Casearia arguta was investigated as part of the ongoing search for synergistic TRAIL (tumor necrosis factor-α-related apoptosis-inducing ligand) sensitizers. As a result of this study, argutins A-H, eight new highly oxygenated clerodane diterpenes were isolated from the plant, Casearia arguta, collected in Guatemala. The modified Mosher ester method was utilized to establish the absolute configuration of argutins A and F. Each of the argutins showed varying levels of synergy with TRAIL. Argutin B showed the highest TRAIL sensitization; the synergistic effect of argutin B and TRAIL together was 3-fold greater than argutin B alone.
Tumor necrosis factor-α-related apoptosis-inducing ligand (TRAIL/Apo2L) belongs to the tumor necrosis factor (TNF) family of cytokines that triggers apoptosis when bound to death domain-containing transmembrane receptors, death receptors 4 and 5 (DR4, DR5).1 TRAIL has gained interest as a promising agent for cancer therapy, as it preferentially induces apoptosis in cancer cells, while showing little to no toxicity in normal cells.1, 2 However, when treating with TRAIL alone, the development of resistance has been widely documented.2-6 There is evidence to suggest that combination chemotherapy regimens may be more effective than traditional cytotoxic mono-chemotherapy.7-9 Based on this supposition, a high throughput screen was developed to identify compounds that could sensitize tumor cells to TRAIL.2 The organic extract of the plant Casearia arguta Kunth. (Flacourtiaceae) showed promising activity in the initial screen. Clerodane diterpenes are prevalent in the genus Casearia and the family Flacourtiaceae, with a wide range of reported activities, including cytotoxic, antimalarial, trypanocidal, antifungal, immunosuppressive, and antiproliferative activity.10-24 Casearia arguta had not been reported previously for natural products, suggesting it would be an attractive source for chemical investigation. As a result, eight new clerodane diterpenes, with varying levels of TRAIL synergism, were isolated from C. arguta.
Results and Discussion
Bioassay guided fractionation of the organic extract of C. arguta, utilizing normal phase chromatography, size exclusion chromatography, and reversed phase HPLC resulted in the isolation of argutins A-H (1-8).
The molecular formula for argutin A (1), C34H48O8, was derived from NMR data and the HRESIMS ion at m/z 607.3253 ([M+Na]+; Δ –1.96 ppm). 1D and 2D NMR studies were utilized
 to establish the structure of argutin A (1). Initial interpretation of the NMR data (Tables 1 and 2) indicated that argutin A (1) contained seven quaternary carbons (three carbonyl), 13 methines, eight methylenes, and six methyls. The NMR data also indicated that 1 contained three methyl singlets (δH 0.77, 1.91, 2.05), one methyl doublet (δH 0.88), two oxymethines (δH 3.74, 5.46), two acetal-acyloxy methines (δH 6.49, 6.68), and a trisubstituted olefin moiety (δH 5.98) attributable to the clerodane diterpenoid core prevalent in compounds isolated from Casearia.12-15, 25 COSY (H-1a/H-2; H-2/H-3; H-6/H-7; H-7b/H-8, H-10/H-1a) and HMBC (H-1 → C-2, C10; H-2 → C-3, C-4; H-3 → C-5, C-18; H-19 → C-4, C-5, C-6; H-6 → C-5, C-7; H-7 → C-8, C-9; Me-17 → C-8; Me-20 → C-9; H-10 → C-5, C-8) correlations further supported the basic clerodane skeleton. A six-carbon conjugated diene side chain, comprised of a terminal olefin (δH 5.04, J = 17.3 Hz; 4.88, J = 10.8 Hz), an olefinic methine (δH 6.26, J = 17.3, 10.8 Hz), an olefinic methyl (δH 1.61) and a trisubstituted olefin (δH 5.36), was also present in 1. HMBC correlations (H-11 → C-12, C13; Me-16 → C-12, C-13, C-14; H-15 → C-13, C-14) confirmed the identity of the six-carbon chain, while the HMBC correlation between Me-20 and C-11 supported the placement at C-9. It was clear that 1 possessed an additional C10 diene side chain, characterized by a Z-olefin (δH 5.62, J = 11.4 Hz; 6.57, J = 11.4 Hz), an E-olefin (δH 7.35, J = 15.2, 11.4 Hz; 6.08, J = 15.2, 7.6, 7.1 Hz), and a triplet methyl (δH 0.84, J = 6.7 Hz). The decadienoate portion of 1, located at C-2, was supported by COSY (H-2′/H-3′; H-3′/H-4′; H-4′/H-5′; H-5′/H-6′; H-6′/H-7′; H-7′/H-8′) and HMBC (H-2 → C-1′; H-2′ → C-1′, C-4′; H-3′ → C-5′; H-4′ → C-6′; H-5′ → C-7′, H-6′ → C-7′, C-8′; H-7′ → C-8′, C-9′; H-10′ → C-8′, C-9′) correlations. The 2Z,4E-decadienoate side chain was identical to that of pitumbin, isolated previously from the seeds of C. pitumba.25
to establish the structure of argutin A (1). Initial interpretation of the NMR data (Tables 1 and 2) indicated that argutin A (1) contained seven quaternary carbons (three carbonyl), 13 methines, eight methylenes, and six methyls. The NMR data also indicated that 1 contained three methyl singlets (δH 0.77, 1.91, 2.05), one methyl doublet (δH 0.88), two oxymethines (δH 3.74, 5.46), two acetal-acyloxy methines (δH 6.49, 6.68), and a trisubstituted olefin moiety (δH 5.98) attributable to the clerodane diterpenoid core prevalent in compounds isolated from Casearia.12-15, 25 COSY (H-1a/H-2; H-2/H-3; H-6/H-7; H-7b/H-8, H-10/H-1a) and HMBC (H-1 → C-2, C10; H-2 → C-3, C-4; H-3 → C-5, C-18; H-19 → C-4, C-5, C-6; H-6 → C-5, C-7; H-7 → C-8, C-9; Me-17 → C-8; Me-20 → C-9; H-10 → C-5, C-8) correlations further supported the basic clerodane skeleton. A six-carbon conjugated diene side chain, comprised of a terminal olefin (δH 5.04, J = 17.3 Hz; 4.88, J = 10.8 Hz), an olefinic methine (δH 6.26, J = 17.3, 10.8 Hz), an olefinic methyl (δH 1.61) and a trisubstituted olefin (δH 5.36), was also present in 1. HMBC correlations (H-11 → C-12, C13; Me-16 → C-12, C-13, C-14; H-15 → C-13, C-14) confirmed the identity of the six-carbon chain, while the HMBC correlation between Me-20 and C-11 supported the placement at C-9. It was clear that 1 possessed an additional C10 diene side chain, characterized by a Z-olefin (δH 5.62, J = 11.4 Hz; 6.57, J = 11.4 Hz), an E-olefin (δH 7.35, J = 15.2, 11.4 Hz; 6.08, J = 15.2, 7.6, 7.1 Hz), and a triplet methyl (δH 0.84, J = 6.7 Hz). The decadienoate portion of 1, located at C-2, was supported by COSY (H-2′/H-3′; H-3′/H-4′; H-4′/H-5′; H-5′/H-6′; H-6′/H-7′; H-7′/H-8′) and HMBC (H-2 → C-1′; H-2′ → C-1′, C-4′; H-3′ → C-5′; H-4′ → C-6′; H-5′ → C-7′, H-6′ → C-7′, C-8′; H-7′ → C-8′, C-9′; H-10′ → C-8′, C-9′) correlations. The 2Z,4E-decadienoate side chain was identical to that of pitumbin, isolated previously from the seeds of C. pitumba.25
Table 1.
1H NMR Data for Argutins A – E (1 -5) (CDCl3, 600 MHz)
| argutin A (1) | argutin B (2) | argutin C (3) | argutin D (4) | argutin E (5) | |
|---|---|---|---|---|---|
| |
|||||
| position | δH mult (J, Hz) | δH mult (J, Hz) | δH mult (J, Hz) | δH mult (J, Hz) | δH mult (J, Hz) |
| 1a | 1.97, m | 2.01, m | 2.02, m; (15.1, 13.2, 5.1)c | 2.07, m | 1.95, m |
| 1b | 1.89,a m | 1.96, m | 1.97, m; (15.1, 3.5, 3.1)c | 1.98, m | 1.91,a m |
| 2 | 5.46, br t (4.1) | 4.41, br t (4.1) | 5.51, br t (4.0) | 4.47, br t (4.0) | 5.45, m (4.3) |
| 3 | 5.98, br d (4.1) | 5.98, br d (4.1) | 6.03, br d (4.0) | 6.01, br d (4.0) | 5.95, dd (4.3, 1.6) |
| 6a | 3.74, dd (12.1, 4.1) | 5.00, dd (12.2, 4.2) | 3.52,b m; (9.8)c | 5.08,b d (10.7) | 2.00, dd (13.5, 4.6) |
| 6b | — | — | — | — | 1.44, dd (13.5, 11.4) |
| 7a | 1.69, m | 1.76, ddd (13.4, 7.6, 4.2) | 3.54,b m; (10.7, 9.8)c | 3.66, br t (10.7) | 3.65, ddd (11.4, 11.2, 4.6) |
| 7b | 1.57, m | 1.62, m (13.4, 12.8, 12.2) | — | — | — |
| 8 | 1.76, m (6.7) | 1.88, ddd (12.8, 7.6, 6.8) | 1.65,a m; (10.7, 6.7)c | 1.77, dd (10.7, 6.7) | 1.54, dd (11.2, 6.7) |
| 10 | 2.27, dd (13.0, 4.0) | 2.37, dd (13.5, 3.7) | 2.30, dd (13.2, 4.0) | 2.37,b m | 2.15, dd (13.9, 4.5) |
| 11a | 2.20,b m | 2.24, dd (17.0, 8.5) | 2.38, br dd (16.8, 8.3) | 2.37,b m | 2.34, dd (16.8, 8.1) |
| 11b | 1.63,a m | 1.72, m (17.0) | 1.61, m (16.8) | 1.61, m | 1.61,a m (16.8) |
| 12 | 5.36, br d (7.5) | 5.41, br d (8.5) | 5.36, br d (8.3) | 5.38, br d (8.1) | 5.38 br d (8.1) |
| 14 | 6.26, dd (17.3, 10.8) | 6.29, dd (17.3, 10.9) | 6.28, dd (17.3, 10.7) | 6.26, dd (17.3, 10.8) | 6.28, dd (17.3, 10.8) |
| 15a | 5.04, d (17.3) | 5.07, d (17.3) | 5.09, d (17.3) | 5.07,b d (17.3) | 5.08, d (17.3) |
| 15b | 4.88, d (10.8) | 4.91, d (10.9) | 4.92, d (10.7) | 4.91, d (10.8) | 4.91, d (10.8) |
| 16 | 1.61, s | 1.64, s | 1.64, s | 1.64, s | 1.64, s |
| 17 | 0.88, d (6.7) | 0.90, d (6.8) | 1.04, d (6.7) | 1.04, d (6.7) | 1.01, d (6.7) |
| 18 | 6.68, t (1.6) | 6.45, t (1.6) | 6.73, t (1.5) | 6.40, t (1.5) | 6.68, t (1.6) |
| 19 | 6.49, br s | 6.59, br s | 6.49, br s | 6.55, s | 6.33, br s |
| 20 | 0.77, s | 0.86, s | 0.84, s | 0.88, s | 0.90, s |
| 2′ | 5.62, d (11.4) | 5.57, d (11.3) | 5.64, d (11.3) | 5.63, d (11.2) | 5.68, d (11.3) |
| 3′ | 6.57, t (11.4) | 6.58, t (11.3) | 6.60, t (11.3) | 6.63, t (11.2) | 6.60, t (11.3) |
| 4′ | 7.35, dd (15.2, 11.4) | 7.32, dd (15.2, 11.3) | 7.38, dd (15.2, 11.3) | 7.30, dd (15.2, 11.2) | 7.43, dd (15.1, 11.3) |
| 5′ | 6.08, ddd (15.2, 7.6, 7.1) | 6.09, ddd (15.2, 7.7, 7.1) | 6.11, ddd (15.2, 7.7, 7.2) | 6.09, ddd (15.2, 7.7, 7.2) | 6.10, ddd (15.1, 7.6, 7.1) |
| 6′ | 2.16,b ddd (14.6, 7.6, 7.2) | 2.17, ddd (14.6, 7.5, 7.1) | 2.18, ddd (14.7, 7.7, 7.2) | 2.14, ddd (14.7, 7.7, 7.3) | 2.20, ddd (14.6, 7.6, 7.2) |
| 7′ | 1.40, ddd (14.6, 7.4, 7.2) | 1.41, ddd (14.6, 7.5, 7.3) | 1.43, ddd (14.7, 7.4, 7.2) | 1.38, ddd (14.8, 7.6, 7.3) | 1.41, ddd (14.6, 7.4, 7.2) |
| 8′ | 1.24,b m | 1.26,b m | 1.28,b m | 1.23,b m | 1.26,b m |
| 9′ | 1.25,b m (6.7) | 1.27,b m (6.8) | 1.28,b m (6.8) | 1.26,b m (6.9) | 1.27,b m (6.8) |
| 10′ | 0.84, t (6.7) | 0.86, t (6.8) | 0.87, t (6.8) | 0.84, t (6.9) | 0.90, t (6.8) |
| 18-OAc | 2.05, s | 2.05, s | 2.09, s | 2.04, s | 2.09, s |
| 19-OAc | 1.91, s | 1.93, s | 1.93, s | 1.91, s | 1.93, s |
Signal obscured by overlapping methyl signal.
Signals overlapped.
Measured in acetone-d6.
Table 2.
13C NMR Data for Argutins A – E (1 -5) (CDCl3, 150 MHz)
| argutin A (1) | argutin B (2) | argutin C (3) | argutin D (4) | argutin E (5) | |
|---|---|---|---|---|---|
| |
|||||
| position | δc, mult | δc, multa | δc, multa | δc, multa | δc, multa |
| 1 | 27.2, CH2 | 29.6 | 27.1 | 30.1 | 26.5 |
| 2 | 66.0, CH | 64.1 | 65.6 | 64.3 | 65.9 |
| 3 | 122.1, CH | 126.4 | 122.4 | 127.6 | 121.3 |
| 4 | 145.2, C | 142.7 | 144.2 | 141.6 | 145.8 |
| 5 | 53.6, C | 52.3 | 52.6 | 52.8 | 49.7 |
| 6 | 73.1, CH | 73.4, CH | 76.7, CH | 76.9, CH | 37.8, CH2 |
| 7 | 37.5, CH2 | 33.4, CH2 | 73.6, CH | 72.8, CH | 69.4, CH |
| 8 | 36.7, CH | 36.4 | 42.2 | 43.9 | 43.8 |
| 9 | 37.8, C | 37.8 | 39.0 | 39.4 | 39.4 |
| 10 | 37.1, CH | 36.6 | 36.5 | 37.1 | 34.6 |
| 11 | 30.6, CH2 | 30.5 | 31.6 | 32.3 | 31.9 |
| 12 | 129.3, CH | 129.4 | 128.6 | 129.5 | 128.7 |
| 13 | 135.8, C | 135.8 | 135.8 | 135.9 | 136.1 |
| 14 | 141.5, CH | 141.4 | 141.2 | 141.7 | 141.4 |
| 15 | 111.1, CH2 | 111.2 | 111.2 | 112.0 | 111.3 |
| 16 | 12.1, CH3 | 12.1 | 12.0 | 12.8 | 12.2 |
| 17 | 15.8, CH3 | 15.7 | 11.0 | 11.9 | 11.0 |
| 18 | 96.0, CH | 95.7 | 95.6 | 95.7 | 94.6 |
| 19 | 97.4, CH | 97.8 | 97.5 | 98.3 | 99.1 |
| 20 | 25.1, CH3 | 25.3 | 25.4 | 26.3 | 25.9 |
| 1′ | 166.2, C | 166.0 | 165.9 | 166.3 | 166.2 |
| 2′ | 115.6, CH | 114.7 | 115.2 | 114.6 | 115.5 |
| 3′ | 146.2, CH | 147.5 | 146.2 | 148.8 | 146.3 |
| 4′ | 127.0, CH | 127.0 | 126.8 | 127.6 | 127.0 |
| 5′ | 146.7, CH | 147.3 | 146.7 | 148.2 | 146.9 |
| 6′ | 33.0, CH2 | 33.2 | 33.1 | 33.7 | 33.3 |
| 7′ | 28.7, CH2 | 28.6 | 28.5 | 28.8 | 28.8 |
| 8′ | 31.6, CH2 | 31.6 | 31.5 | 31.6 | 31.7 |
| 9′ | 22.7, CH2 | 22.7 | 22.5 | 23.3 | 22.7 |
| 10′ | 14.2, CH3 | 14.2 | 14.0 | 14.9 | 14.2 |
| 18-OAc | 170.4, C | 170.3 | 170.3 | 170.1 | 170.5 |
| 21.5, CH3 | 21.5 | 21.3 | 21.7 | 21.5 | |
| 19-OAc | 169.8, C | 169.7 | 169.4 | 169.4 | 169.9 |
| 21.9, CH3 | 22.0 | 21.7 | 22.3 | 21.7 | |
Multiplicities are identical to those of argutin A unless specifically noted.
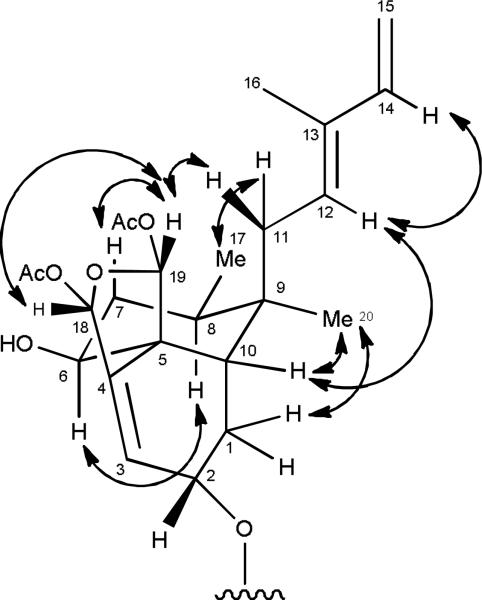
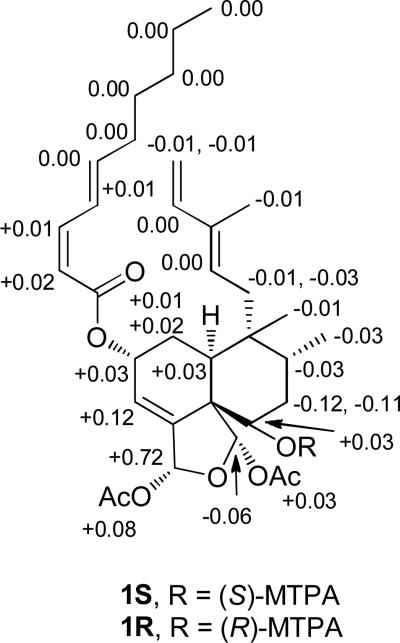
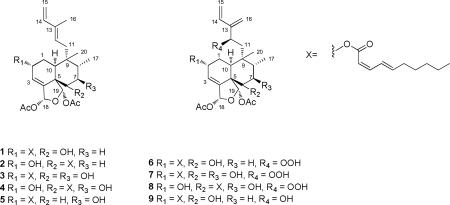
The relative configuration of argutin A (1) was determined on the basis of the coupling constants, ROESY data (Figure 1), and 13C NMR chemical shifts. An ROE observed between H-12 and H-14 supported an E geometry at C-12. The large coupling constant observed between H-6 and H-7b (J = 12.1 Hz) indicated that H-6 was axial, and an ROE observed between H-6 and H-8 implied that H-8 was axial and Me-17 was equatorial. ROE correlations between H-10 and Me-20; H-10 and H-12; and Me-20 and H-1b suggested that Me-20 and H-10 (with respect to the B-ring) were equatorial, and that the A/B ring juncture was cis. ROE correlations were observed between Me-17 and H-11a; H-11b and H-19; and H-19 and H-7b, which indicated H-19 had a β-orientation. The allylic coupling observed between H-3 and H-18 (J = 1.6 Hz) and the ROE correlation observed between H-18 and H-19 suggested that H-18 also had a β-orientation. The 13C NMR chemical shift of C-2 (δC 66.0),13, 19, 26 the narrow multiplicity of H-2 in the 1H NMR spectrum, and the lack of an ROE between H-2 and H-10 were consistent with the C-2 substituent having an α- orientation. The modified Mosher ester method14, 27-29 was utilized to establish the absolute configuration of C-6 and, therefore, the rest of the molecule from the aforementioned data. Reaction of 1 with (R)- and (S)-MTPA chloride yielded the (S)-MTPA ester (1S) and (R)-MTPA ester (1R), respectively. The observed chemical shift differences (ΔδSR = δS – δR) between 1S and 1R indicated that the absolute configuration of C-6 is S (Figure 2). Therefore, argutin A (1) was assigned as (2R,5S,6S,8R,9R,10S,18R,19S)-18,19-diacetoxy-18,19-epoxy-2-[(2′Z,4′E)-decadienoyloxy]-6-hydroxycleroda-3,12(E),14-triene.
Figure 1.
Key ROE correlations supporting the configuration of argutin A (1).
Figure 2.
ΔδSR values (ΔδSR = δS – δR) for the MTPA esters 1S and 1R.
The molecular formula for argutin B (2), C34H48O8, was derived from NMR data and the HRESIMS ion at m/z 607.3240 ([M+Na]+; Δ + 0.21 ppm). Compounds 1 and 2 were isomeric and showed a few disparities in their 1H NMR spectra that suggested their substitution patterns were different. The 2D NMR data confirmed that 2 contained the 6-carbon diene and decadienoate side chains seen in 1. Major variations between H-2 and H-6 in 1 and 2 (Table 1) suggested the decadienoate side chain in 2 was located at C-6. The C-6 location of the decadienoate portion was confirmed based on an HMBC correlation from H-6 to C-1′. The measureable coupling constants in 1 and 2 are virtually indistinguishable and the observed ROE correlations for 2 support argutin B (2) having the same configuration as 1. Thus, from a biosynthetic standpoint, their absolute configurations are the same. Therefore, argutin B (2) was assigned as (2R,5S,6S,8R,9R,10S,18R,19S)-18,19-diacetoxy-18,19-epoxy-2-hydroxy-6-[(2′Z,4′E)-decadienoyloxy]cleroda-3,12(E),14-triene.
The molecular formula for argutin C (3), C34H48O9, derived from the HRESIMS ion at m/z 623.3212 ([M+Na]+; Δ –3.69 ppm), indicated that 3 contained one oxygen atom more than 1 and 2. Comparison of the NMR data of 1 and 3 revealed that 3 contained an additional oxymethine (δH 3.54; δC 73.6) in place of a methylene (Tables 1 and 2). HMBC correlations (H-7 → C-6; H-6 → C-7; Me-17 → C-7) confirmed the location of the additional hydroxy group at C-7. The coupling constants of H-6, H-7, and H-8 could not be readily measured in CDCl3, due to overlap, but were measurable in acetone- d6 (Table 1). H-7 showed 1,2-diaxial coupling (J = 10.7, 9.8 Hz) with H-8 and H-6, respectively. ROEs were observed between H-7/H-19 and H-7/H-11b, which confirmed H-7 was axial. The observed ROE correlations, coupling constants, and biosynthetic precedent supported argutin C (3) having the same absolute configuration as 1. As a result, argutin C (3) was assigned as (2R,5S,6S,7R,8S,9S,10S,18R,19R)-18,19-diacetoxy-18,19-epoxy-2-[(2′Z,4′E)-decadienoyloxy)-6,7-dihydroxy]cleroda-3,12(E),14-triene.
The molecular formula for argutin D (4), C34H48O9, was derived from NMR data and the HRESIMS ion at m/z 623.3207 ([M+Na]+; Δ –2.63 ppm). Compounds 3 and 4 were isomeric, and much like 1 and 2, their substitution patterns appeared to be different. Argutin D (4) showed similarities to the substitution pattern of 2, with the addition of an oxymethine (δH 3.66; δC 72.8) and loss of a methylene. HMBC correlations (H-6 → C-1′; H-7 → C-6; H-6 → C-7; Me-17 → C-7) confirmed the C-6 location of the decadienoate moiety and the C-7 location of the additional hydroxy group. In compound 4, H-7 also exhibited 1,2-diaxial coupling (J = 10.7 Hz) with both H-6 and H-8. As in 3, ROEs were observed between H-7/H-19 and H-7/H-11b, confirming that H-7 was axial. The above evidence, additional observed ROE correlations, similar coupling constants, and biosynthetic precedent substantiated the assignment of argutin D (4) as (2R,5S,6S,7R,8S,9S,10S,18R,19R)-18,19-diacetoxy-18,19-epoxy-2,7-dihydroxy-6-[(2′Z,4′E)-decadienoyloxy]cleroda-3,12(E),14-triene.
Argutin E (5), with the molecular formula, C34H48O8, derived from the HRESIMS ion at m/z 607.3242 ([M+Na]+; Δ –0.16 ppm), was isomeric with 1 and 2. Compound 5 was more similar to 1 than 2, but one of the major distinctions between 1 and 5, made apparent by the HSQC spectra, was a methylene pair; the H-7 methylenes of 1 (δH 1.69, 1.57) were very different from the methylene pair of 5 (δH 2.00, 1.44). A number of variations in the 1H and 13C NMR chemical shifts of argutin E (5) around C-7, as compared to 1, pointed to the hydroxy group residing at C-7 rather than C-6 (Tables 1 and 2). COSY (H-6a/H-7; H-6b/H-7; H-7/H8) and HMBC (H-6a → C-5, C19; H-7 → C-17; Me-17 → C-7) correlations confirmed this notion. The coupling constants of 5 were also resolved, and H-7 showed 1,2-diaxial coupling with both H-6b and H-8. All of these data, in addition to further coupling constants, observed ROE correlations, and biosynthetic precedent supported the assignment of argutin E (5) as (2R,5S,7S,8S,9S,10S,18R,19S)-18,19-diacetoxy-18,19-epoxy-2-[(2′Z,4′E)-decadienoyloxy]-7-hydroxycleroda-3,12(E),14-triene.
The molecular formula for argutin F (6), C34H48O10, derived from the HRESIMS ion at m/z 639.3146 ([M+Na]+; Δ –1.10 ppm), indicated that 6 contained one oxygen atom more than 3 and 4. Initial interpretation of the NMR data (Tables 1 and 2) showed that argutin F (6) contained seven quaternary carbons (3 carbonyl), 13 methines, nine methylenes, and five methyls. The NMR data also indicated that the clerodane core was intact in 6 (three methyl singlets (δH 1.02, 1.97, 2.06), one methyl doublet (δH 1.06), two oxymethines (δH 3.72, 5.46), two acetal-acyloxy methines (δH 6.41, 6.72), and a trisubstituted olefin (δH 5.97). The substitution pattern around the clerodane skeleton was most similar to 1 (C-2 decadienoate, C-6 hydroxy), which suggested the six-carbon side chain at C-9 was oxygenated in 6. The C6 side chain contained two terminal olefins (δH 5.26, 5.16; 5.46, 5.15), and an unusual oxymethine (δH 4.73; δC 83.4) that suggested the presence of a hydroperoxy group. The downfield 1H and 13C NMR shifts of the oxymethine,30-32 the lack of an additional oxymethine signal, and a positive TLC spray test for peroxides (using N,N-dimethyl-1,4-phenylenediammonium dichloride)33 all supported the presence of a hydroperoxy group in 6. COSY (H-12/H11a; H-12/H-11b) and HMBC correlations (H-12 → C-11, C-13, C-14, C-16; H-14 → C-13, C-15) indicated the hydroperoxy was located at C-12, and confirmed the identity of the 6-carbon diene side chain in argutin F (6), as drawn.
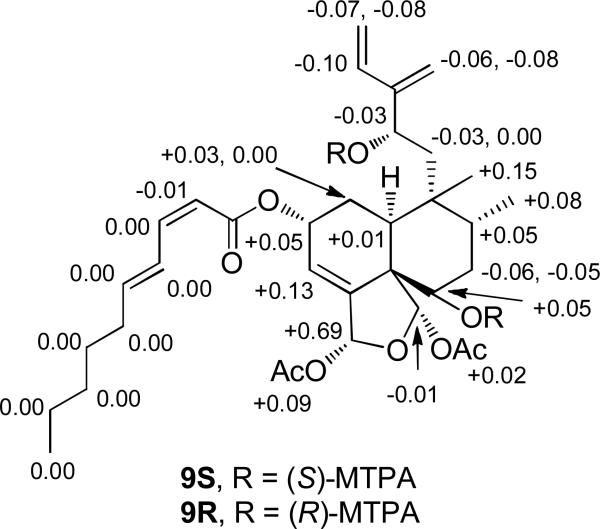
The observed ROE correlations, coupling constants, and biosynthetic precedent supported argutin F (6) having the same relative configuration as 1 in the clerodane ring system (C-2, C-5, C-6, C-8, C-9, C-10, C-18, C-19). In an effort to determine the absolute configuration of C-12, the hydroperoxy functionality in argutin F (6) was reduced using Ph3P to give 9. As with argutin A (1), the modified Mosher ester method14, 27-29 was utilized to establish the absolute configuration of C-12. Reaction of 9 with (R)- and (S)-MTPA chloride yielded the (S)-MTPA diester (9S) and (R)-MTPA diester (9R), respectively. The observed chemical shift differences (ΔδSR = δS – δR) between 9S and 9R indicated that the absolute configuration of C-12 is S (Figure 3). Additionally, the observed chemical shift differences supported the absolute configuration of C-6 as S, as was seen in argutin A (1). Therefore, argutin F (6) was assigned as (2R,5S,6S,8R,9R,10S,12S,18R,19S)-18,19-diacetoxy-18,19-epoxy-2-[(2′Z,4′E)-decadienoyloxy]6-hydroxy-12-hydroperoxycleroda-3,13(16),14-triene.
Figure 3.
ΔδSR values (ΔδSR = δS – δR) for the MTPA esters 9S and 9R.
The molecular formula for argutin G (7), C34H48O11, derived from the HRESIMS ion at m/z 655.3097 ([M+Na]+; Δ –1.42 ppm), revealed that 7 contained an additional oxygen atom more than 6. When compared with the NMR data of 6 (Table 3), it was evident that 7 also contained the allylic hydroperoxide (δH 4.80; δC 82.8), as well as two hydroxy groups. COSY (H-6/H-7; H-7/H-8) and HMBC correlations (H-8 → C-7; Me-17 → C-7) confirmed the additional hydroxy was located at C-7. The coupling constants of 7 were resolved, and revealed 1,2-diaxial coupling between H-7 and both H-6 and H-8. The observed ROE correlations, coupling constants, and biosynthetic precedent supported argutin G (7) having the same absolute configuration as 6. As a result, argutin G (7) was assigned as (2R,5S,6S,7R,8R,9R,10S,12S,18R,19S)-18,19-diacetoxy-18,19-epoxy-2-[(2′Z,4′E)-decadienoyloxy]6,7-dihydroxy-12-hydroperoxycleroda-3,13(16),14-triene.
Table 3.
NMR Data for Argutins F-H (6 -8) (600 MHz, CDCl3)
| argutin F (6) | argutin G (7) | argutin H (8) | ||||
|---|---|---|---|---|---|---|
| |
||||||
| position | δC, mult | δH mult (J, Hz) | δC, multa | δH mult (J, Hz) | δC, multa | δH mult (J, Hz) |
| 1a | 27.5, CH2 | 2.00, m | 27.5 | 2.04, m | 29.5 | 2.09, m |
| 1b | 1.89, m | 1.95,b m | 2.01,c m | |||
| 2 | 66.4, CH | 5.46, br t (4.0) | 65.5 | 5.49,b m (3.7) | 63.6 | 4.45, m (4.0) |
| 3 | 122.2, CH | 5.97, br d (4.0) | 122.3 | 6.02, br d (3.7) | 127.0 | 5.99, br d (4.0) |
| 4 | 145.3, C | — | 146.8 | — | 141.9 | — |
| 5 | 54.0, C | — | 52.8 | — | 53.2 | — |
| 6 | 73.8, CH | 3.72, dd (10.8, 4.3) | 77.0 | 3.54, d (9.7) | 76.3 | 5.06, d (10.2) |
| 7a | 38.1, CH2 | 1.70,b m | 73.2, CH | 3.68, dd (10.2, 9.7) | 71.7, CH | 3.76, dd (10.7, 10.2) |
| 7b | 1.65, m | — | — | |||
| 8 | 36.6, CH | 1.70,b m (6.2) | 42.2 | 1.65, m (10.2, 6.9) | 43.0 | 1.76,b m (10.7, 7.0) |
| 9 | 38.6, C | — | 39.0 | — | 39.4 | — |
| 10 | 42.6, CH | 1.92, dd (13.6, 5.0) | 41.8 | 1.94,b m | 40.8 | 2.01,c m |
| 11a | 38.9, CH2 | 1.78, dd (15.6, 6.8) | 39.7 | 1.80, m | 39.3 | 1.78,b m |
| 11b | 1.22, m (15.6) | 1.27,b | 1.25,b m | |||
| 12 | 83.4, CH | 4.73, dd (6.8, 2.4) | 82.8 | 4.80, dd (6.6, 2.4) | 82.7 | 4.80, dd (7.3, 2.1) |
| 13 | 146.2, C | — | 146.2 | — | 146.1 | — |
| 14 | 135.6, CH | 6.27, dd (17.7, 11.3) | 135.6 | 6.27, dd (17.6, 11.2) | 135.5 | 6.27, dd (17.7, 11.3) |
| 15a | 116.7, CH2 | 5.46, d (17.7) | 116.3 | 5.47,b (17.6) | 116.6 | 5.47, d (17.7) |
| 15b | 5.15, d (11.3) | 5.13, dd (11.2) | 5.14, d (11.3) | |||
| 16a | 117.5, CH2 | 5.26, s | 117.3 | 5.24, s | 117.1 | 5.27, s |
| 16b | 5.16, s | 5.16, s | 5.17, s | |||
| 17 | 16.2, CH3 | 1.06, d (6.2) | 10.7 | 1.20, d (6.9) | 10.7 | 1.20, d (7.0) |
| 18 | 96.0, CH | 6.72, t (1.4) | 95.4 | 6.75, br s | 95.0 | 6.40, br t (1.6) |
| 19 | 98.1, CH | 6.41, br s | 97.9 | 6.44, br s | 98.0 | 6.45, br s |
| 20 | 24.2, CH3 | 1.02, s | 24.6 | 1.04, s | 24.8 | 1.14, s |
| 1′ | 166.2, C | — | 166.1 | — | 166.5 | — |
| 2′ | 115.7, CH | 5.59, d (11.3) | 115.4 | 5.60, d (11.3) | 114.2 | 5.64, d (11.3) |
| 3′ | 146.8, CH | 6.55, t (11.3) | 146.7 | 6.59, t (11.3) | 148.4 | 6.63, t (11.3) |
| 4′ | 127.4, CH | 7.36, dd (15.3, 11.3) | 127.2 | 7.37, dd (15.2, 11.3) | 127.1 | 7.32, dd (15.2, 11.3) |
| 5′ | 147.0, CH | 6.09, ddd (15.3, 7.7, 7.2) | 147.3 | 6.11, ddd (15.3, 7.6, 7.3) | 147.4 | 6.10, ddd (15.2, 7.6, 7.2) |
| 6′ | 33.6, CH2 | 2.18, ddd (14.3, 7.2, 7.0) | 33.3 | 2.19, ddd (14.4, 7.3, 7.1) | 33.5 | 2.15, ddd (14.4, 7.2, 7.0) |
| 7′ | 29.0, CH2 | 1.41, ddd (14.7, 7.4, 7.0) | 28.6 | 1.42, ddd (14.7, 7.4, 7.1) | 28.8 | 1.40, ddd (14.6, 7.4, 7.0) |
| 8′ | 31.9, CH2 | 1.26,b m | 31.7 | 1.28,b m | 31.9 | 1.25,b m |
| 9′ | 22.9, CH2 | 1.28,b m (6.8) | 22.6 | 1.29,b m (6.7) | 23.0 | 1.27,b m (6.9) |
| 10′ | 14.7, CH3 | 0.86, t (6.8) | 14.0 | 0.87, t (6.7) | 13.8 | 0.85, t (6.9) |
| 18-OAc | 171.1, C | — | 170.8 | — | 170.5 | — |
| 21.6, CH3 | 2.06, s | 21.4 | 2.08, s | 21.2 | 2.02, s | |
| 19-OAc | 169.9, C | — | 170.0 | — | 169.8 | — |
| 22.2, CH3 | 1.97, s | 21.7 | 1.97, s | 21.8 | 1.94, s | |
Carbon multiplicities are identical to those of argutin F unless specifically noted.
0 Signals overlapped.
Signal obscured by overlapping methyl signal.
Argutin H (8) was isomeric with 7, with the molecular formula, C34H48O11, derived from the HRESIMS ion at m/z 655.3081 ([M+Na]+; Δ – 1.22 ppm). Argutin H (8) also contained the allylic hydroperoxide, and showed similarities to the substitution pattern of 4 (C-2 and C-7 hydroxyl, C-6 decadienoate). COSY (H-1/H-2; H-2/H-3; H-6/H-7; H-7/H-8) and HMBC correlations (H-6 → C-7; Me-17 → C-7, C-8) confirmed the locations of the decadienoate and hydroxy groups. 1,2-diaxial coupling was also evident between H-6/H-7 and H-7/H-8 (Table 3) in 8. Following the pattern of the other argutins, taking into account the observed ROE correlations and coupling constants, argutin H (8) was assigned as (2R,5S,6S,7R,8R,9R,10S,12S,18R,19S)-18,19-diacetoxy-18,19-epoxy-2,7-dihydroxy-6-[(2′Z,4′E)-decadienoyloxy]-12-hydroperoxycleroda-3,13(16),14-triene.
All of the compounds described herein fall into the “synergistic/toxic” category described previously.2 All have some toxicity alone, but synergize with TRAIL to kill cells (greater than an additive effect in combination). In this regard, the argutins are more potent in the presence of TRAIL than in its absence. ACHN cells are not sensitive to recombinant TRAIL ligand at concentrations of up to 10 μg/mL, but they can be sensitized with pre-exposure to certain chemical sensitizers (e.g. the proteasome inhibitor bortezomib).2 The TRAIL activity of argutins A-H (1-8) is outlined in Table 4. Argutin B (2) showed the highest degree of TRAIL sensitization of the argutins; the synergistic effect of 2 and TRAIL together was 3-fold greater than 2 alone. Argutin H (8) showed the lowest synergistic effect, with the effect of 8 and TRAIL combined being only 1.3 times greater than that of 8 alone. The C-9 side chain, containing a hydroperoxide and two terminal olefin moieties, present in argutins F-H (7-9) negatively affects potency and synergy; argutins A (1), C (3), and D (4) have higher synergistic effects and potencies than argutins F (6), G (7), and H (8). The position of the decadienoate appears to affect potency in ACHN cells; argutins with the decadienoate at C-2 (1, 3, and 7) are more potent (2-5.4×) than comparable argutins with the decadienoate at C-6 (2, 4, and 8). The relationship between the decadienoate position and synergy is harder to define; argutin B (2) has a higher synergistic effect than argutin A (1), but argutins D (4) and G (7) both have lower synergistic effects than argutins C (3) and H (8), respectively. An additional hydroxy group at C-7 makes the synergistic effect lower; argutins A (1) and B (2) have higher synergistic effects than argutins C (3) and D, respectively. A hydroxy group at C-7 rather than C-6 also negatively affects synergy and potency; argutin A (1) has a higher synergistic effect and potency than argutin E (5).
Table 4.
Biological Effect of Argutins A-H (1-8) with and without TRAIL
| Compound | EC50 w/ TRAIL (μM) | EC50 w/o TRAIL (μM) | Synergistic Effecta |
|---|---|---|---|
| argutin A (1) | 2.4 | 5.4 | 2.3 |
| argutin B (2) | 4.6 | 13.8 | 3.0 |
| argutin C (3) | 1.6 | 3.1 | 1.9 |
| argutin D (4) | 8.7 | 14.3 | 1.6 |
| argutin E (5) | 2.2 | 3.3 | 1.5 |
| argutin F (6) | 7.1 | 10.4 | 1.5 |
| argutin G (7) | 2.8 | 4.2 | 1.5 |
| argutin H (8) | 9.8b | 12.6b | 1.3 |
Synergistic effect is defined as the ratio of the two half-maximal (EC50) values obtained from cell killing curves.
EC50 is estimated. Argutin H contains impurities and was isolated in small quantities; further purification would have resulted in loss of material.
Pitumbin is the only other clerodane diterpene that contains the 2Z,4E-decadienoate side chain seen in argutins A-H (1-8).25 However, corymbulosin A contains a 2Z,4Z-decadienoate moiety, and there are several examples of clerodane diterpenes with decanoate groups.12, 26, 34-36 The hydroperoxide-containing side chain present in argutins F-H (6-8) has never been reported, but the reduced form of the side chain, with a hydroxy group at C-12, is present in casearlucins H-K,13 and caseamembrol B.37
Experimental Section
General Experimental Procedures
Optical rotations were measured on a Perkin-Elmer 241 polarimeter. UV spectra were acquired in spectroscopy grade MeOH using a Varian Cary 50 UV-Vis spectrophotometer. NMR data were collected using a Bruker Avance III 600 (1H 600 MHz, 13C 150 MHz) NMR spectrometer (Bruker Biospin) with a 3-mm PATXI probe, referenced to residual solvent (δH 7.24, δC 77.24 for CDCl3). MS spectra were measured with an Agilent Technologies 6510 Q-TOF LC-MS and an Applied Biosystems, Inc. QSTAR XL hybrid triple-quad time-of-flight (QqTOF) mass spectrometer. Initial purification was performed on Diol SPE cartridges (Applied Separations) and Sephadex LH-20 resin (Amersham Biosciences). HPLC was performed on a Rainin SD-1/UV-1 system.
Plant Material
The leaves of C. arguta Kunth. (Flacourtiaceae) were collected near Puerto Barrios, Izabal, Guatemala (March 21, 1988) by N. Marshall of the New York Botanical Gardens under contract to the National Cancer Institute. The plant was identified by N. Trushell, and a voucher specimen (collection number Q65V430) is maintained at the New York Botanical Gardens.
Extraction and Isolation
The leaves of C. arguta (0.91 kg) were extracted successively with DCM/MeOH (1:1) and MeOH.38 The combined extracts were reduced to dryness in vacuo to give 85.05 g of crude extract. A portion of this extract (0.7 g) was separated by six Diol SPE cartridges (2 g resin each), and the equivalent fractions were combined to give five fractions; Fraction A = 9:1 hexanes/DCM, Fraction B = 20:1 DCM/EtOAc, Fraction C = EtOAc, Fraction D = 5:1 EtOAc/MeOH, Fraction E = MeOH. Size exclusion chromatography of the active fraction B on Sephadex LH-20 (2.5 × 90 cm) using hexanes/DCM/MeOH (2:5:1) yielded eight fractions (102A-102H). Fraction 102E was chromatographed on C18 (3.0 × 35 cm) using a step gradient of 70% CH3CN/H2O (+ 0.1% AcOH) to 100% CH3CN in 5 % steps, to yield argutin E (1.5 mg) and 10 other fractions (104B-104K). Fractions 104C and 104D were purified by HPLC using a Rainin Dynamax C18 column (250 × 10 mm) employing a gradient of 78% CH3CN/22% H2O (+0.1% AcOH) to 100% CH3CN at 4.5 mL/min over 15 min, followed by 7 min of 100% CH3CN to yield argutins A (12.0 mg), B (6.1 mg), D (2.5 mg), and F (2.1 mg). Fraction 104B was purified by HPLC utilizing the same method to yield argutin C (12.6 mg), as well as fraction 83B. Fraction 83B was further purified by HPLC using a Rainin Dynamax C18 column (250 × 10 mm) employing a gradient of 75% CH3CN/25% H2O (+0.1% AcOH) to 100% CH3CN at 4.5 mL/min over 15 min to yield argutins G (0.9 mg) and H (0.7 mg).
TRAIL assay
The biological activity of the extract, chromatographic fraction, or pure compound was monitored using the screening assay described previously.2 Briefly, ACHN cell numbers were assessed after 20-24h treatment with varying concentrations of extract, fraction, or pure compound in the absence or presence of 40 ng/mL TRAIL.
Argutin A (1): [α]25D +11.8 (c 0.20, MeOH); UV (MeOH) λmax (log ε) 265 (4.31) 233 (4.42) 205 (4.20) nm; 1H NMR and 13C NMR data, see Table 1 and 2; HRESIMS m/z 607.3253 [M+Na]+ (calcd for C34H48O8Na, 607.3241).
Argutin B (2): [α]25D +9.8 (c 0.20, MeOH); UV (MeOH) λmax (log ε) 267 (3.70) 232 (3.75) 205 (3.49) nm; 1H NMR and 13C NMR data, see Table 1 and 2; HRESIMS m/z 607.3240 [M+Na]+ (calcd for C34H48O8Na, 607.3241).
Argutin C (3): [α]25D +25.5 (c 0.20, MeOH); UV (MeOH) λmax (log ε) 265 (4.26) 232 (4.40) 205 (4.08) nm; 1H NMR and 13C NMR data, see Table 1 and 2; HRESIMS m/z 623.3212 [M+Na]+ (calcd for C34H48O9Na, 623.3191).
Argutin D (4): [α]25D +10.0 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 266 (3.57) 232 (3.65) 205 (3.58) nm; 1H NMR and 13C NMR data, see Table 1 and 2; HRESIMS m/z 623.3207 [M+Na]+ (calcd for C34H48O9Na, 623.3191).
Argutin E (5): [α]25D +6.7 (c 0.20, MeOH); UV (MeOH) λmax (log ε) 265 (3.77) 232 (3.90) 204 (3.75) nm; 1H NMR and 13C NMR data, see Table 1 and 2; HRESIMS m/z 607.3242 [M+Na]+ (calcd for C34H48O8Na, 607.3241).
Argutin F (6): [α]25D +15.0 (c 0.20, MeOH); UV (MeOH) λmax (log ε) 265 (4.00) 226 (3.87) 206 (3.96) nm; 1H NMR and 13C NMR data, see Table 3; HRESIMS m/z 639.3146 [M+Na]+ (calcd for C34H48O10Na, 639.3140).
Argutin G (7): [α]25D +21.7 (c 0.20, MeOH); UV (MeOH) λmax (log ε) 265 (4.15) 225 (4.00) 204 (4.12) nm; 1H NMR and 13C NMR data, see Table 3; HRESIMS m/z 655.3097 [M+Na]+ (calcd for C34H48O11Na, 655.3089).
Argutin H (8): [α]25D +13.3 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 265 (3.78) 226 (3.39) 204 (3.50) nm; 1H NMR and 13C NMR data, see Table 3; HRESIMS m/z 655.3081 [M+Na]+ (calcd for C34H48O11Na, 655.3089).
MTPA Esterification of Argutin A (1)
DMAP (cat.) and (R)-MTPA chloride (3 μL, 16 μmol) were added to a solution of 1 (1.2 mg, 2.1 μmol) in anhydrous pyridine (200 μL), which was allowed to stir under N2 at room temperature overnight. The solution was dried, concentrated in vacuo, and purified by HPLC (C18, 250 × 10 mm) using a gradient of 50% CH3CN/50% H2O (+0.1% AcOH) to 100% CH3CN at 4.5 mL/min over 27 min to yield the (S)-MTPA ester 1S (1.5 mg, 91%). 1H NMR and 13C NMR data, see Supplemental Table 1; HRESIMS m/z 823.3653 [M+Na]+ (calcd for C44H55F3O10Na, 823.3640).
Another aliquot of 1 (1.2 mg, 2.1 μmol) was treated with (S)-MTPA chloride, following the procedure outlined above, to give the (R)-MTPA ester 1R (1.4 mg, 85%). 1H NMR and 13C NMR data, see Supplemental Table 1; HRESIMS m/z 823.3662 [M+Na]+ (calcd for C44H55F3O10Na, 823.3640).
Reduction of Argutin F (6)
PPh3 (1.84 mg, 7.0 μmol) was added to a solution of 6 (1.0 mg, 1.6 μmol) in anhydrous DCM (500 μL), which was allowed to stir under N2 at room temperature overnight. The solution was dried, concentrated in vacuo, and purified by HPLC (C18, 250 × 10 mm) using a gradient of 78% CH3CN/22% H2O (+0.1% AcOH) to 100% CH3CN at 4.5 mL/min over 15 min to yield 9 (0.8 mg, 82%). 1H NMR and 13C NMR data, see Supplemental Table 2; LRESIMS m/z 623.5 [M+Na]+.
MTPA Esterification of Deoxoargutin F (9)
DMAP (cat.) and (R)-MTPA chloride (3 μL, 16 μmol) were added to a solution of 9 (0.4 mg, 0.7 μmol) in anhydrous pyridine (200 μL), which was allowed to stir under N2 at room temperature overnight. The solution was dried, concentrated in vacuo, and purified by HPLC (C18, 250 × 10 mm) using a gradient of 78% CH3CN/22% H2O (+0.1% AcOH) to 100% CH3CN at 4.5 mL/min over 15 min, followed by 7 min of 100% CH3CN to yield the (S)-MTPA ester 9S (0.6 mg, 87%). 1H NMR and 13C NMR data, see Supplemental Table 3; HRESIMS m/z 1055.3993 [M+Na]+ (calcd for C54H62F6O13Na, 1055.3987).
Another aliquot of 9 (0.4 mg, 0.7 μmol) was treated with (S)-MTPA chloride, following the procedure outlined above, to give the (R)-MTPA ester 9R (0.5 mg, 72%). 1H NMR and 13C NMR data, see Supplemental Table 3; HRESIMS m/z 1055.3950 [M+Na]+ (calcd for C54H62F6O13Na, 1055.3987).
Supplementary Material
Acknowledgment
The authors would like to thank N. Marshall (NYBG), N. Trushell (NYBG), D. Newman (NPB) and T. McCloud (NPSG) for collection, identification, contract collection, and extraction of the plant material; the Biophysics Resource of the Structural Biophysics Laboratory for providing technical assistance with Q-TOF LC-MS experiments. This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government. This research was supported in part by the Intramural Research Program of NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Supporting Information Available: 1H NMR spectra of 1-9, 1S, 1R, 9S, and 9R, 13C NMR spectra for 1-5, 9, 1S, and 1R, HSQC NMR spectra for 6-8, 9S and 9R, and a table of assignments for 1S and 1R, 9, and 9S and 9R. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Mahalingam D, Szegezdi E, Keane M, Jong S, Samali A. Cancer Treat. Rev. 2009;35:280–288. doi: 10.1016/j.ctrv.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Booth NL, Sayers TJ, Brooks AD, Thomas CL, Jacobsen K, Goncharova EI, McMahon JB, Henrich CJ. Cancer Immunol. Immunother. 2009;58:1229–1244. doi: 10.1007/s00262-008-0637-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang L, Fang B. Cancer Gene Ther. 2005;12:228–237. doi: 10.1038/sj.cgt.7700792. [DOI] [PubMed] [Google Scholar]
- 4.Van Geelen CM, de Vries EG, de Jong S. Drug Resist. Updat. 2004;7:345–358. doi: 10.1016/j.drup.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 5.O'Kane HF, Watson CJ, Johnston SR, Petak I, Watson RW, Williamson KE. J. Urol. 2006;175:432–438. doi: 10.1016/S0022-5347(05)00160-6. [DOI] [PubMed] [Google Scholar]
- 6.Cheng J, Hylander BL, Baer MR, Chen X, Repasky EA. Mol. Cancer Ther. 2006;5:1844–1853. doi: 10.1158/1535-7163.MCT-06-0050. [DOI] [PubMed] [Google Scholar]
- 7.Huang Y, Sheikh MS. Toxicol. Appl. Pharmacol. 2007;224:284–289. doi: 10.1016/j.taap.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Duiker EW, Mom CH, de Jong S, Willemse PH, Gietema JA, van der Zee AG, de Vries EG. Eur. J. Cancer. 2006;42:2233–2240. doi: 10.1016/j.ejca.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 9.Buchsbaum DJ, Forero-Torres A, LoBuglio AF. Future Oncol. 2007;3:405–409. doi: 10.2217/14796694.3.4.405. [DOI] [PubMed] [Google Scholar]
- 10.Hunter MS, Corley DG, Carron CP, Rowold E, Kilpatrick BF, Durley RC. J. Nat. Prod. 1997;60:894–899. doi: 10.1021/np970141n. [DOI] [PubMed] [Google Scholar]
- 11.De Carvalho PR, Furlan M, Young MC, Kingston DG, Bolzani VS. Phytochemistry. 1998;49:1659–1662. doi: 10.1016/s0031-9422(98)00249-0. [DOI] [PubMed] [Google Scholar]
- 12.Beutler JA, McCall KL, Herbert K, Johnson T, Shoemaker RH, Boyd MR. Phytochemistry. 2000;55:233–236. doi: 10.1016/s0031-9422(00)00281-8. [DOI] [PubMed] [Google Scholar]
- 13.Sai Prakash CV, Hoch JM, Kingston DG. J. Nat. Prod. 2002;65:100–107. doi: 10.1021/np010405c. [DOI] [PubMed] [Google Scholar]
- 14.Kanokmedhakul S, Kanokmedhakul K, Kanarsa T, Buayairaksa M. J. Nat. Prod. 2005;68:183–188. doi: 10.1021/np049757k. [DOI] [PubMed] [Google Scholar]
- 15.Kanokmedhakul S, Kanokmedhakul K, Buayairaksa M. J. Nat. Prod. 2007;70:1122–1126. doi: 10.1021/np070083y. [DOI] [PubMed] [Google Scholar]
- 16.Beutler JA, McCall KL, Herbert K, Herald DL, Pettit GR, Johnson T, Shoemaker RH, Boyd MR. J. Nat. Prod. 2000;63:657–661. doi: 10.1021/np990553r. [DOI] [PubMed] [Google Scholar]
- 17.Oberlies NH, Burgess JP, Navarro HA, Pinos RE, Fairchild CR, Peterson RW, Soejarto DD, Farnsworth NR, Kinghorn AD, Wani MC, Wall ME. J. Nat. Prod. 2002;65:95–99. doi: 10.1021/np010459m. [DOI] [PubMed] [Google Scholar]
- 18.Espindola LS, Vasconcelos Junior JR, de Mesquita ML, Marquie P, de Paula JE, Mambu L, Santana JM. Planta Med. 2004;70:1093–1095. doi: 10.1055/s-2004-832655. [DOI] [PubMed] [Google Scholar]
- 19.Shen YC, Wang CH, Cheng YB, Wang LT, Guh JH, Chien CT, Khalil AT. J. Nat. Prod. 2004;67:316–321. doi: 10.1021/np0303658. [DOI] [PubMed] [Google Scholar]
- 20.Huang DM, Shen YC, Wu C, Huang YT, Kung FL, Teng CM, Guh JH. Eur. J. Pharmacol. 2004;503:17–24. doi: 10.1016/j.ejphar.2004.09.040. [DOI] [PubMed] [Google Scholar]
- 21.Jullian V, Bonduelle C, Valentin A, Acebey L, Guigou AG, Prevost MF, Sauvain M. Bioorg. Med. Chem. Lett. 2005;15:5065–5070. doi: 10.1016/j.bmcl.2005.07.090. [DOI] [PubMed] [Google Scholar]
- 22.Shen YC, Cheng YB, Ahmed AF, Lee CL, Chen SY, Chien CT, Kuo YH, Tzeng GL. J. Nat. Prod. 2005;68:1665–1668. doi: 10.1021/np058063o. [DOI] [PubMed] [Google Scholar]
- 23.Williams RB, Norris A, Miller JS, Birkinshaw C, Ratovoson F, Andriantsiferana R, Rasamison VE, Kingston DG. J. Nat. Prod. 2007;70:206–209. doi: 10.1021/np0605034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vieira GM, Jr., Goncalves Tde O, Regasini LO, Ferreira PM, Pessoa Cdo O, Costa Lotufo LV, Torres RB, Boralle N, Bolzani Vda S, Cavalheiro AJ. J. Nat. Prod. 2009;72:1847–1850. doi: 10.1021/np9004079. [DOI] [PubMed] [Google Scholar]
- 25.Guittet VS, Lallemand JY, Ramiandrasoa F, Kunesch G, Moretti C. Tetrahedron. 1988;44:2893–2901. [Google Scholar]
- 26.Gibbons S, Gray AI, Waterman PG. Phytochemistry. 1996;41:565–570. [Google Scholar]
- 27.Sullivan GR, Dale JA, Mosher HS. J. Org. Chem. 1973;38:2143–2147. [Google Scholar]
- 28.Ohtani I, Kusumi T, Kashman Y, Kakisawa H. J. Am. Chem. Soc. 1991;113:4092–4096. [Google Scholar]
- 29.Rieser MJ, Hui Y, Rupprecht JK, Kozlowski JF, Wood KV, McLauchlin JL, Hanson PR, Zhuang Z, R. HT. J. Am. Chem. Soc. 1992;114:10203–10213. [Google Scholar]
- 30.Corminboeuf O, Overman LE, Pennington LD. Org. Lett. 2003;5:1543–1546. doi: 10.1021/ol034384k. [DOI] [PubMed] [Google Scholar]
- 31.Fattorusso E, Romano A, Taglialatela-Scafati O, Irace C, Maffettone C, Bavestrello G, Cerrano C. Tetrahedron. 2009;65:2898–2904. [Google Scholar]
- 32.Yamada K, Ogata N, Ryu K, Miyamoto T, Komori T, Higuchi R. J. Nat. Prod. 1997;60:393–396. [Google Scholar]
- 33.Knappe E, Peteri D. Fresenius Z. Anal. Chem. 1962;190:386–389. [Google Scholar]
- 34.Itokawa H, Totsuka N, Takeya K, Watanabe K, Obata E. Chem. Pharm. Bull. 1988;36:1585–1588. doi: 10.1248/cpb.36.1585. [DOI] [PubMed] [Google Scholar]
- 35.Itokawa H, Totsuka N, Morita H, Takeya K, Iitaka Y, Schenkel EP, Motidome M. Chem. Pharm. Bull. 1990;38:3384–3388. doi: 10.1248/cpb.38.3384. [DOI] [PubMed] [Google Scholar]
- 36.Morita H, Nakayama M, Kojima H, Takeya K, Itokawa H, Schenkel EP, Motidome M. Chem. Pharm. Bull. 1991;39:693–697. doi: 10.1248/cpb.39.693. [DOI] [PubMed] [Google Scholar]
- 37.Shen YC, Wang LT, Wang CH, Khalil AT, Guh JH. Chem. Pharm. Bull. 2004;52:108–110. doi: 10.1248/cpb.52.108. [DOI] [PubMed] [Google Scholar]
- 38.McCloud TG. Molecules. 2010;15:4526–4563. doi: 10.3390/molecules15074526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.