Abstract
The flavoprotein rotenone-insensitive internal NADH-ubiquinone (UQ) oxidoreductase (Ndi1) is a member of the respiratory chain in Saccharomyces cerevisiae. We reported previously that bound UQ in Ndi1 plays a key role in preventing the generation of reactive oxygen species. Here, to elucidate this mechanism, we investigated biochemical properties of Ndi1 and its mutants in which highly conserved amino acid residues (presumably involved in NADH and/or UQ binding sites) were replaced. We found that wild-type Ndi1 formed a stable charge transfer (CT) complex (around 740 nm) with NADH, but not with NADPH, under anaerobic conditions. The intensity of the CT absorption band was significantly increased by the presence of bound UQ or externally added n-decylbenzoquinone. Interestingly, however, when Ndi1 was exposed to air, the CT band transiently reached the same maximum level regardless of the presence of UQ. This suggests that Ndi1 forms a ternary complex with NADH and UQ, but the role of UQ in withdrawing an electron can be substitutable with oxygen. Proteinase K digestion analysis showed that NADH (but not NADPH) binding induces conformational changes in Ndi1. The kinetic study of wild-type and mutant Ndi1 indicated that there is no overlap between NADH and UQ binding sites. Moreover, we found that the bound UQ can reversibly dissociate from Ndi1 and is thus replaceable with other quinones in the membrane. Taken together, unlike other NAD(P)H-UQ oxidoreductases, the Ndi1 reaction proceeds through a ternary complex (not a ping-pong) mechanism. The bound UQ keeps oxygen away from the reduced flavin.
Keywords: Enzyme Catalysis, Flavoproteins, NADH, Quinones, Reactive Oxygen Species (ROS), Reaction Mechanism, Substrate Binding Site
Introduction
Alternative NADH dehydrogenases (NDH-2)4 catalyze electron transfer from NADH to quinone without energy transduction. They are commonly found in the respiratory chain of bacteria, fungi, and plant mitochondria but not in mammalian mitochondria (1, 2). The NDH-2 enzyme is generally composed of a single polypeptide and can be classified into three groups: A, B, and C. Group A has two adenine dinucleotide (ADP)-binding motifs and is involved in the non-covalent binding of NAD(P)H and FAD. Group B has the same two ADP-binding motifs and an additional conserved EF-hand. Group C has only one conserved ADP-binding motif where flavin is covalently bound (2). The yeast Ndi1 enzyme belongs to group A (2) and catalyzes NADH oxidation on the matrix side of the mitochondrial inner membrane (3) like the energy-transducing NADH dehydrogenase complex I (4).
The physiological electron acceptor of NDH-2 is quinone, which is broadly classified into two types: naphthoquinone (menaquinone) and benzoquinone (ubiquinone (UQ)). Ndi1 utilizes the latter as an electron acceptor (2). Both quinones have a hydrophilic head group and hydrophobic isoprenoid side chains and ensure electron transfer between several membrane-bound respiratory enzymes. Ndi1 does not react with oxygen in the presence of UQ. In the absence of quinone, NADH-reduced Ndi1 reacts with oxygen and produces H2O2, although intrinsic NADH oxidase activity is extremely low (about 300-fold less) compared with NADH-UQ1 oxidoreductase activity (5). Our previous biochemical studies of Ndi1 suggest that Ndi1 has enzyme-bound UQ (bound UQ), which is distinct from the catalytic UQ (electron acceptor). Kinetic studies have indicated the physiological importance of bound UQ in Ndi1. When Ndi1 was incorporated into bovine heart submitochondrial particles, only the UQ-bound form established NADH-linked respiratory activity and did not produce H2O2 (5). In other experiments, NDI1 (which encodes the Ndi1 protein)-transduced cells did not produce detectable levels of reactive oxygen species (ROS) (6, 7). Thus, it was postulated that the presence of bound UQ in Ndi1 is physiologically important in its enzymatic reaction and that Ndi1 bears at least two distinct UQ binding sites: one for bound UQ and another for catalytic UQ (5). However, the function of bound UQ in Ndi1 and the general reaction mechanism of NDH-2 have yet to be elucidated (2, 8–10).
In this study, we mutated highly conserved amino acid residues, which are presumably involved in NADH and/or UQ binding sites based on the x-ray crystal structures of flavin enzymes that have relatively high sequence homology with Ndi1. Using these mutants and/or various inhibitors, we examined the reaction mechanism of Ndi1 and the role of bound UQ in this reaction.
EXPERIMENTAL PROCEDURES
Materials
UQ1, UQ6, and UQ10 were purchased from Sigma. Preparation of the reduced form of UQ1 (UQ1H2) was done according to the method of Rieske (11). Dodecyl β-d-maltoside (DM) was purchased from Anatrace. Materials for PCR product purification, gel extraction, and plasmid preparation were purchased from Promega. Egg yolk lecithin (PL-100M) was kindly provided by Q. P. Corp. All other chemicals were of reagent grade and obtained from commercial sources.
Site-directed Mutagenesis of Ndi1
Amino acid replacement of Ndi1 was performed with the QuikChange II XL site-directed mutagenesis kit (Stratagene) using pET16b(NDI1-m) (5) and synthetic oligonucleotide primers (supplemental Table S1). Pro92, Leu93, Leu94, Thr239, Thr339, Asn341, and Thr392 were replaced with Ala; Leu195 was replaced with Ala, Val, and Ile; Glu242 was replaced with Ala, Gln, and Asp; and Asp383 was replaced with Ala, Asn, Glu, and Gln. Each mutation was confirmed through DNA sequencing (PRISM310, Applied Biosystems).
Expression and Purification of Ndi1
Escherichia coli strain BL21(DE3)pLysS was transformed with mutant pET16b(NDI1-m). The expression and purification procedure of mutant Ndi1 enzyme was basically the same as those for the wild-type enzyme (Triton X-100 (TX) enzyme) (5). Because some mutant enzymes were purified as apoenzyme, 50 μm FAD was added to all buffers after the membrane suspension step. This improved FAD content in purified mutant Ndi1. Purification was performed under aerobic conditions because the enzyme is not sensitive to oxygen.
In addition to the preparation of UQ-free Ndi1 (TX enzyme), UQ-bound Ndi1 (containing endogenous quinone; UQ8 if overexpressed in E. coli) was also prepared by a procedure using DM (5). To further increase the UQ content of purified Ndi1, the ionic strength of the buffers was raised from 200 to 500 mm. We found that at low ionic strength purified Ndi1 with DM was extremely unstable (formed aggregates) compared with the TX enzyme. The enzyme eluted from the nickel-nitrilotriacetic acid column was desalted with a buffer containing 50 mm Mops-KOH (pH 7.0), 500 mm NaCl, 0.1 mm EDTA, 10% glycerol, and 0.02% DM. We called this preparation DM enzyme. Furthermore, to increase enzyme purity and recovery, 0.3% DM was replaced with 0.17% DM plus 0.1% TX from the membrane extraction step. Ndi1 purified by this modified method was called the TX/DM enzyme. The TX/DM enzyme was also unstable at low ionic strength.
Enzyme Assays
NADH-UQ1 oxidoreductase activity was measured as described (5). The decrease in NADH absorption (340 nm) was measured with a Hitachi U-3310 spectrophotometer. Although Ndi1 has NADH oxidase activity, the experiments were carried out under aerobic conditions because NADH oxidase activity is much smaller than NADH-UQ1 oxidoreductase activity, and ROS were not detected in the presence of quinone (data not shown). The enzyme was preincubated with UQ1 for 1 min, and the reaction was initiated by the addition of NADH. For the NADH oxidase activity, UQ1 was omitted.
Quantification of Bound Ubiquinones in Purified Ndi1
As shown in Ref. 5, the UQ content in the purified Ndi1 enzyme was determined using an ultraperformance liquid chromatography system (LaChrome ULTRA, Hitachi). Inertsil ODS-3 (2 μm, 3.0 × 50 mm; GL Sciences) was used, and the flow rate was changed to 0.8 ml/min at 40 °C. Elution was monitored by a diode array detector (L-2455L, Hitachi), and the content (UQ8 and UQ10) was calculated from the peak area by comparison with authentic UQ8 and UQ10. The efficiency of extraction was also verified by the internal standard UQ6 in each measurement.
Proteolytic Digestion
Proteolytic digestion of Ndi1 was carried out in an anaerobic chamber (Plas Labs, Inc.) at room temperature to avoid NAD(P)H oxidation by Ndi1 and oxygen. Purified wild-type enzyme was diluted with a buffer containing 50 mm Tris-HCl (pH 8.0), 1 mm EDTA, 1 mm CaCl2, 200 mm NaCl, and 10% glycerol to obtain a concentration of 1 mg of protein/ml (total, 100 μl). The enzyme was then incubated for 5 min with a substrate (1 mm NADH, 1 mm NADPH, or 100 μm UQ1), 1 mm dithionite, or 5% (v/v) EtOH. After incubation with the substrate, the enzyme was digested by 0.01 mg/ml (>0.3 units/ml) proteinase K from porcine pancreas (proteomics grade, Sigma) for 1–3 h. The reaction was stopped by the addition of 2 mm PMSF. The resulting sample was denatured with the same volume of 2× Laemmli sample loading buffer and subjected to SDS-polyacrylamide gel (9%) electrophoresis. Proteins were then transferred onto the PVDF membrane. The digested peptide fragments were identified by N-terminal protein sequencing (492 Protein Sequencer, Applied Biosystems).
Fusion of UQ10-enriched Liposomes with E. coli Membrane
Unilamellar UQ10-containing liposomes were prepared as described in Ref. 12. Egg yolk lecithin (2.0 g) was mixed with UQ10 (13.3 mg), and the mixture was dissolved in 6 ml of chloroform (7.5 nmol of UQ10/mg of egg yolk lecithin). The solution was concentrated under nitrogen gas and then dried using a centrifugal thickener (TOMY ST-10) for 1 h at 30 °C. A dried lipid/UQ10 mixture was resuspended in 10 ml of a buffer containing 50 mm Tris-Cl (pH 8.0), 200 mm NaCl, 0.1 mm EDTA, and 10% glycerol. After 1 h of hydration, the lipid mixture was sonicated with an ultrasonic cell disruptor (MicrosonTM) at 14 watts for five 10-min cycles at 4 °C.
Fusion of liposome and membrane was performed by a modified method used by Hackenbrock and co-workers (12, 13). 10 ml of liposomes were mixed with 20 ml of E. coli membrane overexpressing Ndi1 (28.9 mg of protein/ml), and the mixture was incubated for 1 h at 30 °C. The mixture was then frozen in liquid nitrogen, thawed, and sonicated at 14 watts for 1 min at 0 °C; this process was repeated five times to improve fusion efficiency. The mixture of liposome and membrane was brought up to 45 ml with the same buffer described above, and the liposome-fused membrane was separated by ultracentrifugation (260,000 × g for 1 h). The pellet (liposome-fused membrane fraction) was suspended at a concentration of 10 mg of protein/ml in 50 mm Tris-HCl (pH 8.0), 500 mm NaCl, 0.1 mm EDTA, and 10% glycerol; quickly frozen in liquid nitrogen; and stored at −80 °C until use.
Other Analytical Procedures
Protein concentration was determined with a protein assay bicinchoninate kit from Nacalai Tesque. FAD content was determined as described in Refs. 14 and 15. SDS-polyacrylamide gel electrophoresis was performed by a modified method used by Laemmli (16). UV and visible absorption spectra were measured on an Olis DW2000 spectrophotometer, a Hitachi U-3310 spectrophotometer, or an ND-1000 spectrophotometer (NanoDrop). Anaerobic experiments were carried out in a controlled anaerobic chamber with a palladium catalyst, which bonds trace oxygen to hydrogen in the anaerobic gas mixture (90% argon and 10% hydrogen), forming water vapor. Any variations from these procedures and other details are described in the figure legends.
RESULTS
Purification of UQ-bound Ndi1
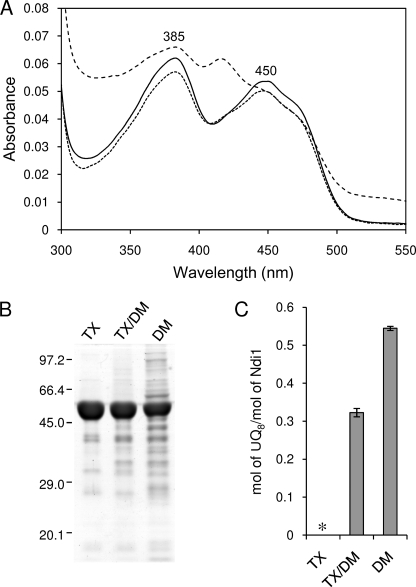
We have reported previously that UQ8 content of Ndi1 extracted by DM was estimated to be 0.17 mol of UQ8/mol of Ndi1 (5). By increasing the salt concentration in buffers, the UQ content of purified Ndi1 (DM enzyme) was increased to 0.54 mol/mol of enzyme (Fig. 1C). However, the purity of the DM enzyme was considerably lower than that of the TX enzyme (Fig. 1, A and B). The additional absorption peak around 420 nm was found in DM enzyme (shown in Fig. 1A), which showed typical characteristics of hemoprotein (data not shown). In contrast, the purity of TX/DM enzyme was almost the same as that of TX enzyme (Fig. 1, A and B), but its UQ content was 0.32 mol/mol, which was a significant amount (Fig. 1C). Therefore, we used TX/DM enzyme as UQ-bound Ndi1 in subsequent experiments.
FIGURE 1.
Effect of detergents on purity and content of bound UQ of Ndi1. A, the absorption spectra of Ndi1, which was purified using TX (5 μm; solid line), TX/DM (5 μm; dotted line), and DM (5 μm; dashed line). The spectra were measured on a Hitachi U-3310 spectrophotometer at 25 °C in a buffer containing 50 mm Mops-KOH (pH 7.0), 0.1 mm EDTA, 10% glycerol, 1 m NaCl, and 0.02% TX (for TX enzyme) or 0.02% DM (for TX/DM and DM enzymes). B, each sample (10 μg) was subjected to SDS-polyacrylamide gel (9%) electrophoresis. C, UQ8 content of each sample was calculated from the results of reverse-phase HPLC. * denotes an undetectable level of UQ8. Each value and vertical bar represent the mean ± S.E.
Redox Properties of Flavin and Bound UQ of Ndi1
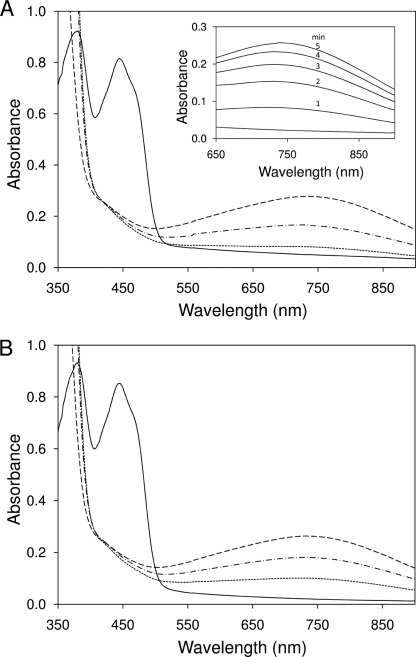
To verify whether the bound UQ of Ndi1 directly participates in the electron transfer reaction, the redox state of FAD of the purified enzyme was examined spectrophotometrically. The experiments were carried out under strict anaerobic conditions to avoid NADH oxidation by Ndi1 and oxygen. The absorptions of oxidized FAD (peaks at 383 and 448 nm) in UQ-free and UQ6-reconstituted Ndi1 were completely lost by the addition of NADH (Fig. 2, A and B, solid lines). However, a small broad band newly appeared around 740 nm when the enzymes were reduced with NADH (Fig. 2, A and B, dotted lines). This band was stable for at least 1 h under anaerobic conditions. This broad absorption band can be regarded as charge transfer (CT) complex because the formation of CT complex has been reported for many flavoproteins (17, 18). Generally, a CT complex is an electron donor-electron acceptor complex characterized by electronic transition to an excited state in which there is a partial transfer of electronic charge from the donor to the acceptor moiety and with optical absorption bands derived from the formation of an electronic transition state. In flavoproteins, a CT complex is usually formed as a result of π-π charge transfer interaction between the parallel stacked isoalloxazine ring of the reduced flavin and the nicotinamide ring of NAD(P) and has a broad absorption band in the long wavelength region (19).
FIGURE 2.
Effect of bound UQ on formation of CT complex in Ndi1. The absorption spectra of UQ-free (100 μm; A) and UQ6-reconstituted Ndi1 (about 1 mol of UQ6/mol of enzyme; 100 μm; B) were measured on an Olis DW2000 spectrophotometer at 25 °C in a buffer containing 50 mm Mops-KOH (pH 7.0), 0.1 mm EDTA, 10% glycerol, and 0.02% DM. The experiments were carried out using an anaerobic chamber (Coy Laboratory Products Inc.) The method of incorporation of UQ6 into the purified Ndi1 was described in Ref. 5. UQ-free Ndi1 (A) and UQ6-reconstituted Ndi1 (B) were reduced by the addition of 1 mm NADH (dotted line), and then 200 μm DBQ was added (dashed-dotted line). After the addition of DBQ, the cuvette was opened under aerobic conditions. The dashed line indicates the maximum height of the broad band. Inset, change of broad band over time after the addition of NADH under aerobic condition (in the absence of DBQ).
In Ndi1, the CT complex was not formed by the addition of NADH alone under anaerobic conditions. In the UQ6-reconstituted Ndi1, the CT complex absorption band was increased 2.5-fold compared with that of UQ-free enzyme (Fig. 2, A and B, dotted lines). Moreover, the addition of excess n-decylbenzoquinone (DBQ) (relatively water-soluble quinone) resulted in the same increase of the CT peak in both UQ-free and UQ6-bound Ndi1 (Fig. 2, A and B, dashed-dotted lines). Then, when the cuvette was exposed to air, the CT complex absorption band progressively increased, reached equilibrium within 5 min (Fig. 2A, inset), and subsequently decreased. This was probably caused by the exhaustion of NADH. The maximum heights of the CT complex absorption band in UQ-free and UQ6-reconstituted Ndi1 were basically the same (Fig. 2, A and B, dashed lines). These results indicate that Ndi1 forms the CT complex with NADH like other flavoproteins. Moreover, quinone (or molecular oxygen) is required as an electron acceptor for the formation of the CT complex. It is noteworthy that the CT complex was not observed after addition of NADPH and that the addition of DBQ or the exposure to air had no effect (supplemental Fig. 1A). This suggests that NADH is specifically required for the CT complex formation.
We also examined the influence of endogenous quinone on the formation of CT complex using UQ8-bound Ndi1 (TX/DM enzyme) (supplemental Fig. 1, B and C) and found that endogenous UQ8 more efficiently formed CT complex compared with exogenous quinone (UQ6) (Fig. 2). This result implies that exogenously bound UQ6 is not bound to exactly the same site as endogenously bound UQ8. Moreover, these results suggest that Ndi1 forms a ternary complex with NADH and UQ in close vicinity of FAD and that UQ can withdraw an electron from reduced flavin, leading to the formation of the CT complex. We thus speculate that quinone can prevent artificial electron transfer from reduced flavin to molecular oxygen.
Conformational Change in Ndi1 by NADH Binding
We reported previously that the bound UQ was dissociated from the enzyme when Ndi1 was reduced with NADH under aerobic conditions, whereas the bound UQ remained attached when Ndi1 was reduced with NADH under anaerobic conditions (see Fig. 7 in Ref. 5). In contrast, the bound UQ was dissociated from the enzyme when Ndi1 was reduced with NADPH under both aerobic and anaerobic conditions (data not shown). These observations suggest that NADH binding might induce conformational change in Ndi1 to prevent the release of bound UQ from Ndi1. Therefore, we examined the susceptibility to protease digestion of Ndi1 in the presence of different substrate analogs to probe conformational changes induced by NADH.
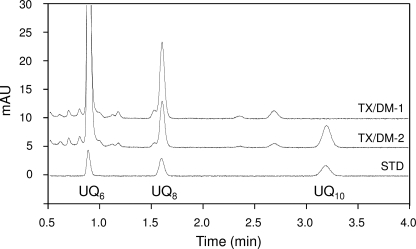
FIGURE 7.
Elution profiles of quinones extracted from Ndi1 purified using TX/DM from control membrane (TX/DM-1) and UQ10-fused membrane (TX/DM-2) on reverse-phase HPLC. Both samples contained UQ6 as an internal standard. Authentic UQ6, UQ8, and UQ10 were used for the standard (STD). mAU, milliabsorbance units.
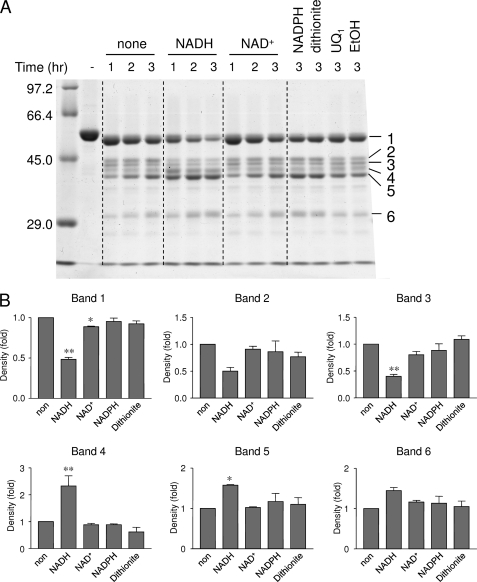
Ndi1 was digested by proteinase K under anaerobic conditions and analyzed using SDS-PAGE (Fig. 3A). Supplemental Fig. 2 confirmed that the amount of loaded protein was the same in all conditions. Ndi1 clearly exhibited much higher sensitivity to proteinase K in the presence of NADH compared with the other substrate analogs (Fig. 3A). Larger peptide bands 1 and 3 were significantly decreased, whereas smaller peptide bands 4 and 5 were significantly increased (Fig. 3B). The presence of NADPH or dithionite did not affect the protease sensitivity of Ndi1 (Fig. 3, A and B). The peptide fragments were identified by N-terminal amino acid sequence and the prediction of molecular mass by SDS-PAGE (supplemental Fig. 3). All of these peptide fragments except the sixth fragment were found to contain a GXGXXG motif for NADH binding. In the control experiment, the digestion pattern of bovine serum albumin by proteinase K was not affected by the addition of NADH (data not shown). These data indicate that the conformational change in Ndi1 was caused by the NADH binding but not by the reduced state of FAD.
FIGURE 3.
Effect of NADH binding on Ndi1 structure. A, Ndi1 (1 mg of protein/ml) was incubated with 1 mm NADH, 1 mm NAD+, 1 mm dithionite, 100 μm UQ1, or 5% EtOH in a buffer containing 50 mm Tris-HCl (pH 8.0), 1 mm EDTA, 1 mm CaCl2, 200 mm NaCl, and 10% glycerol and digested by proteinase K for 1–3 h under strict anaerobic conditions. The reaction was stopped by the addition of 2 mm PMSF. Then the samples were subjected to SDS-polyacrylamide gel (9%) electrophoresis. B, the intensities of each digested peptide band of non-treated (non) protein were compared with those of proteins that were treated under different conditions. The intensities of each peptide band were determined by NIH Image. The band numbers correspond to the number of digested peptides in A. *, p < 0.05; **, p < 0.01 compared with untreated protein. The p value was calculated by unpaired Student's t test. Each value and vertical bar represent the mean ± S.E. (n = 3).
Site-directed Mutagenesis of Ndi1
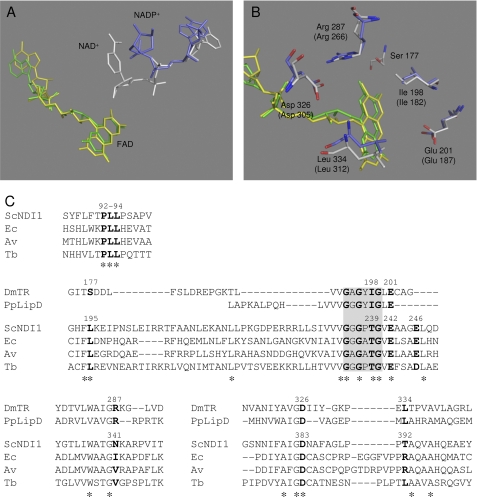
Based on the results in Fig. 2, we speculated that the binding site of “bound UQ” in Ndi1 might be located near the isoalloxazine ring of FAD to directly accept an electron from the reduced FAD. Indeed, the NADH binding site of the group A family of NDH-2 enzymes has been suggested to exist near the FAD binding site (2, 20). In the case of NDH-2 from Yarrowia lipolytica, it was suggested that NADH and the quinone head group interact with the same binding pocket competitively (8). To obtain an understanding of the structure of the binding sites of the substrates in Ndi1, we referred to the x-ray crystal structures of other flavin enzymes: thioredoxin reductase of Drosophila melanogaster (DmTR) (21) and lipoamide dehydrogenase of Pseudomonas putida (PpLipDH) (22). Both enzymes have a similar sequence homology (Protein Data Bank code 2NVK (amino acids 153–349; identity, 25%; similarity, 39%; gap, 24%) and Protein Data Bank code 1LVL (amino acids 165–325; identity, 26%; similarity, 43%; gap, 13%), respectively) with Ndi1 (NCBI accession number CAA89160) by using BLAST of the DNA Data Bank of Japan (DDBJ) against the Protein Data Bank. The nicotinamide-ribose moiety of DmTR is far from the isoalloxazine ring of FAD (Fig. 4A), implying that efficient electron transfer between both molecules will not occur. However, it has been reported that thioredoxin reductase from E. coli causes a conformational change induced by the binding of NADH that moves the nicotinamide-ribose moiety in proximity to the isoalloxazine ring of FAD (17). To achieve efficient electron transfer to the electron acceptor (thioredoxin for DmTR and NAD+ for PpLipDH, which correspond to UQ for Ndi1), these substrates should actually be in closer proximity to FAD. Based on this structural information (Fig. 4, A and B) and the sequence alignment data of DmTR, PpLipDH, and Ndi1 (Fir. 4C), several residues (Leu195, Thr239, Glu242, Glu246, Asn341, Asp383, and Thr392 of Ndi1) that are predicted to be in the surroundings of FAD were chosen for mutational analyses. Especially, Leu195, Thr239, Glu242, and Asn383 were highly conserved in group A of NDH-2 (supplemental Fig. 4). In the three-dimensional modeling of E. coli NDH-2, His51 is located at <5 Å from the isoalloxazine ring and could be a potential hydrogen bond donator for quinone. However, this residue is not conserved in the NDH-2 family (2, 20), and in fact, this residue corresponds to Pro95 in Ndi1. However, a hydrophobic cluster, Pro92-Leu93-Leu94, adjacent to Pro95 is found to be highly conserved in both groups A and B of the NDH-2 family (Fig. 4C and supplemental Fig. 4). We also mutated these amino acid residues in this study.
FIGURE 4.
Predicted UQ binding site of Ndi1. A, FAD (yellow and green) and nicotinamide-ribose (NADP+, blue; NAD+, white) of DmTR and PpLipDH are displayed as a stick model. B, the amino acid residues of DmTR and PpLipDH (in parentheses) surrounding FAD are displayed as a stick model colored in blue and white, respectively. C, amino acid sequence alignment from the group A family of NDH-2, DmTR, and PpLipDH. The GXGXXG consensus motifs are shown with a gray background. Residues marked with an asterisk indicate conserved residues among the NDH-2 enzymes, and those in bold are mutated residues in this study. ScNDI1, S. cerevisiae Ndi1 (NCBI accession number CAA89160); Ec, E. coli (NCBI accession number CAA23586.1); Av, Azotobacter vinelandii (NCBI accession number AAK19737.1); Tb, Trypanosoma brucei (NCBI accession number AAM95239).
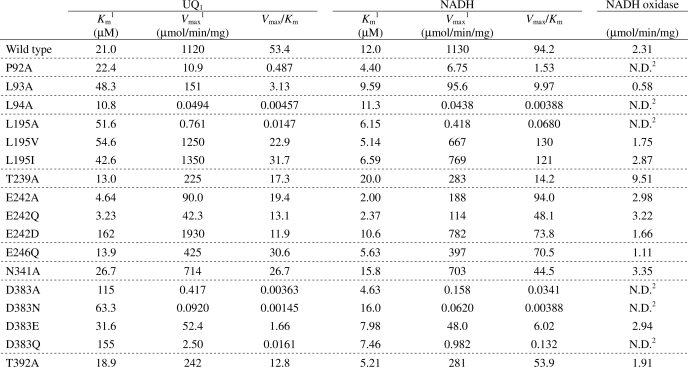
FAD Contents and NADH Oxidase Activities of Mutant Enzymes
Mutations near the FAD binding site affected the stability of Ndi1 enzyme and resulted in the reduction of FAD contents in purified enzyme. FAD contents of wild-type and mutants P92A, L93A, L195I, L195V, D383E, and D383Q were 1.07, 0.41, 0.38, 0.96, 1.07, 0.61, and 0.77 mol of FAD/mol of Ndi1 protein, respectively. These values agreed fairly well with FAD contents calculated from the spectra of mutant enzymes (supplemental Fig. 5). The 383 and 448 nm peaks derived from the oxidized FAD of wild-type enzyme did not shift in mutant Ndi1 enzymes (supplemental Fig. 5). Other mutants showed values similar to the wild-type level except mutants L195A, D383A, and D383N for which expression levels were too low to obtain enough materials for FAD estimation. These data suggest that residues Pro92 and Leu93 are located in close proximity to FAD.
NADH oxidase activities of purified Ndi1 were largely unaffected by mutations (within a 2-fold difference) except for T239A when enzyme activities were normalized by the FAD content in mutant enzymes. For instance, the NADH oxidase activities of L93A and D383E were 0.58 and 2.94 μmol/min/mg, respectively (Table 1). Because FAD contents of L93A and D383E mutants contained only 0.38 and 0.61 mol/mol of protein, the NADH oxidase activities should be equivalent to 1.53 and 4.81 μmol/min/mg of holoenzyme, respectively. On the other hand, the NADH-UQ1 oxidoreductase activities varied to a greater extent depending on the mutations (Table 1). These results indicate that NADH oxidase activity of Ndi1 is not influenced by changes in the NADH and UQ binding sites but rather is controlled by other unknown factors.
TABLE 1.
Kinetic parameters of wild type and mutant Ndi1
1 Apparent Km and Vmax values of wild type and mutant enzymes for both NADH and UQ1 were determined from double reciprocal plots at fixed concentration of UQ1 (60 μm) and NADH (100 μm), respectively.
2 N.D., not detected.
Kinetic Analyses of Mutant Ndi1 Enzymes
The kinetic parameters of wild-type and mutant enzymes for both NADH and UQ1 were measured at fixed concentrations of 60 μm UQ1 and 100 μm NADH, respectively (Table 1). The Km value of UQ1 was 21 μm in the wild type and moderately increased in L93A, L195A, L195V, and L195I by 2.0–2.5-fold and greatly increased in E242D, D383A, D383N, and D383Q by 3.3–7.7-fold. The Vmax values decreased from 1120 μmol/min/mg (wild type) to less than 10% in L195A, E242A, E242Q, D383A, D383N, D383E, and D383Q. Amino acid residues Leu195, Glu242, and Asp383 are expected to participate in UQ binding sites (Fig. 4). The kinetic parameters of these mutants were especially varied depending on the property of substituted amino acids. In Leu195, the Km value was almost the same in all mutants, but the Vmax value of the Ala mutant was drastically lower than that of Val and Ile mutants, suggesting that the hydrophobicity of Leu195 is essential for NADH-UQ oxidoreductase activity. In Asp383, the Km value of the Glu mutant was 2–5 times smaller than that of Ala, Asn, and Gln mutants, and the Vmax value was at least 20-fold higher, suggesting that the carboxyl group of Asp383 is essential for UQ binding and NADH-UQ oxidoreductase activity. Both Km and Vmax values of the Asp mutant in Glu242 were higher than that of Ala and Gln mutants, suggesting that the carboxyl group of Glu242 is essential for NADH-UQ oxidoreductase activity and that the carboxyl group is located in close vicinity of the UQ binding site. These results indicate that these highly conserved residues (Leu93, Leu195, Glu242, and Asp383) are important for UQ binding compared with the non-conserved residues (Glu246, Asn341, and Thr392) in group A of NDH-2 (supplemental Fig. 4). UQ contents of L93A, L195V, E242D, and D383Q mutants extracted by TX/DM were 0.19, 0.19, 0.29, and 0.20 mol of UQ8/mol of Ndi1 protein, respectively. It is noteworthy that UQ contents of these mutants were lower than that of wild-type enzyme (0.32 mol of UQ8/mol of enzyme).
In contrast to the Km values for UQ1, the Km values for NADH of mutants were basically similar (0.2–1.3-fold) except for T239A for which the Km value for NADH increased 1.7-fold. Thr239 is possibly involved in NADH binding because Tyr181 of PpLipDH interacts with NAD+ by hydrogen bonding; Tyr181 locates in the same position as Pro238 in Ndi1 (Fig. 4A) (the neighbor residue of the conserved Thr239) (18). This result indicates that the amino acids contributing to UQ binding, such as Leu93, Leu195, Glu242, and Asp383, are not involved in NADH binding.
Inhibition Mode of Ubiquinol and Reactive Blue-2
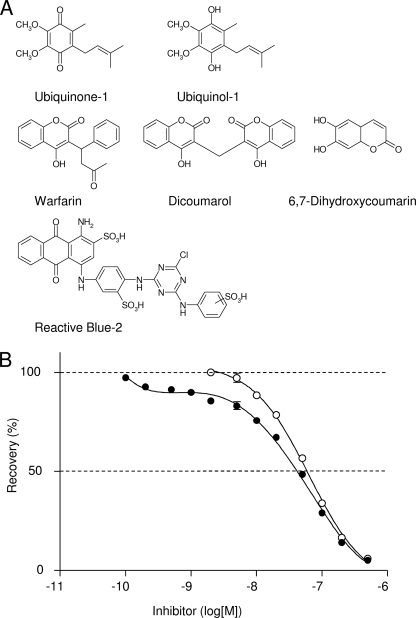
Various compounds were screened in the search for potent inhibitors against DT-diaphorase (NAD(P)H-quinone oxidoreductase 1 (NQO1)). NQO1 is a soluble flavoprotein that catalyzes nicotinamide nucleotide-dependent reductions of quinones, cyclohexa-3,5-diene-1,2-diylidenediamine (quinoneimines), azo dyes, and nitro group (23, 24). Studies of NQO1 structure and function have shown that the NAD(P)H and the quinone binding sites of enzymes have a significant overlap (25), and the enzymatic mechanism is a typical ping-pong reaction. In this study, two coumarin derivatives (warfarin and 6,7-dihydroxycoumarin) (26, 27), dicoumarol (28), and the reactive dye Cibacron Blue (Reactive Blue-2 (RB-2)) (29, 30), which compete with NAD(P)H for binding to NQO1, were tested to see whether these compounds inhibit Ndi1. The structures of these inhibitors are shown in Fig. 5A.
FIGURE 5.
Inhibitory effects of Reactive Blue-2 and ubiquinol on NADH-UQ1 reductase activity of Ndi1. A, structures of UQ1, UQ1H2, and several inhibitors for NQO1. B, NADH-UQ1 reductase activities were measured in the presence of increasing concentrations of RB-2 (closed circle). The NADH-UQ1 reductase activity was corrected for nonspecific inhibition using the value in the presence of 1 nm RB-2 as 100% activity (open circle). 100% activity was 900 μmol of NADH oxidized/min/mg of Ndi1. Each value and vertical bar represent the mean ± S.E.
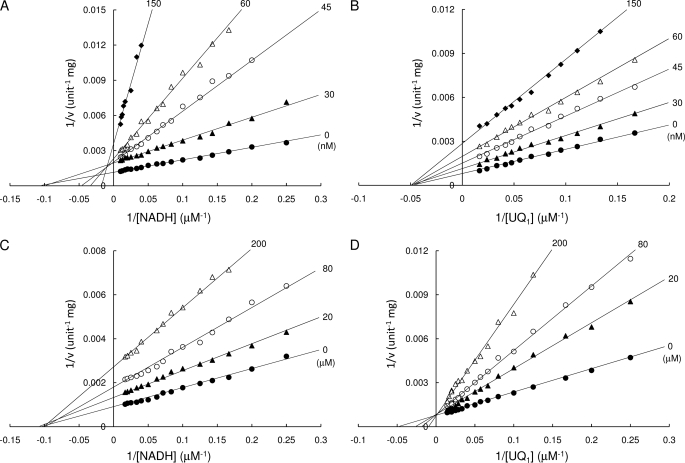
The IC50 values against Ndi1 for these inhibitors, including the most potent competitive inhibitor for NQO1, dicoumarol (Ki of 1–10 nm) (28), were higher than 10 μm, suggesting that the substrate binding site of NQO1 and Ndi1 are different. In contrast, the IC50 of RB-2 against Ndi1 was relatively low, 45 nm, and the inhibition curve was biphasic (Fig. 5B). The inhibition mode of RB-2 against NADH was competitive (Ki = 145 nm) (Fig. 6A) at high concentrations ([RB-2] > 2 nm), similar to the result of NQO1 (31). In contrast, RB-2 behaved as a non-competitive inhibitor with respect to UQ1 (Ki = 61 nm) (Fig. 6B). The non-competitive inhibition with NADH at low RB-2 concentrations (Fig. 6A) is likely due to nonspecific binding of this agent to the enzyme because non-competitive inhibition constituted less than 15% of the total inhibition (Fig. 5B). Taken together, these results indicate that RB-2 is competitive against NADH but non-competitive against UQ1 with respect to inhibition of Ndi1. It is noteworthy that we could not determine the inhibition mode of NAD+ against both NADH and UQ1 because NAD+ inhibition was extremely weak; less than 10% of total activity was inhibited at 20 mm. This result is consistent with the previous observation that NAD+ did not inhibit Ndi1 (9).
FIGURE 6.
Inhibition mode of RB-2 and UQ1HinfMACROS BELOW ARE FOR THE VISUAL2. Inhibition kinetics for NADH (A and C; 60 μm UQ1) and UQ1 (B and D; 100 μm NADH) were measured in the presence of RB-2 (A and B) and UQ1H2 (C and D). The concentrations of RB-2 and UQ1H2 are as indicated in the figure.
On the other hand, the IC50 for ubiquinol (UQ1H2) against NADH-UQ1 oxidoreductase activity was 120 μm. Ubiquinol clearly showed competitive inhibition with respect to UQ1 (Ki = 52 μm; Fig. 6D) but non-competitive inhibition with respect to NADH (Ki = 115 μm; Fig. 6C). These data strongly suggest that NADH and UQ1 do not bind to the same binding pocket and that both ubiquinol and NADH can coincidentally bind to the enzyme, pointing to the existence of the ternary complex, which conflicts with the ping-pong mechanism.
Behavior of Bound UQ in Ndi1 in Membrane
It is not yet clear whether bound UQ of Ndi1 is firmly attached or can reversibly dissociate from the enzyme in the membrane. To investigate this point, UQ10-enriched liposomes were fused with the Ndi1-overexpressing E. coli membrane containing endogenous UQ8. It was expected that if bound UQ of Ndi1 can reversibly dissociate from the enzyme in the membrane then bound endogenous UQ8 of Ndi1 will be partly replaced by UQ10. After the fusion, UQ8 and UQ10 contents of UQ10-fused membrane were determined to be 9.71 and 7.15 nmol/mg of protein, respectively (Table 2), whereas NADH oxidase activity was the same as before the fusion. However, when Ndi1 was purified from the UQ10-fused membrane with TX/DM, it contained exogenous UQ10 (0.144 ± 0.002 mol/mol of enzyme) in addition to endogenous UQ8 (0.189 ± 0.002 mol/mol of enzyme) (Fig. 7). The ratio of UQ10 per total UQ in the purified enzyme (0.43) was nearly identical to that in UQ10-fused membrane (0.42). This result suggests that the bound UQ in Ndi1 reversibly dissociates from the enzyme and equilibrates with the quinone pool unlike other quinone-binding enzymes, such as E. coli membrane-bound glucose dehydrogenase (32) and cytochrome bo-type quinol oxidase (33).
TABLE 2.
Comparison of UQ contents and NADH oxidase activities in control membrane and UQ10-fused membrane
| UQ8a | UQ10a | UQ10/UQtotal | NADH oxidaseb (+KCN)c | |
|---|---|---|---|---|
| Control membraned | 9.35 ± 0.01 | NDe | 6.02 (0.11) | |
| UQ10-fused membraned | 9.71 ± 0.02 | 7.15 ± 0.03 | 0.42 | 6.06 (0.14) |
a nmol of UQ/mg of protein.
b μmol of NADH/min/mg of protein.
c Activities were measured in the presence of 20 mm KCN.
d Each membrane was prepared from Ndi1-overexpressing cells.
e Not detected.
DISCUSSION
In this study, we uncovered the catalytic mechanism of Ndi1 and the function of bound UQ against ROS production. Contrary to our previous assumption, our current data strongly suggest that the Ndi1 reaction proceeds through a ternary complex mechanism under physiological conditions. This was an unexpected result because it is generally accepted that NDH-2, including Ndi1, operates by a ping-pong mechanism (8–10).
Quinones can be a one- or two-electron acceptor. In general, flavin-dependent quinone reductases, including NDH-2, afford the strict two-electron reduction of the quinone to its hydroquinone form and avoid the generation of the semiquinone anion, which is prone to react with molecular oxygen, leading to the generation of ROS (34). However, it is not yet clear how NDH-2 interacts with their hydrophobic substrate quinones. To be more precise, in Ndi1, the bound UQ seems to play a key role in the prevention of ROS generation (5), but the mechanism underlying ROS suppression by the bound UQ has not been disclosed.
We found that Ndi1 forms a stable CT complex with NADH and UQ under anaerobic conditions, displaying a fairly broad absorbance band between 500 and 900 nm with a peak at around 740 nm. The formation of the CT complex itself is common among many flavin enzymes. It is known that the pyridine nucleotide-disulfide oxidoreductase family, such as DmTR and PpLipDH, forms the CT complex under aerobic conditions (35, 36). Recently, it was reported that the mitochondrial flavoprotein apoptosis-inducing factor, which shares relatively high sequence homology with Ndi1, also produces air-stable FADH2-NAD(P) CT complexes (37). The CT complex of apoptosis-inducing factor was similar to that of Ndi1, showing an absorption maximum at 770 nm, which is much longer than that usually observed in the CT complex of other flavin enzymes. The greater the interaction between the reacting molecules, the longer the wavelength of the CT complex absorption band (19), suggesting that the interaction among NADH, FAD, and UQ in Ndi1 is very strong.
Bound UQ accepts an electron from reduced FAD in the CT complex of Ndi1. Therefore, the formation of the CT complex might result in the formation of a semiquinone radical. The semiquinone radical does not react with molecular oxygen because the oxygen molecule may not enter the catalytic site of Ndi1 when the ternary complex, in which the substrate binding sites are completely occupied with NADH and UQ, is formed. This is supported by the previous observation that hydrogen peroxide was not produced during the catalysis when UQ-bound Ndi1, but not UQ-free Ndi1, was incorporated into bovine heart submitochondrial particles (see Fig. 8 in Ref. 5). In addition, NDI1-transfected cells, in which Ndi1 harboring endogenously bound UQ is overexpressed, did not produce reactive oxygen species as control non-transfected cells (6, 7). Furthermore, the electron transfer from flavin radical to UQ radical may occur on a millisecond time scale based on the report that in E. coli membrane-bound glucose dehydrogenase the electron transfer rate between two semiquinone radicals of pyrroloquinoline quinone (which corresponds to FAD in Ndi1) and UQ was 1.2 × 103 s−1 (38). Thus, it is highly likely that a semiquinone radical formed in the ternary complex will be immediately converted into a fully reduced form before reacting with molecular oxygen.
We strongly believe that our data shown in this study are all important circumstantial evidence to support a ternary complex mechanism for the Ndi1 reaction, which cannot be explained by a ping-pong mechanism. First, Ndi1 forms a CT complex with only NADH (Fig. 2). The specificity to NADH was also evident by the proteolytic digestion experiment (Fig. 3). Under anaerobic conditions, the bound UQ remained attached when Ndi1 was reduced with NADH but not with NADPH (see Fig. 7 in Ref. 5). Second, the kinetic study with inhibitors (RB-2 and UQ1H2) clearly indicated there is no overlap between NADH and UQ binding sites (Fig. 6). Especially, the inhibition mode of ubiquinol clearly showed a non-competitive inhibition with respect to NADH (Fig. 6 C), allowing the formation of the ternary complex with NADH and ubiquinol. This piece of information itself is against a ping-pong mechanism. Third, in the oxidized state of Ndi1, the bound UQ can reversibly dissociate and be replaced with other quinones in the membrane (Fig. 7). Fourth, our proteolytic digestion analysis supports the conformational change induced by NADH (Fig. 3), which is essential for the formation of the ternary complex.
Consequently, a question immediately arises. Why was it concluded that the reaction of Ndi1 with NADH and UQ was a ping-pong mechanism in our previous study? This was partly because we did not consider the reaction environment of purified Ndi1. Our previous data showed a typical ping-pong reaction in the hydrophilic artificial reaction environment where Ndi1 is simply surrounded by detergent micelle and UQ1 molecules in a disorganized way. This is completely different from the physiological, hydrophobic membrane environment where Ndi1 provides an ordered and limited entrance for quinone to access the catalytic pocket. UQ1 is much more hydrophilic and smaller than physiological UQ6 and can easily enter through the entry port for NADH. In this case, NADH and UQ1 compete and enter alternately. Therefore, even though NADH and UQ binding sites in Ndi1 are different, apparent bisubstrate kinetics could give consistent results that support a ping-pong reaction. The same behavior of UQ1 was also reported recently in mitochondrial complex I (39, 40). Hydrophilic quinines, such as UQ1, can be reduced by the flavin mononucleotide cofactor in the active site for NADH oxidation of complex I through a non-energy-transducing pathway (which is insensitive to quinone binding site inhibitors, such as rotenone and piericidin A). This inhibitor-insensitive UQ reduction occurs by a ping-pong type mechanism (39, 40). In addition, we have further experimental evidence that the reaction environment could drastically affect the Ndi1 catalytic properties and mode of action. The apparent rate constant for the release of bound UQ (UQ6) from the purified Ndi1 in the aqueous solution was extremely slow (∼0.14 s−1) compared with the turnover rate of the NADH oxidase reaction of Ndi1 incorporated into submitochondrial particles, which contain endogenous UQ10 (∼8.3 s−1) (5). A possible explanation is that the dissociation of UQ6 from the purified enzyme becomes very slow in an aqueous environment because UQ6 cannot freely diffuse into the hydrophilic solvent due to its hydrophobicity. This can lead to underestimation of the rate constant of the release of bound UQ6. The reaction environment is extremely important and should not be undermined for the mechanistic study of membrane-bound respiratory enzymes that involves quinone.
We previously postulated that Ndi1 bears two different UQ (bound UQ and catalytic UQ) binding sites (5). This speculation was made based on the ping-pong mechanism of Ndi1 (5) in which the binding of substrate UQ to the catalytic site occurs only after a sequence of events related to NADH binding and subsequent NAD+ release. However, because the reaction mechanism of Ndi1 is now considered a ternary complex mechanism (described above), it became unnecessary to assume two different sites for bound UQ and catalytic UQ. In fact, all of our results from previous and current studies favor only one UQ binding site in Ndi1. (i) The bound UQ is freely replaced with other quinones available in the membrane and also during the turnover. (ii) The kinetic data with competitive inhibitors (RB-2 and UQ1H2) clearly indicated that there is no additional UQ binding site in Ndi1. (iii) In fact, the C-terminal 26 amino acids (Gly374–Glu399) containing Asp383 was identified recently as the sole UQ binding site by a photoaffinity labeling experiment (41). All of these data lead us to the more realistic conclusion that the bound UQ site in Ndi1 is identical to the catalytic UQ site (41).
In summary, our current study revealed that Ndi1 exists as a UQ-bound form in the membrane. The UQ-bound form prevents oxygen from accessing the FAD site in Ndi1. At the same time, the bound UQ facilitates the formation of a ternary complex with NADH and safely accepts two electrons from the NADH-reduced FAD. After the resulting ubiquinol leaves Ndi1, the UQ binding site is immediately occupied with UQ from the quinone pool. This whole reaction mechanism contributes to the suppression of ROS generation from Ndi1.
Supplementary Material
Acknowledgments
We thank Dr. Kazunobu Matsushita and Dr. Mamoru Yamada (Yamaguchi University, Yamaguchi, Japan), Kimitoshi Sakamoto (University of Tokyo, Tokyo, Japan), and Hiromi Yoshida (Kagawa University, Kagawa, Japan) for valuable discussions. The DNA sequencer, protein sequencer, and anaerobic chamber used here were maintained and supported by the Division of Instrument and Equipment, Kagawa University.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM033712 (to T. Yagi). This work was also supported by Japan Society for the Promotion of Science Grant-in-aid for Young Scientists (Start-up) 19870016 (to T. Yamashita) and by the fund for Kagawa University Young Scientists 2007 (to T. Yamashita).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. 1–5.
- NDH-2
- alternative NADH dehydrogenase
- complex I
- mitochondrial proton-translocating NADH-quinone oxidoreductase
- CT
- charge transfer
- DBQ
- n-decylbenzoquinone
- DM
- dodecyl β-d-maltoside
- PpLipDH
- lipoamide dehydrogenase of P. putida
- Ndi1
- internal rotenone-insensitive NADH-ubiquinone oxidoreductase from S. cerevisiae
- NQO1
- NAD(P)H-quinone oxidoreductase 1 or DT-diaphorase
- ROS
- reactive oxygen species
- DmTR
- thioredoxin reductase of D. melanogaster
- TX
- Triton X-100
- UQ
- ubiquinone
- RB-2
- Reactive Blue-2.
REFERENCES
- 1. Yagi T., Di Bernardo S., Nakamaru-Ogiso E., Kao M. C., Seo B. B., Matsuno-Yagi A. (2004) in Respiration in Archaea and Bacteria (Zannori D. ed) pp. 15–40, Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 2. Melo A. M., Bandeiras T. M., Teixeira M. (2004) Microbiol. Mol. Biol. Rev. 68, 603–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Moller I. M. (2001) Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 561–591 [DOI] [PubMed] [Google Scholar]
- 4. Yagi T., Seo B. B., Nakamaru-Ogiso E., Marella M., Barber-Singh J., Yamashita T., Kao M. C., Matsuno-Yagi A. (2006) Rejuvenation Res. 9, 191–197 [DOI] [PubMed] [Google Scholar]
- 5. Yamashita T., Nakamaru-Ogiso E., Miyoshi H., Matsuno-Yagi A., Yagi T. (2007) J. Biol. Chem. 282, 6012–6020 [DOI] [PubMed] [Google Scholar]
- 6. Seo B. B., Marella M., Yagi T., Matsuno-Yagi A. (2006) FEBS Lett. 580, 6105–6108 [DOI] [PubMed] [Google Scholar]
- 7. Marella M., Seo B. B., Matsuno-Yagi A., Yagi T. (2007) J. Biol. Chem. 282, 24146–24156 [DOI] [PubMed] [Google Scholar]
- 8. Eschemann A., Galkin A., Oettmeier W., Brandt U., Kerscher S. (2005) J. Biol. Chem. 280, 3138–3142 [DOI] [PubMed] [Google Scholar]
- 9. Velázquez I., Pardo J. P. (2001) Arch. Biochem. Biophys. 389, 7–14 [DOI] [PubMed] [Google Scholar]
- 10. Yano T., Li L. S., Weinstein E., Teh J. S., Rubin H. (2006) J. Biol. Chem. 281, 11456–11463 [DOI] [PubMed] [Google Scholar]
- 11. Rieske J. S. (1967) Methods Enzymol. 10, 239–245 [Google Scholar]
- 12. Schneider H., Lemasters J. J., Hackenbrock C. R. (1982) J. Biol. Chem. 257, 10789–100793 [PubMed] [Google Scholar]
- 13. Schneider H., Lemasters J. J., Höchli M., Hackenbrock C. R. (1980) J. Biol. Chem. 255, 3748–3756 [PubMed] [Google Scholar]
- 14. Massey V., Swoboda B. E. (1963) Biochem. Z. 338, 474–484 [PubMed] [Google Scholar]
- 15. Yagi T. (1986) Arch. Biochem. Biophys. 250, 302–311 [DOI] [PubMed] [Google Scholar]
- 16. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 17. Williams C. H., Jr. (1995) FASEB J. 9, 1267–1276 [DOI] [PubMed] [Google Scholar]
- 18. Nonaka Y., Fujii S., Yamano T. (1986) J. Biochem. 99, 803–814 [DOI] [PubMed] [Google Scholar]
- 19. Sakurai T., Hosoya H. (1966) Biochim. Biophys. Acta 112, 459–468 [Google Scholar]
- 20. Schmid R., Gerloff D. L. (2004) FEBS Lett. 578, 163–168 [DOI] [PubMed] [Google Scholar]
- 21. Eckenroth B. E., Rould M. A., Hondal R. J., Everse S. J. (2007) Biochemistry 46, 4694–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mattevi A., Obmolova G., Sokatch J. R., Betzel C., Hol W. G. (1992) Proteins 13, 336–351 [DOI] [PubMed] [Google Scholar]
- 23. Ernster L., Navazio F. (1957) Biochim. Biophys. Acta 26, 408–415 [DOI] [PubMed] [Google Scholar]
- 24. Brunmark A., Cadenas E., Lind C., Segura-Aguilar J., Ernster L. (1987) Free Radic. Biol. Med. 3, 181–188 [DOI] [PubMed] [Google Scholar]
- 25. Li R., Bianchet M. A., Talalay P., Amzel L. M. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 8846–8850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hosoda S., Nakamura W., Hayashi K. (1974) J. Biol. Chem. 249, 6416–6423 [PubMed] [Google Scholar]
- 27. Garten S., Wosilait W. D. (1971) Biochem. Pharmacol. 20, 1661–1668 [DOI] [PubMed] [Google Scholar]
- 28. Asher G., Dym O., Tsvetkov P., Adler J., Shaul Y. (2006) Biochemistry 45, 6372–6378 [DOI] [PubMed] [Google Scholar]
- 29. Chen S., Wu K., Zhang D., Sherman M., Knox R., Yang C. S. (1999) Mol. Pharmacol. 56, 272–278 [DOI] [PubMed] [Google Scholar]
- 30. Prestera T., Prochaska H. J., Talalay P. (1992) Biochemistry 31, 824–833 [DOI] [PubMed] [Google Scholar]
- 31. Prochaska H. J. (1988) Arch. Biochem. Biophys. 267, 529–538 [DOI] [PubMed] [Google Scholar]
- 32. Elias M. D., Nakamura S., Migita C. T., Miyoshi H., Toyama H., Matsushita K., Adachi O., Yamada M. (2004) J. Biol. Chem. 279, 3078–3083 [DOI] [PubMed] [Google Scholar]
- 33. Mogi T., Sato-Watanabe M., Miyoshi H., Orii Y. (1999) FEBS Lett. 457, 61–64 [DOI] [PubMed] [Google Scholar]
- 34. Kozlov A. V., Nohl H., Gille L. (1998) Bioorg. Chem. 26, 334–344 [DOI] [PubMed] [Google Scholar]
- 35. Schirmer R. H., Schulz G. E. (1987) in Coenzymes and Cofactors (Dolphin D., Poulson R., Avramovic O. eds) Vol. IIB, pp. 333–379, Wiley, New York [Google Scholar]
- 36. Williams C. H., Jr. (1992) in Chemistry and Biochemistry of Flavoenzymes (Müller F. ed) Vol. III, pp. 121–211, CRC Press, Boca Raton, FL [Google Scholar]
- 37. Churbanova I. Y., Sevrioukova I. F. (2008) J. Biol. Chem. 283, 5622–5631 [DOI] [PubMed] [Google Scholar]
- 38. Kobayashi K., Mustafa G., Tagawa S., Yamada M. (2005) Biochemistry 44, 13567–13572 [DOI] [PubMed] [Google Scholar]
- 39. King M. S., Sharpley M. S., Hirst J. (2009) Biochemistry 48, 2053–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Birrell J. A., Yakovlev G., Hirst J. (2009) Biochemistry 48, 12005–12013 [DOI] [PubMed] [Google Scholar]
- 41. Murai M., Yamashita T., Senoh M., Mashimo Y., Kataoka M., Kosaka H., Matsuno-Yagi A., Yagi T., Miyoshi H. (2010) Biochemistry 49, 2973–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.