Abstract
Escherichia coli K-12, the most widely used laboratory bacterium, does not secrete proteins into the extracellular medium under standard growth conditions, despite possessing chromosomal genes encoding a putative type II secretion machinery (secreton). We show that in wild-type E.coli K-12, divergent transcription of the two operons in the main chromosomal gsp locus, encoding the majority of the secreton components, is silenced by the nucleoid-structuring protein H-NS. In mutants lacking H-NS, the secreton genes cloned on a moderate-copy-number plasmid are expressed and promote efficient secretion of the endogenous, co-regulated endochitinase ChiA. This is the first time that secretion of an endogenous extracellular protein has been demonstrated in E.coli K-12.
Keywords: chitinase/H-NS/promoters/protein secretion/secreton
Introduction
In most Gram-negative bacteria, the Sec pathway that allows proteins to cross the cytoplasmic membrane is extended by one or more terminal branches that permit extracellular secretion or assembly of surface organelles (Pugsley, 1993a). One such terminal branch of this general secretory pathway (GSP) is the secreton or type II secretion machinery that specifically recognizes proteins in a folded state and facilitates their extrusion through a putative large gated pore in the outer membrane (Pugsley, 1993a). Proteins secreted via this pathway include virulence determinants such as cholera toxin in Vibrio cholerae (Sandkvist et al., 1997), the pore-forming toxin aerolysin in Aeromonas hydrophila (Howard et al., 1993) and pectate lyases and cellulases in the plant pathogenic Erwinia species (Andro et al., 1984).
The genetic organization and the number and primary sequence of individual secreton components are well conserved among Gram-negative bacteria. However, secretons show at least some degree of specificity, since some components are not interchangeable (Possot et al., 2000; Sandkvist et al., 2000) and proteins secreted by one bacterial species are not always secreted by the secreton in another species (de Groot and Tommassen, 1991; Lindeberg et al., 1996; Possot et al., 2000).
An active secreton is composed of 12–15 different proteins, many of which are known or are predicted to be located in the cytoplasmic membrane (Possot et al., 2000). In one of the best studied secretons, the Klebsiella oxytoca pullulanase (Pul) secreton, two proteins, PulD and PulS, form a complex that is presumed to function as a gated outer membrane channel to permit secretion of the enzyme pullulanase (Nouwen et al., 1999, 2000). This second step in pullulanase secretion is dependent on the proton gradient across the cytoplasmic membrane (Possot et al., 1997). In addition, Pul secreton component PulE, which is anchored to the inner face of the cytoplasmic membrane through an interaction with secreton component PulL (Possot et al., 2000; Sandkvist et al., 2000), shares characteristics with known ATP binding proteins (Possot and Pugsley, 1994). By analogy with the type IV piliation system, which includes many homologues of Pul secreton components (Turner et al., 1993), PulE is presumed to be required for the assembly of surface-exposed pili that we have recently observed in bacteria expressing the secreton genes at high levels (Sauvonnet et al., 2000b).
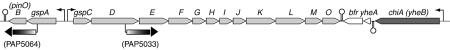
Escherichia coli K-12 does not secrete endogenous proteins, although it contains genes (gsp) that are homologous to those encoding other secretons (Whitchurch and Mattick, 1994; Stojiljkovic et al., 1995; Francetic and Pugsley, 1996; Blattner et al., 1997). The principal gsp locus at min 74.5 is composed of two divergent operons, gspCDEFGHIJKLMO (gspC-O) and gspAB (formerly gspA-pinO) (Figure 1) (Blattner et al., 1997). Another putative secreton gene, yacC (gspS), homologous to the K.oxytoca secreton gene pulS (d’Enfert and Pugsley, 1989), is located at min 2.95 of the E.coli K-12 chromosome (Blattner et al., 1997). Together, these genes account for ∼0.5% of the coding capacity of the E.coli genome. The gsp genes are transcriptionally silent under standard laboratory conditions (Francetic and Pugsley, 1996). In the present study we attempted to find conditions allowing their expression, as a prerequisite to determining whether the E.coli Gsp secreton is functional and to identifying its substrates.
Fig. 1. Schematic representation of the gsp locus at min 74.5 of the E.coli K-12 chromosome. The gsp genes (drawn to approximate scale) are represented by light grey boxes and chiA is dark grey. The positions of the lacZYA reporter insertions are indicated and the names of the resulting strains are given in parentheses. Terminators are represented by the stem–loop structures and arrowheads indicate the direction of transcription.
Results
gsp gene transcription in E.coli K-12 is silenced by H-NS
To probe the expression of the two divergent operons of the main gsp locus, we inserted the promoterless lacZYA operon into the gspA and gspD genes using an R6K-based suicide vector (Figure 1). Growth of the resulting strains, PAP5033 (gspD-lacZYA) and PAP5064 (gspA-lacZYA), was indistinguishable from that of the parent strain, indicating that the gsp genes are not essential. Both strains were Lac–, consistent with the previously demonstrated poor expression of the gsp genes (Francetic and Pugsley, 1996). Transcription of the gspAB operon was ∼10-fold lower than that of gspC-O (Table I). Both operon fusions were more highly expressed at 30°C than at 37°C (Table I) and after the end of exponential growth (data not shown), but none of a large variety of other growth conditions tested (anaerobiosis, variations in carbon source, addition of serum or tissue culture medium, iron starvation) seemed to affect gsp transcription.
Table I. Expression of the gsp-lacZYA operon fusions.
| Strain | Relevant genotype | Growth temperature (°C) | β-galactosidase activity (Miller units) |
|---|---|---|---|
| PAP5033 | gspD-lacZYA | 30 | 57.6 ± 0.8 |
| PAP5033 | gspD-lacZYA | 37 | 33.4 ± 3.7 |
| PAP5064 | gspA-lacZYA | 37 | 2.4 ± 1.0 |
| PAP5108 | gspA-lacZYA hns-1001 | 37 | 8.5 ± 1.9 |
| PAP5090 | gspD-lacZYA hns-F2 | 37 | 152.3 ± 36.6 |
| PAP5048 | gspD-lacZYA hns-1001 | 37 | 147.4 ± 26.2 |
Cells were grown in LB medium to mid-exponential phase. β-galactosidase activity was measured according to Miller (Miller, 1972). The values presented are means from at least three independent measurements with standard deviations.
To identify putative repressors of gsp expression, we generated a library of 20 000 random insertions in the Lac– strain PAP5033 (gspD-lacZYA), using transposon Tn10 (Way et al., 1984), and screened for mutations conferring a Lac+ phenotype on MacConkey lactose agar. Two classes of mutants were obtained in which the Lac+ phenotype was caused by the Tn10 insertion. In the eight class I mutants, Tn10 was cotransducible with the gspD-lacZYA fusion. Sequencing showed that one of these Tn10 insertions was within the gspC gene, resulting in gspD-lacZYA transcription from the Tn10 pout promoter (data not shown). The pleiotropic phenotype caused by the Tn10 insertion in the four independent class II mutants (mucoidy and serine sensitivity) was similar to that caused by mutations inactivating the transcriptional silencer protein H-NS (Atlung and Ingmer, 1997). Indeed, all four Tn10 insertions were 100% linked to the hns-1001 knockout mutation (Bertin et al., 1994) in P1 transduction experiments. Furthermore, the hns-1001 mutation, like the Tn10 insertions (e.g. hns-F2), led to >3-fold derepressed transcription of both gsp operons (Table I).
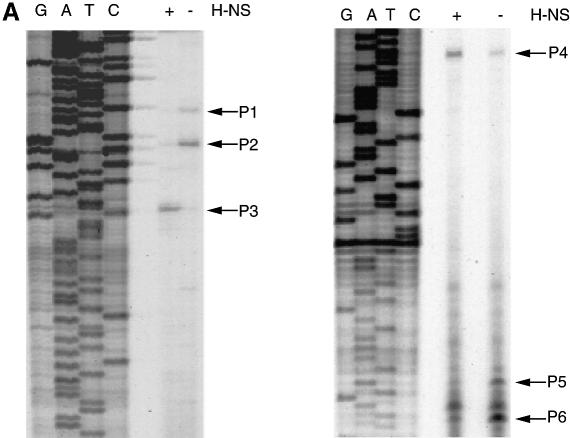
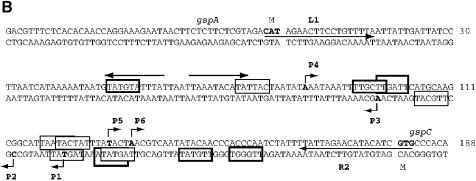
Primer extension experiments indicated that each gsp operon was expressed from three promoters (Figure 2). The gspA transcription start sites P1 and P2 corresponded to overlapping σ70-type promoters with relatively well conserved –35 and –10 regions (thick and thin boxes in Figure 2B, respectively) and a consensus 17 bp distance between them (TTGGGT-N17-TAGTAT for P1 and TTGTAT-N17-TAGTAT for P2; consensus nucleotides in bold). The fact that the P1 and P2 promoters initiate at an unconventional pyrimidine base (Jeong and Kang, 1994) could explain why the gspAB operon is expressed at lower levels than the gspC-O operon. The P3 start site had a putative –10 box (TTGCAT), while the corresponding –35 region (AGTATA) was very different from the consensus. The overlapping gspC promoters P5 and P6 belong to the σ70 type with the sequences TTGCTT-N17-TAATAC for P5 and TTGATT-N17-TACTAT for P6. Promoter P4, like P3, has a well conserved putative –10 box (TATTAC) and a poorly conserved –35 box (TATGTA). The positions of the gspC transcription start sites were confirmed using a more distal primer (data not shown). Promoters P1, P2, P5 and P6 were derepressed in the hns mutants, whereas transcription from the P3 and P4 promoters was down-regulated (Figure 2A).
Fig. 2. (A) Primer extension mapping of the gspA and gspC promoters. Left panel, primer L1 (see B) was used to map the transcription initiation sites of the gspAB operon (P1–P3). Right panel, promoters driving gspC-O operon transcription (P4–P6) were mapped using primer R2 (see B). (B) DNA sequence of the gsp promoter region. Base +1 is the first base after the ATG codon of gspA. Primers L1 and R2 hybridized with the sequences marked by arrows. Transcription start sites (P1–P6), as mapped by primer extension, are marked in bold and the arrowheads indicate the direction of transcription. The promoter –10 and –35 boxes are framed with thin and thick lines, respectively. Divergent arrows indicate the palindromic sequence (see Discussion).
To examine the possibility of direct silencing of the gsp promoters by H-NS, we used a competitive gel retardation assay to analyse the in vitro binding of purified H-NS to the gsp promoter region. A 185 bp intergenic region between the gspA and gspC genes was amplified by PCR and mixed in an equimolar ratio with DNA fragments generated by digesting pBR322 with the restriction endonucleases SspI and TaqI. The binding of purified H-NS protein to this DNA mixture was analysed on a polyacrylamide gel as described in Materials and methods (Figure 3). Migration of the gsp promoter fragment was specifically retarded at 20 nM H-NS, while retardation of the bla promoter fragment, known to bind H-NS avidly (Francetic et al., 2000), required 50 nM H-NS in this experiment. These results indicate that H-NS probably acts directly to repress gspA and gspC promoter activity.
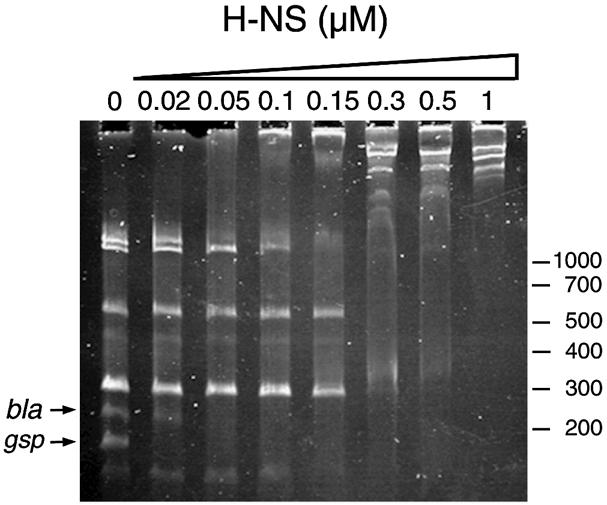
Fig. 3. Competitive gel retardation experiment to assay binding of H-NS protein to the gsp promoter. Purified H-NS protein was incubated at the indicated concentrations with the mixture of DNA fragments as described in Materials and methods and analysed on a 7.5% acrylamide gel in TBE buffer. The positions of the gsp- and bla promoter-containing fragments are indicated by arrows. Molecular size markers are given in bp.
Production of GspG protein under conditions of derepressed gsp gene expression
We next assessed the steady-state levels of GspG, which is homologous to the major pseudopilin in other secretons (Pugsley, 1993b; Sauvonnet et al., 2000b), under conditions allowing maximal gsp expression (Figure 4). Whole-cell extracts of wild-type E.coli K-12 or the hns mutants did not contain sufficient GspG for detection by immunoblotting (Figure 4, lanes 5 and 6), but processed GspG could be detected in hns mutant strains containing the gspC-O operon cloned into the pACYC-based plasmid pACKJ195-4 (Figure 4, lanes 1 and 4). GspG production was stimulated by growth at 30°C (Figure 4), consistent with the higher gsp transcription observed at this temperature (Table I). Interestingly, cloning of the gspAB operon into the same plasmid with gspC-O (pCHAP4278) caused a marked increase in GspG levels and trace amounts of unprocessed preGspG could also be detected (Figure 4, compare lanes 1 with 2 and 4 with 7). This suggested that GspA and/or GspB might play a regulatory role, possibly at the level of transcription or secreton function (e.g. by affecting the stability of one or more of the secreton components). Primary sequence comparisons indicate that GspB, previously designated PinO and implicated in chromosomal DNA replication (Guzman et al., 1991), belongs to the PulB/OutB/ExeB family of proteins, which might or might not be required for secretion (d’Enfert and Pugsley, 1989; Howard et al., 1996; Condemine and Shevchik, 2000).
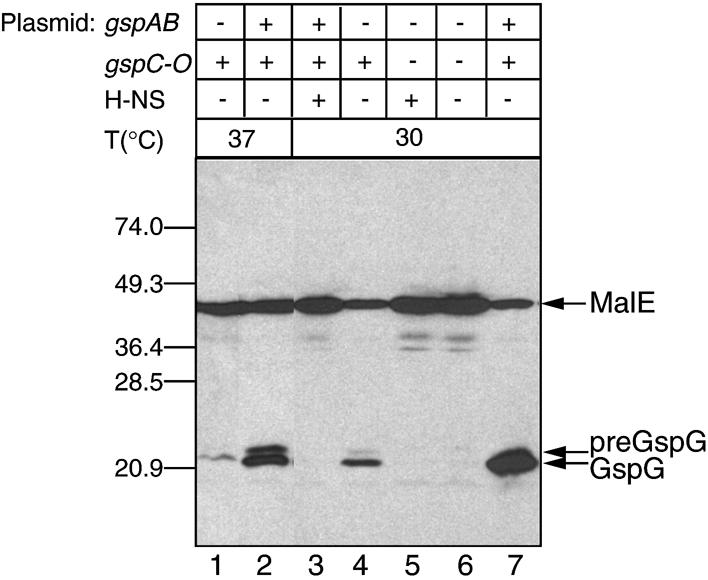
Fig. 4. Steady-state levels of the secreton protein GspG in different strain backgrounds. The gspC-O operon is carried by pACKJ194-5, and pCHAP4278 carries both gspAB and gspC-O. Lane 1, strain PAP5066 (gspC-lacZYA hns-118) (pACKJ194-5); lane 2, strain PAP5066 (pCHAP4278); lane 3, strain PAP5033 (pCHAP4278); lane 4, PAP5066 (pACKJ194-5); lane 5, MC4100; lane 6, MC4100 hns-1001; lane 7, strain PAP5066 (pCHAP4278). The bacteria were grown in LB medium at 37 (lanes 1 and 2) or 30°C (lanes 3–7). Proteins were analysed by SDS–PAGE and immunoblotting using rabbit antiserum generated against a MalE–GspG hybrid protein. The positions of MalE, preGspG, GspG and molecular size markers (in kDa) are indicated.
Chitinase secretion under conditions of derepressed gsp gene expression
To test the functionality of the E.coli K-12 secreton, we attempted to identify cognate substrates of the Gsp secretion machinery. Since genes for secreton substrates are often linked to the secreton genes, we paid particular attention to proteins encoded in the region around the gsp operons. The codon adaptation index of the entire gsp locus and downstream genes, bfr, yheA and yheB (Figure 1), is lower than that of the upstream ribosomal protein operons, indicating that this entire region might represent a more recent acquisition to the E.coli genome (Blattner et al., 1997). Recently, we reported that yheB, which we renamed chiA (Figure 1), encodes a periplasmic endochitinase (Francetic et al., 2000). Endochitinases degrade chitin, an N-acetylglucosamine polymer that is the main component of fungal cell walls and crustacean exoskeletons (Cohen-Kupiec and Chet, 1998). To access these insoluble substrates, endochitinases are secreted to the cell surface and, in at least one case, secretion occurs via a type II machinery (Connel et al., 1998). Interestingly, chiA, like the gsp genes, is cryptic, up-regulated at 30°C and derepressed by mutations in hns (Francetic et al., 2000), suggesting that ChiA could be a substrate of the E.coli secreton.
To investigate this possibility, we introduced plasmid pCHAP4288 (pBGS18::chiA) into strain PAP5066 (PAP5033 hns-118) carrying either pCHAP4278 (encoding the intact gsp locus) or its deletion derivative pCHAP4280 (ΔgspDE). As shown in Figure 5, lane 4, the supernatant fraction of the secretion-proficient strain contained a single ∼100 kDa protein. Immunoblot analysis confirmed that this protein, secreted with high efficiency, was ChiA (Figure 5, lower panel). No other protein was found in the culture medium. Strain PAP5066 (pCHAP4280) did not secrete the chitinase (Figure 5, lane 6), demonstrating the requirement for the intact gsp operon. Only very low levels of ChiA secretion were detected in the absence of gspAB operon (plasmid pACKJ195-4, Figure 5, lane 2).
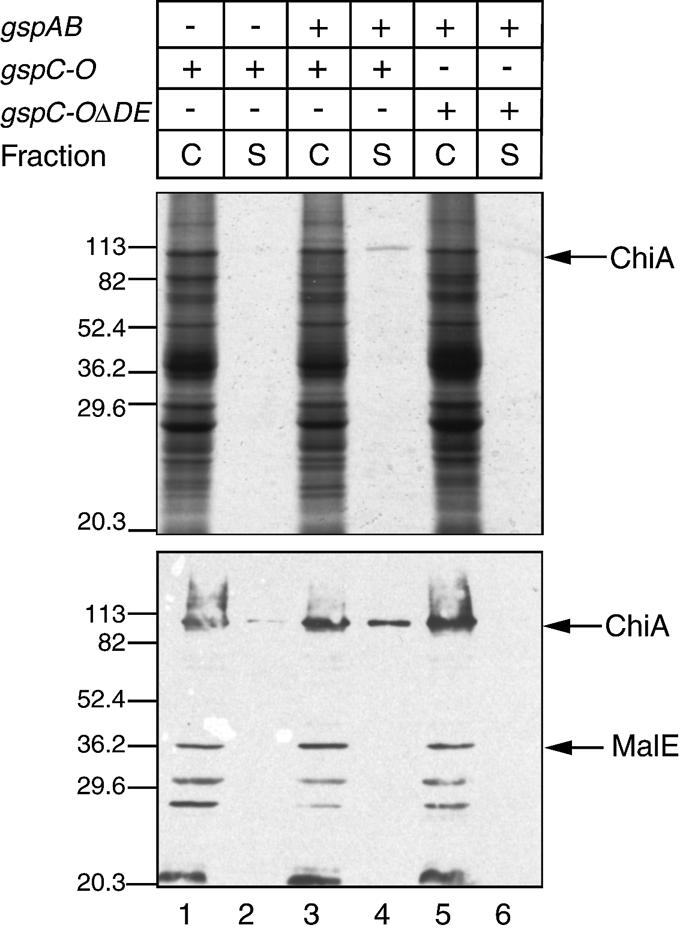
Fig. 5. The E.coli chitinase ChiA is secreted via the Gsp secreton. Cells were grown to mid-exponential phase at 30°C and cell (C) and supernatant (S) fractions from 0.1 A600 of cultures were analysed by SDS–PAGE and stained by Coomassie Brilliant Blue R (upper panel), or by immunoblotting using anti-MalE–ChiA antiserum (Francetic et al., 2000) (lower panel). Lanes 1 and 2, strain PAP5066 (pACJK194-5; gspC-O); lanes 3 and 4, strain PAP5066 (pCHAP4278; gspAB and gspC-O); lanes 5 and 6, strain PAP5066 (pCHAP4280; gspAB and gspC-O with a gspDE deletion). The arrows indicate the positions of MalE and ChiA. Molecular size standards are given in kDa.
Discussion
The results presented demonstrate that, contrary to current dogma, E.coli K-12 does possess a functional pathway for the secretion of extracellular proteins. The gsp genes, as well as the chiA gene encoding the secreton substrate protein, are cryptic under standard laboratory conditions and might provide a new model for studying gene silencing as well as the evolutionary maintenance of cryptic genes in the bacterial chromosome. The best studied example of cryptic genes in E.coli is the bgl operon, involved in transport and utilization of β-glucosides such as arbutin (Lopilato and Wright, 1990). Like the gsp and chiA genes, the bgl operon has remained fully functional, despite the apparent absence of selective pressure. As suggested by Bender, the fact that the cryptic genes have remained functional despite genetic drift could reflect selective pressures exerted outside the laboratory (Bender, 1996). Consistent with this idea, the bgl operon is transcribed in mouse liver during septicaemic infections caused by E.coli (Khan and Isaacson, 1998).
Another common feature shared by the gsp and chiA genes and the bgl operon is the fact that their transcription is silenced by H-NS (Defez and De Felice, 1981). Genes repressed by H-NS often need a specific activator for maximal expression, even in an hns background (Atlung and Ingmer, 1997). This might explain why chitinase secretion was only observed when the derepressed gsp operons were present on a multiple-copy-number plasmid. Specific regulators of the gsp genes and environmental conditions and signalling required for their induction have so far remained elusive, but their identification will be facilitated by the fact that at least one protein is now known to be secreted via the Gsp secreton.
At least two types of promoters seem to drive gsp transcription. One class (P1, P2, P5 and P6) resembles σ70 promoters and is silenced by H-NS. The increased transcription from these promoters observed in the absence of H-NS correlates with the increased expression of the gspA-lacZ and gspD-lacZ operon fusions (Table I). The second class of promoters, P3 and P4, are apparently activated by H-NS, probably indirectly, and could depend on an alternative σ factor. Further studies will be required to understand the molecular mechanism of H-NS repression of the gsp promoters as well as to identify other transcription factors involved in gsp regulation. We suggested previously that the palindromic sequence present in the promoter region (Figure 2B) could represent a binding site for a putative repressor (Francetic and Pugsley, 1996). Consistent with this idea, a deletion within this sequence leads to 3-fold derepression of transcription of both gsp operons without affecting the repression by H-NS (O.Francetic, unpublished data).
In addition, the gspAB operon itself appears to play a regulatory role, since its absence reduces the level of GspG and presumably other secreton components, leading to a drastic reduction in secretion efficiency. A similar genetic organization is found in A.hydrophila, where a homologous operon, exeAB, is transcribed divergently from the exeC-M operon (Howard et al., 1996). The role of ExeA is not very well understood but it contains a consensus nucleotide binding motif and, unlike secreton components encoded by the exeC-M operon, its elimination reduced secretion without affecting the assembly of outer membrane proteins (Howard et al., 1996). ExeA interacts with ExeB, and both are predicted to be cytoplasmic membrane proteins. The level of identity between GspA and ExeA is ∼22%, while GspB and ExeB are ∼19% identical. Homologues of exeA and exeB were found in the recently sequenced V.cholerae genome (Heidelberg et al., 2000) but their role in secretion has not been investigated.
We identified one natural substrate of the E.coli secreton pathway, the endochitinase ChiA, but additional proteins might also be secreted via the Gsp secreton. For example, the V.cholerae eps secreton, besides allowing secretion of an endogenous chitinase, is able to promote secretion of the cholera toxin and the closely related heat-labile enterotoxin (LT) of E.coli (Neill et al., 1983). It would be interesting to test whether the same proteins are secreted in E.coli strains containing the active Gsp secreton and whether the secretion of LT by certain strains of enteropathogenic E.coli (Fleckenstein et al., 2000) is gsp dependent.
Another candidate substrate of the Gsp secreton is a putative laccase encoded by the E.coli yacK gene, transcribed in the opposite direction from yacC (gspS), with which it shares a common regulatory region. Laccases, polyphenol oxidases with broad substrate specificity (Alexandre and Zhulin, 2000), are usually secreted by the producing organism and are of considerable biotechnological interest, being used in the degradation of lignin, effluent detoxification or transformation of steroid compounds.
Besides protein secretion, secretons are involved in outer membrane and capsule biogenesis, filamentous phage assembly and type IV piliation (Howard et al., 1993; Ali et al., 2000; Davis et al., 2000; Sauvonnet et al., 2000b). The involvement of the gsp genes in all of these processes deserves further investigation, as does the role of the related type IV piliation determinants that are also present in the E.coli chromosome (Francetic et al., 1998; Sauvonnet et al., 2000a).
Materials and methods
Bacterial strains and growth conditions
All E.coli K-12 strains used or constructed in this study were derived from strain MC4100 (Silhavy et al., 1984). Luria Bertani (LB) broth, 2× YT broth and LB agar were prepared as described (Miller, 1972), and MacConkey lactose agar was from Difco. Antibiotics were added as required at the following concentrations: ampicillin (Ap), 50 or 100 µg/ml; tetracycline (Tc), 16 µg/ml; kanamycin (Km), 30 µg/ml; chloramphenicol (Cm), 15 or 34 µg/ml; streptomycin (Sm), 100 µg/ml; spectinomycin (Sp), 100 µg/ml. Expression of genes under lacZ promoter control was induced with 1 mM isopropyl-β-d-thiogalactopyranoside. Generalized transduction using P1 phage was performed as described (Miller, 1972). Transposon mutagenesis using defective lambda phage λNK55 carrying transposon Tn10 was performed according to Way et al. (1984).
Molecular biology techniques and plasmid constructions
DNA manipulation, plasmid purification, DNA electrophoresis and bacterial transformation were as described (Sambrook et al., 1989). DNA sequencing from double-stranded DNA templates was performed according to Zagursky et al. (1985) using Sequenase (USB) or T7 polymerase (Pharmacia). PCR amplifications were carried out for 30 cycles (1 min at 94°C, 1 min at 48 or 52°C and 1–4 min at 72°C) with Taq polymerase (Perkin Elmer). Amplified DNA was purified using Qiagen PCR purification columns.
In pACKJ195-4, a 25.6 kb BamHI fragment from strain KJ195 (Bost and Belin, 1995), containing a Tn10 that is 90–95% linked to secY, was cloned into the BamHI site of pDB185, a TcS derivative of pACYC184 obtained by deletion of the AvaI–SalI fragment. The BamHI fragment contains 6140 bp of the right end of Tn10, inserted in gspA (position 1158, DDBJ/EMBL/GenBank accession No. AE000409), and the entire gspC-O operon; the insert ends at the BamHI site of rpsG (position 7304, AE000410).
pCHAP4278 containing the entire gsp locus was constructed as follows. A gspAB DNA fragment generated by PCR using primers GspBL (5′-CAATTGCTGCAGTACTTAACCACCGAACGC-3′) and GspCR (5′-CTTGAGGATCCGCATGATTTTCGTTACGCC-3′), flanked by PstI and BamHI sites, was cloned into pBluescript SK+, giving pCHAP4047. The same fragment, flanked with EcoRV and SphI sites was then cloned into pACYC184, giving pCHAP4277. The gspC-O operon from pACKJ195-4 was cloned as a SphI–SalI fragment into pCHAP4277, generating a complete gsp locus clone, pCHAP4278.
pCHAP4280 was made by deleting the 1644 bp BglII fragment containing the gspD and gspE genes from pCHAP4278. pCHAP4288 was made by cloning the BamHI–HindIII fragment containing the chiA gene from pCHAP4180 (Francetic et al., 2000) into pBGS18.
To construct the MalE–GspG hybrid protein (where MalE is maltose binding protein), the gspG gene fragment encoding the hydrophilic C-terminal part of GspG was amplified by PCR using primers GspG4 (5′-CGAATCGGATCCCCTAACCTAATGGGCAAT-3′) and OLIG2 (5′-CTCGAGAAGCTTATCTCCAGCAGGGTAAAC-3′). This fragment was cloned into pMal-c2 (New England Biolabs) and digested with BamHI and HindIII, giving pCHAP4088.
Construction of lacZ operon fusions
To generate chromosomal lacZYA fusions in the gsp locus, two plasmids were made using the suicide vector pCHAP4079 (Francetic et al., 2000): pCHAP4105 containing a 1063 bp ScaI fragment from the gspD gene, and plasmid pCHAP4162 containing a 924 bp SspI fragment internal to the gspA gene. These plasmids were mobilized from strain SM10 λpir into the MC4100 recipient, and colonies resistant to ampicillin and streptomycin that were pale blue on plates containing 5-bromo-3-chloro-3-indolyl-β-d-galactopyranoside were selected. Their correct chromosomal integration in the resulting strains [PAP5033 (gspD- lacZYA) and PAP5064 (gspA-lacZYA), respectively] was confirmed by P1 transduction (Miller, 1972) and by PCR.
Primer extension
For primer extension, total E.coli RNA was prepared using the Qiagen RNAeasy kit, from mid-exponential cultures of strain MC4100 or MC4100 hns-1001, carrying the gsp promoter cloned on plasmid pCHAP4051. The oligonucleotides L1 (5′-CATAGAACTTCCTGTTTTA-3′) and R2 (5′-CACGATGTATGTTCTAATA-3′) (5 pmol/µl) (Figure 2B) were labelled at their 5′ ends with [γ-32P]ATP (4500 Ci/mmol) using T4 kinase (Pharmacia) in a buffer containing 50 mM Tris–HCl pH 7.6, 10 mM MgCl2, 5 mM dithiothreitol (DTT), 0.1 mM spermidine and 0.1 mM EDTA for 1 h at 37°C. The γ-32P-labelled primers (0.5 pmol) were annealed with 5 µg of total RNA by incubating for 3 min at 70°C and 10 min at 42°C in the reaction mixtures that contained 50 mM Tris–HCl pH 8.3, 75 mM KCl, 3 mM MgCl2, 10 mM DTT and 0.5 mM dNTPs. M-MLV reverse transcriptase (BRL) was added (200 U) and the mixtures were incubated for 1 h at 40°C, followed by the addition of 0.15 M NaOAc and 12.5 mM EDTA. The reaction mixtures were precipitated with cold ethanol, centrifuged for 30 min in a microfuge at 15 000 r.p.m., washed with 70% ethanol and resuspended in formamide– EDTA sample buffer. The samples were separated on 6% polyacrylamide–8 M urea gels, which were then dried and exposed to autoradiography film at –80°C.
Competitive gel retardation experiments
Competitive gel retardation assays to detect binding of H-NS protein to the gsp promoter were performed according to Lucht et al. (1994). DNA fragments obtained from the digestion of plasmid pBR322 with SspI and TaqI endonucleases were mixed with the 185 bp PCR fragment containing the intergenic region between gspA and gspC, generated using primers L1 and R2. The DNA was pre-incubated with the purified H-NS protein at the indicated concentrations for 15 min at room temperature in buffer containing 40 mM HEPES pH 8.0, 8 mM Mg-aspartate, 60 mM K-glutamate, 0.3 mg/ml bovine serum albumin, 0.05% NP-40 and 2 mM DTT. Protein–DNA complexes were resolved on a 7.5% acrylamide–0.25% bisacrylamide gel in Tris–borate–EDTA buffer at 20 V/cm and stained with ethidium bromide.
Purification of MalE–GspG hybrid protein and antibody production
Plasmid pCHAP4088 carrying the DNA fragment encoding the last 118 amino acids of GspG was introduced into strain PAP5023 (MC4100 ΔmalE444/F′lacIQ). The cells were harvested and disrupted using a French press. Total soluble proteins from a 1 l culture were extracted and loaded onto an amylose–agarose affinity column (New England Biolabs). The column was washed extensively and specifically bound proteins were eluted according to the manufacturer’s instructions and used to immunize a rabbit.
SDS–PAGE and immunoblotting
Total cellular fractions or culture supernatants were prepared and analysed by SDS–PAGE. Proteins were separated in 10 or 12.5% acrylamide gels, stained with Coomassie Blue or transferred to nitrocellulose by semi-dry electroblotting. Immunoblots were stained with Ponceau S and incubated for 1 h in a 5% solution of non-fat dry milk in TBST buffer (10 mM Tris–HCl pH 8, 150 mM NaCl, 0.05% Tween-20). The primary antibodies, anti-MalE–GspG and anti-ChiA–MalE (Francetic et al., 2000), were diluted 1:1000. The secondary antibody was horseradish peroxidase-linked anti-rabbit IgG (Amersham) diluted 1:40 000. Immunoblots were developed by enhanced chemiluminescence using an ECL kit (Amersham) and detected using Kodak XAR-2 film.
Acknowledgments
Acknowledgements
Carol Kumamoto and Guy Tran Van Nhieu are thanked for critical reading of the manuscript, Roy Williams for suggesting that gsp might be regulated by H-NS, Sylvie Rimsky for advice on H-NS, Philippe Bertin for advice and the gift of strains, Evelyne Richet for helpful comments, Dominique Vidal for computer assistance and all members of the Pugsley laboratory for their help and support. This work was supported by the European Union (Training and Mobility in Research grant numbers FMRX-CT96-0004 and HPRN-CT-2000-00075) and by the Swiss National Science Foundation. During the course of this work, O.F. was supported by the Institut Pasteur, by the Fondation pour la Recherche Médicale, by the Fondation Mérieux and by Aventis-Pasteur. C.B. is a recipient of an MENRT fellowship.
References
- Alexandre G. and Zhulin,I. (2000) Laccases are widespread in bacteria. Trends Biotechnol., 18, 41–42. [DOI] [PubMed] [Google Scholar]
- Ali A., Johnson,J.A., Franco,A.A., Metzger,D.J., Connell,T.D., Morris,J.G.,Jr and Sozhamannan,S. (2000) Mutations in the extracellular protein secretion pathway genes (eps) interfere with rugose polysaccharide production and motility of Vibrio cholerae. Infect. Immun., 68, 1967–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andro T., Chambost,J.-P., Kotoujansky,A., Cattaneo,J., Bertheau,Y., Barras,F., van Gijsegem,F. and Coleno,A. (1984) Mutants of Erwinia chrysanthemi defective in secretion of pectinase and cellulase. J. Bacteriol., 160, 1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlung T. and Ingmer,H. (1997) H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol., 24, 7–17. [DOI] [PubMed] [Google Scholar]
- Bender R. (1996) Variations on a theme by Escherichia. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella. Cellular and Molecular Biology, 2nd edn, Vol. 1. ASM Press, Washington, DC, pp. 4–9. [Google Scholar]
- Bertin P., Terao,E., Lee,E.H., Lejeune,P., Colson,C., Danchin,A. and Collatz,E. (1994) The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J. Bacteriol., 176, 5537–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner F.R. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1462. [DOI] [PubMed] [Google Scholar]
- Bost S. and Belin,D. (1995) A new genetic selection identifies essential residues in SecG, a component of the Escherichia coli protein export. EMBO J., 14, 4412–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Kupiec R. and Chet,I. (1998) The molecular biology of chitin digestion. Curr. Opin. Biotechnol., 9, 270–277. [DOI] [PubMed] [Google Scholar]
- Condemine G. and Shevchik,V.E. (2000) Overproduction of the secretin OutD suppresses the secretion defect of an Erwinia chrysanthemi outB mutant. Microbiology, 146, 639–647. [DOI] [PubMed] [Google Scholar]
- Connel T.D., Metzger,D.J., Lynch,J. and Folster,J.P. (1998) Endo chitinase is transported to the extracellular milieu by the eps-encoded general secretory pathway of Vibrio cholerae. J. Bacteriol., 180, 5591–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B.M., Lawson,E.H., Sandkvist,M., Sozhamannan,S., Ali,A. and Waldor,M.K. (2000) Convergence of the secretory pathways for cholera toxin and the filamentous phage, CTXφ. Science, 288, 333–335. [DOI] [PubMed] [Google Scholar]
- Defez R. and De Felice,M. (1981) Cryptic operon for β-glucoside metabolism in Escherichia coli K12: genetic evidence for a regulatory protein. Genetics, 97, 11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot A. and Tommassen,J. (1991) Conservation of xcp genes, involved in the two-step protein secretion process, in different Pseudomonas species and other Gram-negative bacteria. Mol. Gen. Genet., 229, 278–284. [DOI] [PubMed] [Google Scholar]
- d’Enfert C. and Pugsley,A.P. (1989) Klebsiella pneumoniae pulS gene encodes an outer membrane lipoprotein required for pullulanase secretion. J. Bacteriol., 171, 3673–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckenstein J., Lindler,L., Elsinghorst,E. and Dale,J. (2000) Identification of a gene within a pathogenicity island of enterotoxigenic Escherichia coli H10407 required for maximal secretion of the heat-labile enterotoxin. Infect. Immun., 68, 2766–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francetic O. and Pugsley,A.P. (1996) The cryptic general secretory pathway (gsp) operon of Escherichia coli K-12 encodes functional proteins. J. Bacteriol., 178, 3544–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francetic O., Lory,S. and Pugsley,A.P. (1998) A second prepilin peptidase gene in Escherichia coli K-12. Mol. Microbiol., 27, 763–775. [DOI] [PubMed] [Google Scholar]
- Francetic O., Badaut,C., Rimsky,S. and Pugsley,A.P. (2000) The ChiA (YheB) protein of Escherichia coli K-12 is an endochitinase whose gene is negatively controlled by the nucleoid-structuring protein H-NS. Mol. Microbiol., 35, 1506–1517. [DOI] [PubMed] [Google Scholar]
- Guzman E.C., Pritchard,R.H. and Jiménez-Sánchez,A. (1991) A calcium binding protein that may be required for the initiation of chromosome replication in Escherichia coli. Res. Microbiol., 142, 137–140. [DOI] [PubMed] [Google Scholar]
- Heidelberg J.F. et al. (2000) DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature, 406, 477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard S.P., Critch,J. and Bedi,A. (1993) Isolation and analysis of eight exe genes and their involvement in extracellular protein secretion and outer membrane assembly in Aeromonas hydrophila. J. Bacteriol., 175, 6695–6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard S.P., Meiklejohn,H.C., Shivak,D. and Jahagirdar,R. (1996) A TonB-like protein and a novel membrane protein containing an ATP-binding cassette function together in exotoxin secretion. Mol. Microbiol., 22, 595–604. [DOI] [PubMed] [Google Scholar]
- Jeong W. and Kang,C. (1994) Start site selection at lacUV5 promoter affected by the sequence context around the initiation site. Nucleic Acids Res., 22, 4667–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.A. and Isaacson,R.E. (1998) In vivo expression of the β-glucoside (bgl) operon of Escherichia coli occurs in mouse liver. J. Bacteriol., 180, 4746–4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg M., Salmond,G.P.C. and Collmer,A. (1996) Complementation of deletion mutations in a cloned functional cluster of Erwinia chrysanthemi out genes with Erwinia carotovora out homologs reveals OutC and OutD as candidate gatekeepers of species-specific secretion of proteins via the Type II pathway. Mol. Microbiol., 20, 175–190. [DOI] [PubMed] [Google Scholar]
- Lopilato J. and Wright,A. (1990) Mechanism of activation of the cryptic bgl operon. In Drlica,K. and Riley,M. (eds), The Bacterial Chromosome. American Society of Microbiology, Washington, DC, pp. 439–444. [Google Scholar]
- Lucht J.M., Dersch,P., Kempf,B. and Bremer,E. (1994) Interactions of the nucleoid-associated DNA-binding protein H-NS with the regulatory region of the osmotically controlled proU operon of Escherichia coli. J. Biol. Chem., 269, 6578–6586. [PubMed] [Google Scholar]
- Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Neill R.J., Ivins,B.E. and Holmes,R.K. (1983) Synthesis and secretion of the plasmid-coded heat-labile enterotoxin of Escherichia coli in Vibrio cholerae. Science, 22, 289–291. [DOI] [PubMed] [Google Scholar]
- Nouwen N., Ranson,N., Saibil,H., Wolpensinger,B., Engel,A., Ghazi,A. and Pugsley,A.P. (1999) Secretin PulD: association with pilot protein PulS, structure and ion-conducting channel formation. Proc. Natl Acad. Sci. USA, 96, 8173–8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouwen N., Stahlberg,H., Pugsley,A.P. and Engel,A. (2000) Domain structure of secretin PulD revealed by limited proteolysis and electron microscopy. EMBO J., 19, 2229–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possot O. and Pugsley,A.P. (1994) Molecular characterization of PulE, a protein required for pullulanase secretion. Mol. Microbiol., 12, 287–299. [DOI] [PubMed] [Google Scholar]
- Possot O., Letellier,L. and Pugsley,A.P. (1997) Energy requirement for pullulanase secretion by the main terminal branch of the general secretory pathway. Mol. Microbiol., 24, 457–464. [DOI] [PubMed] [Google Scholar]
- Possot O., Vignon,G., Bomchil,N., Ebel,F. and Pugsley,A.P. (2000) Multiple interactions between pullulanase secreton components involved in stabilization and cytoplasmic membrane association of PulE. J. Bacteriol., 182, 2142–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley A.P. (1993a) The complete general secretory pathway in Gram-negative bacteria. Microbiol. Rev., 57, 50–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley A.P. (1993b) Processing and methylation of PulG, a pilin-like component of the general secretory pathway of Klebsiella oxytoca. Mol. Microbiol., 9, 295–308. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sandkvist M., Michel,L.O., Hough,L.P., Morales,V.M., Bagdasarian,M., Koomey,M., DiRita,V.J. and Bagdasarian,M. (1997) General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J. Bacteriol., 179, 6994–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandkvist M., Keith,J.M., Bagdasarian,M. and Howard,S.P. (2000) Two regions of EpsL involved in species-specific protein–protein interactions with EpsE and EpsM of the general secretion pathway in Vibrio cholerae. J. Bacteriol., 182, 742–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvonnet N., Gounon,P. and Pugsley,A.P. (2000a) PpdD type IV pilin of Escherichia coli K-12 can be assembled into pili in Pseudomonas aeruginosa. J. Bacteriol., 182, 848–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvonnet N., Vignon,G., Pugsley,A.P. and Gounon,P. (2000b) Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J., 19, 2221–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy T.J., Berman,M.L. and Enquist,L.W. (1984) Experiments with Gene Fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Stojiljkovic I., Schönherr,R. and Kusters,J.G. (1995) Identification of the hopG gene, a component of Escherichia coli K-12 type II export system and its conservation among different pathogenic Escherichia coli and Shigella isolates. J. Bacteriol., 177, 1892–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner L.R., Cano-Lara,J., Nunn,D.N. and Lory,S. (1993) Mutations in the consensus ATP-binding sites of XcpR and PilB eliminate extracellular protein secretion and pilus biogenesis in Pseudomonas aeruginosa. J. Bacteriol., 175, 4962–4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way J.C., Davis,M.A., Morisato,D., Roberts,D.E. and Kleckner,N. (1984) New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene, 32, 369–379. [DOI] [PubMed] [Google Scholar]
- Whitchurch C.B. and Mattick,J.S. (1994) Escherichia coli contains a set of genes homologous to those involved in protein secretion, DNA uptake and the assembly of type-4 fimbriae in other bacteria. Gene, 150, 9–15. [DOI] [PubMed] [Google Scholar]
- Zagursky R., Baumeister,N., Lomax,N. and Berman,M. (1985) Rapid and easy sequencing of large linear double stranded DNA and supercoiled plasmid DNA. Gene Anal. Tech., 2, 89–94. [Google Scholar]