Abstract
The turnover of tumor suppressor p53 is critical for its role in various cellular events. However, the pathway that regulates the turnover of the Drosophila melanogaster DMP53 is largely unknown. Here, we provide evidence for the first time that the E2 ligase, Drosophila homolog of Rad6 (dRad6/Dhr6), plays an important role in the regulation of DMP53 turnover. Depletion of dRad6 results in DMP53 accumulation, whereas overexpression of dRad6 causes enhanced DMP53 degradation. We show that dRad6 specifically interacts with DMP53 at the transcriptional activation domain and regulates DMP53 ubiquitination. Loss of dRad6 function in transgenic flies leads to lethalities and altered morphogenesis. The dRad6-induced defects in cell proliferation and apoptosis are found to be DMP53-dependent. The loss of dRad6 induces an accumulation of DMP53 that enhances the activation of apoptotic genes and leads to apoptosis in the presence of stress stimuli. In contrast to that, the E3 ligase is the primary factor that regulates p53 turnover in mammals, and this work demonstrates that the E2 ligase dRad6 is critical for the control of DMP53 degradation in Drosophila.
Keywords: Drosophila, p53, Protein Degradation, Transcription, Ubiquitination, Rad6
Introduction
The tumor suppressor p53 (1, 2) is characterized as a “cellular gatekeeper” or “the guardian of the genome” (3, 4). It is a sequence-specific DNA-binding protein (5, 6) that functions as a node for organizing whether the cell responds to various types of stress. p53 also plays a critical role in cell apoptosis, cell cycle regulation, senescence, DNA repair, and other cellular events (7). Mutations in p53 or its altered function have been found in more than 50% of cancer cases (8–10).
p53 stabilization and activation are associated with post-transcriptional modifications in mammals (7, 11). For instance, the ubiquitin-proteosome degradation pathway is critical in maintaining a low cellular level of p53 in normal cells (12, 13). Ubiquitin-conjugating enzymes (E2s) and ubiquitin ligases (E3s) confer substrate specificity during the ubiquitination reaction (14). The E3 ligase mouse double minute protein 2 (MDM2) is believed to be the primary factor that regulates p53 turnover (15–17). MDM2 regulates p53 through mono- or polyubiquitination and may act in concert with a number of other molecules to participate in the turnover process in mammalian cells. However, other studies suggest that p53 is degraded also through an MDM2-independent pathway (14, 18–21).
The pathway that regulates the stabilization of the Drosophila melanogaster DMP53 (22, 23) is not known. The DMP53 gene encodes three DMP53 protein isoforms as follows: DMP53L (495 amino acids), DMP53 (385 amino acids), and DMP53n (110 amino acids) (24). Previous studies have suggested that Drosophila DMP53 may be regulated differently from mammalian p53 (25). First, the mammalian p53 can induce cell cycle arrest either in the G1 phase or at the G2/M checkpoint (26), whereas DMP53 is required for damage-induced apoptosis but not for cell cycle arrest (22, 27). Second, p53 in mammals is regulated by binding to MDM2, which in turn binds to p19AFR (28). However, MDM2 and p19AFR homologs have not been detected in D. melanogaster nor have the residues that are critical for p53 binding to MDM2 been found in DMP53 (10). Given that Drosophila DMP53 is one of the most remote representatives of the p53 family (25), the study of DMP53 regulation will likely provide important insights into the ancestry of this gene family.
Rad6 is an E2 enzyme (29) that repairs DNA damage and regulates gene transcription by catalyzing the ubiquitination of different target proteins in yeast and mammalian cells (30–34). In Saccharomyces cerevisiae, Rad6 interacts with Rad18 to catalyze the monoubiquitination of proliferating cell nuclear antigen on lysine 164 (Lys-164), thereby promoting the error-prone DNA damage repair pathway (35–37). However, a separate mechanism exists in response to DNA damage wherein a complex containing Ubc13-MMS2-Rad5/Rad18-Rad6 promotes the polyubiquitination of proliferating cell nuclear antigen and activates the error-free repair pathway (31, 32). Rad6 also regulates gene transcription. In both yeast and mammals, Rad6 determines the monoubiquitination of H2B on Lys-123 (yeast) or Lys-120 (mammals) and controls the methylation of histone H3 at lysine 4 (H3K4) and lysine 79 (H3K79) by interacting with the E3 ligase Bre1 (34, 38–42). Rad6 also participates in the protein degradation process by cooperating with different E3 ligases (43–45). This interaction is required for sporulation in S. cerevisiae (9). In addition, Rad6/HHR6 is overexpressed in some cancer cell lines and tumors (46, 47). Mutations in the catalytic site of Rad6 confer hypersensitivity to a variety of DNA damage agents (43, 48). Whereas the Drosophila homolog of yeast Rad6, dRad6/Dhr6, was identified in 1991 (49), its function remains less well understood.
In this work, we demonstrate for the first time that Drosophila dRad6 negatively regulates DMP53 turnover. We show that dRad6 forms a complex with DMP53 through specific interactions and thereby regulates DMP53 ubiquitination. The loss of dRad6 inhibits DMP53 degradation, leading to altered development and morphogenesis in Drosophila. dRad6-depleted cells exhibit enhanced stimulation-induced apoptosis in a DMP53-dependent manner. The downstream targets of dRad6 and DMP53 were also investigated, and it was found that Drosophila dRad6 regulates gene expression through two types of mechanisms. Therefore, the data in this work provide important clues for understanding the function of dRad6 in development as well as how dRad6 influences DMP53-induced apoptosis and transcription.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
Drosophila S2 cells were cultured at 25 °C in Schneider's Drosophila medium (Invitrogen) supplemented with 10% fetal calf serum and antibiotics (penicillin and streptomycin). Transfection of constructs into S2 cells was performed with StarFect (Genstar) according to the manufacturer's standard protocol.
Plasmid Constructs
A pIB/V5-His-TOPO (Invitrogen) plasmid expressing DsRed2 (pIB/DsRed2) was constructed by cloning DsRed2 PCR products that were amplified from pDsRed2-N1 (Clontech) into the pIB vector. The pIB/V5-His-TOPO plasmid expressing a GFP (pIB/GFP) tag was constructed previously (50). Plasmids expressing H2A, H2B, dRad6, and DMP53 were constructed by cloning the coding sequences for H2A (fused with a Myc tag at its 5′-terminal region), H2B (fused with a Myc tag at its 5′-terminal region), dRad6 (fused with an HA tag at its 5′-terminal region), and DMP53 amplified from D. melanogaster cDNA into pIB/V5-His TOPO (H2A and H2B), pIB/DsRed2 (HA-dRad6), or pIB/GFP (DMP53) expression vectors. The primer sequences are provided.
RNAi in S2 Cells
The coding sequences of Drosophila dRad6 and DMP53 were first amplified with primers containing the gene sequence plus the sequence of a T7 promoter at the 5′-terminal end. dsRNA2 were prepared using a MEGAscript T7 kit (Ambion) according to the manufacturer's standard protocol. The products were quantified and stored at −80 °C. RNAi in Drosophila S2 cells was performed according to the protocol of the Dixon laboratory and Ni et al. (50).
Co-immunoprecipitation Analysis
S2 cells were transfected with HA-tagged dRad6 and Myc-tagged H2A or H2B with StarFect (Genstar). After 72 h, cells were harvested, washed with ice-cold PBS, resuspended in BC300 buffer (containing 20 mm Tris-Cl, pH 8.0, 300 mm NaCl, 0.2 mm EDTA, 10% glycerol, 0.2 mm PMSF, and 0.2% Tween 20), and sonicated 10 times, for 3 s each time, on ice with 30% efficiency. The cell lysates were incubated with normal mouse IgG (Santa Cruz Biotechnology, as a negative control), anti-HA, or anti-Myc antibodies (Santa Cruz Biotechnology) at 4 °C overnight. Protein A/G-agarose beads were then added and incubated for another 3 h, followed by centrifugation to harvest the agarose beads after they were washed five times with BC300 buffer. The precipitated proteins were released by boiling in BC300 buffer and resolved by SDS-PAGE (15%) and immunoblot analysis with antibodies against ubiquitin or p53, as indicated.
Western Blot Analysis
S2 cells were lysed in BC300 buffer (containing 20 mm Tris-Cl, pH 8.0, 300 mm NaCl, 0.2 mm EDTA, 10% glycerol, 0.2 mm PMSF, and 0.2% Tween 20). The protein concentration of the supernatant was measured with a BCA assay kit (Calbiochem). Then 15% SDS-PAGE was performed to resolve the protein compounds. Different amounts of total proteins were loaded in each experiment to facilitate the detection of different target proteins as follows: 150 μg/lane for p53, 70 μg/lane for dRad6, 30 μg/lane for tubulin and H3, and 100–120 μg/lane for histone modifications. After electrophoresis, proteins were transferred onto PVDF membranes (Amersham Biosciences) and then hybridized with primary antibodies at the following dilutions: DMP53 (Santa Cruz Biotechnology; 1:2,000); dRad6 (Santa Cruz Biotechnology; 1:2,000); tubulin (Santa Cruz Biotechnology; 1:3,000); H3 (Upstate; 1:3,000); histone modifications (AbCAM; H3K4Me3, H3K79Me3, H3S10p, and H3K9Ac, 1:1,000);and Myc and HA tag (Zhongshan Golden Bridge; 1:2,000). The HRP-labeled secondary antibodies (Zhongshan Golden Bridge) were all used at the dilution of 1:2,000. An ECL detection system (Amersham Biosciences) was used to detect signals on the membranes.
Immunofluorescence Staining
Immunofluorescence staining was performed according to Ni et al. (50). The primary antibodies used were anti-dRad6 (1:50), anti-DMP53 (Santa Cruz Biotechnology; 1:50) and anti-tubulin (Santa Cruz Biotechnology; 1:100). DAPI (Sigma) was used at a concentration of 1 × 10−4 μg/μl. Secondary antibodies coupled with FITC (1:100) and Texas Red (1:100) were purchased from Zhongshan Co., China. Images were photographed with a laser scanning confocal microscope (Leica) with a 100× oil-immersion objective.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed according to the published protocols of Upstate and Ni et al. (50).
RT-PCR Assay
A total of 4 × 106 S2 cells were lysed to isolate total RNA using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. The reverse transcription was performed according to Ni et al. (50). Total RNA (5 μg) was reverse-transcribed to synthesize cDNA in a volume of 20 μl (reverse transcriptase Moloney murine leukemia virus, Takara). For each 25-μl PCR, 1 μl of cDNA was used for 20–25 cycles. PCR products (10 μl) were loaded onto a 2% agarose gel, stained with ethidium bromide, and photographed. The primer sequences used for RT-PCR are provided in the supplemental material.
Microarray Analysis
Two independent populations of S2 cells were treated with double dsRNA encoding GFP (as a control), dRad6, or dRad6/DMP53 for 6 days. Total RNA was extracted with TRIzol reagent (Invitrogen) according to a standard protocol. The total RNA samples were then sent to CapitalBio Corp. for Chip (Affymetrix) assay.
TUNEL Staining
TUNEL staining was performed according to the manufacturer's instruction (Roche Applied Science) and Arama E and Steller H (51).
Apoptosis Assay
S2 cell apoptosis assays were performed according to the manufacturer's standard protocol (KeyGen). In general, 5 × 105 S2 cells were harvested and resuspended in 500 μl of binding buffer. Resuspended cells were treated with 5 μl each of annexin V-FITC and propidium iodide solutions. Cells were then incubated for 5 min in the dark and subjected to flow cytometric analysis. The annexin V-positive cells are defined as the apoptotic cells.
RESULTS
dRad6 Is Essential in Regulating DMP53 Degradation
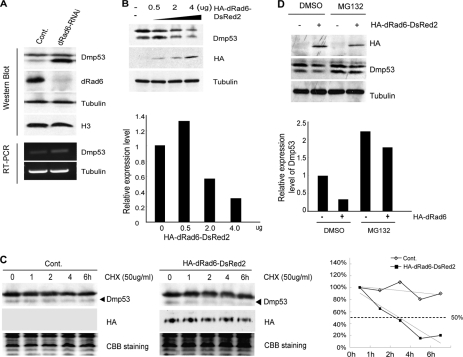
In mammals, p53 ubiquitination requires the E3 ubiquitin ligase MDM2 (52). However, no MDM2 homolog has been identified in D. melanogaster. As such, the mechanism by which DMP53 degradation is controlled in D. melanogaster remains unclear. A previous study showing an interaction between p53 and Rad6 in mammalian cells (53) indicates a possible role for dRad6 in the regulation of DMP53. Therefore, we tested whether depletion of dRad6 from S2 cells affects DMP53 protein levels. Indeed, the results show that dRad6 depletion led to a dramatic accumulation of DMP53 (Fig. 1A).
FIGURE 1.
dRad6 regulates DMP53 degradation through the 26 S proteasome pathway. A, depletion of dRad6 results in the accumulation of DMP53 protein. S2 cells treated with dsRNA encoding for GFP control (Cont) or dRad6 (dRad6-RNAi) are indicated. The cell extracts were subjected to Western blot analysis using antibodies against DMP53, antibodies against dRad6, and antibodies against histone H3 and tubulin. The DNA gels at the bottom show the RT-PCR analysis of transcription level of dmp53 and tubulin. B, overexpression of dRad6 decreases the protein level of DMP53. S2 cells transfected with increasing amounts of HA-dRad6-DsRed2 plasmid (0, 0.5, 2, and 4 μg) are indicated. The immunoblot was stained with anti-DMP53 or anti-HA, and anti-tubulin antibodies are shown. The quantification of band densities based on these gels is shown to the right. C, dRad6-induced degradation of DMP53 is 26 S proteasome-dependent. S2 cells transfected with (+) or without (−)HA-dRad6-DsRed2 in a 6-well plate and treated with DMSO or MG132 are indicated. The immunoblot was stained with antibodies against the HA tag (to visualize HA-dRad6-DsRed2) and DMP53. Anti-tubulin was used as an internal control. The quantification of band densities is shown on the right. CBB, Coomassie Brilliant Blue. D, overexpression of dRad6 decreases the half-life of DMP53 in S2 cells. S2 cells transfected with or without HA-dRad6-DsRed2 and then treated with cycloheximide (CHX) for the indicated time are indicated. Immunoblot was stained with antibodies against DMP53 or the HA tag (to visualize HA-dRad6-DsRed2). A representative control experiment, performed under the same conditions, is shown at the bottom. The quantification of band densities is shown on the right.
To confirm the correlation between Drosophila dRad6 and the down-regulation of DMP53, we next overexpressed various levels of HA/DsRed2-tagged dRad6 proteins in S2 cells and examined whether DMP53 protein levels were altered. Indeed, the level of DMP53 was decreased in response to increased levels of dRad6 protein expression in S2 cells (Fig. 1B).
To test whether the above results were due to Drosophila DMP53 degradation, we employed a chase analysis. Control cells and transfected cells, expressing higher levels of dRad6, were treated with 50 μg/ml cycloheximide for the indicated times. Cells were then lysed and subjected to Western blot analysis. From the results in Fig. 1C, we calculated the half-life time in both treatments, and they are more than 6 h in the control group and about 3 h in dRad6 overexpression groups, respectively. So, we can conclude that overexpression of dRad6 decreased the half-life of DMP53 (Fig. 1C), confirming that dRad6 affects DMP53 degradation.
DMP53 Degradation Proceeds through the 26 S Proteasome Pathway
We wondered whether the degradation of DMP53 by dRad6 occurs via the 26 S proteasome pathway. We therefore examined this mechanism in S2 cells using the 26 S proteasome-specific inhibitor, MG132. DMP53 protein accumulated after 8–12 h of treatments with 25 μm MG132 (data not shown). Therefore, we conclude that Drosophila DMP53 degradation is mediated via the 26 S proteasome pathway.
We next overexpressed HA/DsRed2-tagged dRad6 in control cells and cells treated with MG132. Cells were lysed and subjected to Western blot analysis. As indicated in Fig. 1D, treatment with MG132 prevented the decrease in DMP53 protein expected under conditions of dRad6 overexpression. This suggests that dRad6 regulates DMP53 protein levels via the 26 S proteasome pathway.
dRad6 Regulates DMP53 Degradation through Ubiquitination
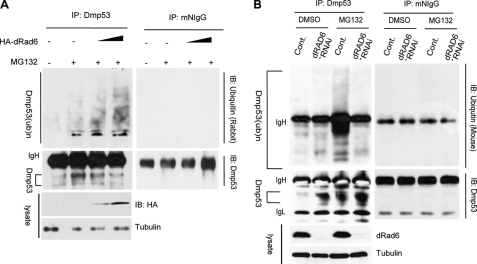
Because Rad6 is involved in the ubiquitination of various target proteins in both yeast and mammals (31–34), it may also regulate the protein level of DMP53 through the ubiquitination pathway. We analyzed this possibility using an in vivo ubiquitination assay. S2 and Kc cells were transfected with DsRed2 control plasmid or different amounts of HA-dRad6-DsRed2 plasmids, together with DMSO or 25 μm MG132 treatment for 8 h; at this time point, the cells were lysed and subjected to immunoprecipitation (IP) with anti-DMP53 antibody under denaturing conditions. IP lysates were then immunoblotted with anti-ubiquitin or anti-DMP53 antibodies. When the proteasome was inhibited by MG132, the endogenous ubiquitinated DMP53 was not degraded (data not shown) and therefore could be detected in these assays. The results show that the overexpression of dRad6 promotes DMP53 ubiquitination in a dosage-dependent manner (Fig. 2A and supplemental Fig. S1).
FIGURE 2.
dRad6 regulates DMP53 ubiquitination in vivo. A, dRad6 overexpression promotes DMP53 ubiquitination in S2 cells. S2 cells, transfected without (−) or with HA-dRad6-DsRed2 plasmid with increased amounts and then treated with DMSO (−) or MG132 (25 μm), are indicated. The immunoblot (IB) was stained with anti-ubiquitin antibodies to visualize the ubiquitinated form of DMP53 (upper panel). The anti-DMP53 antibody was used to visualize the amount of precipitated DMP53 (middle panel). The gel on the right shows an identical control IP using mouse IgG (mIgG). The expression of HA-dRad6 (HA) is shown in the lower panel. B, effect of dRad6 depletion on the ubiquitination of DMP53. S2 cells treated with dsRNA encoding GFP (Cont) or dRad6, followed by treatment with DMSO (as a control) or MG132, are indicated. Immunoblot analysis was performed with a monoclonal anti-ubiquitin antibody to visualize the ubiquitinated form of DMP53 (upper panel). Anti-DMP53 antibody was used to visualize the level of DMP53 precipitated (middle panel). The efficiency of dRad6-RNAi is shown in the lower panel.
To further test the role of dRad6 in the ubiquitination of DMP53, we also performed an RNAi assay. S2 cells were transfected with dsRNA encoding GFP (control) or dRad6 6 days prior to analysis. Cells were also treated with DMSO or MG132 for an additional 8 h before lysis. Cell extracts were subjected to IP with an anti-DMP53 antibody and then immunoblotted with an anti-ubiquitin antibody. The result shows that the ubiquitinated DMP53 was detectable in the MG132-treated cells; however, it disappeared in the MG132-treated cells when dRad6 was depleted (compare 1st and 4th lanes with 3rd lane, Fig. 2B). Thus, the depletion of dRad6 inhibited the ubiquitination of DMP53, which further confirms that dRad6 is required for DMP53 ubiquitination in vivo.
dRad6 Interacts with DMP53 Both in Vitro and in Vivo
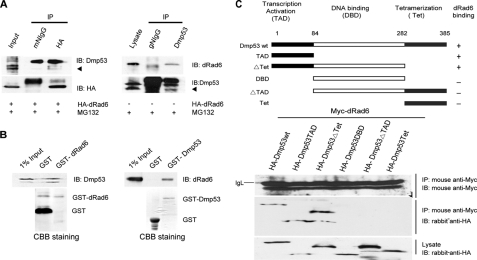
According to our model, DMP53 ubiquitination will require an interaction between DMP53 and dRad6. To determine whether these two molecules interact in vivo, we performed co-immunoprecipitation (co-IP) assays using S2 cells transfected with or without HA/DsRed2-tagged dRad6 in the absence of MG132. Transfected S2 cells were lysed and subjected to co-IP with antibodies directed against the HA tag. Precipitated proteins were then detected by Western blot analysis with an anti-DMP53 antibody. In reverse, transfected S2 cells were lysed and subjected to co-IP with an anti-DMP53 antibody, followed by immunoblot analysis with an anti-dRad6 antibody. The results show that no obvious interaction could be detected between these two molecules (data not shown). However, treating transfected S2 cells with 25 μm MG132, which inhibits the degradation of DMP53, permitted the interaction between dRad6 and DMP53 proteins to be observed in co-IP assays, as expected (Fig. 3A). In addition, we compared the subcellular localization of dRad6 and DMP53 in S2 cells with or without MG132 treatment by immunofluorescence microscopy. These results demonstrate that the localization of these two molecules is dynamic and that dRad6 partially co-localizes with DMP53 in a subset of cells in normal status (data not shown), although a better co-localization can be detected under MG132 treatment (data not shown). Thus, a dynamic interaction between dRad6 and DMP53 occurs in vivo.
FIGURE 3.
dRad6 interacts with DMP53 both in vitro and in vivo. A, dRad6 interacts with DMP53 in vivo in the presence of MG132. S2 cells transfected with HA-dRad6-DsRed2 plasmid (left) and treated with MG132 are indicated. The immunoblot (IB) was stained with anti-DMP53 antibody or anti-HA antibody to visualize HA-dRad6-DsRed2. The result shows that DMP53 can be pulled down by anti-HA antibody. The gels on the right show that dRad6 can be pulled down by anti-DMP53. Antibodies against DMP53 and dRad6 are all indicated. B, GST pulldown assay shows that dRad6 interacts with DMP53 in vitro. The immunoblot analysis was performed with antibodies against DMP53 (left) and dRad6 (right). The positions of DMP53 (left upper panel) and dRad6 (right upper panel) are indicated. The GST fusion proteins were visualized by Coomassie (CBB) staining (lower panel). C, domains of DMP53 that are required for its interaction with dRad6. Myc-tagged dRad6 and different HA-tagged DMP53 fragments co-transfected into the H1299 cell line are shown. Mouse anti-Myc and rabbit anti-HA antibodies are indicated.
To assess the interaction between dRad6 and DMP53 in vitro, we employed a GST pulldown assay. S2 cell lysates were subjected to pulldown assay using GST-fused dRad6 or DMP53 that had been immobilized on Sepharose 4B beads. DMP53 was pulled down by GST-dRad6 protein, and in reverse, dRad6 was pulled down by GST-DMP53 (Fig. 3B). Together, these results support the notion that dRad6 interacts directly with DMP53.
Transcription Activation Domain of DMP53 Interacts with dRad6
To map the domain of DMP53 that interacts with dRad6, we prepared constructs expressing either the HA-tagged wild type DMP53 or the fragments of DMP53, including a fragment encoding the transcriptional activation domain of DMP53 (amino acids 1–84), a fragment encoding both the transcriptional activation domain and the DNA-binding domain of DMP53 (amino acids 1–282), a fragment encoding the DNA-binding domain of DMP53 alone (amino acids 84–282), a fragment encoding both the DNA-binding domain and the domain required for tetramerization (amino acids 84–385), and a fragment encoding only the domain required for tetramerization (amino acids 282–385). These constructs, together with a construct encoding for a Myc-tagged wild type dRad6 protein, were co-transfected into the human lung carcinoma cell line H1299, a cell line without endogenous p53. Co-immunoprecipitation experiments were performed using a mouse anti-Myc antibody (indicating dRad6). Immunoblotting was performed with an antibody against HA (indicating DMP53 and its fragments). The interaction between dRad6 and DMP53 mainly occurred at the N terminus of DMP53, between amino acids 1 and 84, which corresponds to the transcriptional activation domain (Fig. 3C).
Loss of dRad6 in Transgenic Flies Results in Developmental Defects
Because the loss of dRad6 led to an accumulation of DMP53, we wondered whether the depletion of dRad6 would affect development. Therefore, we produced transgenic flies expressing dsRNAs of the D. melanogaster dRad6 using a Gal4-induced RNAi system (54).
The full cDNAs encoding dRad6 were cloned into the pUAST vector between two inverted upstream activating sequence (Gal4-binding sequence elements). When GAL4 binds these sequence elements, it activates transcription of the target from opposite directions and therefore produces dsRNA that silences the endogenous target gene. The transgenic flies were first crossed with an act-Gal4 transgenic line (yw;;act-Gal4/TM6B) that expresses Gal4 ubiquitously during development. Most progeny failed to survive to the adult stage. Further analysis suggested that lethality was most pronounced at the larval and pupal stages (data not shown).
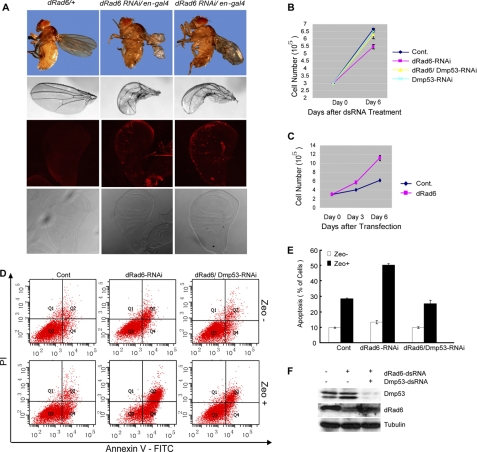
We next crossed the transgenic flies with a line carrying an en-Gal4 driver, in which the Gal4 is mainly activated in the posterior compartment of developing wings. At the adult stage, more than 50% of flies showed different degrees of curled wings (Fig. 4A), suggesting that dRad6 is essential in wing morphogenesis. Furthermore, staining of the wing imaginal disc of 3rd-instar larva with the TUNEL method to detect apoptotic cells showed that the depletion of dRad6 caused an increase in apoptotic cell staining in the posterior compartment (Fig. 4A).
FIGURE 4.
dRad6 is required for normal developmental and cell proliferation. A, dRad6 is required for the normal development of D. melanogaster. The adult fly and wing were photographed by a stereomicroscope (upper two panels). The wing discs from 3rd-instar larva were stained with TUNEL method (in red, lower two panels). B and C, dRad6 regulates S2 cell proliferation. B, up-regulation of DMP53 protein level induced by the deletion of dRad6 results in decreased cell number. S2 cells treated with GFP (control (Cont)), dRad6, combined dRad6/DMP53, or DMP53 dsRNAs are indicated. C, overexpression of dRad6 resulted in an increase in the number of total cells. Three independent experiments were performed for each assay. D, dRad6 depletion-induced apoptosis is dramatically enhanced by the presence of Zeocin. S2 cells, treated with dsRNA of GFP (control, left panel), dRad6 (middle panel), or dRad6/DMP53 (right panel), then treated with (Zeo+) or without (Zeo−) Zeocin, are indicated. Cells were harvested and doubly labeled with annexin V-FITC and propidium iodide (PI) before analysis by flow cytometry. The data are quantified in the histogram on the right. The efficiency of DMP53 and dRad6 RNAi as indicated by Western blot analysis is provided below the histogram. E, quantified histogram according to the data from D. F, RNAi efficiency in D is determined by Western blot assay. S2 cells were treated as in D. The cell extracts were then prepared and subjected to Western blot assay with specific antibodies as indicated.
dRad6 Depletion-induced Change in Cell Proliferation and Apoptosis Correlates with DMP53 Dosage
We further investigated the effect of dRad6 on proliferation and apoptosis using the Drosophila embryonic cell line S2. The depletion of dRad6 resulted in a reduction in S2 cell proliferation (Fig. 4B). This reduction was partially rescued by dRad6 and DMP53 double RNAi, suggesting that the inhibitory effects of dRad6 depletion on cellular proliferation are at least partially due to the accumulation of DMP53. Consistent with this notion, overexpression of dRad6 in S2 cells promoted proliferation (Fig. 4C).
Because DNA damage triggers the activation of DMP53 and induces apoptosis (27), we wondered whether apoptosis is enhanced in cells depleted of dRad6. Therefore, S2 cells were transfected for 6 days with dsRNA of GFP (control), dRad6 alone, or dRad6 and DMP53 in the presence or absence of Zeocin, an antibiotic known to produce DNA double-stranded breaks. Flow cytometric analysis revealed that dRad6-RNAi indeed promoted apoptosis in the presence of Zeocin and that this increase could be partially rescued by co-treating with dRad6 and DMP53 RNAi (Fig. 4, D–F). These results support a model wherein dRad6-induced apoptosis, in the presence of a stress stimulus, is largely mediated by DMP53.
dRad6 Depletion Leads to Altered Gene Transcription in Both a DMP53-dependent and -independent Manner
To address the relationship between dRad6 and DMP53 in the regulation of gene transcription, we performed genome-wide gene expression profile analysis using S2 cells cultured in the presence or absence of dRad6. To separate the downstream gene targets of dRad6 from those regulated by an accumulation of DMP53, we included a sample in which cells were treated with both dRad6 and DMP53 dsRNAs, as well as the control dsRNAs, GFP, and dRad6 alone.
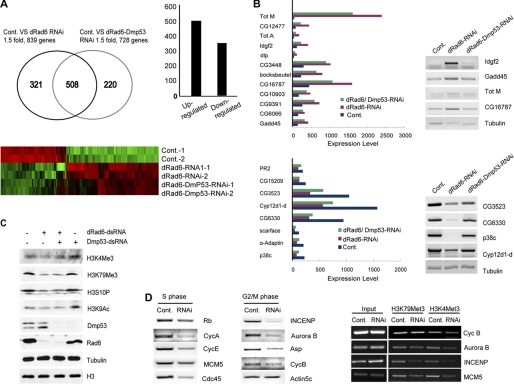
The array analyses show that depletion of dRad6 affects a large number of genes (Fig. 5A). In the absence of dRad6, 829 genes were altered by at least 1.5-fold. Among the affected genes, ∼60% were up-regulated. The depletion of both dRad6 and DMP53 resulted in 728 genes with altered transcription. Between the two different experiments, dRad6 depletion alone and the depletion of both dRad6 and DMP53, over 500 genes were consistently identified. Furthermore, the depletion of DMP53 in dRad6 RNAi-treated cells resulted in the altered transcription pattern of a few specific genes (Fig. 5A) that are likely targets of DMP53. The observed up-regulation of the genes returned to control levels upon DMP53 depletion, confirming that they are specific targets of DMP53. Altered transcription of these genes was further verified by semi-quantitative RT-PCR (Fig. 5B).
FIGURE 5.
Role of dRad6 and DMP53 in gene transcriptional regulation. A, Drosophila microarray analysis. The number of genes specifically affected (over 1.5-fold) in dRad6-depleted cells (dRad6-RNAi) and in dRad6/DMP53 double-depleted cells (dRad6-DMP53 RNAi) are indicated in the Venn diagram (top left). The exact number of genes down-regulated and up-regulated after dRad6 depletion are shown in the histogram (top right). Cluster analysis indicates the differentially expressed genes in these treatment groups (red, up-regulated genes; green, down-regulated genes). Cont, control. B, genes up-regulated by dRad6 RNAi and down-regulated by DMP53 are shown in the histogram at the top panel. These results were validated by semi-quantitative RT-PCR (shown on the right). The genes down-regulated by dRad6 RNAi, but up-regulated by DMP53, are shown in the histogram in the bottom panel. The CG numbers of each gene are indicated to the right of the gels. Tubulin was used as an internal control. C, loss of histone H3 modifications after dRad6 depletion was not due to the up-regulation of DMP53 protein level. S2 cells treated with (+) or without (−) different dsRNAs are indicated. Cells were lysed for Western blot analysis using antibodies against different histone modifications, as indicated. Antibodies against DMP53, dRad6, tubulin, and histone H3 are also indicated. D, subset of cell cycle-related genes are down-regulated after depletion of dRad6 (left and middle panels). RT-PCR analysis to assess the expressional level of cell cycle-related genes, as indicated. The CHIP analysis (panel on the right) shows that the depletion of dRad6 resulted in decreased levels of H3K4me3 and H3K79me3 in the promoter regions of these cell cycle genes. The names of the genes analyzed are labeled next to the gels.
The loss of dRad6 may affect genes via a reduction in H3K4 and H3K79 trimethylation. H3K4 methylation is associated with transcriptionally active genes (55, 56). Furthermore, Western blot analysis showed that the dRad6 depletion-induced reduction in H3K4 and H3K79 trimethylation was not affected by the depletion of DMP53, which ruled out the possibility that core histone modifications were due to the accumulation of DMP53 (Fig. 5C). To test whether a change in H3K4 and H3K79 methylation occurred specifically on chromatin of the affected genes, we performed ChIP analysis of the genes down-regulated following dRad6 depletion. Several genes associated with cell cycle progression, such as INCENP, Aurora B, CycA, CycB, CycD, and CycE, that were down-regulated by loss of dRad6 (Fig. 5D) were selected. Indeed, ChIP analysis confirmed that the loss of H3K4 and H3K79 trimethylation occurred at sites corresponding to the promoter regions of these genes (Fig. 5D). However, genes that were down-regulated in a DMP53-dependent manner showed a different pattern of histone H3K4 and H3K79 methylation than the affected cell cycle regulators (data not shown), suggesting that distinct mechanisms may be utilized.
Loss of function mutations in INCENP or Aurora B cause abnormal chromosomal segregation at metaphase as well as certain cytokinesis defects (57, 58). Indeed, a variety of chromosomal defects were observed among the dRad6-depleted cells (data not shown). Thus, dRad6-induced defects in proliferation (Fig. 4B) may be in part due to defective cell cycle progression.
DMP53 Accumulation Activates Apoptotic Genes under Stress Conditions
A few p53-targeted genes, such as reaper, grim, sickle, and hid, are known to play critical roles in the regulation of apoptosis in Drosophila (25, 28, 59–61). We wondered whether the transcriptional regulation of these apoptotic genes would be up-regulated in the absence of dRad6. However, our results show that the transcription of these genes was not affected under nonstress conditions (Fig. 6A), which also implies that the observed reduction in S2 cell proliferation in the absence of dRad6 (Fig. 4B) cannot be attributed to changes in these cell death regulators. Because depletion of DMP53 in the dRad6-depleted cells resulted in a partial rescue of proliferation (Fig. 4B), it is likely that other DMP53 functions may be involved.
FIGURE 6.
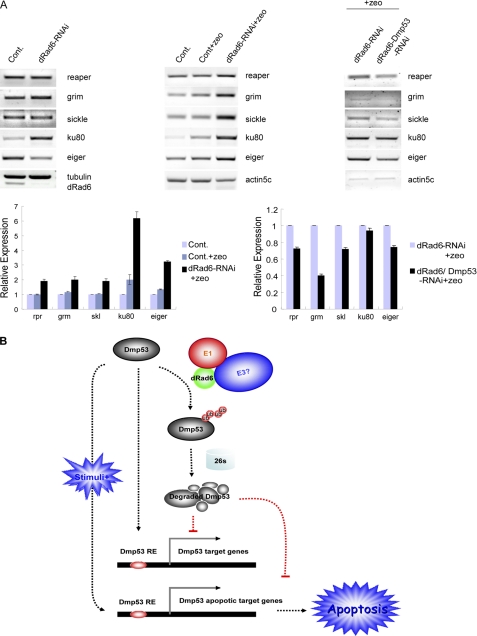
Role of dRad6 and DMP53 in the transcriptional regulation of DMP53 apoptotic target genes. A, activation of DMP53 apoptotic target genes is regulated by the presence of stress stimuli. RT-PCR analysis to check the expressional level of DMP53 target genes using total RNA from control (Cont), dRad6-RNAi, and dRad6-DMP53 double RNAi cells treated with (+Zeo) is shown. The RNA used in experiments shown in the left panel was from cells that had not been treated with Zeocin. The gels in the middle panel show that apoptotic genes were activated following treatment with Zeocin. The gels on the right panel show that the activation of these apoptotic genes is DMP53-dependent. The gene names are indicated next to the gels. The gene expression level under different conditions is quantified at the bottom. B, pathways through which dRad6 regulates DMP53 stability and DMP53-regulated gene transcription and apoptosis. Under normal conditions, Drosophila dRad6 maintains a low level of DMP53 protein through ubiquitination and the 26 S proteasome pathway. This mechanism is executed by dRad6 and other ubiquitination enzymes (E1 and unknown E3s) to inhibit the expression of DMP53 target genes under nonstress conditions (e.g. in the absence of Zeocin). However, DMP53-regulated apoptotic genes are only activated in the presence of stress-inducing stimuli (e.g. Zeocin). As a consequence of this activation, apoptosis occurs. DMP53 RE refers to the DMP53 regulatory sequence element.
The presence of stress stimuli and the accumulation of DMP53 in dRad6 RNAi-treated cells clearly enhanced apoptosis (Fig. 4D). Therefore, we wondered whether the induction of a stress response would promote the transcription of apoptotic genes. Total RNA was isolated from control cells or cells that had been depleted of dRad6, with or without Zeocin treatment. Our semi-quantitative RT-PCR assay demonstrated that the transcription of apoptotic regulators, including reaper, grim, and sickle, was only slightly changed in control cells in the presence of Zeocin. However, the transcription of these genes was increased by up to 5-fold in dRad6-depleted cells under the same conditions (Fig. 6A). This result is consistent with the increased apoptosis observed in Fig. 4. Further depletion of DMP53 in these cells rescued the expression pattern of these genes (Fig. 6A), supporting a direct role of DMP53 in the transcriptional regulation of the apoptotic genes and apoptosis. Overall, these results suggest that the dRad6-induced accumulation of DMP53 is not sufficient to induce transcriptional activation of apoptotic regulators unless additional environmental stress signals are present.
DISCUSSION
The mechanism of the p53 regulation has been a central subject in p53 biology for the past 30 years. Previous studies have greatly enhanced our understanding of this molecule. p53 stabilization appears to be the critical step in the different p53 degradation models proposed (7). It is clear that MDM2 is the predominant E3 ligase for p53 in mammals and is capable of catalyzing both mono- and polyubiquitination on p53 (14, 50). However, the lack of MDM2 homologs in Drosophila and the fact that MDM2-binding sites are apparently lacking in DMP53 (10) raise the question of how DMP53 stability is controlled in Drosophila, especially considering the ∼550 million years of evolutionary distance between insects and vertebrates.
We show in this work that the Drosophila homolog dRad6 plays a critical role in DMP53 stabilization, as supported by the following evidence: 1) the depletion of Drosophila dRad6 protein led to an immediate accumulation of DMP53; 2) dRad6 overexpression caused DMP53 degradation; 3) DMP53 specifically interacts with dRad6; and 4) dRad6 regulates DMP53 ubiquitination. The dRad6 depletion-induced accumulation of DMP53 was not due to changes in either the transcriptional level of the gene or its translation (Fig. 1). The chase experiment also confirmed that dRad6 regulates the level of DMP53 through protein degradation. Consistent with this, dRad6 is localized mainly in the cytoplasm and is present at only low levels in the nucleus under nonstress conditions. Presumably, this subcellular localization ensures the degradation of DMP53 when it is not needed, a property similar to that of the E3 ligase, MDM2, in mammals.
We also noticed that DMP53, but not the larger DMP53L isoform, seemed to be sensitive to dRad6 depletion. The level of DMP53L remained mostly unchanged in the absence of dRad6. However, dRad6 overexpression caused reductions in both DMP53 and DMP53L. We also confirmed that both isoforms correspond to the endogenous DMP53 because transfection of cells with dsRNA of DMP53 resulted in the loss of both isoforms. Our results imply that the loss of dRad6 has major effects on DMP53, but not on the DMP53L form, although the exact mechanism for this difference is not clear.
Our transgenic models showing that dRad6 is essential for normal development in Drosophila, together with the loss-of-function analysis of Rad6 homologs in mouse (62, 63), support an essential role for Rad6 in development. The lethality observed in the dRad6 RNAi flies is likely due to the DMP53-induced apoptosis and/or defective proliferation observed in the absence of dRad6. DMP53 overexpression induced widespread apoptosis in the developing Drosophila retina (22), which supports this hypothesis. However, the analysis of S2 cells following dRad6 depletion demonstrated only a partial increase in apoptosis, implying that the function of DMP53 may be different between various tissues and/or cell types. DMP53 RNA levels are abundant until the cellularization of the blastoderm, after which DMP53 RNA levels become progressively more restricted and fall dramatically in all tissues except the germ cells and a small patch of hindgut cells (22). This expression pattern supports a potential tissue-specific and/or developmental stage-specific function of DMP53. This is also supported by our microarray analysis of dRad6/DMP53-depleted S2 cells, which showed that over 800 genes were affected by the loss of dRad6. In contrast, only a small number of genes appear to be regulated by DMP53 in embryonic S2 cells. Among these dRad6-regulated genes, the transcription of apoptotic genes, such as reaper, grim, sickle, and hid (23, 64, 65), were clearly not affected under nonstress conditions and therefore are unlikely contribute to defects in proliferation. The expression of genes essential for cell cycle progression was reduced after dRad6 depletion (Fig. 5). Indeed, our cell cycle analysis in dRad6-depleted S2 cells found defective cell cycle progression at both metaphase and anaphase (data not shown). In contrast, DMP53 accumulation may directly contribute to the defects in cell proliferation. DMP53 overexpression in larval eye discs alters the entry into and/or duration of M phase, although no effect on the transition of cells from G1 to S phase was observed (22). Whether this is the case in embryonic cells remains unclear.
The study described herein supports a model whereby Drosophila dRad6 regulates gene expression through two types of mechanisms (Fig. 6B). The first mechanism is a DMP53-dependent mechanism, as supported by the fact that a small set of genes affected by loss of dRad6 appears to be rescued by further depletion of DMP53. Clearly, this set of genes is activated or repressed by DMP53 without stress stimulation. Apoptotic genes such as reaper, grim, or sickle, which all contain the DMP53 DNA-binding motif in their promoter regions, do not appear to respond to the accumulation of DMP53 under nonstress conditions. Because of the specificity of the DMP53 antibody, we could not confirm whether DMP53 directly binds to their promoters or whether the binding is enhanced under conditions of stress. However, these genes appear to be activated in the presence of Zeocin. The DMP53 RNAi experiment confirmed that the increased transcription of these genes was due to the increased DMP53 expression because further depletion of DMP53 led to a reversal in their transcription level. A stress-inducing stimulation may be required for p53 to bind to DNA and activate or suppress gene transcription (66–68). Nonetheless, it is not clear whether DMP53, similar to its mammalian p53 ortholog, is constitutively bound to the promoter regions of target genes under nonstress conditions (69, 70) or whether DMP53 remains largely inactive until the induction of a stress response (71).
The second mechanism by which dRad6 controls gene expression involves the histone H3 methylation pathway, as has been demonstrated in a number of studies. The monoubiquitination of histone H2B by the Rad6-Bre1 complex is required for H3K4 methylation by COMPASS (Complex of Protein Associated with Set1) (40, 58, 72, 73) and is highly conserved from yeast to humans (72, 74–76). Rad6-mediated regulation of H2B monoubiquitination is also required for proper H3K79 methylation by Dot1 (39, 77). A recent study revealed that the Cps35 subunit of COMPASS mediates the cross-talk between H2B monoubiquitination and H3 methylation by COMPASS (34). COMPASS can mono-, di-, and tri-methylate lysine 4 of histone H3 (78–82), a marker often associated with actively transcribed genes. Here we show, by Western blot analysis, that the depletion of Drosophila dRad6 results in the loss of H2B mono-ubiquitination, H3K4 and H3K79 trimethylation, which is consistent with the established relationship between Rad6 and histone H3 methylation (supplemental Fig. S2). Our ChIP analysis also confirmed that the reduction in H3K4 and H3K79 methylation was associated with reduced transcription of cell cycle regulators, specifically those genes that are regulated by dRad6.
In summary, we demonstrate a novel role for the E2 ligase, dRad6, in the stabilization of DMP53 in D. melanogaster. It is not clear whether the pathway by which dRad6 controls DMP53 degradation is conserved in other organisms. Further study of p53 regulation in different organisms will likely yield important clues concerning the ancestry of this molecule throughout evolution.
Supplementary Material
Acknowledgment
We thank Dr. Vanda Pogacic for help in the early stages of this work.
This work was supported by National Grant “Jie-Chu-Qing-Nian-Ke-Xue Fund” from the Chinese National Science Foundation 30625017, National “973” Grants from the Ministry of Science and Technology 2011CB965300, 2007CB948101, and 2009CB825603, and a start-up grant from the Yu-Yuan Foundation and the Wu-Shun-De Foundation of Tsinghua University.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Results,” Figs. S1 and S2, Table 1, and additional references.
- dsRNA
- double-stranded RNA
- IP
- immunoprecipitation.
REFERENCES
- 1. Baker S. J., Fearon E. R., Nigro J. M., Hamilton S. R., Preisinger A. C., Jessup J. M., vanTuinen P., Ledbetter D. H., Barker D. F., Nakamura Y., White R., Vogelstein B. (1989) Science 244, 217–221 [DOI] [PubMed] [Google Scholar]
- 2. Finlay C. A., Hinds P. W., Levine A. J. (1989) Cell 57, 1083–1093 [DOI] [PubMed] [Google Scholar]
- 3. Lane D. P. (1992) Curr. Biol. 2, 581–583 [DOI] [PubMed] [Google Scholar]
- 4. Levine A. J. (1997) Cell 88, 323–331 [DOI] [PubMed] [Google Scholar]
- 5. Bargonetti J., Friedman P. N., Kern S. E., Vogelstein B., Prives C. (1991) Cell 65, 1083–1091 [DOI] [PubMed] [Google Scholar]
- 6. Kern S. E., Kinzler K. W., Bruskin A., Jarosz D., Friedman P., Prives C., Vogelstein B. (1991) Science 252, 1708–1711 [DOI] [PubMed] [Google Scholar]
- 7. Kruse J. P., Gu W. (2009) Cell 137, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kastan M. B., Radin A. I., Kuerbitz S. J., Onyekwere O., Wolkow C. A., Civin C. I., Stone K. D., Woo T., Ravindranath Y., Craig R. W. (1991) Cancer Res. 51, 4279–4286 [PubMed] [Google Scholar]
- 9. Vousden K. H., Lu X. (2002) Nat. Rev. Cancer 2, 594–604 [DOI] [PubMed] [Google Scholar]
- 10. Allton K., Jain A. K., Herz H. M., Tsai W. W., Jung S. Y., Qin J., Bergmann A., Johnson R. L., Barton M. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 11612–11616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu X. (2005) Curr. Opin. Genet. Dev. 15, 27–33 [DOI] [PubMed] [Google Scholar]
- 12. Brooks C. L., Gu W. (2006) Mol. Cell 21, 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Michael D., Oren M. (2003) Semin. Cancer Biol. 13, 49–58 [DOI] [PubMed] [Google Scholar]
- 14. Brooks C. L., Gu W. (2004) Cell Cycle 3, 895–899 [PubMed] [Google Scholar]
- 15. Honda R., Tanaka H., Yasuda H. (1997) FEBS Lett. 420, 25–27 [DOI] [PubMed] [Google Scholar]
- 16. Haupt Y., Maya R., Kazaz A., Oren M. (1997) Nature 387, 296–299 [DOI] [PubMed] [Google Scholar]
- 17. Kubbutat M. H., Jones S. N., Vousden K. H. (1997) Nature 387, 299–303 [DOI] [PubMed] [Google Scholar]
- 18. Leng R. P., Lin Y., Ma W., Wu H., Lemmers B., Chung S., Parant J. M., Lozano G., Hakem R., Benchimol S. (2003) Cell 112, 779–791 [DOI] [PubMed] [Google Scholar]
- 19. Dornan D., Wertz I., Shimizu H., Arnott D., Frantz G. D., Dowd P., O'Rourke K., Koeppen H., Dixit V. M. (2004) Nature 429, 86–92 [DOI] [PubMed] [Google Scholar]
- 20. Chen J., Ruan H., Ng S. M., Gao C., Soo H. M., Wu W., Zhang Z., Wen Z., Lane D. P., Peng J. (2005) Genes Dev. 19, 2900–2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ringshausen I., O'Shea C. C., Finch A. J., Swigart L. B., Evan G. I. (2006) Cancer Cell 10, 501–514 [DOI] [PubMed] [Google Scholar]
- 22. Ollmann M., Young L. M., Di, Como C. J., Karim F., Belvin M., Robertson S., Whittaker K., Demsky M., Fisher W. W., Buchman A., Duyk G., Friedman L., Prives C., Kopczynski C. (2000) Cell 101, 91–101 [DOI] [PubMed] [Google Scholar]
- 23. Brodsky M. H., Nordstrom W., Tsang G., Kwan E., Rubin G. M., Abrams J. M. (2000) Cell 101, 103–113 [DOI] [PubMed] [Google Scholar]
- 24. Bourdon J. C., Fernandes K., Murray-Zmijewski F., Liu G., Diot A., Xirodimas D. P., Saville M. K., Lane D. P. (2005) Genes Dev. 19, 2122–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nordstrom W., Abrams J. M. (2000) Cell Death. Differ. 7, 1035–1038 [DOI] [PubMed] [Google Scholar]
- 26. May P., May E. (1999) Oncogene 18, 7621–7636 [DOI] [PubMed] [Google Scholar]
- 27. Sogame N., Kim M., Abrams J. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4696–4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Steller H. (2000) Nat. Cell Biol. 2, E100–E102 [DOI] [PubMed] [Google Scholar]
- 29. Jentsch S., McGrath J. P., Varshavsky A. (1987) Nature 329, 131–134 [DOI] [PubMed] [Google Scholar]
- 30. Koken M. H., Reynolds P., Jaspers-Dekker I., Prakash L., Prakash S., Bootsma D., Hoeijmakers J. H. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 8865–8869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. (2002) Nature 419, 135–141 [DOI] [PubMed] [Google Scholar]
- 32. Pfander B., Moldovan G. L., Sacher M., Hoege C., Jentsch S. (2005) Nature 436, 428–433 [DOI] [PubMed] [Google Scholar]
- 33. Lee K. Y., Myung K. (2008) Mol. Cells 26, 5–11 [PMC free article] [PubMed] [Google Scholar]
- 34. Lee J. S., Shukla A., Schneider J., Swanson S. K., Washburn M. P., Florens L., Bhaumik S. R., Shilatifard A. (2007) Cell 131, 1084–1096 [DOI] [PubMed] [Google Scholar]
- 35. Bailly V., Lamb J., Sung P., Prakash S., Prakash L. (1994) Genes Dev. 8, 811–820 [DOI] [PubMed] [Google Scholar]
- 36. Bailly V., Lauder S., Prakash S., Prakash L. (1997) J. Biol. Chem. 272, 23360–23365 [DOI] [PubMed] [Google Scholar]
- 37. Bailly V., Prakash S., Prakash L. (1997) Mol. Cell. Biol. 17, 4536–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yamashita K., Shinohara M., Shinohara A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11380–11385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ng H. H., Xu R. M., Zhang Y., Struhl K. (2002) J. Biol. Chem. 277, 34655–34657 [DOI] [PubMed] [Google Scholar]
- 40. Sun Z. W., Allis C. D. (2002) Nature 418, 104–108 [DOI] [PubMed] [Google Scholar]
- 41. Shema E., Tirosh I., Aylon Y., Huang J., Ye C., Moskovits N., Raver-Shapira N., Minsky N., Pirngruber J., Tarcic G., Hublarova P., Moyal L., Gana-Weisz M., Shiloh Y., Yarden Y., Johnsen S. A., Vojtesek B., Berger S. L., Oren M. (2008) Genes Dev. 22, 2664–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim J., Roeder R. G. (2009) J. Biol. Chem. 284, 20582–20592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sung P., Berleth E., Pickart C., Prakash S., Prakash L. (1991) EMBO J. 10, 2187–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Watkins J. F., Sung P., Prakash S., Prakash L. (1993) Genes Dev. 7, 250–261 [DOI] [PubMed] [Google Scholar]
- 45. Dohmen R. J., Madura K., Bartel B., Varshavsky A. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 7351–7355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shekhar M. P., Lyakhovich A., Visscher D. W., Heng H., Kondrat N. (2002) Cancer Res. 62, 2115–2124 [PubMed] [Google Scholar]
- 47. Shekhar M. P., Gerard B., Pauley R. J., Williams B. O., Tait L. (2008) Cancer Res. 68, 1741–1750 [DOI] [PubMed] [Google Scholar]
- 48. Lyakhovich A., Shekhar M. P. (2004) Oncogene 23, 3097–3106 [DOI] [PubMed] [Google Scholar]
- 49. Koken M., Reynolds P., Bootsma D., Hoeijmakers J., Prakash S., Prakash L. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 3832–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ni J. Q., Liu L. P., Hess D., Rietdorf J., Sun F. L. (2006) Genes Dev. 20, 1959–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Arama E., Steller H. (2006) Nat. Protoc. 1, 1725–1731 [DOI] [PubMed] [Google Scholar]
- 52. Li M., Brooks C. L., Wu-Baer F., Chen D., Baer R., Gu W. (2003) Science 302, 1972–1975 [DOI] [PubMed] [Google Scholar]
- 53. Lyakhovich A., Shekhar M. P. (2003) Mol. Cell. Biol. 23, 2463–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu L. P., Ni J. Q., Shi Y. D., Oakeley E. J., Sun F. L. (2005) Nat. Genet. 37, 1361–1366 [DOI] [PubMed] [Google Scholar]
- 55. Ruthenburg A. J., Allis C. D., Wysocka J. (2007) Mol. Cell 25, 15–30 [DOI] [PubMed] [Google Scholar]
- 56. Berger S. L. (2007) Nature 447, 407–412 [DOI] [PubMed] [Google Scholar]
- 57. Giet R., Glover D. M. (2001) J. Cell Biol. 152, 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mackay A. M., Ainsztein A. M., Eckley D. M., Earnshaw W. C. (1998) J. Cell Biol. 140, 991–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. White K., Tahaoglu E., Steller H. (1996) Science 271, 805–807 [DOI] [PubMed] [Google Scholar]
- 60. Srinivasula S. M., Datta P., Kobayashi M., Wu J. W., Fujioka M., Hegde R., Zhang Z., Mukattash R., Fernandes-Alnemri T., Shi Y., Jaynes J. B., Alnemri E. S. (2002) Curr. Biol. 12, 125–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen P., Nordstrom W., Gish B., Abrams J. M. (1996) Genes Dev. 10, 1773–1782 [DOI] [PubMed] [Google Scholar]
- 62. Baarends W. M., Wassenaar E., Hoogerbrugge J. W., van Cappellen G., Roest H. P., Vreeburg J., Ooms M., Hoeijmakers J. H., Grootegoed J. A. (2003) Mol. Cell. Biol. 23, 1151–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Roest H. P., Baarends W. M., de Wit J., van Klaveren J. W., Wassenaar E., Hoogerbrugge J. W., van Cappellen W. A., Hoeijmakers J. H., Grootegoed J. A. (2004) Mol. Cell. Biol. 24, 5485–5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. McCall K., Steller H. (1997) Trends Genet. 13, 222–226 [DOI] [PubMed] [Google Scholar]
- 65. Abrams J. M. (1999) Trends Cell Biol. 9, 435–440 [DOI] [PubMed] [Google Scholar]
- 66. Hupp T. R., Meek D. W., Midgley C. A., Lane D. P. (1992) Cell 71, 875–886 [DOI] [PubMed] [Google Scholar]
- 67. Cain C., Miller S., Ahn J., Prives C. (2000) J. Biol. Chem. 275, 39944–39953 [DOI] [PubMed] [Google Scholar]
- 68. Kim E., Rohaly G., Heinrichs S., Gimnopoulos D., Meissner H., Deppert W. (1999) Oncogene 18, 7310–7318 [DOI] [PubMed] [Google Scholar]
- 69. Liu X., Yue P., Khuri F. R., Sun S. Y. (2004) Cancer Res. 64, 5078–5083 [DOI] [PubMed] [Google Scholar]
- 70. Kaeser M. D., Iggo R. D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 95–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Szak S. T., Mays D., Pietenpol J. A. (2001) Mol. Cell. Biol. 21, 3375–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wood A., Schneider J., Dover J., Johnston M., Shilatifard A. (2003) J. Biol. Chem. 278, 34739–34742 [DOI] [PubMed] [Google Scholar]
- 73. Shilatifard A. (2006) Annu. Rev. Biochem. 75, 243–269 [DOI] [PubMed] [Google Scholar]
- 74. Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D. (2006) Cell 125, 703–717 [DOI] [PubMed] [Google Scholar]
- 75. Kim J., Hake S. B., Roeder R. G. (2005) Mol. Cell 20, 759–770 [DOI] [PubMed] [Google Scholar]
- 76. Zhu B., Zheng Y., Pham A. D., Mandal S. S., Erdjument-Bromage H., Tempst P., Reinberg D. (2005) Mol. Cell 20, 601–611 [DOI] [PubMed] [Google Scholar]
- 77. Wood A., Schneider J., Dover J., Johnston M., Shilatifard A. (2005) Mol. Cell 20, 589–599 [DOI] [PubMed] [Google Scholar]
- 78. Nislow C., Ray E., Pillus L. (1997) Mol. Biol. Cell 8, 2421–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Miller T., Krogan N. J., Dover J., Erdjument-Bromage H., Tempst P., Johnston M., Greenblatt J. F., Shilatifard A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Roguev A., Schaft D., Shevchenko A., Pijnappel W. W., Wilm M., Aasland R., Stewart A. F. (2001) EMBO J. 20, 7137–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nagy P. L., Griesenbeck J., Kornberg R. D., Cleary M. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Krogan N. J., Dover J., Khorrami S., Greenblatt J. F., Schneider J., Johnston M., Shilatifard A. (2002) J. Biol. Chem. 277, 10753–10755 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.