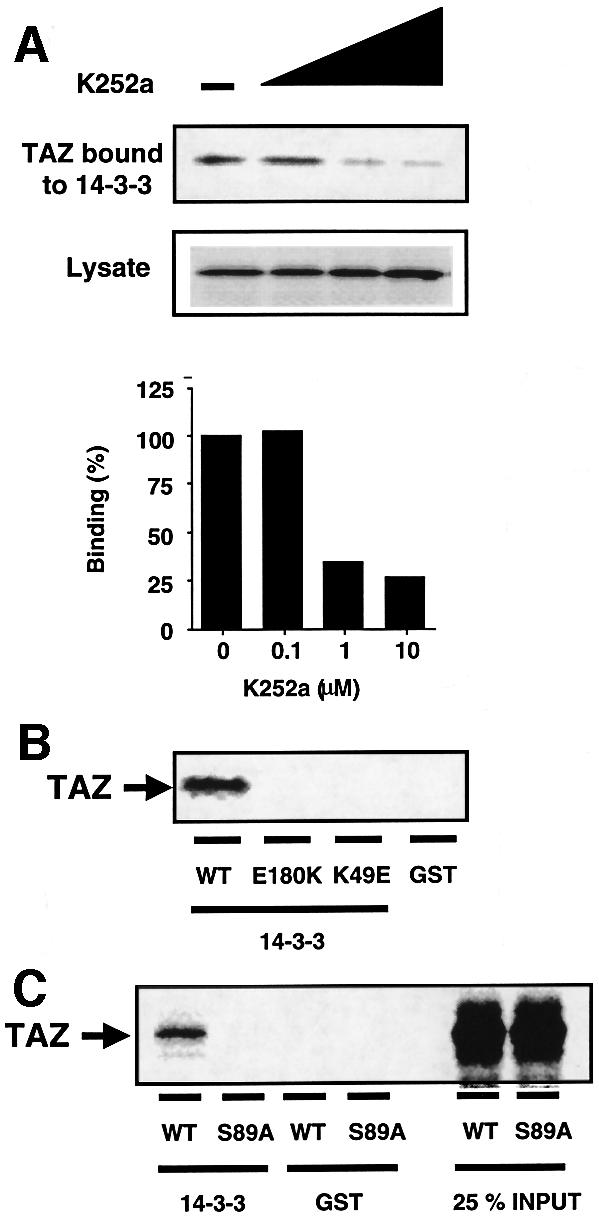
Fig. 2. Phosphorylation of TAZ is required for the binding to 14-3-3. (A) [35S]methionine-labeled TAZ was synthesized by in vitro translation in the presence of various concentrations of the protein kinase inhibitor K252a (0, 0.1, 1 and 10 µM). The amount of TAZ present in the lysate (middle) and bound by GST–14-3-3 (top) was quantitated (bottom). (B) 35S-labeled TAZ was incubated with GST–14-3-3ε wild-type, the E180K mutant, the K49E mutant or GST control beads and analyzed by autoradiography. (C) Ser89 of TAZ is the binding site of 14-3-3 in vitro. 35S-labeled TAZ (wild-type or S89A mutant) was pulled-down with GST–14-3-3 beads or GST control beads.
We previously identified two different optimal phosphoserine motifs, RSXpSXP and RXY/FXpSXP, which are recognized by all 14-3-3 isotypes (Yaffe et al., 1997). There is a perfect match to the first consensus motif within TAZ, centered on Ser89 (RSHSSP), and a near-optimal motif at the same position in YAP (RAHSSP). Furthermore, the 14-3-3-binding motif is conserved in all TAZ and YAP orthologs (Figure 1B). To verify that this site was responsible for the phosphorylation-dependent binding of TAZ to 14-3-3, Ser89 in TAZ was mutated to alanine and binding to 14-3-3 was examined following in vitro translation. Replacement of Ser89 with alanine completely abolished the binding of TAZ to 14-3-3 (Figure 2C).
To confirm that the interaction between TAZ and 14-3-3 was phosphorylation dependent, cell lysates were treated with alkaline phosphatase, incubated with GST–14-3-3 beads, and analyzed by immunoblotting for endogenous TAZ using an anti-TAZ antibody. Phosphatase treatment dramatically reduced the amount of TAZ binding to 14-3-3, and this effect was blocked when phosphatase inhibitors were present during the reaction (Figure 3A). To investigate whether TAZ binds to endogenous 14-3-3 in a cellular context, Flag-tagged versions of wild-type and the S89A mutant were introduced into cells. Immunoprecipitation followed by immunoblotting demonstrated that wild-type TAZ, but not the S89A mutant, interacted with endogenous 14-3-3 in these cells (Figure 3B). Thus, TAZ interacts with 14-3-3 in vivo and in vitro, and this interaction requires S89 phosphorylation. We also found that YAP binds to 14-3-3 and this interaction requires phosphorylation of the homologous serine residue (S112) in the YAP 14-3-3-binding motif (data not shown).
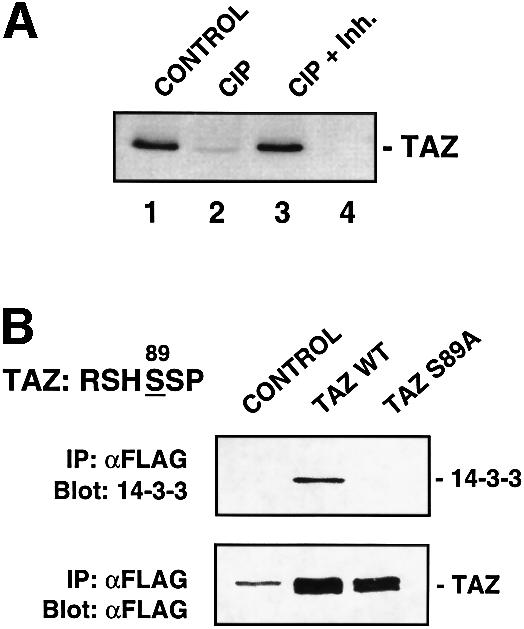
Fig. 3. TAZ associates with 14-3-3 through the phosphoserine-binding motif RSHpSSP in vivo. (A) 293T cell lysates (30 µg) were incubated with or without calf intestine alkaline phosphatase (CIP) [lanes 1 and 4, no phosphatase; lane 2, 200 U of CIP; lane 3, 200 U of CIP and a mixture of phosphatase inhibitors (Inh.)] and pulled-down with GST–14-3-3ε beads (lanes 1, 2 and 3) or GST–14-3-3ε (K49E) beads (lane 4). Proteins bound to beads were eluted, fractionated by SDS–PAGE and immunoblotted with an anti-TAZ antibody. No TAZ was detected in pull-downs using GST alone (data not shown). (B) 293T cells were transfected with Flag-TAZ wild-type (WT) or mutant (S89A). Lysates were immunoprecipitated with anti-Flag and immunoblotted for 14-3-3 (top). Blots were reprobed using anti-Flag (bottom). Cells transfected with pEF-BOS were used as a control. The band in the control lane and the upper band in the sample lanes is the cross-reacting immunoglobulin heavy chain.
TAZ functions as a transcriptional co-activator by binding to PPXY motifs on transcription factors via its WW domain
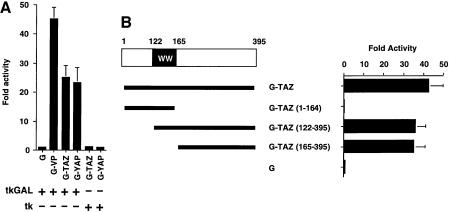
Yagi et al. found that the PPXY motif in Runx transcription factors bound to the WW domain of YAP, allowing YAP to function as a transcriptional co-activator of Runx-mediated gene transcription (Yagi et al., 1999). To investigate whether TAZ contains a similar transcription activation function, we first fused full-length TAZ to the minimum GAL4 DNA-binding domain, GAL4(1–93), and examined the effect of the fusion protein using a luciferase reporter gene construct containing a GAL4 DNA-binding site. The TAZ fusion protein strongly stimulated GAL4-binding site-dependent transcription at a level similar to that attained by GAL4-YAP (Figure 4A). For comparison, G-VP, a plasmid encoding the potent transcriptional activation domain of VP16 fused to the GAL4(1–93) DNA-binding domain, was used as a positive control in these experiments. The region of TAZ containing the transcriptional activation domain was mapped using deletion constructs (Figure 4B). N-terminal deletions of TAZ up to residue 165 had only minimal effect, whereas C-terminal deletions of TAZ beginning at residue 165 completely abolished the transcriptional stimulatory activity. These results indicate that the region C-terminal to the WW domain (amino acids 165–395) of TAZ has a strong intrinsic ability to activate transcription.
Fig. 4. TAZ contains a strong intrinsic transactivation domain in its C-terminus. (A) NIH-3T3 cells were transfected with 100 ng of tk-GALpx3-LUC (tkGAL) or tk-LUC (tk), 1 ng of pRL-EF, 500 ng of pEF-BOS backbone vector and 100 ng of the plasmid expressing the indicated GAL4(1–93) fusion protein. The luciferase activities relative to those obtained with GAL4(1–93) are shown. Results are the mean ± SD from three separate experiments. (B) Schematic illustration of mouse TAZ and its deletion constructs, and the transcriptional activity of each fragment fused to GAL4(1–93). NIH-3T3 cells were transfected with 100 ng of tk-GALpx3-LUC, 1 ng of pRL-EF, 500 ng of pEF-BOS and 100 ng of the plasmid expressing the indicated GAL4–TAZ fusion protein. The luciferase activities relative to those of GAL4(1–93) are indicated. Results are the mean ± SD from three separate experiments.
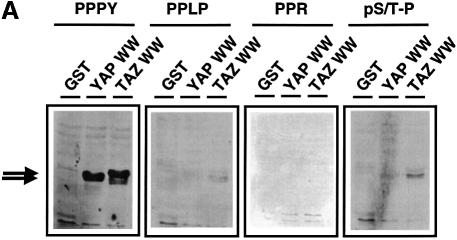
Since the WW domain of YAP has been shown to bind to proteins with Pro-Pro-Xxx-Tyr motifs, we investigated the binding specificity of the TAZ WW domain. Oriented peptide library screening (Table I) revealed that the TAZ WW domain selects for the optimal binding sequence Leu/Pro-Pro-Xxx-Tyr, similarly to the motif found for the WW domain of YAP (Chen and Sudol, 1995; Bedford et al., 2000b). Direct verification of PPXY selection was obtained using blot overlay experiments (Figure 5A). Both the TAZ and YAP WW domains demonstrated strong binding to peptides containing the sequence PPPY, but failed to bind peptides containing the sequences PPLP, phosphoS/T-P (pS/T-P) or PPR, which are recognized by other WW domains (Espanel and Sudol, 1999; Lu et al., 1999; Bedford et al., 2000b). The selection of similar PPXY motifs by the WW domains of TAZ and YAP is in agreement with a structural analysis of the YAP WW domain–peptide complex (Macias et al., 1996), since key residues involved in PPXY recognition (Leu175 and His177 in the mYAP sequence in Figure 1B) are conserved in all TAZ and YAP isoforms.
Table I. Motif selection for the TAZ WW domain determined by peptide library screening.
| Position |
–1 |
Pro |
+1 |
+2 |
| L (1.3) | P | X | Y (2.3) | |
| P (1.3) |
The WW domain of TAZ was screened using a proline-oriented degenerate peptide library of general sequence XXXXPXXXX (theoretical degeneracy = 1.7 × 1010), where X indicates all amino acids except cysteine, as described previously (Bedford et al., 2000). Bound peptides were eluted and sequenced on a Procise protein sequencer. Preference values for amino acid selection were determined by comparing the relative abundance of each amino acid at a particular cycle of Edman sequencing (i.e. mol%) in the recovered peptides with that of each amino acid in the original peptide library mixture at the same position. Significant selection was only observed in the P–1, and P+2 positions, with the selectivity values shown in parentheses. Values ≥1.3 represent moderate selection, and values ≥2 represent strong selection.
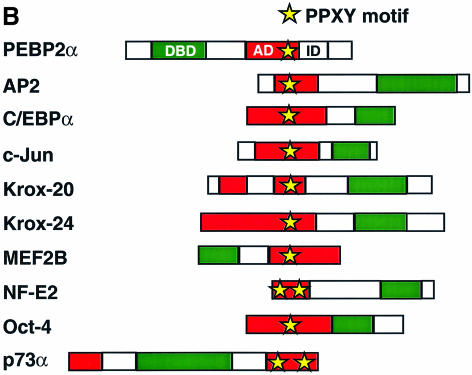
Fig. 5. The TAZ WW domain binds to a PPXY motif. (A) Blot overlay analysis verifies PPXY motif selection for the TAZ WW domain. GST or GST fusion proteins containing the WW domains of YAP and TAZ were analyzed by SDS–PAGE, transferred to membranes and probed using biotin-labeled peptides corresponding to the WW domain-binding motifs PPXY, PPLP, PPR and pS/T-P. In control experiments, the PPLP peptide bound strongly to the FBP11 WW domain, and the PPR peptide bound strongly to the FBP30 WW domains (data not shown). (B) The activation domains of many transcription factors contain a PPXY motif. DBD denotes the DNA-binding domain, AD the activation domain, and ID the autoinhibitory domain of PEBP2α.
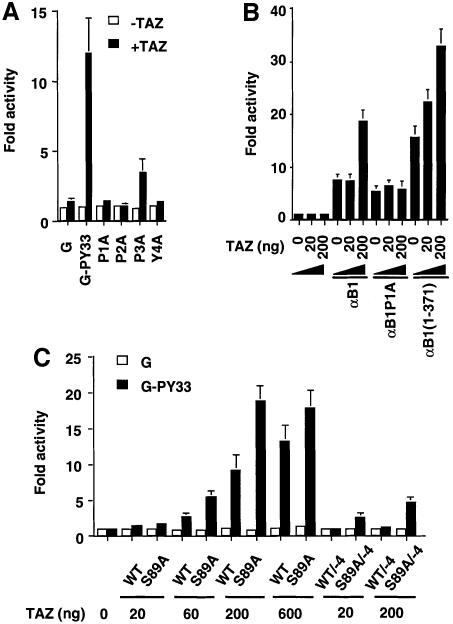
The transcriptional activation domains of many transcription factors, such as Runx/PEBP2α, AP2, C/EBPα, c-Jun, Krox-20, Krox-24, MEF2B, NF-E2, Oct-4 and p73α, contain proline-rich sequences with one or more copies of the PPXY motif (Figure 5B). Deletion of this proline-rich region abrogates transcriptional stimulation for several of these factors (Williams and Tjian, 1991; Vesque and Charnay, 1992; Mosser et al., 1998; Takada et al., 1999; Yagi et al., 1999). To determine whether TAZ, like YAP, can regulate transcription through WW domain–PPXY motif interactions, we investigated whether TAZ could function as a transcriptional co-activator for Runx/PEBP2α (Figure 6). In these experiments, Jurkat cells (which lack TAZ and YAP) were transfected with vectors expressing TAZ, a GAL4 DNA-binding domain construct fusion containing the 33 amino acid region having the PPXY motif from Runx1/PEBP2αB/AML1(G-PY33) and the GAL4-luciferase reporter. Co-expression of increasing amounts of TAZ led to a dramatic increase in the transactivation activity of G-PY33, up to 12-fold in a dose-dependent manner (Figure 6A and C). Furthermore, point mutations P1A, P2A or Y4A within the PPXY motif of G-PY33 completely abrogated TAZ-dependent transcriptional co-activation, whereas the P3A mutant showed a diminished response (Figure 6A). These results suggest that TAZ can function as a transcriptional co-activator through binding to the PPXY motifs present on transcription factors.
Fig. 6. TAZ functions as a transcriptional co-activator through its WW domain in a manner that is regulated by 14-3-3 and PDZ domain binding. (A) Jurkat cells were transfected with 100 ng of tk-GALpx3-LUC, 1 ng of pRL-EF, 200 ng of pEF-TAZ-N-Flag and 100 ng of plasmids expressing either GAL(1–93) (G) or GAL4(1–93) fused to a 33 amino acid region from PEBP2αB/AML1 (G-PY33) having wild-type or mutated versions of the PPXY motif. The luciferase activities relative to those obtained with GAL(1–93) are shown. Results are the mean ± SD from three separate experiments. (B) P19 cells were transfected with 200 ng of pFL56–3, 1 ng of pRL-EF, 200 ng of a pEF-based plasmid expressing full-length PEBP2αB1, PEBP2αB1(P1A) or PEBP2αB1(1–371), and the indicated amount of pEF-TAZ-N-Flag. Luciferase activities relative to those obtained without effector plasmids are shown. Results are the mean ± SD from three separate experiments. (C) Jurkat cells were transfected with 100 ng of tk-GALpx3-LUC, 1 ng of pRL-EF and 100 ng of GAL4(1–93) (open bars) or GAL4(1–93)-PY33 (filled bars) together with the indicated amounts of TAZ constructs. WT, pEF-TAZ-N-Flag; S89A, pEF-TAZ-N-Flag (S89A); WT/–4, pEF-TAZ-N-Flag/–4; S89A/–4, pEF-TAZ-N-Flag (S89A)/–4. Results are the mean ± SD from three separate experiments.
To test whether TAZ could co-activate transcription of an endogenous PEBP2-responsive element through the natural PPXY-binding site present in full-length Runx1, we employed a reporter construct containing three tandem copies of the transforming growth factor-β (TGF-β)-responsive element from the Ig Cα promoter, which contains a natural PEBP2-binding site (Hanai et al., 1999). In this system, co-transfection of TAZ stimulated the activity of Runx1 in a dose-dependent manner, and stimulation of PEBP2αB1 activity by TAZ was eliminated when the PPXY motif was mutated (αB1P1A) (Figure 6B). Stimulation of the reporter activity by TAZ was enhanced further when the ‘inhibitory domain’ of Runx1 was removed (Figure 6B). Thus, TAZ can stimulate transcription of PEBP2-responsive elements by binding to the PPXY motif of Runx1 in a cellular context.
Transcriptional co-activation is negatively regulated by 14-3-3 binding and requires an intact PDZ-binding motif
To investigate the role of 14-3-3 in TAZ-mediated transcriptional co-activation, Jurkat cells were transfected with increasing amounts of wild-type TAZ or the S89A mutant construct in which the 14-3-3-binding site is disrupted, along with GAL4 DNA-binding domain constructs and the GAL4-luciferase reporter. In every instance, the S89A mutant demonstrated a 1.5- to 2-fold increase in co-activation activity (Figure 6C), suggesting that binding of endogenous 14-3-3 to TAZ inhibits its ability to transactivate gene expression. As shown in Figure 1B and C, TAZ also contains a highly conserved PDZ-binding motif at its C-terminus. Unexpectedly, we found that disruption of this motif by deleting the last four C-terminal residues of TAZ eliminated TAZ-mediated transcriptional co-activation (Figure 6C, WT/–4 lanes). A small amount of co-activation activity could be restored, however, by disrupting the PDZ-binding motif together with the 14-3-3-binding site (Figure 6C, S89A/–4 lanes). These data suggest that the transcriptional co-activation by TAZ is balanced through positive regulation involving binding to PDZ domain proteins and negative regulation through sequestration with 14-3-3.
Subcellular localization of TAZ
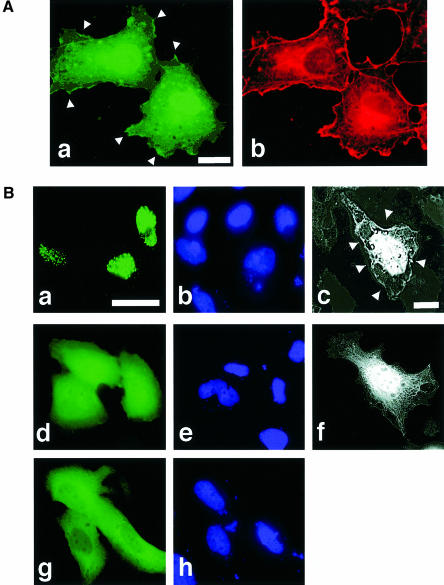
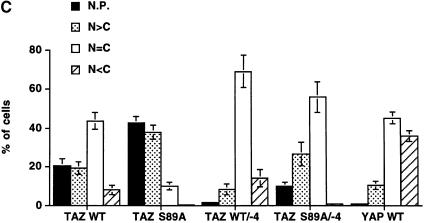
We investigated whether the differential regulation of the co-activation activity of TAZ through PDZ- and 14-3-3 binding was mediated through alterations in intracellular localization. This was accomplished using N-terminal green fluorescent protein (GFP)-tagged fusions containing either wild-type TAZ (GFP–TAZ), a TAZ mutant lacking the 14-3-3-binding site (GFP–TAZ S89A), a TAZ mutant lacking the PDZ-binding motif (GFP–TAZ WT/–4), a double mutant (GFP–TAZ S89A/–4) or YAP. In cells expressing high levels of GFP–TAZ, 50% demonstrated diffuse fluorescence distributed equally between the cytoplasm and nucleus (Figure 7A,a and C), while 40% showed signal limited entirely to the nuclear compartment (Figure 7B,a and C). In half of the cells showing nuclear-dominant staining, GFP–TAZ accumulated in discrete nuclear punctate bodies (Figure 7A,a, B,a and C). Some appearance of GFP–TAZ at discrete sites along the plasma membrane was also observed (Figure 7A,a and B,c, arrowheads). GFP–TAZ S89A was found much more frequently in the nucleus compared with wild-type, suggesting that 14-3-3 binding retains TAZ in the cytosol. Interestingly, deletion of the PDZ-binding motif of TAZ eliminated the punctate nuclear pattern and also eliminated the binding to the plasma membrane (Figure 7B,d and f). This mutant (GFP–TAZ/–4) also had reduced nuclear localization compared with the wild-type TAZ (Figure 7C). The double mutant of TAZ (GFP–TAZ S89A/–4) showed a partial recovery of nuclear accumulation, though the staining pattern was predominantly diffuse (Figure 7C). There was a striking correlation between the ability of each construct to localize in punctate nuclear bodies and its ability to activate transcription (compare Figure 6C, G-PY33 bars with Figure 7C, N.P. bars), suggesting that these two processes are linked via a PDZ domain-mediated interaction. In contrast to the results obtained with TAZ, in >80% of cells expressing GFP–YAP, the fluorescent signal was distributed diffusely within the cytoplasm (Figure 7B,g and C), with ∼10% of cells showing a primarily nuclear staining pattern with no punctate nuclear staining whatsoever. These results for YAP are in agreement with those reported by others (Mohler et al., 1999; Yagi et al., 1999).
Fig. 7. Subcellular localization of TAZ. (A) COS7 cells transiently expressing GFP–TAZ (a) and corresponding F-actin staining with rhodamine–phalloidin (b). Bar = 10 µm. In addition to cytoplasmic staining, TAZ is concentrated along specific portions of the plasma membrane (arrowheads) and within the nucleus. The nuclear staining of the upper cell in (a) is diffuse, while that of the lower cell is punctate, though these details are obscured in this exposure. (B) HeLa cells expressing GFP–TAZ (a), GFP–TAZ lacking the C-terminal four amino acids (d) or GFP–YAP (g). (b), (e) and (h) are the corresponding DAPI staining. (c) and (f) are Z-mode scanning of 293T cells expressing GFP–TAZ (c) and GFP–TAZ/-4 (f). Removal of the PDZ-binding motif eliminated punctate nuclear staining and plasma membrane staining (arrowheads). Bar = 10 µm. (C) Quantitative analysis of TAZ localization. The subcellular distribution of the various GFP fusion proteins was scored according to whether it was higher in the nucleus (N>C), evenly distributed between the nucleus and cytoplasm (N=C) or higher in the cytoplasm (N<C). The percentage of cells showing punctate nuclear staining (N.P.) was also quantitated. Results are the mean ± SD from three separate experiments.
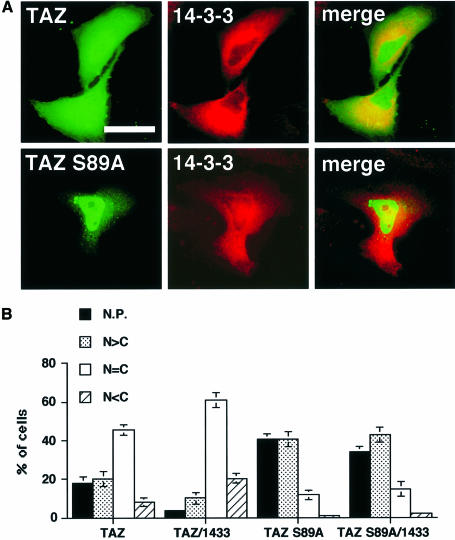
We investigated the effect of co-expression of 14-3-3 with GFP–TAZ wild type or GFP–TAZ S89A (Figure 8). 14-3-3 significantly increased the cytoplasmic localization of GFP–TAZ wild type; however, 14-3-3 had no effect on the predominantly nuclear localization of the S89A mutant (Figure 8A and B), demonstrating that binding to 14-3-3 promotes the cytoplasmic retention and/or nuclear exclusion of wild-type TAZ.
Fig. 8. Co-expression of 14-3-3 localizes TAZ to the cytoplasm. (A) HeLa cells expressing GFP–TAZ wild-type (upper) or GFP–TAZ S89A (lower) together with Xpress-tagged 14-3-3 were stained with an anti-Xpress antibody. TAZ and 14-3-3 were visualized by green (left) or red (middle) fluorescence, respectively. Merged images are shown in the right panels. Bar = 10 µm. (B) Quantitative localization of GFP–TAZ WT or S89A in cells expressing 14-3-3 was performed as described in Figure 7. Results are the mean ± SD from three separate experiments.
TAZ binds to the first PDZ domain of E3KARP/NHERF-2
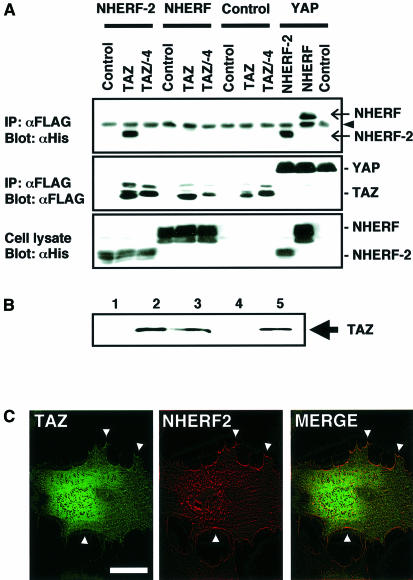
Mohler et al. (1999) recently reported that YAP could localize to the apical plasma membrane in confluent airway epithelial cells through association between the C-terminal PDZ-binding motif in YAP and the second PDZ domain of EBP50/NHERF (ERM-binding phosphoprotein 50, also called Na+/H+ exchanger regulatory factor). To investigate whether TAZ could participate in similar signaling complexes, co-immunoprecipitation studies on cells transfected with TAZ or YAP and NHERF or its homolog E3KARP/NHERF-2 were performed (Figure 9A). TAZ specifically bound to NHERF-2, whereas YAP interacted with both NHERF and NHERF-2. Removal of the last four C-terminal amino acids of TAZ (TAZ/–4) eliminated the NHERF-2 interaction. Pull-down assays revealed that TAZ binds to both full-length NHERF-2 (Figure 9B, lane 2) and the N-terminal PDZ domain (lane 3), but not to the C-terminal PDZ domain (lane 4). The TAZ/–4 mutant was unable to bind to any of the NHERF-2 constructs (data not shown). Thus, the TAZ–NHERF-2 interaction occurs through binding of the TAZ C-terminus to the first PDZ domain in NHERF-2, in contrast to YAP which binds to the second PDZ domain in NHERF (Mohler et al., 1999).
Fig. 9. TAZ interacts specifically with the N-terminal PDZ domain of NHERF-2. (A) 293T cells were transfected with Flag-tagged YAP or TAZ constructs and His-tagged NHERF or NHERF-2, as indicated. Cell lysates were immunoprecipitated with anti-Flag followed by blotting with anti-His (upper) to detect NHERF and NHERF-2 (arrows). A cross-reacting immunoglobulin band is indicated by an arrowhead. The same blot was reprobed with anti-Flag (middle) to show TAZ or YAP expression. The amount of His-tagged NHERF or NHERF-2 within the total cell lysates is shown (lower). (B) TAZ interacts with PDZ1 of NHERF-2. [35S]methionine-labeled TAZ was pulled-down with beads containing GST (lane 1), GST–NHERF-2 (lane 2), GST–NHERF-2N (containing the PDZ1) (lane 3) or GST–NHERF-2C (containing the PDZ2) (lane 4). Ten percent of the input is also shown (lane 5). (C) Z-mode scanning of HeLa cells expressing both GFP–TAZ (left panel) and RFP–NHERF-2 (middle panel). A merged image is shown in the right panel. Co-localization at the plasma membrane is indicated by arrowheads. Bar = 10 µm.
To determine whether NHERF-2 is the PDZ domain protein responsible for the nuclear punctate staining pattern observed for TAZ, GFP–TAZ and a red fluorescent protein (RFP) fusion of NHERF-2 (RFP–NHERF-2) were transfected into HeLa cells and assayed for subcellular localization. As observed previously, GFP–TAZ displayed both diffuse cytoplasmic staining and punctate nuclear staining, along with some accumulation at discrete sites along the plasma membrane (Figure 9C, left panel). RFP–NHERF-2 stained diffusely throughout the entire cell, with some accumulation at the plasma membrane (Figure 9C, middle panel), in agreement with the results of Yun et al. (1998). Merging the images revealed that NHERF-2 co-localized with a fraction of TAZ at discrete sites along the plasma membrane (Figure 9C, right panel). However, TAZ and NHERF-2 did not co-localize within punctate nuclear bodies, suggesting that NHERF-2, while able to bind and target TAZ to the membrane, is not the PDZ domain protein responsible for TAZ accumulation in punctate nuclear bodies.
Discussion
We identified TAZ, a close relative of YAP, as a 14-3-3-binding partner. TAZ functions as a co-activator for transcription factors having PPXY motifs, and its activity is regulated by interactions with 14-3-3 and with PDZ domain-containing proteins. The discovery of TAZ as a relative of YAP fits with an expanding recognition of a role for WW domain-containing proteins in cell signaling events. The ability of WW domains to recognize a variety of distinct proline-rich sequence motifs facilitates ligand-specific targeting of WW domain-containing proteins, particularly when fused to other modular signaling domains (Sudol, 1998). To date, TAZ appears to be the only protein with a domain organization similar to that of YAP. Despite the similarities between TAZ and YAP, the proteins probably subserve separate but perhaps overlapping functions. TAZ mRNA is expressed at the highest levels within the kidney, and the endogenous protein is detected at high levels in cell lines of renal origin, whereas we found that YAP mRNA is expressed at the highest levels in the lung and is reported to also be high in placenta, prostate, ovary and testis (Sudol et al., 1995). YAP contains a proline-rich region through which it binds to Yes, a feature that is lacking in TAZ, and the interactions of YAP and TAZ with the NHERF and NHERF-2 PDZ domains are different. Furthermore, TAZ accumulates in punctate nuclear bodies via its C-terminal PDZ-binding motif, whereas YAP shows only diffuse nuclear staining. In addition, we repeatedly have been unable to obtain stable cell lines overexpressing TAZ, though in the same experiments we have been able routinely to obtain such cell lines overexpressing YAP, suggesting that in contrast to YAP, high level expression of TAZ may impair growth or survival.
We have shown that TAZ, like YAP, can function as a transcriptional co-activator of Runx transcription factors through specific interactions between the PPXY motif of Runx and the TAZ WW domain. In addition, the PPXY motif is found within the transcription activation domains of numerous other transcription factors (Figure 5B) including c-Jun, AP-2, C/EBPα, NF-E2, KROX-20, KROX-24, Oct-4 and MEF2B, along with the p53 homolog p73. Which of these transcription factors are physiologically regulated through TAZ and YAP binding remains to be explored.
A role for 14-3-3 in regulating TAZ-mediated transcription
Our identification of TAZ as a transcriptional co-activator in a search for novel 14-3-3-binding proteins is consistent with a general role for 14-3-3 proteins in regulating cell signaling events through directed subcellular localization. 14-3-3 inhibited the transcriptional co-activation function of TAZ by localizing TAZ in the cytoplasmic compartment. YAP also bound to 14-3-3, demonstrating that 14-3-3 binding is a general regulator of molecules belonging to this protein family of transcriptional co-activators. A variety of other molecules involved in transcriptional control also appear to be regulated through 14-3-3 binding, including FKHRL (Brunet et al., 1999), MSN2 and MSN4 (Beck and Hall, 1999), p53 (Waterman et al., 1998), histone acetyltransferase (Imhof and Wolffe, 1999) and histone deacetylase (Grozinger and Schreiber, 2000). Thus, 14-3-3 proteins probably play a major role in regulating gene transcription through the nuclear– cytoplasmic partitioning of ligands like TAZ.
Transcriptional co-activation by TAZ seems to involve an unknown PDZ domain protein
Unexpectedly, the ability of TAZ to function as a transcriptional co-activator paralleled its ability to localize into punctate nuclear bodies. Both events require an intact PDZ-binding motif, suggesting that an as yet unidentified PDZ domain protein plays a critical role in this process. Most PDZ domain proteins localize to the plasma membrane or cytoplasm. A few nuclear proteins, however, contain PDZ domains. One of these, a splice variant of NHERF-2 named SIP-1, was identified as a nuclear factor binding to the SRY that acts as a trigger for male sex development (Poulat et al., 1997). The SIP-1–SRY interaction involves either PDZ domain of SIP-1 and the C-terminal YSHWTKL sequence of SRY that bears some similarity to the C-terminus of TAZ. Furthermore, immunostaining of endogenous SIP-1 revealed a nuclear punctate staining pattern similar to the one we observed for TAZ. We cannot exclude the possibility that TAZ interacts with NHERF-2/SIP-1 to localize into punctate nuclear bodies, though our inability to demonstrate nuclear co-localization of both proteins argues against this.
Does TAZ transmit a signal from the plasma membrane to the nucleus?
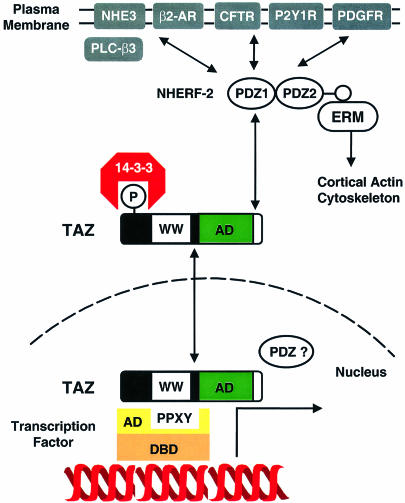
NHERF and a related homolog E3KARP/NHERF-2 were identified originally as regulatory factors involved in protein kinase A-mediated regulation of the Na+/H+ exchanger (Reczek et al., 1997; Yun et al., 1997; Lamprecht et al., 1998; Weinman et al., 1998). Both proteins contain two PDZ domains through which they bind the β2-adrenergic receptor (Hall et al., 1998b), the cystic fibrosis transmembrane conductance regulator (CFTR) (Hall et al., 1998a; Short et al., 1998), the P2Y1 purinergic receptor (Hall et al., 1998a), the platelet-derived growth factor (PDGF) receptor (Seedorf and Ullrich, unpublished data; DDBJ/EMBL/GenBank accession No. Z50150), as well as the Na+/H+ exchanger 3 (NHE-3) (Yun et al., 1998), and a C-terminal sequence that binds directly to the N-terminus of the actin-binding protein ezrin (Bretscher, 1999; Figure 10). In addition, non-integral membrane proteins also bind to NHERF and NHERF-2. Phospholipase C-β3, for example, binds to the second PDZ of NHERF-2 with increased enzymatic activity upon binding (Hwang et al., 2000). It is conceivable that TAZ participates directly in a subset of these complexes to propagate signal transduction events across and along the plasma membrane.
Fig. 10. Model of TAZ-mediated transcriptional regulation. TAZ functions as a co-activator through binding PPXY motifs within the activation domains of transcription factors via its WW domain. 14-3-3 retains phosphorylated TAZ in the cytosol, negatively regulating this activity. TAZ also binds to the N-terminal PDZ domain of NHERF-2 at the plasma membrane. NHERF-2 interacts with ion channels, receptors and cytosolic signaling proteins, tethering them to the actin cytoskeleton through the ezrin–radixin–moesin (ERM) proteins. Thus, TAZ may link events at the plasma membrane and cytoskeleton to nuclear transcription.
TAZ joins a growing list of proteins that distribute between the nucleus and the cell membrane. The ability of TAZ and YAP to interact with both cortical cytoskeletal proteins in epithelial cells and with nuclear transcription factors invites parallels between TAZ/YAP and molecules such as β-catenin and CASK/LIN-2. β-catenin participates in both cadherin-mediated cell–cell contact and in transcriptional co-activation downstream of Wnt family members (Peifer and Polakis, 2000), while CASK functions both as a structural girder of cell–cell junctions in epithelial cells and translocates into the nucleus as a transcriptional co-activator of Tbr-1, a T-box transcription factor (Hsueh et al., 2000). A deeper understanding of the normal physiological functions subserved by TAZ, along with whether TAZ plays a role in regulating gene transcription in response to signals from the plasma membrane and cortical cytoskeleton should be discernable by studying the endogenous protein in cell lines, primary cells and tissues.
Materials and methods
In vitro expression cloning
Preparation of cDNA pools and in vitro translation reactions were performed as described (Lustig et al., 1997). A HeLa human cell line cDNA library (Invitrogen) was fractionated into small pools containing ∼100 different clones each. Each pool was used to express 35S-labeled proteins, using a coupled transcription–translation system (Promega) and [35S]methionine (New England Nuclear). Each 35S-labeled pool was incubated with bead-immobilized GST–14-3-3 proteins (a mixture of β, γ, ε, η, σ, ζ and τ isoforms) or GST–control beads in buffer I [20 mM Tris–HCl pH 7.5, 150 mM NaCl, 10% glycerol, 1% NP-40, 10 mM EDTA, 200 mM sodium orthovanadate, 10 mM NaF, 10 mM tetrasodium pyrophosphate, 4 µg/ml each of pepstatin, leupeptin and aminoethylbenzenesulfonyl fluoride (AEBSF)] at 4°C for 2 h. Following incubation, the beads were washed and analyzed by 10% SDS–PAGE/autoradiography.
Plasmids
The murine TAZ cDNA was subcloned into pcDNA3.1/His C (Invitrogen) and pEF-BOS lacking the SV40 origin (Mizushima and Nagata, 1990) to generate pcDNA3.1/His-TAZ and pEF-TAZ. A Flag tag was inserted into the N-terminus of mTAZ cDNA by PCR, and cloned into pEF-BOS to create pEF-TAZ-N-Flag. Murine TAZ and YAP cDNA were cloned into pEGFP-C1 or C2 (Clontech), to generate pEGFP-TAZ and pEGFP-YAP. The TAZ (S89A) point mutant and TAZ/–4 construct lacking the final four amino acids were constructed using the Transformer Site-Directed Mutagenesis Kit (Clontech). For expression of the minimal GAL4 DNA-binding domain fusion proteins, the corresponding fragments of mTAZ were amplified and cloned into pGN (Yagi et al., 1999). Fragments coding for the mTAZ WW domain (amino acids 122–164 and 79–164) were subcloned into pGEX4T2 (Amersham Pharmacia Biotech) for bacterial expression. DNAs for full-length NHERF-2 (residues 1–337), the N-terminus of NHERF-2 containing PDZ1 (residues 1–148) or the C-terminus of NHERF-2 containing PDZ2 (residues 149–337) were cloned into pGEX4T1 to create pGEX-NHERF-2, pGEX-NHERF-2N and pGEX-NHERF-2C, respectively. GST fusion proteins were prepared as described previously (Rittinger et al., 1999). Rat 14-3-3ε cDNA was inserted into pcDNA3.1/HisC, to create pcDNA3.1/His-14-3-3ε. Full-length cDNA of NHERF-2 was cloned into pDsRed1-C1 (Clontech) to create pRFP-NHERF-2. Plasmids pMT3/NHERF-HS and pMT/E3KARP-HS, which express His-tagged EBP50/NHERF and E3KARP/NHERF-2, and pEF-αB1, pEF-αB1(1–371), pEF-αB1(P1A), pEF-YAP-N-Flag, pG-YAP, pG-VP16, pG-PY33 and mutants within the PPXY motif have been described previously (Lamprecht et al., 1998; Yagi et al., 1999). Reporter plasmids included ptk-GALpx3-LUC (Kanno et al., 1998), ptk-luc (Yagi et al., 1999) and pFL56-3 (Hanai et al., 1999) which express firefly luciferase, and pRL-EF (Kim et al., 1999), which expresses Renilla luciferase as an internal control. All constructs and mutants were verified by DNA sequencing
Cells and anti-TAZ antibodies
HeLa cells and human embryo kidney 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM)/10% fetal calf serum (FCS), NIH-3T3 cells were cultured in DMEM/10% calf serum (CS), and Jurkat T lymphocytes were cultured in RPMI 1640/10% FCS. P19 cells were maintained in α-minimum essential medium containing 7.5% CS and 2.5% FCS, and type II MDCK cells were maintained in DMEM/5% FCS. Antibodies to TAZ were obtained by immunizing rabbits with the keyhole limpet hemocyanin (KLH)-coupled peptide CVESVLNKS EPFLTWL corresponding to the C-terminus of mouse TAZ (Research Genetics).
Northern blot analysis
A mouse multiple-tissue northern blot (Clontech) was probed with [α-32P]dCTP-labeled mouse TAZ cDNA (amino acids 184–265). The same blot was reprobed with 32P-labeled mouse YAP cDNA (amino acids 213–251). A human multiple-tissue northern blot (Clontech) was probed with 32P-labeled human TAZ cDNA (amino acids 183–266).
Chromosome mapping of TAZ
The GeneBridge 4 Radiation Hybrid Panel (Research Genetics) was screened using the primer set (5′-CTTGGATGTAGCCATGACCTT-3′ and 5′-TCAATCAAAACCAGGCAATG-3′) designed in the 3′-untranslated region of hTAZ cDNA. The amplification profile consisted of one cycle at 94°C for 2 min, and 37 cycles at 94°C for 30 s, 61°C for 30 s, 72°C for 30 s, and the amplified products were separated by 1.5% agarose gels. Each hybrid clone was scored as positive (1), negative (0) or uncertain (2) according to the presence of the fragment band. The following data vector was generated: 100000001001000001211000102110010010010100001000111110101000011100000000000001110000101010110, and analyzed by two-point maximum-likelihood analysis software through the URL at http://www.sanger.ac.uk/Software/RHserver/RHserver.shtml. This revealed that the human TAZ gene is located on the long arm of chromosome 3, 2.33 cR telomeric of AFM234wa1 (LOD = 18.364) and 1.41 cR centromeric of RH42049 (LOD = 19.558).
In vitro binding assay
pcDNA3.1/His-TAZ (wild-type or S89A) was translated in vitro using [35S]methionine. Reaction products (2 µl) were incubated with 20 pmol of bead-immobilized GST–14-3-3 or GST for 2 h at 4°C, and the beads washed and analyzed by SDS–PAGE/autoradiography. Where indicated, the Ser/Thr kinase inhibitor K252a (Biomol) was included in the in vitro translation reaction and the amount of 14-3-3-bound TAZ quantitated using a Molecular Imager System. For the analysis of interaction between TAZ and NHERF-2, in vitro labeled TAZ was pulled-down with beads containing GST, GST fusions containing full-length NHERF-2, or the N- or C-terminal NHERF-2.
Blot overlay analysis
Blot overlay was performed as described (Bedford et al., 2000a). Biotinylated peptides corresponding to the following WW-binding motifs were used: biotin-SGSGGTPPPPYTVG (PPXY); biotin-SGSGAPPTPPLPP (PPLP); biotin-PPGMRPPPPGMRRGPPPPGMRPPRP (PPR); and biotin-SGSGEQPL-phosphoT-PVTDL (pS/T-P). A 25 mg aliquot of each peptide was pre-complexed to 20 ml of streptavidin–horseradish peroxidase (HRP) (30 min, 4°C). GST or GST fusion proteins containing the WW domains of TAZ and YAP were separated by SDS–PAGE and transferred onto PVDF membranes (Millipore). Blots were blocked in TBST/1% milk (1 h, 22°C), incubated with the pre-complexed peptide (overnight, 4°C) and the HRP signal detected by enhanced chemiluminescence.
Immunoprecipitation and immunoblotting
To detect endogenous TAZ, 50 µg of total protein from cell lysates was resolved on 10% SDS–gels and transferred to PVDF. Blots were blocked in TBST/5% milk and incubated with rabbit anti-TAZ antibodies (1:500, 2 h, 22°C). After washing, blots were incubated with HRP-conjugated goat anti-rabbit antibody (1 h, 22°C) and visualized using enhanced chemiluminescence. For analysis of the interaction between TAZ and 14-3-3, 293T cells transfected with pEF-TAZ-N-Flag or pEF-TAZ-N-Flag (S89A) using Superfect (Qiagen) were lysed after 24 h using buffer I and immunoprecipitated using anti-Flag M2 affinity gel (Sigma). The immunoprecipitates were washed, and bound proteins were analyzed by SDS–PAGE and immunoblotted using an anti-14-3-3 antibody (K19, Santa Cruz). For analysis of the interaction between TAZ, YAP and NHERFs, 293T cells were transfected with the Flag-tagged TAZ or YAP together with pMT/E3KARP-HS or pMT3/NHERF-HS. After 24 h, lysates were immunoprecipitated with anti-Flag antibody (Sigma) and blotted with anti-His antibody (Qiagen). Phosphatase treatment of cell lysates was performed by adding 30 µg of 293T cell lysate to 250 µl of reaction buffer [20 mM Tris pH 8.0, 150 mM NaCl, 1 mM MgCl2, 1 mM dithiothreitol (DTT)] with or without 200 U of calf intestinal alkaline phosphatase (New England Biolabs), followed by incubation at 37°C for 2 h. Where indicated, the inhibitors NaF (50 mM) and Na3VO4 (1 mM) were added. The lysates were then incubated with GST–14-3-3ε wild-type or K49E mutant beads followed by immunoblotting with the anti-TAZ antibody.
Luciferase assays
Cells in 24-well plates were transfected with the plasmids indicated in the figure legends using FuGene6 (Roche) and harvested 24 h later. Total transfected DNA was fixed by adding corresponding amounts of backbone plasmid. Firefly and Renilla luciferase activities were assayed with the dual luciferase assay system (Promega) and firefly luciferase activity normalized with respect to Renilla luciferase activity. All experiments were performed at least three times.
Immunofluorescence microscopy
293T cells and COS7 cells on glass coverslips were transfected with pEGFP-TAZ using Superfect. After 24 h, cells were fixed with 3% paraformaldehyde. F-actin was visualized by staining with rhodamine–phalloidin (Sigma). HeLa cells on glass coverslips were transfected with pEGFP-TAZ or pEGFP-TAZ (S89A) together with pcDNA3.1/His-14-3-3ε, or with pEGFP-TAZ and pRFP-NHERF-2 using FuGene6. After 24 h, the cells were fixed, permeabilized with 0.2% Triton X-100, blocked in phosphate-buffered saline (PBS)/5% bovine serum albumin (BSA) and, where indicated, stained with anti-Xpress antibody (1:500) (Invitrogen) and Cy3-conjugated goat anti-mouse antibody (1:200) (Jackson Immunoresearch laboratories). Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma) to visualize the nucleus. Images were acquired using a Nikon Diaphot 300 microscope equipped with a Sensys CCD digital camera (Photometrics, Tucson, Arizona) and processed using Vaytek Imaging Software (Fairfield, Iowa). Z-mode (confocal-like) analysis, in which sequential images were taken at 0.2 µm intervals throughout the cell body, was used in some experiments.
Acknowledgments
Acknowledgements
We thank Marius Sudol (New York University) and Mark Bedford (Harvard Medical School) for providing reagents, and Sharon L.Milgram (University of North Carolina), Akira Tanigami (Otsuka GEN Research Institute) and the members of the Division of Signal Transduction for discussions. F.K. was supported in part by a fellowship from Toyobo Biotechnology Foundation and by a fellowship from Uehara Memorial Foundation. This work was supported by NIH grants HL03601 and GM59281 to M.B.Y., GM56203 to L.C.C., an HMS/Affiliated Hospital Collaborative Award and a Burroughs-Wellcome Career Development Award (M.B.Y.).
Note added in proof
Accession numbers for mouse and human TAZ are AJ299430 and AJ299431, respectively.
References
- Aitken A. (1996) 14-3-3 and its possible role in co-ordinating multiple signalling pathways. Trends Cell Biol., 6, 341–347. [DOI] [PubMed] [Google Scholar]
- Beck T. and Hall,M.N. (1999) The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature, 402, 689–692. [DOI] [PubMed] [Google Scholar]
- Bedford M.T., Frankel,A., Yaffe,M.B., Clarke,S., Leder,P. and Richard,S. (2000a) Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J. Biol. Chem., 275, 16030–16036. [DOI] [PubMed] [Google Scholar]
- Bedford M.T., Sarbassova,D., Xu,J., Leder,P. and Yaffe,M.B. (2000b) A novel pro-Arg motif recognized by WW domains. J. Biol. Chem., 275, 10359–10369. [DOI] [PubMed] [Google Scholar]
- Bretscher A. (1999) Regulation of cortical structure by the ezrin–radixin–moesin protein family. Curr. Opin. Cell Biol., 11, 109–116. [DOI] [PubMed] [Google Scholar]
- Brunet A. et al. (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell, 96, 857–868. [DOI] [PubMed] [Google Scholar]
- Chang H.C. and Rubin,G.M. (1997) 14-3-3ε positively regulates Ras-mediated signaling in Drosophila. Genes Dev., 11, 1132–1139. [DOI] [PubMed] [Google Scholar]
- Chen H.I. and Sudol,M. (1995) The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc. Natl Acad. Sci. USA, 92, 7819–7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal S.N., Schweitzer,C.M., Gan,J. and DeCaprio,J.A. (1999) Cytoplasmic localization of human cdc25C during interphase requires an intact 14-3-3 binding site. Mol. Cell. Biol., 19, 4465–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espanel X. and Sudol,M. (1999) A single point mutation in a group I WW domain shifts its specificity to that of group II WW domains. J. Biol. Chem., 274, 17284–17289. [DOI] [PubMed] [Google Scholar]
- Fanning A.S. and Anderson,J.M. (1999) PDZ domains: fundamental building blocks in the organization of protein complexes at the plasma membrane. J. Clin. Invest., 103, 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Subramanian,R.R. and Masters,S.C. (2000) 14-3-3 proteins: structure, function and regulation. Annu. Rev. Pharmacol. Toxicol., 40, 617–647. [DOI] [PubMed] [Google Scholar]
- Grozinger C.M. and Schreiber,S.L. (2000) Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl Acad. Sci. USA, 97, 7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R.A., Ostedgaard,L.S., Premont,R.T., Blitzer,J.T., Rahman,N., Welsh,M.J. and Lefkowitz,R.J. (1998a) A C-terminal motif found in the β2-adrenergic receptor, P2Y1 receptor and cystic fibrosis transmembrane conductance regulator determines binding to the Na+/H+ exchanger regulatory factor family of PDZ proteins. Proc. Natl Acad. Sci. USA, 95, 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R.A. et al. (1998b) The β2-adrenergic receptor interacts with the Na+/H+ exchanger regulatory factor to control Na+/H+ exchange. Nature, 392, 626–630. [DOI] [PubMed] [Google Scholar]
- Hanai J. et al. (1999) Interaction and functional cooperation of PEBP2/CBF with Smads. Synergistic induction of the immunoglobulin germline Cα promoter. J. Biol. Chem., 274, 31577–31582. [DOI] [PubMed] [Google Scholar]
- Hsueh Y.P., Wang,T.F., Yang,F.C. and Sheng,M. (2000) Nuclear translocation and transcription regulation by the membrane-associated guanylate kinase CASK/LIN-2. Nature, 404, 298–302. [DOI] [PubMed] [Google Scholar]
- Hwang J.I., Heo,K., Shin,K.J., Kim,E., Yun,C., Ryu,S.H., Shin,H.S. and Suh,P.G. (2000) Regulation of phospholipase C-β 3 activity by Na+/H+ exchanger regulatory factor 2. J. Biol. Chem., 275, 16632–16637. [DOI] [PubMed] [Google Scholar]
- Imhof A. and Wolffe,A.P. (1999) Purification and properties of the Xenopus Hat1 acetyltransferase: association with the 14-3-3 proteins in the oocyte nucleus. Biochemistry, 38, 13085–13093. [DOI] [PubMed] [Google Scholar]
- Kanno T., Kanno,Y., Chen,L.F., Ogawa,E., Kim,W.Y. and Ito,Y. (1998) Intrinsic transcriptional activation–inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor α subunit revealed in the presence of the β subunit. Mol. Cell. Biol., 18, 2444–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.Y., Sieweke,M., Ogawa,E., Wee,H.J., Englmeier,U., Graf,T. and Ito,Y. (1999) Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J., 18, 1609–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht G., Weinman,E.J. and Yun,C.H. (1998) The role of NHERF and E3KARP in the cAMP-mediated inhibition of NHE3. J. Biol. Chem., 273, 29972–29978. [DOI] [PubMed] [Google Scholar]
- Lu P.J., Zhou,X.Z., Shen,M. and Lu,K.P. (1999) Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science, 283, 1325–1328. [DOI] [PubMed] [Google Scholar]
- Lupas A., Van Dyke,M. and Stock,J. (1991) Predicting coiled coils from protein sequences. Science, 252, 1162–1164. [DOI] [PubMed] [Google Scholar]
- Lustig K.D., Stukenberg,P.T., McGarry,T.J., King,R.W., Cryns,V.L., Mead,P.E., Zon,L.I., Yuan,J. and Kirschner,M.W. (1997) Small pool expression screening: identification of genes involved in cell cycle control, apoptosis and early development. Methods Enzymol., 283, 83–99. [DOI] [PubMed] [Google Scholar]
- Macias M.J., Hyvonen,M., Baraldi,E., Schultz,J., Sudol,M., Saraste,M. and Oschkinat,H. (1996) Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature, 382, 646–649. [DOI] [PubMed] [Google Scholar]
- Mizushima S. and Nagata,S. (1990) pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res., 18, 5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler P.J., Kreda,S.M., Boucher,R.C., Sudol,M., Stutts,M.J. and Milgram,S.L. (1999) Yes-associated protein 65 localizes p62(c-Yes) to the apical compartment of airway epithelia by association with EBP50. J. Cell Biol., 147, 879–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser E.A., Kasanov,J.D., Forsberg,E.C., Kay,B.K., Ney,P.A. and Bresnick,E.H. (1998) Physical and functional interactions between the transactivation domain of the hematopoietic transcription factor NF-E2 and WW domains. Biochemistry, 37, 13686–13695. [DOI] [PubMed] [Google Scholar]
- Muslin A.J., Tanner,J.W., Allen,P.M. and Shaw,A.S. (1996) Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell, 84, 889–897. [DOI] [PubMed] [Google Scholar]
- Peifer M. and Polakis,P. (2000) Wnt signaling in oncogenesis and embryogenesis—a look outside the nucleus. Science, 287, 1606–1609. [DOI] [PubMed] [Google Scholar]
- Peng C.Y., Graves,P.R., Thoma,R.S., Wu,Z., Shaw,A.S. and Piwnica-Worms,H. (1997) Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science, 277, 1501–1505. [DOI] [PubMed] [Google Scholar]
- Poulat F., Barbara,P.S., Desclozeaux,M., Soullier,S., Moniot,B., Bonneaud,N., Boizet,B. and Berta,P. (1997) The human testis determining factor SRY binds a nuclear factor containing PDZ protein interaction domains. J. Biol. Chem., 272, 7167–7172. [DOI] [PubMed] [Google Scholar]
- Reczek D., Berryman,M. and Bretscher,A. (1997) Identification of EBP50: a PDZ-containing phosphoprotein that associates with members of the ezrin–radixin–moesin family. J. Cell Biol., 139, 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittinger K., Budman,J., Xu,J., Volinia,S., Cantley,L.C., Smerdon,S.J., Gamblin,S.J. and Yaffe,M.B. (1999) Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell, 4, 153–166. [DOI] [PubMed] [Google Scholar]
- Schultz J., Copley,R.R., Doerks,T., Ponting,C.P. and Bork,P. (2000) SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res., 28, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short D.B., Trotter,K.W., Reczek,D., Kreda,S.M., Bretscher,A., Boucher,R.C., Stutts,M.J. and Milgram,S.L. (1998) An apical PDZ protein anchors the cystic fibrosis transmembrane conductance regulator to the cytoskeleton. J. Biol. Chem., 273, 19797–19801. [DOI] [PubMed] [Google Scholar]
- Sudol M. (1994) Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene, 9, 2145–2152. [PubMed] [Google Scholar]
- Sudol M. (1998) From Src Homology domains to other signaling modules: proposal of the ‘protein recognition code’. Oncogene, 17, 1469–1474. [DOI] [PubMed] [Google Scholar]
- Sudol M., Bork,P., Einbond,A., Kastury,K., Druck,T., Negrini,M., Huebner,K. and Lehman,D. (1995) Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module, the WW domain. J. Biol. Chem., 270, 14733–14741. [DOI] [PubMed] [Google Scholar]
- Takada N., Ozaki,T., Ichimiya,S., Todo,S. and Nakagawara,A. (1999) Identification of a transactivation activity in the COOH-terminal region of p73 which is impaired in the naturally occurring mutants found in human neuroblastomas. Cancer Res., 59, 2810–2814. [PubMed] [Google Scholar]
- Thomas M.K., Yao,K.M., Tenser,M.S., Wong,G.G. and Habener,J.F. (1999) Bridge-1, a novel PDZ-domain coactivator of E2A-mediated regulation of insulin gene transcription. Mol. Cell. Biol., 19, 8492–8504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesque C. and Charnay,P. (1992) Mapping functional regions of the segment-specific transcription factor Krox-20. Nucleic Acids Res., 20, 2485–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M.A., Spillett,D.J., Thomas,P., Weissenbach,J. and Goodfellow,P.N. (1994) A method for constructing radiation hybrid maps of whole genomes. Nature Genet., 7, 22–28. [DOI] [PubMed] [Google Scholar]
- Waterman M.J., Stavridi,E.S., Waterman,J.L. and Halazonetis,T.D. (1998) ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nature Genet., 19, 175–178. [DOI] [PubMed] [Google Scholar]
- Weinman E.J., Steplock,D., Tate,K., Hall,R.A., Spurney,R.F. and Shenolikar,S. (1998) Structure–function of recombinant Na/H exchanger regulatory factor (NHE-RF). J. Clin. Invest., 101, 2199–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T. and Tjian,R. (1991) Analysis of the DNA-binding and activation properties of the human transcription factor AP-2. Genes Dev., 5, 670–682. [DOI] [PubMed] [Google Scholar]
- Yaffe M.B., Rittinger,K., Volinia,S., Caron,P.R., Aitken,A., Leffers,H., Gamblin,S.J., Smerdon,S.J. and Cantley,L.C. (1997) The structural basis for 14-3-3:phosphopeptide binding specificity. Cell, 91, 961–971. [DOI] [PubMed] [Google Scholar]
- Yagi R., Chen,L.F., Shigesada,K., Murakami,Y. and Ito,Y. (1999) A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J., 18, 2551–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C.H., Oh,S., Zizak,M., Steplock,D., Tsao,S., Tse,C.M., Weinman,E.J. and Donowitz,M. (1997) cAMP-mediated inhibition of the epithelial brush border Na+/H+ exchanger, NHE3, requires an associated regulatory protein. Proc. Natl Acad. Sci. USA, 94, 3010–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C.H., Lamprecht,G., Forster,D.V. and Sidor,A. (1998) NHE3 kinase A regulatory protein E3KARP binds the epithelial brush border Na+/H+ exchanger NHE3 and the cytoskeletal protein ezrin. J. Biol. Chem., 273, 25856–25863. [DOI] [PubMed] [Google Scholar]
- Zhang L., Wang,H., Liu,D., Liddington,R. and Fu,H. (1997) Raf-1 kinase and exoenzyme S interact with 14-3-3ζ through a common site involving lysine 49. J. Biol. Chem., 272, 13717–13724. [DOI] [PubMed] [Google Scholar]