Abstract
Neuronal precursor cell-expressed developmentally down-regulated 4 (Nedd4) proteins are ubiquitin ligases, which attach ubiquitin moieties to their target proteins, a post-translational modification that is most commonly associated with protein degradation. Nedd4 ubiquitin ligases have been shown to down-regulate both potassium and sodium channels. In this study, we investigated whether Nedd4 ubiquitin ligases also regulate Cav calcium channels.
We expressed three Nedd4 family members, Nedd4-1, Nedd4-2, and WWP2, together with Cav1.2 channels in tsA-201 cells. We found that Nedd4-1 dramatically decreased Cav whole-cell currents, whereas Nedd4-2 and WWP2 failed to regulate the current. Surface biotinylation assays revealed that Nedd4-1 decreased the number of channels inserted at the plasma membrane. Western blots also showed a concomitant decrease in the total expression of the channels. Surprisingly, however, neither the Cav pore-forming α1 subunit nor the associated Cavβ and Cavα2δ subunits were ubiquitylated by Nedd4-1. The proteasome inhibitor MG132 prevented the degradation of Cav channels, whereas monodansylcadaverine and chloroquine partially antagonized the Nedd4-1-induced regulation of Cav currents. Remarkably, the effect of Nedd4-1 was fully prevented by brefeldin A. These data suggest that Nedd4-1 promotes the sorting of newly synthesized Cav channels for degradation by both the proteasome and the lysosome. Most importantly, Nedd4-1-induced regulation required the co-expression of Cavβ subunits, known to antagonize the retention of the channels in the endoplasmic reticulum. Altogether, our results suggest that Nedd4-1 interferes with the chaperon role of Cavβ at the endoplasmic reticulum/Golgi level to prevent the delivery of Cav channels at the plasma membrane.
Keywords: Calcium Channels, Lysosomes, Membrane Proteins, Trafficking, Ubiquitylation, Cav Beta Subunits, Proteasome
Introduction
Voltage-dependent calcium channels (Cav)3 constitute a major pathway for the entry of calcium into excitable cells. There are three main classes of Cav channels (Cav1–3), which expression varies according to tissue and cell type (1). The main pore-forming Cavα1 subunit carries the main biophysical and pharmacological properties of the channel. Cav1α1 and Cav2α1 subunits are associated with accessory subunits Cavβ and Cavα2δ. Both Cavβ and Cavα2δ play a dual role in regulating both the biophysical properties and trafficking of Cav channels (2–4). In contrast, Cav3 channels are thought to be solely constituted of the pore-forming Cavα1 subunit (1).
In addition to their voltage dependence, the channels are regulated by signaling pathways. Previously, we have shown that phosphorylation of Cavβ2 regulates the expression of Cav1 and Cav2 channels at the plasma membrane (4). A growing number of studies show that, like phosphorylation, ubiquitylation plays a major role in regulating the surface density of membrane proteins. In many cases, ubiquitylation acts as a signal for endocytosis of plasma membrane proteins, which are subsequently degraded by the lysosome or the proteasome (5). Ubiquitylation also occurs at the exit of the endoplasmice reticulum (ER) (5) and the trans-Golgi network (TGN) (6, 7). One family of ubiquitin ligases, Nedd4 (8), has been shown to ubiquitylate and down-regulate a wide range of ion channels, including KCNQ1 potassium (9) and Nav1.5 sodium channels (10, 11) in heterologous expression systems. In native tissues, ENaC epithelial sodium channels (12), Kv1.3 (13), and ClC-5 channels (14) have also been shown to be down-regulated by Nedd4-like enzymes. However, the potential role of Nedd4 ubiquitin ligases on Cav channels has not yet been examined.
Our study focuses on dihydropyridine-sensitive Cav1.2 channels, which are ubiquitously expressed in excitable cells, and are the predominant isoform expressed in the cardiovascular system. We used mammalian cell lines to investigate whether Nedd4 ligases regulate Cav1.2α1 channels expressed together with Cavβ2 and Cavα2δ-1. Of the three family members tested (Nedd4-1, Nedd4-2, and WWP2), only Nedd4-1 was found to alter Cav whole-cell currents. Overexpressing Nedd4-1 reduced both Cav currents and the surface expression of Cav1.2α1. The Nedd4-1-induced down-regulation of Cav channels at the plasma membrane was associated with a decrease in the total expression of Cav1.2α1, Cavβ2, and Cavα2δ-1. Immunoprecipitation and pulldown assays of ubiquitylated proteins failed to show any increase in the ubiquitylation level of either Cav subunit, suggesting that the ubiquitin ligase acts indirectly on the channel. Evidence obtained using a variety of tools, interfering with either the internalisation or the forward trafficking of membrane proteins, suggests that Nedd4-1 promotes the targeting of Cav channels directly from the ER-Golgi network toward the proteasomal and lysosomal compartments. Finally, Cav1.2α1/α2δ-1 (without Cavβ subunit) channels failed to be regulated by Nedd4-1, suggesting that the accessory Cavβ subunit is essential for the Nedd4-1-induced degradation of newly synthesized Cav channels.
EXPERIMENTAL PROCEDURES
cDNA Constructs
Rabbit Cav1.2α1 (cP15381.1), Cavβ2 (P54288), and Cavα2δ-1 (P13806) cDNAs, inserted into pCARDHE, pBH17, and pCA1S respectively, were gifts from Dr. G. S. Pitt (Division of Cardiology, Department of Medicine, Duke University Medical Center, Durham, NC). Rabbit Cav2.2α1 (D14157), Cav3.2α1, rat Cavβ1 (X61394), β3 (M88751), and β4 (L02315) cDNAs inserted into pMT2 were gifts from professor A. C. Dolphin (Department of Neuroscience, Physiology, and Pharmacology, University College London, London, UK). Human Nedd4-1 (KIAA0093) and Nedd4-2 (KIAA0439) cDNA subcloned into pcDNA3.1 (Invitrogen) were gifts from Dr. T. Nagase (Kazusa Institute, Kazusa, Japan). Human WWP2 cDNA (U96114) was amplified by PCR from a human heart cDNA library (Matchmaker, NT 91-2769; Clontech, Basel, Switzerland), cloned into EcoRI-digested pcDNA3.1. CD8 cDNA was subcloned into EBOpcD-Leu2. Human dynamin I wild-type and mutant (K44R) cDNAs inserted into pcDNA3.1 were gifts from Pr. O. Staub (Department of Pharmacology and Toxicology, Lausanne, Switzerland). A GST fusion of the ubiquitin binding proteasomal subunit S5A (GST-S5A) was obtained from a S5A cDNA cloned into pGEX-4T1 (Amersham Biosciences, Otelfingen, Switzerland). Cavβ2Y221A and Nedd4-1C867S mutant were generated using the QuikChange XL II mutagenesis kit (Stratagene) and verified by sequencing. Myc-Nedd4-1 was generated by PCR, using 5′-GTC CAT AAG CTT GCC ACC ATG GAG CAG AAG TTG ATT AGC GAG GAG GAT TTG GCA ACT TGC GCG GTG GAG GTG-3′ and 5′-GTC CAT CTC GAG CTA ATC AAC TCC ATC AAA-3′ primers.
Transfections
For electrophysiology experiments, tsA-201 cells were transiently transfected with 7.5 μl of transfection reagent FuGENE 6® (Roche Diagnostics) and 0.3 μg cDNA of each Cav channel subunit (Cav1.2α1, Cavβ, and Cavα2δ-1; ratio 1:1:1) together with 1.0 μg of either Nedd4, WWP2, or dynamin constructs, per 25 cm2 flask. Empty pcDNA3.1 vector was used as a control. In addition, 0.5 μg of cDNA encoding CD8 antigen was added to all transfections as a reporter gene. At 24 h post-transfection, cells were split at low density (3% of one 25 cm2 flask per dish). Anti-CD8 beads (Dynal®, Oslo, Norway) were used to identify transfected cells. For biochemistry experiments, 10-cm dishes of HEK-293 cells were transfected using 10 μg of cDNAs and 30 μl of Lipofectamine LTX® (Invitrogen). The ratio of the different constructs was similar as for the patch clamp experiments. Cells were used 48 h after transfection.
Electrophysiology
Whole-cell currents were measured at room temperature (22–23 °C) using an Axopatch200B amplifier (Axon Instruments, Union City, CA). The internal pipette solution was composed of the following: 60 mm CsCl, 70 mm cesium-aspartate, 1 mm MgCl2, 10 mm HEPES, 11 mm EGTA, and 5 mm Mg-ATP, pH 7.2, with CsOH. The external solution contained the following: 130 mm NaCl, 5.6 mm KCl, 1–20 mm BaCl2, 1 mm MgCl2, 10 mm HEPES, and 11 mm d-glucose, pH 7.4, with NaOH. Current records are shown after leak and capacitance current subtraction (P/4 protocol). Data were analyzed using pClamp (version 9.2, Axon Instruments, Union City, California) and Origin (version 7.5, OriginLab® corporation, Northampton, MD) software. Barium current densities in picoamperes/picofarads (pA/pF) were calculated by dividing the peak current by the cell capacitance. The I-V relationship (IV) was fitted with the following equation, y = (Gmax(Vh − Vrev,Ca))/(1 + exp((Vh − V50)/k)), in which y is the normalized peak current (pA/pF) at a given holding potential (Vh), V50 is the voltage at which half of the channels are activated, k is the slope factor, and Gmax is the maximum conductance. Activation curves and steady-state inactivation curves were fitted with the following single Boltzmann equation, y = 1/(1 + exp((Vh − V50)/k)), in which y is the normalized conductance (activation) or peak current (steady-state inactivation) at a given holding potential (Vh), V50 is the voltage at which half of the channels are activated (V50,act) or inactivated (V50,inact) respectively, and k is the slope factor.
Western Blots
10-cm HEK-293 cells dishes or whole C57BL/6 mouse heart mechanically dissociated were lysed in 1.0 ml of lysis buffer (50 mm HEPES, pH 7.4, 150 mm NaCl, 10% glycerol, 1% Triton, 1 mm EGTA supplemented with 10 mm N-ethyl maleimide, and protease inhibitors). Soluble fractions were recovered in supernatants after 15 min of centrifugation at 13,000 × g at 4 °C.
Immunoprecipitations
One mg of HEK-293 cells or C57BL/6 mouse heart lysate were incubated for 2 h at 4 °C with appropriate antibody previously linked to Dynabeads A (Invitrogen) according to the manufacturer's instructions. Negative control for mouse heart immunoprecipitation was done with unspecific rabbit antibodies (Santa Cruz Biotechnology, Santa Cruz, CA).
Pulldown Assays
One mg of HEK-293 cell lysate was added to either 50 μg of GST-S5A beads or 50 μg of beads from the ubiQapture kit (Enzo Life Science, Lausanne, Switzerland) and incubated for 2 h at 4 °C. Precipitated proteins were eluted with sample buffer (Invitrogen).
Surface Biotinylation Assays
Cells were treated for 30 min at 4 °C with 4 ml of biotin per 10-cm dish (1 mg/ml; EZ link sulfo-NHS-SS-biotin; Pierce), washed three times with cold PBS containing 200 mm glycine, and lysed with 1-ml/dish of lysis buffer. Fifty μl of streptavidin Sepharose beads (GE Healthcare Europe, Glattbrugg, Switzerland) were added to 1 mg of HEK-293 cell lysate and incubated 2 h on a wheel at 4 °C. The beads were washed five times with lysis buffer and resuspended in sample buffer (Invitrogen). Eluted proteins were analyzed by Western blot.
Antibodies
Anti-ubiquitin monoclonal antibody FK2 (Enzo Life Science, Lausanne, Switzerland) was used at a dilution of 1:500. Antibodies against Cav1.2α1 (ACC003; Alomone, Jerusalem, Israel) and Cavβ2 (ab54920; Abcam, Cambridge, UK) were used at a dilution 1:200 and Cavα2δ-1 (ab2864; Abcam) was used at a dilution of 1:1000. Nedd4 antibody, used at a dilution of 1:1000, was a gift from professor O.Staub (Department of Pharmacology and Toxicology, University of Lausanne, Lausanne, Switzerland). Monoclonal antibodies raised against actin and Myc were purchased from Sigma-Aldrich (Sigma-Aldrich Chemie, Postfach, Switzerland) and used at a dilution of 1:2000.
Drugs
Monodansylcadaverine (MDC), brefeldin A (BFA), MG132, and chloroquine diphosphate were purchased from Sigma-Aldrich (Sigma-Aldrich, Dorset, UK). Chloroquine was dissolved in water. MDC, BFA, and MG132 were dissolved in DMSO. DMSO did not exceed 0.1% in the final solution.
Statistical Analysis
Data are presented as means ± standard error of the mean. Unpaired, two-tailed Student's t tests or one-way analysis of variance with a Bonferroni post-test were used to compare the means; p < 0.05 was considered significant.
RESULTS
Nedd4-1 Down-regulates Cav1.2 Channels
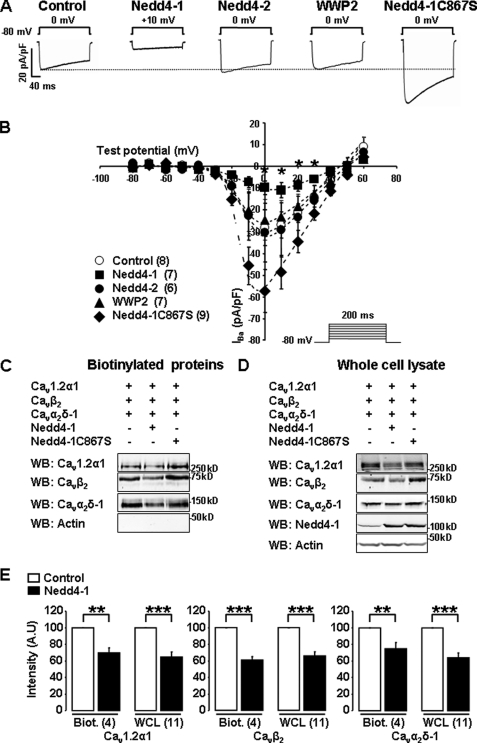
We expressed Cav1.2α1/β2/α2δ-1 channels together with Nedd4-1, Nedd4-2, or their close relative WWP2. Fig. 1A shows that Nedd4-1 dramatically reduced Cav currents by 67 ± 10% (n = 7), whereas neither Nedd4-2 nor WWP2 showed significant effects (+8 ± 47%; n = 6 and −13 ± 34%; n = 7). Conversely, expression of a catalytically inactive Nedd4-1 mutant (Nedd4-1C867S) significantly increased Cav1.2 current densities by 90 ± 33% (n = 9; Fig. 1, A and B), suggesting that Cav1.2 channels may be constitutively regulated by an endogenous pool of Nedd4-1. Nedd4-1 also induced a minor shift of the current-voltage relationship and steady-state inactivation curves (supplemental Table 1). Interestingly, it is known that in the absence of Cavβ, Cav currents are also dramatically reduced, and their biophysical parameters are shifted in the same direction as with Nedd4-1, although the amplitude of the shifts is significantly more pronounced (2) (supplemental Table 1). To examine whether Nedd4-1 may reduce the current by disrupting the interaction of Cavβ with the pore of the channel, we performed co-immunoprecipitation experiments (supplemental Fig. 1). These showed that the same proportion of Cavβ2 was associated with Cav1.2α1 in control and Nedd4-1-transfected cells. Hence, there is no evidence that Nedd4-1 interferes with the binding of Cavβ to the pore subunit of the channel.
FIGURE 1.
Nedd4-1 down-regulates Cav1.2 channels. A, representative whole-cell current traces. B, I-V relationships recorded from tsA-201 cells transfected with Cav1.2α1/Cavβ2/Cavα2δ-1 channels alone (Control, ○) or together with either Nedd4-1 (■), Nedd4-2 (●), WWP2 (▲), or the catalytically inactive mutant Nedd4-1C867S (♦). The number of cells recorded is indicated in parentheses. *, p < 0.05 when compared with control cells transfected with Cav1.2α1/Cavβ2/Cavα2δ-1 channels alone. C and D, surface biotinylation assays were performed in control, Nedd4-1- and Nedd4-1C867S-transfected HEK-293 cells. Western blots (WB) showing Cav channels detected in the biotinylated fraction (C) and corresponding whole-cell lysates (D). E, bar graphs illustrating the down-regulation of Cav1.2α1, Cavβ2, and Cavα2δ-1 subunits mediated by Nedd4-1 in biotinylation assays (Biot.) and in Western blot on whole-cell lysates (WCL). The number of independent experiments is indicated in parentheses. A.U., arbitrary units. **, p < 0.01 and ***, p < 0.001 when compared with control cells transfected with Cav1.2α1/β2/α2δ-1 channels only.
Because Nedd4 ligases have been shown to promote the internalization of other ion channels (5, 12, 15, 16), we examined whether the decrease of Cav currents may be correlated to a decrease in the expression of Cav channels at the plasma membrane. We first performed immunocytochemistry experiments to examine the subcellular localization of Cav1.2α1. The distribution pattern of Cav1.2 channels was similar in control and Nedd4-1-transfected cells. As reported previously in other transfected cell lines (4), there was little or no clear accumulation of Cav channels at the plasma membrane of control cells (supplemental Fig. 2). Nonetheless, we observed a reduction in the total fluorescence of 30 ± 9% in Nedd4-1-transfected cells compared with control, which hinted that this ubiquitin ligase may alter the overall expression of the channels. To quantify and compare the insertion of Cav channels in the two conditions, we used biochemistry techniques to measure the total expression and the membrane localization of the channels. We performed surface biotinylation assays, which detect only the proteins inserted in the plasma membrane. In control cells, transfected with the channels only, we were able to detect both Cav1.2α1 and Cavα2δ-1 subunits, as well as the cytosolic Cavβ2 (Fig. 1C). This was expected as Cavβ subunits interact with Cav1.2α1 and co-immunoprecipitate with this pore-forming subunit (supplemental Fig. 1) (1, 2). Another cytosolic protein, actin, which does not co-immunoprecipitate with Cav channels, was not detected in the surface biotinylated fraction, confirming that our protocol respected the integrity of the plasma membrane of the cells. We found that cells overexpressing Nedd4-1 showed a reduction in both the number of Cav1.2 channels inserted at the plasma membrane (Fig. 1, C and E) and in the total amount of channels expressed in the cells (Fig. 1, D and E). Interestingly, both Cavβ2 and Cavα2δ-1 were down-regulated in similar proportion as Cav1.2α1 in cells overexpressing Nedd4-1 (Fig. 1, C–E), indicating that Nedd4-1 regulates the whole channel complex rather than the sole pore-forming Cav1.2α1. In agreement with its effect on Cav currents, the catalytically inactive Nedd4-1C867S increased both the total amount of Cav1.2α1 subunits (Fig. 1D) and their expression at the plasma membrane (Fig. 1C). These findings suggest that the Nedd4-1-induced decrease of Cav current is caused by a reduction in the availability of channels at the plasma membrane rather than a functional inhibition of the channels.
Nedd4-1 Does Not Ubiquitylate Cav1.2 Channels
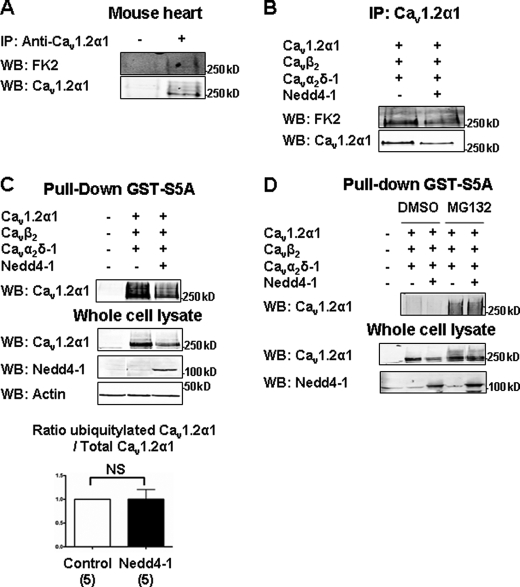
We next examined whether Cav1.2α1 was ubiquitylated by Nedd4-1. We assessed the ubiquitylation level of immunoprecipitated Cav1.2α1 in Western blots, using FK2 antibodies that recognize polyubiquitylated proteins. We found that both native Cav1.2α1 in cardiac lysate (Fig. 2A) and Cav1.2α1 expressed in HEK-293 cells (Fig. 2, B–D) were ubiquitylated in basal conditions. In agreement with the Nedd4-1-induced reduction of Cav1.2α1 shown in Fig. 1D, the amount of immunoprecipitated Cav1.2α1 (Fig. 2B) was ∼45% smaller in HEK-293 cells co-transfected with Nedd4-1 versus the channels only. We compared the amount of ubiquitylated Cav1.2α1, detected with the FK2 antibody, with the total amount of immunoprecipitated Cav1.2α1 detected with the anti-Cav1.2α1 antibody. This ratio, displayed in Fig. 2C, shows that overexpressing Nedd4-1 failed to significantly increase the amount of ubiquitylated channels. Pulldown experiments performed using a GST-fused peptide corresponding to the protein S5A, which recognizes polyubiquitylated proteins (17), also failed to show any Nedd4-1-induced ubiquitylation of Cav1.2α1 (Fig. 2C). Likewise, the ubiquitylation level of Cav1.2α1 was unchanged with the inactive mutant Nedd4-1C867S (supplemental Fig. 3).
FIGURE 2.
The basal ubiquitylation of Cav1.2α1 is not regulated by Nedd4-1. A, native cardiac Cav1.2α1 subunits were immunoprecipitated (IP) from mouse heart lysates, and their ubiquitylation state was determined by Western blotting (WB) with anti-ubiquitin FK2 antibodies. The first lane (−) shows a control experiment performed with unspecific rabbit antibodies. The second lane (+) shows the immunoprecipitated Cav1.2α1. B, Western blots showing that Nedd4-1 reduced both the amount of immunoprecipitated Cav1.2α1 and the corresponding FK2 signal in HEK-293 cells. C, ubiquitylated proteins were pulled down using ubiquitin binding GST-S5A. Western blots show that Nedd4-1 decreased the amount of Cav1.2α1 in pulled down fractions and in whole-cell lysates. Bar graph shows the ratio of pulled down ubiquitylated Cav1.2α1 versus total Cav1.2α1, in five independent experiments. This ratio was calculated as the intensity of the smear containing several bands corresponding to ubiquitylated Cav1.2α1 channels, divided by the intensity of the band corresponding to the total pool of Cav1.2α1 subunits (ubiquitylated and nonubiquitylated) shown in the whole cell lysate. D, Western blots showing that the proteasome inhibitor MG132 (10 μm for 8 h) antagonized the effect of Nedd4-1 and increased the amount of Cav1.2α1 both in GST-S5A pulled down fractions and the corresponding whole-cell lysates. NS, not significant.
Kawaguchi et al. (18) had previously shown that ubiquitylation of Cav1.2α1 was increased in the presence of the proteasome inhibitor MG132. Moreover, degradation of Cav channels by the proteasome has previously been reported (19). To make sure that a possible ubiquitylation by Nedd4-1 was not occulted by the concomitant degradation of the channels by the proteasome, we repeated GST-S5A pulldown experiments after treatment with MG132. As expected, the proteasome inhibitor increased the amount of ubiquitylated Cav1.2 channels recovered in both control and Nedd4-1-transfected cells. Interestingly, MG132 also prevented the Nedd4-1-induced decrease of Cav1.2α1 protein. However, and in agreement with our other results, Cav1.2α1 were not further ubiquitylated by Nedd4-1 (Fig. 2D).
Altogether, these results indicate that Cav1.2α1 is not ubiquitylated by Nedd4-1. The effect of MG132 also suggests that proteasome-dependent degradation contribute to the inhibitory effect of Nedd4-1 on Cav channels.
Cavβ Subunits Are Essential for Regulation of Cav Channels by Nedd4-1
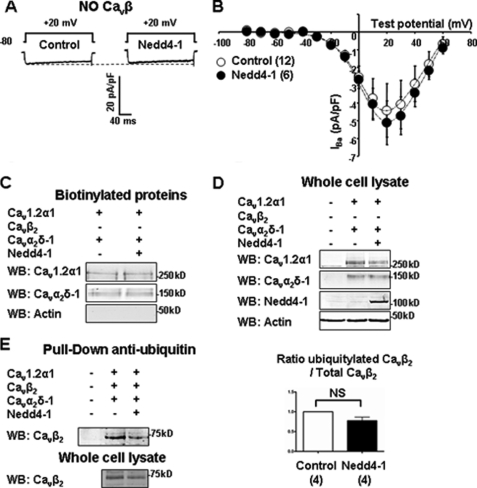
Because Cav1.2α1 does not appear to be the direct target of Nedd4-1, we examined the possibility that Cav accessory subunits may be involved. The concentration of charge carrier (Barium) was increased to 20 mm to compensate for the reduced Cav trafficking observed in the absence of Cavα2δ or Cavβ (2, 3). Whereas Cav channels expressed without Cavα2δ were still sensitive to Nedd4-1 (−53 ± 13%; n = 14), those expressed without Cavβ failed to be regulated (Fig. 3, A and B). Accordingly, neither the total amount (Fig. 3D) nor the biotinylated fraction of Cav channels (Fig. 3C) was affected by Nedd4-1 in the absence of Cavβ. Fig. 3E shows that Cavβ2 subunits were ubiquitylated in control conditions; however, there was no further ubiquitylation with Nedd4-1. Likewise, Cavα2δ-1, which are also ubiquitylated in basal conditions, were not further ubiquitylated by Nedd4-1 (supplemental Fig. 4). Interestingly, all four isoforms (β1–β4) of Cavβ subunits supported the effect of Nedd4-1 on Cav currents (supplemental Table 1).
FIGURE 3.
The effect of Nedd4-1 is Cavβ-dependent. A, representative current traces. B, current-voltage relationships of Cav1.2α1/Cavα2δ-1 channels expressed without Cavβ in control and Nedd4-1-transfected cells. C and D, surface biotinylation assays were performed in cells transfected with Cav1.2α1/Cavα2δ-1 alone or together with Nedd4-1. Representative Western blots (WB) showing that Nedd4-1 did not affect the amount of Cav1.2α1 and Cavα2δ-1 subunits detected in the biotinylated fraction (C; n = 4) or corresponding whole-cell lysates (D; n = 4). E, anti-ubiquitin pulldown assays were performed in Cav1.2α1/Cavβ2/Cavα2δ-1-transfected cells. Western blots show that Nedd4-1 decreased the amount of Cavβ2 subunits in pulled down fractions and whole-cell lysates. Bar graph shows that the ratio of ubiquitylated Cavβ2 versus total Cavβ2 from four independent experiments. NS, not significant.
Hence, although they are not the direct target of Nedd4-1, Cavβ subunits are essential for the regulation of Cav1.2 channels by Nedd4-1. Furthermore, the effect of Nedd4-1 is not Cavβ isoform-specific and may occur with any combination of Cavβ.
Nedd4-1 Does Not Promote Dynamin-dependent Cav Down-regulation
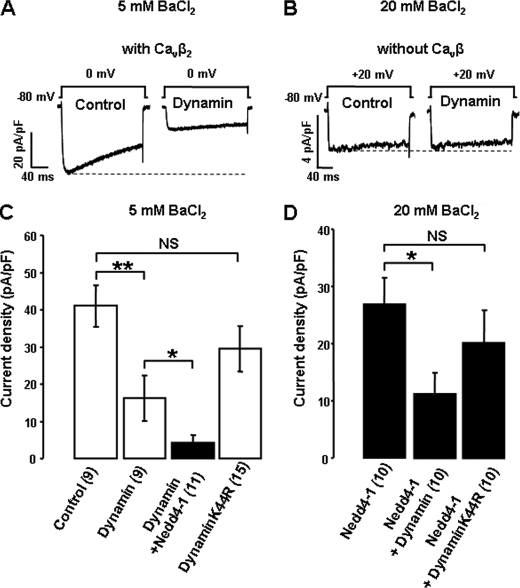
Cavβ subunits have been shown to bind to dynamin, a small GTPase involved in the formation of membrane vesicles, resulting in the internalisation of Cav channels inserted at the plasma membrane (20). We found that overexpressing dynamin alone was sufficient to decrease Cav1.2α1/β2/α2δ-1 currents by 61 ± 15% (n = 10) (Fig. 4A) but, as previously reported (20), there was no effect of dynamin on Cav1.2α1/α2δ-1 channels lacking Cavβ (Fig. 4B), confirming the essential role of Cavβ in the dynamin-dependent retrieval of Cav channels. In contrast, the catalytically inactive dynamin K44R, which cannot hydrolyze GTP and has been shown to interfere with the internalization of membrane proteins (21), did not significantly alter Cav current densities (Fig. 4C), indicating that there is little or no constitutive down-regulation of Cav channels by endogenous dynamin in our model. In addition, Cav current densities were similar in cells transfected with dynamin K44R and Nedd4-1 versus Nedd4-1 alone (Fig. 4D). The magnitude of the Nedd4-1-induced reduction in Cav current was also similar in the absence or presence of dynamin and reciprocally for the effect of dynamin in the absence or presence of Nedd4-1 (Fig. 4, C and D). Therefore, although both Nedd4-1 and dynamin act through Cavβ subunits to regulate Cav channels, these two pathways are independent, and Nedd4-1 does not reduce Cav currents by promoting dynamin-dependent internalization of Cav channels.
FIGURE 4.
Dynamin requires Cavβ to down-regulate Cav channels but is not involved in Nedd4-1-induced regulation. A and B, representative whole-cell current traces in control and dynamin-transfected cells expressing Cav1.2α1/Cavβ2/Cavα2δ-1 (A) and Cav1.2α1/Cavα2δ-1 (B) channels. C and D, effect of dynamin and dynamin K44R on Cav1.2α1/Cavβ2/Cavα2δ-1 current densities in control (white bars) and Nedd4-1-transfected cells (black bars), recorded in 5 mm BaCl2 (C) or 20 mm BaCl2 (D). NS, not significant. Error bars are standard errors of the mean. *, p < 0.05, **, p < 0.01 when compared with cells not transfected with either dynamin constructs.
Nedd4-1 Prevents Targeting of Newly Synthesized Cav Channels to Plasma Membrane
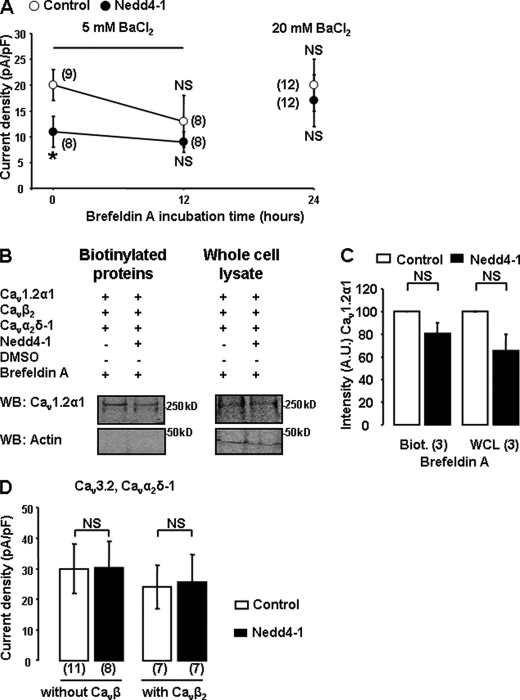
Cavβ subunits have also been shown to promote the exit of Cavα1 pore-forming subunits from the ER/Golgi (22). Therefore, we examined whether Nedd4-1 affected the forward trafficking of Cav channels. We used BFA, which impairs Golgi trafficking. 12 h of treatment with BFA was sufficient to significantly reduce Cav currents by 35 ± 25% (n = 8) in control cells (Fig. 5A). After 24 h of BFA treatment, and despite a further 12-h expression, the amplitude of the residual Cav currents recorded in 5 mm BaCl2 was too small to reliably measure an effect of Nedd4-1(Fig. 5A); hence, we proceeded using 20 mm BaCl2. We found that the inhibitory effect of Nedd4-1 was no longer apparent when cells were treated with BFA (for 12 or 24 h; Fig. 5A), suggesting that Nedd4-1 acts by interfering with the exit of Cav channels from the Golgi. This result was supported by surface biotinylation assays, which showed no significant change in the amount of Cav1.2α1 channels inserted at the plasma membrane in BFA-treated cells (Fig. 5B).
FIGURE 5.
Nedd4-1 targets newly-synthesized Cav channels. A, effect of BFA on Cav1.2α1/Cavβ2/Cavα2δ-1 current densities in control (○) or Nedd4-1-transfected cells (●). Cells were incubated with BFA (50 ng/ml) 24 h post-transfection. *, p < 0.05 when compared with corresponding controls. B, representative Western blots (WB) showing that, upon treatment with BFA, Nedd4-1 did not affect the amount of Cav1.2 channels detected in the biotinylated fraction (n = 3) or corresponding whole-cell lysates (WCL; n = 3). C, quantification is shown in the bar graph. NS, not significant; Biot., biotinylated. The number of independent experiments is indicated in parentheses. A.U., arbitrary units. D, lack of effect of Nedd4-1 on Cav3.2α1/Cavα2δ-1 current densities expressed without or with Cavβ2 subunits.
Recently, further roles have emerged for Cavβ subunits that may also act independently of Cav channels (3). To ensure that the Cavβ-dependent effect of Nedd4-1 was not due to an overall inhibition of membrane trafficking, we examined the effect of Nedd4-1 on Cav3.2 T-type channels. Whether Cavβ interacts with Cav3α1 subunit is still controversial (1, 23); however, it is generally accepted that Cav3 channels do not require the co-expression of auxiliary subunits to be efficiently targeted to the plasma membrane (23, 24). In agreement with this, the amplitude of Cav3.2 currents was similar in the absence or presence of Cavβ (Fig. 5C). We found that Nedd4-1 did not affect the current densities of Cav3.2 channels expressed either with or without Cavβ, indicating that trafficking of Cav3.2 to the plasma membrane was not affected by the ubiquitin ligase. Additional experiments performed on cloned neuronal Cav2.2 channels, which (like Cav1.2) depend on Cavβ for efficient membrane targeting (2), showed that these channels are also regulated by Nedd4-1 (supplemental Fig. 5). This result suggests that this regulation is not due to a specific interaction of Cavβ2 with Cav1.2 and further highlights the broad physiological relevance of this regulation.
Nedd4-1 Targets Cav Channels for Degradation by Lysosome
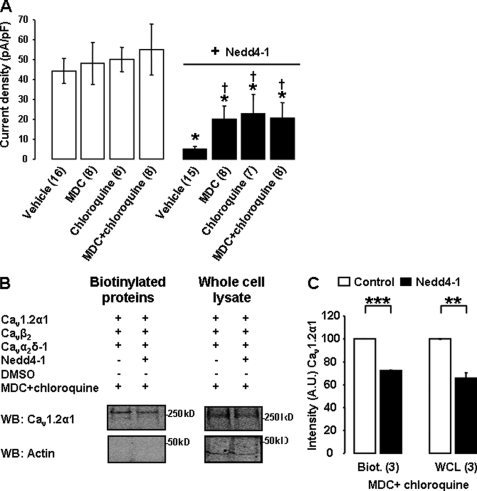
The inhibitory effect of MG132, shown in Fig. 2D, suggested that the proteasome was involved in Nedd4-1-induced degradation of Cav channels. To determine whether other degradation pathways were implicated, we pretreated the cells for 15 h (36 h post-transfection) with the lysosome inhibitor chloroquine. Chloroquine had no effect on basal Cav current densities; however, it partially prevented the effect of Nedd4-1 (by 45 ± 24%; n = 7; Fig. 6A), suggesting that Nedd4-1 directs Cav channels for lysosomal degradation. The partial effect of chloroquine was consistent with an additional contribution of the proteasome.
FIGURE 6.
Nedd4-1-induced regulation of Cav channels involves the lysosome. A, effect of MDC (100 μm for 15 h) and chloroquine (50 μm for 15 h) on Cav1.2α1/Cavβ2/Cavα2δ-1 current densities recorded in control (white bars) and Nedd4-1-transfected cells (black bars). †, p < 0.05 when compared with Nedd4-1-transfected cells treated with vehicle only; *, p < 0.05 when compared with corresponding control cells. B, surface biotinylation assays were performed in control and Nedd4-1-transfected cells treated with MDC and chloroquine. Western blots (WB) and associated bar graph, in C, show that the treatment failed to prevent the down-regulation of Cav1.2 channels in Nedd4-1-transfected cells. A.U., arbitrary units. The number of independent experiments is indicated in parentheses. **, p < 0.01, ***p < 0.001 when compared with control cells transfected with Cav1.2α1/Cavβ2/Cavα2δ-1 channels only. Biot., biotinylated; WCL, whole-cell lysate.
One way to transport cargo proteins from the TGN to other membrane compartments involves clathrin-coated post-Golgi carriers (25). To assess whether Nedd4-1 targeted Cav channels-containing post-Golgi carriers, we treated the cells with MDC, a drug that inhibits the formation of clathrin-coated pits (26). MDC was applied for 15 h, 36 h post-transfection. MDC did not significantly affect control Cav currents, suggesting that Cavα1 channels are trafficked to the plasma membrane via clathrin-independent pathways. MDC partially antagonized the effect of Nedd4-1 on Cav currents (by 38 ± 17%; n = 8; Fig. 6A). The extent of the inhibition produced by MDC was similar to the effect of chloroquine, and there was no additional effect when both drugs were added concomitantly (40 ± 19%; n = 8; Fig. 6A), suggesting that these drugs inhibited a common pathway and that Nedd4-1 recruits clathrin-coated vesicles to transport Cavα1 to the lysosome. This partial inhibition could not be seen on surface biotinylation and whole-cell lysate Western blots (Fig. 6B). However, this could be attributed to the lack of sensitivity of the method. Most importantly, these experiments suggest that Nedd4-1 also induces the degradation of Cav channels via a lysosome-independent pathway, in agreement with the involvement of the proteasome suggested in Fig. 2D.
Tyrosine 221 on Cavβ2 Subunit Is Required for Sorting of Cav Channels to Lysosome
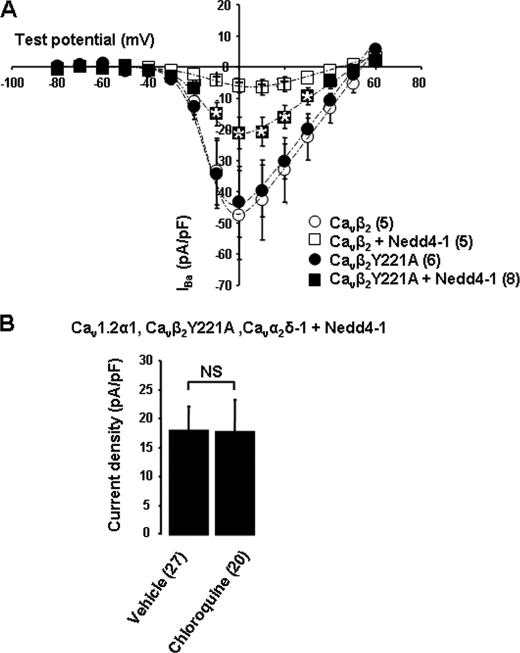
We next investigated the role of Cavβ in this regulation. Because Cavβ is cytosolic, it is conceivable that they act as a bridge to enable interaction between Cavα1 and the trafficking machinery. Several sorting signals for selection into clathrin-coated vesicles have been identified in the cytoplasmic tails of trans-membrane proteins. The best characterized is the YXXØ signal (Ø, bulky hydrophobic amino acid; X, any amino acid), which binds to the μ-subunits of the adaptor protein AP-1 (27). We identified a conserved YDVV motif in the SH3 domain of all four isoforms of Cavβ. Substitution of tyrosine 221 with an alanine in Cavβ2 (mutant Cavβ2Y221A) did not affect the basal current density (Fig. 7A) nor the biophysical properties of Cav channels (supplemental Table 1; Fig. 7A). However, the Y221A mutation partially reduced the effect of Nedd4-1 on Cav currents by 36%± 11 (n = 8) (Fig. 7A). Most importantly, there was no further effect of chloroquine when Cav channels were expressed with Cavβ2Y221A instead of Cavβ2 (Fig. 7B), confirming the involvement of this residue in the lysosome-dependent down-regulation of Cav channels.
FIGURE 7.
Cavβ tyrosine 221 is involved in Nedd4-1-induced degradation of Cav channels by the lysosome. A, current-voltage relationships showing the effect of Nedd4-1 on Cav1.2α1/Cavα2δ-1 channels expressed together with Cavβ2 or Cavβ2Y221A. Statistical analysis compared Cav1.2α1/Cavβ2Y221A/Cavα2δ-1/Nedd4-1 current densities with control cells (*, p < 0.05) and with Cav1.2α1/Cavβ2/Cavα2δ-1/Nedd4-1-transfected cells (†, p < 0.05). B, Nedd4-1-transfected cells were treated with either vehicle (H2O) or chloroquine (50 μmol/liter) for 15 h at 37 °C. The bar graph shows that chloroquine had no significant (NS) effect on Cav1.2α1/Cavβ2Y221A/Cavα2δ-1 current densities.
DISCUSSION
In this study, we uncovered a new Cavβ- and Nedd4-1-dependent mechanism controlling the sorting of newly synthesized Cav channels. Our data suggest that Nedd4 ubiquitin ligases promote the degradation of Cavβ-containing channels by both the proteasome and the lysosomes. Misfolded Cav channels have previously been reported to be degraded via endoplasmic reticulum-associated degradation (19). Most ubiquitin ligases involved in endoplasmic reticulum-associated degradation are yet to be identified (5), and one could suggest that endoplasmic reticulum-associated degradation account for the Nedd4-1-induced degradation of Cavα1 by the proteasome. Nonetheless, this is unlikely to fully explain the effect of Nedd4-1 for three reasons: (i) the effect of Nedd4-1 is dependent on Cavβ, which associates with Cavα1 at the exit of the ER (22), (ii) Nedd4-1 down-regulates both Cavβ and Cavα2δ-1 together with Cav1.2α1, and (iii) Nedd4-1 did not alter the proportion of Cavβ co-immunoprecipitated with Cav1.2α1. These three points suggest that the Nedd4-1-regulated channels are correctly folded and processed beyond the ER until fully assembled at the exit of the TGN. Furthermore, we showed that treatment with either brefeldin A or chloroquine counteracted the effect of Nedd4-1 on Cav currents. Altogether, these data provide the first evidence that fully assembled Cav channels can be sorted directly from the TGN to the lysosome, without transiting to the plasma membrane. This finding is original because, to our knowledge, there is only one other example of such a regulation; in yeast, newly synthesized Gap1 (general amino acid permease 1) is targeted to lysosomes instead of the plasma membrane (6), when the cell senses Gap1 is not required at the plasma membrane (28).
In contrast with Gap1 and other channels directly regulated by Nedd4, we found that Nedd4-1 does not ubiquitylate Cav1.2α1 channels nor their associated subunits Cavβ2 and Cavα2δ-1. Nonetheless, the increase in Cav current induced by Nedd4-1C867S (Fig. 1) indicates that Nedd4-1 acts through its ubiquitin ligase activity. Interestingly, Nedd4 has previously been shown to indirectly regulate other proteins. For example, Myat et al. (29) have reported that Drosophila roundabout receptors were down-regulated upon ubiquitylation of their binding partner Commisureless by DNedd4, rather than by ubiquitylation of the receptor itself. In mammalian cells, Nedd4-1 has also been shown to promote the translocation of the lysosomal transmembrane protein LAPTM5 without ubiquitylating LAPTM5 (30). Instead, Nedd4-1 ubiquitylates Golgi-associated, γ-adaptin homologue, Arf-binding 3 (GGA3), which selects LAPTM5 at the TGN for translocation to the lysosomes, where LAPTM5 exerts its function. Likewise, Nedd4-1 may target signaling partners associated with the trafficking of Cav channels.
One important clue revealed by our study is the specific requirement of Cavβ subunits, which suggests Cavβ-interacting proteins as possible targets of Nedd4-1. Cavβ has been shown to interact with numerous proteins involved in signal transduction or protein trafficking. We tested the possible involvement of dynamin (20) which, like Nedd4-1, induces a Cavβ-dependent reduction in Cav currents. However, we found no evidence that Nedd4-1 either required or modified the interaction of dynamin with Cavβ. Cavβ subunits are also targeted by many signaling effectors, such as Akt serine/threonine kinases (4) and small G proteins (31). Akt promotes the insertion of Cav channels at the plasma membrane (4), whereas small G proteins of the RGK Ras-related family bind and sequestrate Cavβ subunits in the nucleus, preventing the surface expression of Cav channels (31). However, so far, there is no evidence suggesting a direct link between these candidates and Nedd4-1. We show that Cav currents are regulated by Nedd4-1 but not Nedd4-2 nor WWP2. Interestingly, it has recently been found that the μ1 subunit of AP-1 specifically interacts with Nedd4-1 and not Nedd4-2 nor WWP2, albeit the ubiquitylation of AP-1 itself has not been investigated (32). Furthermore, AP-1 has also been shown to interact with GGA1, -2, and -3 via their γ subunits (33), and GGA3 is an identified target of Nedd4-1 (30). Since the regulation of Cav channels is also specific to Nedd4-1 and because the clathrin inhibitor MDC partially antagonizes its effect on Cav currents, one attractive possibility is that the effect of Nedd4-1 may be mediated by the GGA-AP1 complex. One could speculate that ubiquitylation of GGA3 by Nedd4-1 may recruit Cav channels at the TGN, via their common interaction with AP-1. In support of this hypothesis, we provided preliminary data showing that the mutation of Cavβ on a putative μ1 binding motif prevented the lysosome-dependent down-regulation of Cav currents. Additional studies would be required to fully investigate this potential mechanism. The role of Cavβ in mediating the additional proteasome-dependent regulation of Cav channels by Nedd4-1 should also be further investigated.
Remarkably, the residual Cav channels that escaped Nedd4-1-induced degradation exhibited slightly different gating properties than in controls, depending on the isoform of Cavβ co-expressed with Cav1.2α1 (supplemental Table 1). Phosphorylation of Cavβ has been shown to regulate Cav biophysical properties (34, 35). Hence, it is possible that Nedd4-1 modifies the gating of the channels by targeting one of the many kinases regulating Cavβ. It is also important to point out that Cavβ are composed of two main domains: a guanylate kinase domain and an SH3 domain, which promotes protein-protein interaction (2, 3). Changes in the interaction of the guanylate kinase and SH3 domains with each other, and with Cavα1, have been shown to affect both the current-voltage relationship and steady-state inactivation of the channels (3, 36). Hence, the observed changes in gating properties may reflect the Nedd4-1-induced association of an intermediate partner binding the Cavβ SH3 and altering its interaction with Cav1.2α1. In agreement with this hypothesis, the mutation Y221A on Cavβ2 SH3 domain antagonized the effect of Nedd4-1 both on whole-cell current densities and gating properties.
Nedd4-1 is expressed in many tissues (8), including the cardiovascular system. One role for Nedd4-1 may be to adjust Cav1.2 channels availability at the membrane to meet the cellular demand. Cav1.2 channels are responsible for the plateau of the action potential in ventricular cardiomyocytes (37, 38, 39), whereas in vascular smooth muscle cells, calcium entry via Cav1.2 channels regulates vascular tone, making these channels a primary target for the treatment of cardiovascular diseases (1). It has been shown that Cav currents decrease in response to atrial fibrillation (40), and Cav density is reduced in cardiac hypertrophy (41); however, the underlying mechanisms are still unknown. Whether Nedd4-1 is involved in these adaptative mechanisms and how the action of Nedd4-1 on Cav channels may be regulated remain to be investigated.
Altogether, our results show that Nedd4-1 ubiquitin ligases control the availability of Cav1.2 channels at the plasma membrane and reveal a new role for the Cavβ subunit, permitting Nedd4-1-dependent degradation of Cav channels at their exit from the ER/Golgi network.
Supplementary Material
Acknowledgments
We thank Professor Annette C. Dolphin (Department of Neurosciences, Physiology, and Pharmacology, University College London, London, UK) for ongoing support and access to laboratory facilities. We also thank Sophie Roy (Department of Pharmacology and Toxicology, University of Lausanne, Lausanne, Switzerland) for excellent technical assistance.
This work was supported by the Wellcome Trust (to J.-S. R. and P. V.) and Swiss National Science Foundation Grant 310030_120707 (to H. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Table 1, and Figs. 1–5.
- Cav
- voltage-dependent calcium channel
- Nedd4
- Neuronal precursor cell-expressed developmentally down-regulated 4
- ER
- endoplasmic reticulum
- TGN
- trans-Golgi network
- MDC
- monodansylcadaverine
- BFA
- brefeldin A.
REFERENCES
- 1. Catterall W. A., Perez-Reyes E., Snutch T. P., Striessnig J. (2005) Pharmacol. Rev. 57, 411–425 [DOI] [PubMed] [Google Scholar]
- 2. Dolphin A. C. (2003) J. Bioenerg. Biomembr. 35, 599–620 [DOI] [PubMed] [Google Scholar]
- 3. Dolphin A. C. (2009) Curr. Opin. Neurobiol. 19, 237–244 [DOI] [PubMed] [Google Scholar]
- 4. Viard P., Butcher A. J., Halet G., Davies A., Nürnberg B., Heblich F., Dolphin A. C. (2004) Nat. Neurosci. 7, 939–946 [DOI] [PubMed] [Google Scholar]
- 5. Abriel H., Staub O. (2005) Physiology 20, 398–407 [DOI] [PubMed] [Google Scholar]
- 6. Scott P. M., Bilodeau P. S., Zhdankina O., Winistorfer S. C., Hauglund M. J., Allaman M. M., Kearney W. R., Robertson A. D., Boman A. L., Piper R. C. (2004) Nat. Cell Biol. 6, 252–259 [DOI] [PubMed] [Google Scholar]
- 7. Puertollano R., Bonifacino J. S. (2004) Nat. Cell Biol. 6, 244–251 [DOI] [PubMed] [Google Scholar]
- 8. Rotin D., Kumar S. (2009) Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 9. Jespersen T., Membrez M., Nicolas C. S., Pitard B., Staub O., Olesen S. P., Baró I., Abriel H. (2007) Cardiovasc. Res. 74, 64–74 [DOI] [PubMed] [Google Scholar]
- 10. Abriel H., Kamynina E., Horisberger J. D., Staub O. (2000) FEBS Lett. 466, 377–380 [DOI] [PubMed] [Google Scholar]
- 11. Rougier J. S., van Bemmelen M. X., Bruce M. C., Jespersen T., Gavillet B., Apothéloz F., Cordonier S., Staub O., Rotin D., Abriel H. (2005) Am. J. Physiol. Cell Physiol. 288, C692–701 [DOI] [PubMed] [Google Scholar]
- 12. Rotin D., Kanelis V., Schild L. (2001) Am. J. Physiol. Renal Physiol. 281, F391–399 [DOI] [PubMed] [Google Scholar]
- 13. Henke G., Maier G., Wallisch S., Boehmer C., Lang F. (2004) J. Cell. Physiol. 199, 194–199 [DOI] [PubMed] [Google Scholar]
- 14. Hryciw D. H., Ekberg J., Lee A., Lensink I. L., Kumar S., Guggino W. B., Cook D. I., Pollock C. A., Poronnik P. (2004) J. Biol. Chem. 279, 54996–55007 [DOI] [PubMed] [Google Scholar]
- 15. van Bemmelen M. X., Rougier J. S., Gavillet B., Apothéloz F., Daidié D., Tateyama M., Rivolta I., Thomas M. A., Kass R. S., Staub O., Abriel H. (2004) Circ. Res. 95, 284–291 [DOI] [PubMed] [Google Scholar]
- 16. Kamynina E., Debonneville C., Bens M., Vandewalle A., Staub O. (2001) FASEB J. 15, 204–214 [DOI] [PubMed] [Google Scholar]
- 17. Deveraux Q., Ustrell V., Pickart C., Rechsteiner M. (1994) J. Biol. Chem. 269, 7059–7061 [PubMed] [Google Scholar]
- 18. Kawaguchi M., Minami K., Nagashima K., Seino S. (2006) J. Biol. Chem. 281, 13015–13020 [DOI] [PubMed] [Google Scholar]
- 19. Page K. M., Heblich F., Davies A., Butcher A. J., Leroy J., Bertaso F., Pratt W. S., Dolphin A. C. (2004) J. Neurosci. 24, 5400–5409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gonzalez-Gutierrez G., Miranda-Laferte E., Neely A., Hidalgo P. (2007) J. Biol. Chem. 282, 2156–2162 [DOI] [PubMed] [Google Scholar]
- 21. Damke H., Baba T., Warnock D. E., Schmid S. L. (1994) J. Cell Biol. 127, 915–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bichet D., Cornet V., Geib S., Carlier E., Volsen S., Hoshi T., Mori Y., De Waard M. (2000) Neuron 25, 177–190 [DOI] [PubMed] [Google Scholar]
- 23. Dubel S. J., Altier C., Chaumont S., Lory P., Bourinet E., Nargeot J. (2004) J. Biol. Chem. 279, 29263–29269 [DOI] [PubMed] [Google Scholar]
- 24. Dolphin A. C., Wyatt C. N., Richards J., Beattie R. E., Craig P., Lee J. H., Cribbs L. L., Volsen S. G., Perez-Reyes E. (1999) J. Physiol. 519, 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luini A., Mironov A. A., Polishchuk E. V., Polishchuk R. S. (2008) Histochem. Cell Biol. 129, 153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Davies P. J., Davies D. R., Levitzki A., Maxfield F. R., Milhaud P., Willingham M. C., Pastan I. H. (1980) Nature 283, 162–167 [DOI] [PubMed] [Google Scholar]
- 27. Robinson M. S., Bonifacino J. S. (2001) Curr. Opin. Cell Biol. 13, 444–453 [DOI] [PubMed] [Google Scholar]
- 28. De Craene J. O., Soetens O., Andre B. (2001) J. Biol. Chem. 276, 43939–43948 [DOI] [PubMed] [Google Scholar]
- 29. Myat A., Henry P., McCabe V., Flintoft L., Rotin D., Tear G. (2002) Neuron 35, 447–459 [DOI] [PubMed] [Google Scholar]
- 30. Pak Y., Glowacka W. K., Bruce M. C., Pham N., Rotin D. (2006) J. Cell Biol. 175, 631–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Béguin P., Nagashima K., Gonoi T., Shibasaki T., Takahashi K., Kashima Y., Ozaki N., Geering K., Iwanaga T., Seino S. (2001) Nature 411, 701–706 [DOI] [PubMed] [Google Scholar]
- 32. Camus G., Segura-Morales C., Molle D., Lopez-Vergès S., Begon-Pescia C., Cazevieille C., Schu P., Bertrand E., Berlioz-Torrent C., Basyuk E. (2007) Mol. Biol. Cell 18, 3193–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Doray B., Ghosh P., Griffith J., Geuze H. J., Kornfeld S. (2002) Science 297, 1700–1703 [DOI] [PubMed] [Google Scholar]
- 34. Puri T. S., Gerhardstein B. L., Zhao X. L., Ladner M. B., Hosey M. M. (1997) Biochemistry 36, 9605–9615 [DOI] [PubMed] [Google Scholar]
- 35. Fitzgerald E. M. (2002) J. Physiol. 543, 425–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen Y. H., He L. L., Buchanan D. R., Zhang Y., Fitzmaurice A., Yang J. (2009) FEBS Lett. 583, 1969–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Takimoto K., Li D., Nerbonne J. M., Levitan E. S. (1997) J. Mol. Cell Cardiol. 29, 3035–3042 [DOI] [PubMed] [Google Scholar]
- 38. Marionneau C., Couette B., Liu J., Li H., Mangoni M. E., Nargeot J., Lei M., Escande D., Demolombe S. (2005) J. Physiol. 562, 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Seisenberger C., Specht V., Welling A., Platzer J., Pfeifer A., Kühbandner S., Striessnig J., Klugbauer N., Feil R., Hofmann F. (2000) J. Biol. Chem. 275, 39193–39199 [DOI] [PubMed] [Google Scholar]
- 40. Ming Z., Nordin C., Siri F., Aronson R. S. (1994) J. Mol. Cell Cardiol. 26, 1133–1143 [DOI] [PubMed] [Google Scholar]
- 41. Chen H., Polo S., Di Fiore P. P., De Camilli P. V. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14908–14913 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.