Abstract
Human alkyladenine DNA glycosylase (AAG) locates and excises a wide variety of structurally diverse alkylated and oxidized purine lesions from DNA to initiate the base excision repair pathway. Recognition of a base lesion requires flipping of the damaged nucleotide into a relatively open active site pocket between two conserved tyrosine residues, Y127 and Y159. We have mutated each of these amino acids to tryptophan and measured the kinetic effects on the nucleotide flipping and base excision steps. The Y127W and Y159W mutant proteins have robust glycosylase activity toward DNA containing 1,N6-ethenoadenine (εA), within 4-fold of that of the wildtype enzyme, raising the possibility that tryptophan fluorescence could be used to probe the DNA binding and nucleotide flipping steps. Stopped-flow fluorescence was used to compare the time-dependent changes in tryptophan fluorescence and εA fluorescence. For both mutants, the tryptophan fluorescence exhibited two-step binding with essentially identical rate constants as were observed for the εA fluorescence changes. These results provide evidence that AAG forms an initial recognition complex in which the active site pocket is perturbed and the stacking of the damaged base is disrupted. Upon complete nucleotide flipping, there is further quenching of the tryptophan fluorescence with coincident quenching of the εA fluorescence. Although these mutations do not have large effects on the rate constant for excision of εA, there are dramatic effects on the rate constants for nucleotide flipping that result in 40 to 100-fold decreases in the flipping equilibrium relative to wildtype. Most of this effect is due to an increased rate of unflipping, but surprisingly the Y159W mutation causes a 5-fold increase in the rate constant for flipping. The large effect on the equilibrium for nucleotide flipping explains the greater deleterious effects that these mutations have on the glycosylase activity toward base lesions that are in more stable base pairs.
Human alkyladenine DNA glycosylase (AAG) recognizes a wide variety of structurally diverse deaminated and alkylated purine lesions. This enzyme initiates the base excision DNA repair pathway by catalyzing the hydrolysis of the N-glycosidic bond and releasing the lesion base. Subsequent action of an abasic-site specific endonuclease, a 5’-deoxyribose phosphate lyase, a DNA polymerase, and a DNA ligase are required to restore the correct DNA sequence, using the intact strand as a template. AAG is the only human glycosylase known to recognize a wide variety of alkylated and deaminated purines (1-4). The expression of AAG confers increased cell survival in the presence of exogenous alkylating agents (5, 6), however its glycosylase activity has been shown to be responsible for retinal degeneration (apoptosis) in mice (7) and the overexpression of AAG leads to enhanced sensitivity to DNA alkylating agents in certain contexts (8, 9). Furthermore, increased levels of AAG have been linked to microsatellite instability (10, 11). Thus, it is important that cells have the correct balance of AAG activity.
Crystal structures of AAG in complex with DNA revealed that this enzyme uses nucleotide flipping to gain access to the N-glycosidic bond of a damaged nucleotide (12, 13). We previously used the intrinsic fluorescence of εA to characterize the minimal kinetic mechanism for the recognition, flipping, and excision of this lesion by wildtype AAG (Scheme 1; (14)). Rapid mixing experiments indicated that an initial recognition complex rapidly and reversibly formed in which the εA lesion is partially unstacked. Given the excess of undamaged sites on the oligonucleotide substrate, and our previous finding that AAG uses facilitated diffusion to search DNA (15), we infer that nonspecific binding is as fast or faster than the apparent association rate constant of ~2×108 M-1s-1 that we observe for the first change in fluorescence of the εA lesion (14). We hypothesized that the further change in the fluorescence of the εA lesion is indicative of the capture of the lesion in the active site of AAG. In order to test this hypothesis we sought to observe conformational changes in the protein directly, as similar approaches have been informative for other DNA glycosylases (16-19).
Scheme 1.
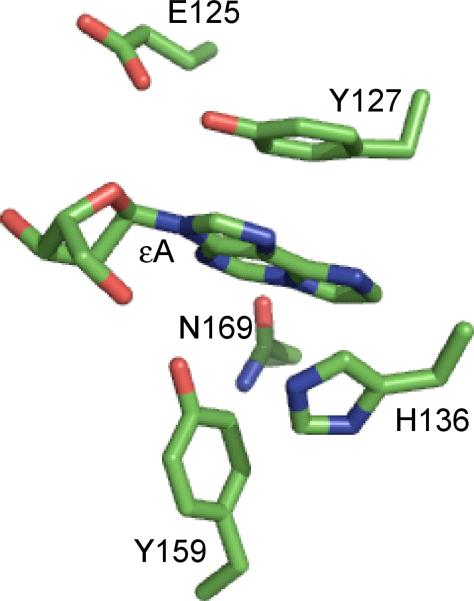
The catalytic domain of AAG (residues 80-298) contains 6 tyrosine and 3 tryptophan residues. The three native tryptophan residues are all found near the surface on the opposite side of the protein from the active site and the DNA binding interface, suggesting that they would not change their environment upon DNA binding. Five of the tyrosine residues line either the binding pocket or the DNA-protein interface, but the absorbance of tyrosine is very close to the absorbance of DNA and it is unlikely that they would serve as useful reporters. Therefore, we took the approach of introducing tryptophan residues into the binding pocket of AAG, substituting either Y127 or Y159, the two tyrosine residues that sandwich the bound lesion in the crystal structures (12, 13). The Y127 side chain stacks on top of the εA lesion, whereas the Y159 side chain makes a T-shaped interaction with the lesion (Figure 1). Comprehensive analysis of aromatic interactions in protein structures suggests that T-type interactions are common and presumably contribute similar binding energy as stacked interactions (20). It has been suggested that tyrosine to tryptophan mutations are relatively conservative (21), but these two positions in AAG are invariably found to be tyrosine in organisms ranging from bacteria to vertebrates (22).
Figure 1.
Active site contacts with the flipped out εA from the published crystal structure of AAG bound to εA-DNA. The coordinates are from 1EWN (13) and the image was rendered with Pymol (http://www.pymol.org). Only the immediate residues surrounding the εA base are shown.
We found that substitution of tyrosine residues with tryptophan at position 127 and 159 in the AAG active site had minimal effects on the excision of εA, however, there were much larger effects on the rate and equilibrium constant for nucleotide flipping. These results are consistent with the simple model that bulky tryptophan residues in the active site destabilize the flipped out conformation and accelerate the rate constant for unflipping. However, it is surprising that the Y159W mutation resulted in a 5-fold faster rate constant for εA flipping. Although flipping is perturbed in these mutants, they follow the same mechanism as the wildtype enzyme with formation of two distinct intermediates prior to N-glycosidic bond cleavage. The observation of identical rate constants for time-dependent changes in either εA or tryptophan fluorescence, demonstrates that there are already changes in the active site environment prior to full nucleotide flipping, and that the most strongly quenched intermediate previously observed is indeed the extrahelical specific recognition complex (14).
MATERIALS AND METHODS
Purification of Wildtype and Mutant AAG Proteins
The catalytic domain of human AAG that lacks the first 79 amino acids was expressed in E. coli and purified as previously described (23). The Y127W and Y159W mutants were constructed by site-directed mutagenesis and purified using the same protocol (23). The concentration of AAG was determined by fluorescent titration of εA-DNA, as described below, and the concentration was found to be within 2-fold of the concentration determined from the calculated extinction coefficient of the wildtype or mutant proteins.
Synthesis and Purification of Oligodeoxynucleotides
The 25mer oligonucleotides were synthesized by Integrated DNA Technologies or by the W.M. Keck Facility at Yale University (see Supporting Information for DNA sequences). The oligonucleotides were desalted using Sephadex G-25 and purified using denaturing polyacrylamide gel electrophoresis as previously described (15). Oligonucleotides for gel-based assays were labeled on the lesion containing strand with a 5’-fluorescein (6-fam) label. The concentrations of the single stranded oligonucleotides were determined from the absorbance at 260 nm, using the calculated extinction coefficients for all oligonucleotides except those containing εA. For oligonucleotides containing εA, the extinction coefficient was calculated for the same sequence with an A in place of the εA and corrected by subtracting 9400 M-1cm-1 to account for the weaker absorbance of εA as compared to A (24). The lesion-containing oligonucleotides were annealed with a 1.2-fold excess of the complement by heating to 90°C and cooling slowly to 4°C. Two different sequence contexts were used and these are referred to by the central 3 nucleotides (i.e., TEC has a central εA lesion flanked by a 5’T and a 3’C; See Supporting Information for sequences).
Steady-State Fluorescence Measurements
Fluorescence emission spectra were collected with a PTI QuantaMaster fluorometer controlled by FeliX software. For εA fluorescence, an excitation wavelength of 320 nm (6 nm band-pass) was used and the total fluorescence was measured at emission wavelengths from 350-500 nm (6 nm band-pass). Samples (300 μL) of 400 nM εA-containing DNA were prepared in the standard buffer (50 mM NaMES (pH 6.5), 100 mM NaCl, 1 mM EDTA, 1 mM DTT) and spectra were recorded at 25°C. To determine the steady-state fluorescence of εA-containing DNA bound to AAG, the spectra were recorded within 1 minute. No significant excision of εA occurs during this time. Three independent titrations were averaged, and the data were fit by a quadratic equation assuming tight binding by AAG (eq 1), in which Frel is the relative fluorescence, A is the fractional quenching, and Kd is the dissociation constant for the εA-DNA.
| (1) |
Single Turnover Excision of εA Monitored by Fluorescence
Experiments with mutant and wildtype AAG were performed as previously described (14). Since εA is strongly quenched in duplex DNA and when bound to AAG, we were able to follow the release of εA into solution by the increase in fluorescence. Single turnover glycosylase assays were performed in the standard buffer at 25°C with 400 nM DNA duplex and 1.2–2.4 μM AAG. A full emission spectrum was recorded at various times between 2 and 120 minutes. The greatest change in fluorescence occurred at 405 nm, therefore this wavelength was chosen to follow the release of εA into solution. The fluorescence at 405 nm was used to determine the fraction of εA product (Fraction Product = (Ft-F0)/Fmax; Ft is the fluorescence at time = t, F0 is the initial fluorescence at time = 0 and Fmax is the maximal fluorescence change). This normalization gives fraction product values between 0 and 1. These data were fit by a single exponential equation using nonlinear least squares regression with Kaleidagraph (Synergy Software), in which kobs is the rate constant, t is the time, and A is the amplitude (eq 2). In all cases, the same rate constant was measured at two different concentrations of AAG. This demonstrates that AAG was saturating and that the maximal single turnover rate constant was determined (See Supporting Information). Under these conditions, the observed rate constant is equal to the maximal single turnover rate constant (kobs = kmax).
| (2) |
Gel-based Glycosylase Assay
We also measured single turnover glycosylase activity with the standard glycosylase activity assay that utilizes abasic site cleavage by NaOH followed by DNA separation on a denaturing PAGE gel (15, 23). Fluorescein-labeled DNA substrates (50 nM) containing either εA or inosine (I) were prepared in the standard buffer. The reactions were initiated with the addition of 75 nM – 5 μM AAG and incubated at 25°C. At various time points, a sample from the reaction was removed and quenched in 2 volumes of 0.3 M NaOH, giving a final hydroxide concentration of 0.2 M. The abasic sites were cleaved by heating at 70°C for 10 minutes. Samples were mixed with an equal volume of formamide/EDTA loading buffer before loading onto a 15% polyacrylamide gel. Gels were scanned with a Typhoon Imager (GE Healthcare) to detect the fluorescein label by exciting at 488 nm and measuring emission with a 520BP40 filter. The gel bands were quantified using ImageQuant TL (GE Healthcare). The data was converted to fraction product [Fraction Product = product / (product + substrate)] and then fit by a single exponential (eq 2). The observed rate constant for the single turnover reaction was independent of the concentration of AAG, indicating that the maximal rate constant was measured (kobs = kmax; see Supporting Information).
Assignment of Single Turnover Rate Constants
Glycosylases require a nucleotide flipping step to gain access to the labile bond and therefore a saturating single turnover rate constant could be limited by either nucleotide flipping or by hydrolysis of the N-glycosidic bond. This reaction scheme is shown below (Scheme 2) and the contribution of individual rate constants is given in eq 3. For wildtype AAG-catalyzed excision of εA we have previously established that the flipping rate constant (kflip) is much faster than the rate of N-glycosidic bond hydrolysis (kchem). Furthermore, we found that the equilibrium constant for nucleotide flipping (Kflip; eq 4) is highly favorable (14). Therefore, for AAG-catalyzed excision of εA the observed single turnover rate constant is simply kchem. The analysis herein suggests that this is true for the Y127W and Y159W mutants as well. However, for AAG-catalyzed excision of hypoxanthine (Hx), the rates of flipping are unknown and the inverse correlation between base pair stability and excision rate strongly suggests that the equilibrium for nucleotide flipping is unfavorable for this lesion (3, 25). Therefore, the value of kmax for wildtype and mutant AAG should be considered a lower limit for the value of kchem (eq 5).
| (3) |
| (4) |
| (5) |
Scheme 2.
Stopped-Flow Kinetics
Pre-steady state kinetic experiments were performed on a Hi-Tech SF-61DSX2, controlled by Kinetic Studio (TgK Scientific). The fluorescence of εA was measured using an excitation wavelength of 320 nm and a WG360 long-pass emission filter as previously described (14). The fluorescence of tryptophan was measured using an excitation wavelength of 295 nm and a 330BP20 band-pass emission filter. Due to the weak fluorescence of εA and tryptophan, and the fast association rate constant, we monitored binding with equimolar concentrations of εA-DNA and protein. Under these conditions the time-dependent changes in fluorescence was fit by eq 6, in which F is εA fluorescence as a function of time, C is the fluorescence of the free DNA, Y and Z are the changes in fluorescence of the first and second intermediates relative to the free DNA, E0 is the starting concentration of AAG and DNA ([AAG] = [DNA]), k1 is the bimolecular rate constant for binding (kon; Scheme 2), k2, obs is the observed unimolecular rate constant for the subsequent flipping step, and t is the time (14). For an approach to equilibrium, the observed rate constant is equal to the sum of the rate constants for the forward and reverse flipping steps (eq 7). However, in the case of a favorable equilibrium constant for flipping (kflip >> kunflip), the observed rate constant for the forward reaction is approximately equal to the flipping rate constant (eq 8). Dissociation experiments were performed for the mutant proteins to validate this assignment (see below).
| (6) |
| (7) |
| (8) |
Several different concentrations between 0.125 μM and 1.0 μM (all concentrations are given after mixing) gave the same rate constants, confirming that the concentration chosen was saturating for the mutant and wildtype proteins (see Supporting Information and data not shown). At least three traces were averaged together for each concentration in two independent experiments, and the averaged data was fit by eq 6 using Kaleidagraph.
Pulse-Chase Assay to Measure Substrate Dissociation
The macroscopic rate constant for dissociation of wildtype and mutant AAG from εA-containing DNA was measured by pulse-chase in the standard reaction buffer at 25°C as previously described for wildtype AAG (14). Briefly, in 20 μL reactions, 50 nM fluorescein-labeled TEC DNA was mixed with 75 nM or 150 nM AAG for 20 seconds, and then a chase of 10 μM unlabeled TEC DNA was added. At various time points, a sample from the reaction was removed and analyzed as described for “Gel-based glycosylase assay”. Base excision catalyzed by AAG results in fluorescein-labeled product, whereas dissociation releases unreacted fluorescein-labeled substrate. The partitioning between hydrolysis and dissociation can be determined from either the exponential rate constant or by the change in burst amplitude. If AAG dissociates from the labeled DNA before the chemical cleavage step and then binds to the unlabeled DNA, less of the reaction will occur during the single turnover part of the curve as compared to the same experiment without chase. The data was converted to fraction product (fraction product = product/(product + substrate)) and fit by a single exponential (eq 2). The observed rate constant was independent of the concentration of AAG.
According to the two step binding mechanism described in Scheme 2, two different partitioning equations can be written (26). All labeled substrate is initially bound, and therefore the fraction of product formed is given by the fraction that goes on to react. This is indicated by eq 9, in which A is the burst amplitude (the fraction of product formed in the burst phase of the experiment), kmax is the maximal single turnover rate constant for formation of product, and koff,obs is the macroscopic rate constant for dissociation from the flipped out complex. This expression can be rearranged to solve for the desired dissociation rate constant (eq 10). Similarly, for branched pathways, the observed rate constant for the burst phase of the pulse-chase experiment is given by the sum of the rate constants for the competing pathways, formation of product is given by kmax and the macroscopic dissociation of substrate is designated koff-obs (eq 11). Solving for koff,obs gives eq 12.
| (9) |
| (10) |
| (11) |
| (12) |
Control reactions in which no chase was added provided the single turnover rate constant, kmax, and confirmed that these concentrations of AAG were saturating. From these values, the dissociation rate constant, koff, for AAG dissociating from εA-DNA was calculated by two different methods (eqs 10 and 12). Both methods gave very similar values for koff,obs and we report the average of the results obtained with both methods.
Since the εA-DNA binds in two steps, the observed rate constant for dissociation of substrate (koff,obs) could be limited by the unflipping rate (kunflip) or dissociation from nonspecific DNA (koff). According to Scheme 2, and assuming that the flipped out complex is stable (i.e., kflip >> kunflip), this observed dissociation rate constant can be expressed in terms of the microscopic rate constants [eq 13; (26)]. Stopped flow fluorescence suggests that dissociation from the initial AAG•DNA complex is rapid (koff ~30 s-1) relative to the forward flipping rate constant kflip = 2.6 s-1, and therefore the observed rate constant for substrate dissociation from the εA-DNA•AAG complex is approximately equal to the reverse rate constant for flipping (eq 14).
| (13) |
| (14) |
Direct measurement of the macroscopic rate for substrate dissociation
Stopped-flow double-mixing experiments were performed to measure unflipping and dissociation of εA-DNA directly. To rapidly form the flipped-out AAG-DNA complex, 2 μM AEA DNA was mixed with 2.8 μM AAG. After an aging time of 1 second, 18 μM pyrrolidine-DNA was added as a competitor. The final concentrations after mixing were 500 nM AEA DNA, 700 nM AAG, and 9 μM pyrrolidine-DNA. The fluorescence of εA was measured using an excitation wavelength of 320 nm and a WG360 long-pass emission filter. The reaction was followed for 100 seconds; no significant excision of εA occurs during this time. Three traces were averaged together for each mutant, and the normalized fluorescence was fit by a single exponential equation (eq 2). As described above for the pulse-chase assay, the observed rate constant for dissociation is described by eq 13 and simplifies to eq 14 under the conditions of koff >> kflip.
Free Energy Reaction Profile
The energetic effects of the Y127W and Y159W mutations can be illustrated by constructing a free energy reaction profile for the binding, flipping, and excision of an εA lesion by wildtype and mutant AAG. A standard state of 25 °C, pH 6.5, 1 nM enzyme, and 1 μM εA-DNA was chosen. The E•S state was arbitrarily set to ΔG=0 for all enzymes and the individual heights of barriers were determined by eq 15, in which R is the gas constant (1.987×10-3 kcal/molK), T is the temperature (298 K), k is the rate constant, h is Planck's constant (1.58×10-37 kcal s), and kB is the Boltzmann constant (33×10-27 kcal/K).
| (15) |
RESULTS
Tryptophan Fluorescence of Wildtype AAG
The catalytic domain of AAG (residues 80-298) has almost identical glycosylase activity as the full-length protein (23, 27, 28), but is considerably more stable. Therefore, we used this construct for all of the experiments described below. To gain insight into the steps involved in nucleotide flipping we sought to establish fluorescent probes that could report on ligand binding and conformational changes in AAG. Although is is possible to use the natural fluorescence of εA to characterize the interaction of AAG with this alkylative lesion, the majority of substrates recognized by AAG are not fluorescent (14). The catalytic domain of AAG contains three tryptophan residues, raising the possibility that the native protein might undergo changes in its intrinsic fluorescence upon binding to DNA. The crystal structure of AAG bound to εA-DNA shows that all three of these residues are located near the surface on the opposite side of the enzyme from the DNA binding surface ((12); see Supporting Information). We initially tried to remove these tryptophan residues to simplify the fluorescence spectra when additional tryptophan residues were added to the protein. Each individual tryptophan could be mutated singly to phenylalanine and full glycosylase activity was maintained, however attempts to combine these into a triple mutant resulted in insoluble protein in E. coli (data not shown). Before embarking on more extensive mutagenesis studies, we tested whether the native protein undergoes any changes in tryptophan fluorescence upon binding to εA-containing DNA. Whereas binding could be readily detected by changes in the εA fluorescence (excitation at 320 nm and emission at 400 nm), parallel experiments in which tryptophan fluorescence was measured gave no detectable changes in fluorescence (excitation at 295 nm and emission at 330 nm; see Supporting Information). This suggests that the native tryptophan residues do not change their environment upon DNA binding and nucleotide flipping. Therefore, the changes in tryptophan fluorescence for the Y127W and Y159W mutants simply reflect changes in the local environment of the active site tryptophan residue.
Interaction of Y127W and Y159W AAG with εA-DNA
We prepared two different mutants of AAG in which one of the active site tyrosines was mutated to tryptophan. These tyrosine residues, Y127 and Y159, interact with the flipped out base in the crystal structure of AAG bound to εA-DNA (13). As tyrosine to tryptophan substitutions are often tolerated (21), we investigated the possibility that these mutants might exhibit tryptophan fluorescence that reports on the nucleotide flipping step. This would enable us to correlate changes in the fluorescence of εA-DNA with protein fluorescence. It was previously shown that εA-DNA of the sequence AEA shows a large decrease in fluorescence upon binding to wildtype AAG (See Materials and Methods; (14)). Fluorescence titrations were performed with a fixed concentration of εA-DNA and increasing concentration of mutant AAG proteins to determine the concentration of active AAG (Figure 2; See Materials and Methods). The active concentration determined by this method was within 60% of the value calculated from the UV-absorbance of the mutant proteins and this corrected concentration was used for all of the experiments described below.
Figure 2.
Titration of εA-DNA with AAG mutant proteins. Samples of 400 nM εA-DNA were incubated with increasing concentration of either Y127W (◇) or Y159W (○) AAG and the εA fluorescence was monitored. Three independent titrations were performed and the average and standard deviation are shown. The normalized fluorescence was fit by the quadratic equation under conditions of tight binding (eq 1). These fits indicate that the number of εA-binding sites is 70% of the expected value for Y127W and 60% of the expected value for Y159W.
Glycosylase Activity of AAG Mutants Toward εA-DNA
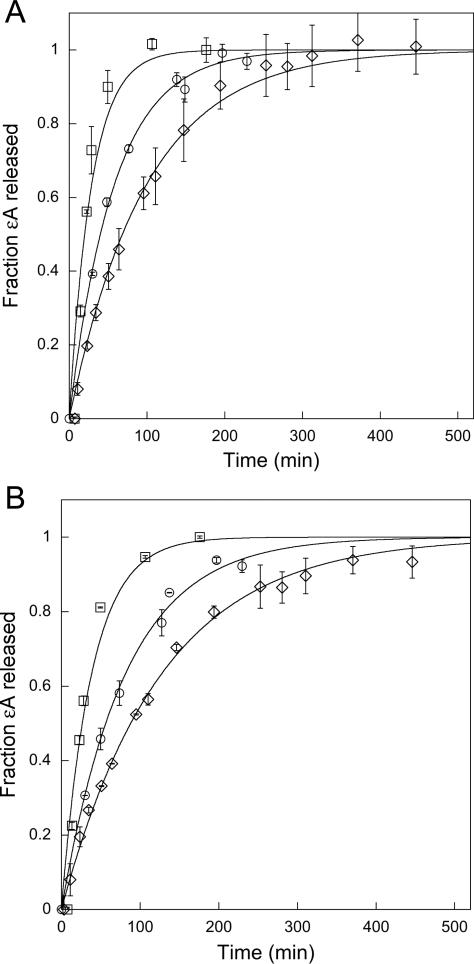
We next measured the single turnover glycosylase activity of the wildtype and mutant proteins. Since εA is strongly quenched in duplex DNA, and becomes fluorescent upon its release into solution, we monitored the fluorescence of εA-DNA incubated with either wildtype or mutant forms of AAG under single turnover conditions, with enzyme in excess over substrate and far above the Kd for substrate binding (Figure 3). Two different sequence contexts were tested, a heterogenous sequence (TEC) and a polyA sequence (AEA; see Supporting Information; (14)). The analysis of εA excision from the TEC duplex by wild-type AAG gave a single turnover rate constant of 0.048 min-1, similar to the previously reported value of 0.034 min-1 (14). The Y159W mutant is 2.7-fold slower than wildtype and the Y127W mutant is 4-fold slower (Table 1). These effects are similar to the respective 2.7-fold and 8-fold effects of these mutations on the single turnover excision of εA measured by abasic site formation under different reaction conditions (23). Similar results were obtained for the AEA sequence context with 3.4-fold and 4.9-fold decreases in the single turnover rate constant for Y159W and Y127W, relative to wildtype AAG. The relatively modest decreases in the single turnover glycosylase activity raised the possibility that these introduced tryptophan residues could serve as useful reporters on the rates of conformational changes within the protein. For wildtype AAG, the maximal single turnover rate constant is likely to be N-glycosidic bond hydrolysis since the equilibrium constant for flipping is highly favorable. However, the maximal single turnover rate constant could be limited by either the nucleotide flipping step or the N-glycosidic bond cleavage step for these mutants (Scheme 2).
Figure 3.
Single turnover excision of εA by wildtype and mutant AAG. Since εA is strongly quenched in duplex DNA and when bound to AAG, the release of εA into solution results in an increase in fluorescence that can be monitored by exciting at 320 nm and recording the fluorescence at 400 nm. The reaction time course for wildtype (□), Y127W (◇), and Y159W (○) AAG are shown for two different sequences, TEC (A) and AEA (B). The concentration of DNA was 400 nM and the concentration of AAG was either 1.2 or 2.4 μM. The two concentrations of AAG gave identical reaction curves, indicating that AAG is saturating under these conditions. Each data point reflects the average and standard deviation of three independent reactions and the lines indicate the best fits to a single exponential (eq 2).
Table 1.
Kinetic parameters for flipping and excision of damaged bases by wildtype and mutant AAGa
| Wildtype | Y159W | Y127W | |
|---|---|---|---|
| kon (M-1s-1) | 2.0×108b | 3.1×108 | 6.8×107 |
| kflip (s-1) | 2.6b | 13.6 | 1.1 |
| kunflip (s-1) | 0.002b | 0.37 | 0.12 |
| Kflipc | 1300b | 40 | 9 |
| | |||
| kchem εA (s-1)d | 8.0×10-4 | 3.0×10-4 | 2.0×10-4 |
| kmax Hx (s-1)e | 4.9×10-2 | 2.8×10-4 | 8.7×10-5 |
The binding and flipping rate constants were determined from stopped-flow fluorescence using the TEC oligonucleotide. The values shown are from the changes in εA fluorescence, but the tryptophan fluorescence of Y159W and Y127W gave almost identical values (see Supporting Information). The standard conditions were 25 °C, 50 mM NaMES, pH 6.5, 100 mM NaCl, 1 mM EDTA, 1 mM DTT.
The values for wildtype AAG were previously reported (14).
The equilibrium constant for flipping is given by the ratio of the flipping and unflipping rate constants (Kflip = kflip/kunflip).
The rate constants for single turnover excision of εA from TEC DNA are from Figure 3A, and reflect the rate of N-glycosidic bond cleavage.
Stopped-Flow Fluorescence of εA
To evaluate whether the Y127W or Y159W mutants alter the rate constants for DNA binding or nucleotide flipping, we performed rapid mixing experiments with εA-DNA. We used equimolar concentrations of DNA and protein far above the dissociation constant for DNA binding to monitor fluorescence changes in εA, as previously described for wildtype AAG (14). This experiment provides optimal signal for both binding and flipping steps and is necessary given the fast binding of AAG and the weak fluorescence of εA. Representative data are shown in Figure 4 (A & C). Both mutants exhibit transient increases and subsequent decreases in εA fluorescence similar to the two-step binding that was observed for wildtype AAG. We previously showed that AAG uses nonspecific DNA binding to efficiently locate the site of damage (Scheme 1), however these searching steps are very fast relative to the nucleotide flipping step (14, 15). Therefore, a simplified kinetic mechanism is sufficient to explain the transient changes in fluorescence (Scheme 2). We previously found that the first phase of the binding reaction is concentration-dependent, and reflects the initial binding of AAG to the site of damage (E•S). The second phase of the binding reaction is independent of DNA concentration and corresponds to the flipping of the εA lesion into the active site pocket (E•S′).
Figure 4.
Stopped-flow fluorescence to measure binding and nucleotide flipping by Y127W and Y159W mutants of AAG. Equimolar mixtures of AAG and εA-DNA were mixed in the stopped flow (1 μM after mixing) and either εA fluorescence or tryptophan fluorescence was measured as described in the Materials and Methods. The fluorescence is reported in arbitrary units. (A) εA fluorescence for Y127W. (B) Tryptophan fluorescence for Y127W. (C) εA fluorescence for Y159W. (D) Tryptophan fluorescence for Y159W. The data is the average of 3 independent reactions and the lines indicate the best fits of eq 3. The rate constants are given in Table 1.
For the Y127W mutant, the initial binding step yielded an observed rate constant that is similar to the rate constant for wildtype AAG under these conditions (Table 1). In contrast, the second step is approximately 3-fold slower for the mutant than for wildtype AAG. This observed rate constant is equal to the sum of the rate constants for the flipping and unflipping steps, because it is an approach to equilibrium (eq 7). However, the results from dissociation experiments suggest that the rate constant for flipping is significantly faster than the rate constant for unflipping (see below). This indicates that the observed rate constant for the second phase of the reaction is approximately equal to the microscopic rate constant for flipping (kflip). Therefore, the Y127W mutation decreases the rate constant for flipping by a factor of ~3-fold.
The Y159W mutant showed a lower level of εA fluorescence in the transient formation of the initial recognition complex (E•S; Scheme 2). Given this decrease in signal it was more difficult to accurately measure the rate constant for the initial phase of the binding reaction, but the best fits yielded a binding rate constant that is also similar to that of wildtype and the Y127W mutant (Table 1). Remarkably, the observed rate constant for the second phase of the reaction (flipping) is more than 5-fold faster for the Y159W mutant than for the wildtype protein. This places an upper limit on the rate constant for flipping, but does not provide sufficient information to establish whether or not this mutation increases the rate constant for nucleotide flipping. This faster observed rate constant could also be explained by a 6000-fold increase in the reverse rate constant for flipping with no change in the forward rate constant for flipping. Therefore, additional experiments were performed to resolve this ambiguity and to establish that the Y159W mutation does increase the rate constant for nucleotide flipping (see below).
Stopped-Flow Fluorescence of Tryptophan
We also followed the fluorescence of the tryptophan during the rapid mixing experiments, and the results are shown in Figure 4 (B & D). Control reactions in which the tryptophan fluorescence was recorded for wildtype AAG mixed with εA-DNA (295 nm excitation, 330/20 nm emission) established that none of the three tryptophan residues present in AAG are sensitive to DNA binding. When the Y127W and Y159W mutants were mixed with εA-DNA, two phases of decreasing fluorescence were observed (Figure 4B & 4D). These data were fit by the same equation used to monitor εA fluorescence (eq 6), and yielded almost identical rate constants as were observed for εA fluorescence changes. These experiments reveal that tryptophan at position 127 and 159 in the active site pocket are perturbed in the initial lesion recognition complex, relative to when AAG is free in solution. The tryptophan quenching in the first phase, corresponding to formation of an initial recognition complex, is coincident with the increased fluorescence of εA, suggesting that the protein changes its local conformation as fast or faster than the DNA conformational change that is monitored by the εA fluorescence. In the second step, tryptophan residues at either position 127 or 159 are quenched in the flipped out complex with the same rate constant as the εA is quenched. This correlation is consistent with the simple model that the εA lesion interacts with the tyrosine or tryptophan residues at positions 127 and 159 (Figure 1).
Pulse-Chase Experiments to Determine the Rate of Substrate Dissociation
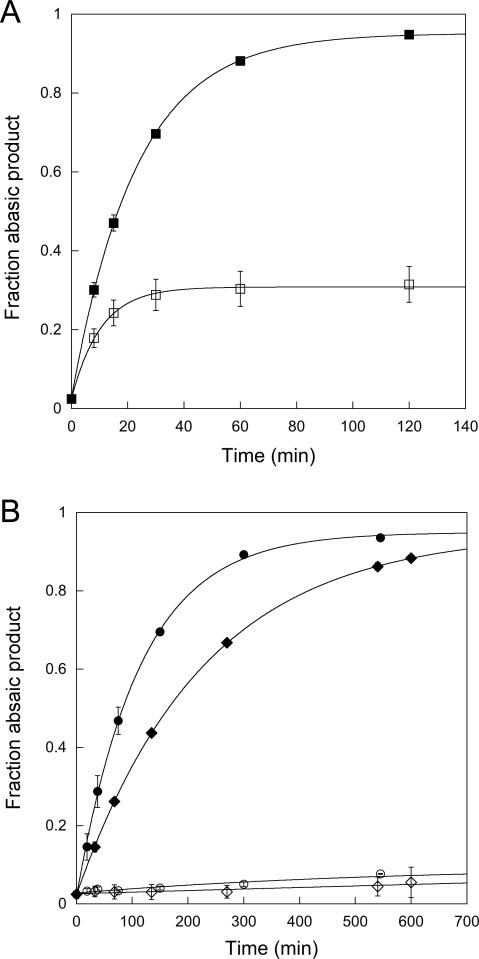
As the nucleotide flipping step is an approach to an equilibrium, it is important to measure the kinetics of the reaction in the reverse direction. Therefore, we performed pulse-chase experiments in which wildtype or mutant AAG proteins were initially mixed with fluorescently labeled εA-DNA and then chased with excess unlabeled εA-DNA (Figure 5). This experiment allows the partitioning between dissociation and base excision to be directly determined (see Materials and Methods). The wildtype protein gave the same results as previously reported, with 30% of the substrate partitioning forward to product and ~70% dissociating. The dissociation of AAG from nonspecific DNA is relatively fast, and therefore the observed rate constant is simply the rate constant for unflipping [kobs = kunflip = 1.6×10-3 s-1; (14)]. In contrast, the addition of chase completely blocked the reaction by the Y127W and Y159W mutants (Figure 5B). This indicates that the rate of substrate dissociation is much faster than the rate of N-glycosidic bond cleavage and places a lower limit on the rate of εA dissociation of ~20-fold faster than the rate of N-glycosidic bond cleavage, assuming that 5% abasic product could have been detected. However, a much bigger effect on the unflipping rate could have been masked. Therefore, we adopted an alternative approach to measure the rate of substrate dissociation, and to determine the rate constant for the unflipping step with these mutant enzymes.
Figure 5.
Pulse-chase experiments to measure dissociation of εA-DNA. The partitioning between the forward reaction (base excision) and substrate dissociation were measured for wildtype, Y127W, and Y159W AAG. Either wildtype (A) or mutant (B) AAG was mixed with fluorescein-labeled εA-DNA in the presence (open symbols) or absence (closed symbols) of excess unlabeled εA-DNA as described in the Materials and Methods. The fraction of fluorescent abasic DNA product was quantified using the standard gel-based glycosylase assay. For the wildtype enzyme approximately 30% of the εA-DNA is committed to excision. In contrast, essentially all of the εA-DNA dissociated from both Y127W (◇) and Y159W (○) mutant proteins under these conditions, indicating the kunflip is much greater than kchem (Scheme 2).
Double-mixing Experiments to Measure Unflipping and Dissociation of εA-DNA
We used double-mixing experiments to rapidly form the flipped-out AAG-DNA complex with the AEA substrate, and then challenged it with a tight-binding pyrrolidine-DNA competitor. Pyrrolidine is positively charged at the pH of these reactions and binds tightly to AAG as a transition state analog (12, 29). The change in fluorescence is adequately fit by a single exponential increase for both the Y127W and Y159W mutant proteins (Figure 6). We interpret these data as a rate-limiting unflipping step, followed by rapid dissociation of the nonspecific DNA. The small deviations from a simple exponential could be explained by a small amount of rebinding of εA-DNA. Consistent with this interpretation, experiments performed with a lower concentration of pyrrolidine-DNA chase resulted in a larger deviation from single exponential behavior (data not shown). The values of 0.12 s-1 for Y127W and 0.4 s-1 for Y159W are 75-fold and 250-fold faster than the rate of εA-unflipping for the wildtype protein (Table 1). Nevertheless, these unflipping rate constants are significantly slower than the observed rate constants for flipping, indicating that the observed rate constant for εA and tryptophan quenching that we have assigned to the nucleotide flipping step is approximately equal to the microscopic rate constant for flipping (eq 8). The ratio of the rate constants for flipping and unflipping yields the equilibrium constant for nucleotide flipping (eq 4; Table 1).
Figure 6.
Direct measurement of εA-DNA dissociation from Y127W and Y159W AAG. Double-mixing experiments were performed using stopped-flow fluorescence. The AAG•DNA complex was formed, aged for one second, and then chased with an excess of pyrrolidine-DNA as a competitor. The normalized fluorescence was obtained by dividing the observed fluorescence signal by the fluorescence of the sample after the dissociation reaction was complete. The traces reflect the average of 3 separate mixing experiments and the solid lines depict the best fit of a single exponential. Only one out of every twenty data points is shown for clarity. The observed rate constants for dissociation are 0.12 s-1 for Y127W (◇) and 0.37 s-1 for Y159W (○) mutant proteins.
Single Turnover Excision of Hx
The decreased stability of the flipped-out εA-DNA complex for the Y127W and Y159W mutants suggests that these mutations are likely to have a much larger deleterious effect on the flipping of more stably paired lesions. Therefore, we examined the single turnover excision of Hx from the most favorable Hx•T context. Wildtype AAG catalyzes the excision of Hx with a rate constant that is 60-fold faster than the single turnover excision of εA when T is the opposing base (Figure 7A; Table 1). This is very similar to the 54-fold difference that was reported previously at a higher temperature (3). In contrast, the single turnover rate constants for the AAG mutant proteins are essentially identical for excision of both εA and Hx (Figure 7B; Table 1). We believe this is a coincidence, because the single turnover rate constants report on different steps for the excision of εA and Hx. For εA excision, the N-glycosidic bond cleavage step is likely to be rate-limiting. In the case of Hx excision, the observed rate constant is significantly smaller than the rate constant for the chemical step because there is an unfavorable equilibrium for nucleotide flipping (eq 13). In the absence of direct measurements of nucleotide flipping for this lesion, it is also possible that the nucleotide flipping step becomes rate-limiting for these mutants. Importantly, the Y127W and Y159W mutations have severe (>100-fold) reductions in the observed rate constant for excision of Hx (Table 1).
Figure 7.
Single-turnover excision of Hx. The reaction progress curves are shown for wildtype (A), Y127W (◆) and Y159W (●) AAG (B). Note that the time scales are different for experiments with the wildtype and mutant proteins. Saturating amount of enzyme (1,5 μM) was always in excess over fluorescein-labeled DNA (50 nM). Reactions were followed in triplicate and the average values are shown; errors bars indicate one standard deviation. The lines indicate the best fit of a single exponential and the rate constants are reported in Table 1 (See Materials and Methods for details).
DISCUSSION
We mutated the two tyrosine residues in the AAG active site pocket to tryptophan in order to gain insight into the mechanism by which AAG recognizes DNA damage. Tryptophan was chosen as a conservative replacement with the intent of being able to directly monitor conformational changes and binding of base lesions in the active site. For both the Y127W and the Y159W mutant proteins, the changes in tryptophan fluorescence were coincident with the changes in εA fluorescence, supporting the previous assignments of the εA fluorescence changes to an initial recognition complex and to an excision-competent extrahelical complex (14). Although these mutants are efficient catalysts for the excision of εA lesions, the full kinetic analysis demonstrates that these mutations severely impair the equilibrium for flipping by AAG. The large perturbation of the equilibrium constant for nucleotide flipping explains why these mutations have a much larger deleterious effect on the excision of other lesions, such as Hx.
Functional Consequences of the Y127W and Y159W Mutations
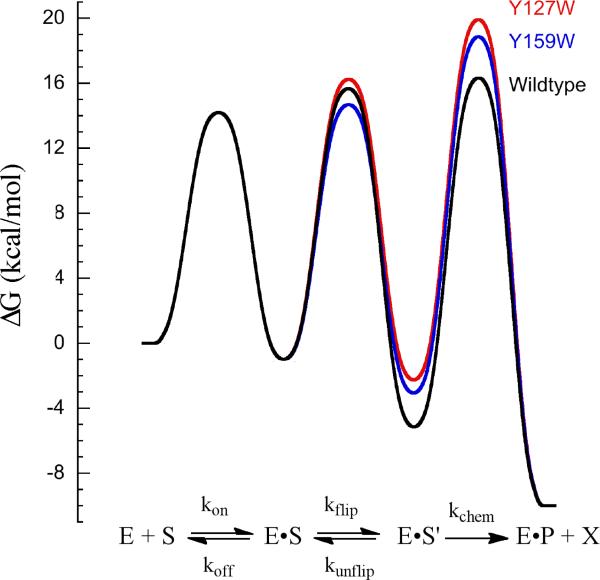
We performed a pre-steady state kinetic analysis of the interaction of these mutant proteins with εA-DNA under the same conditions that were previously used to characterize wildtype AAG. These experiments provide an unambiguous kinetic characterization of the binding, flipping, and base excision steps (Scheme 2). The resulting microscopic rate constants are reported in Table 1, and can be used to calculate the free energy profile for the reactions catalyzed by wildtype and mutant forms of AAG (Figure 8). From this analysis it is apparent that the substitution of either tyrosine with tryptophan significantly destabilizes the active complex with the flipped out εA lesion.
Figure 8.
Free energy profile to illustrate the effects of the Y127W and Y159W mutations on the AAG-catalyzed excision of εA. The relative barrier heights for wildtype (black), Y127W (red), and Y159W (blue) were calculated as described in the Materials and Methods. Whereas there are relatively large effects on the nucleotide flipping step and the stability of the flipped out nucleotide, similar destabilization of the transition state results in much more modest effects on the rate of N-glycosidic bond hydrolysis (kchem).
Perhaps the most remarkable finding is that the Y159W mutation conferred a 5-fold increase in the rate constant for flipping (Table 1). This could be due to a favorable interaction with the side chain of W159 that stabilizes the transition state for flipping of εA. The crystal structure of the wildtype protein shows a T-shaped interaction between Y159 and the εA lesion, and it may be possible for the slightly larger tryptophan to contact the lesion before it fully enters the nucleobase binding pocket (Figure 1). This faster flipping is countered by a much greater increase in the unflipping rate constant (~200-fold; Table 1) and the overall equilibrium for base flipping is perturbed by ~30-fold. This destabilization may be due to the loss of the hydroxyl moiety or to the slightly increased side chain volume of tryptophan relative to tyrosine. In contrast, there is only a subtle effect on the rate constant for εA excision (~3-fold; Table 1). As the equilibrium constant for flipping remains favorable, this suggests that the Y159W mutation has a very similar destabilizing effect in the ground state of the flipped out complex as in the transition state for N-glycosidic bond cleavage (Figure 8).
The Y127W mutation destabilizes the flipped out complex to a greater extent than the Y159W mutation. Whereas there is a modest 2.4-fold decrease in the rate constant for flipping, the unflipping rate constant is increased by 60-fold, resulting in ~140-fold decrease in the stability of the flipped out complex. The Y127W mutant has only a 4-fold decreased rate constant for εA excision, relative to wildtype, which is very similar to the ~3-fold deleterious effect of the Y159W mutation (Table 1). We conclude that the increase in the size of Y127 from tyrosine to tryptophan does not greatly alter the transition state for flipping, but destabilizes the flipped out εA-DNA complex by a similar extent in both the ground state and the transition state for N-glycosidic bond cleavage.
Wildtype AAG has a highly favorable equilibrium for flipping of εA, and therefore the Y127W and Y159W mutant proteins retain favorable equilibrium constants for flipping despite the deleterious effects of these substitutions. This results in relatively small effects on the overall rates of εA excision by the Y127W and Y159W mutants. It is likely that the inability of εA to form hydrogen bonds with an opposing base facilitates this highly favorable flipping equilibrium. In contrast to εA, most of the lesions recognized by AAG, including 7-methylguanine and Hx, do not appear to be bound stably in the flipped out conformation by AAG (3, 25, 30). Consistent with this model, the Y127W and Y159W mutations have much larger deleterious effects of 520- and 140-fold for the excision of Hx from a Hx•T wobble pair (Table 1). These defects are larger by a factor of ~4 than the defects in the flipping equilibrium for εA (Table 1), suggesting that the tryptophan mutations specifically compromise the positioning of Hx in the transition state for N-glycosidic bond cleavage.
Insight Into the Mechanism of AAG-catalyzed Nucleotide Flipping
Although the Y127W and Y159W mutations were more disruptive than expected, the characterization of these mutant proteins provided valuable insight into the mechanism of AAG-catalyzed nucleotide flipping. In spite of the large decreases in the stability of the flipped out, pre-hydrolysis complex, the stopped-flow fluorescence experiments showed that these mutant proteins exhibit the same two-state binding model as was observed for wildtype AAG. The same rate constants were observed for tryptophan and εA fluorescence with both mutants (Figure 4). These observations indicate the εA lesion and tryptophan side chains located in the AAG active site both report on the initial recognition complex and on the fully flipped, active complex. Structural and biochemical studies of two distinct DNA glycosylases, uracil DNA glycosylase (UNG) and 8-oxoguanine DNA glycosylase (OGG1) have suggested that these glycosylases have a pre-binding site for prospective damaged bases that has been referred to as the exosite (31-34). The finding that two different fluorescent reporters in the AAG active site report on an intermediate that forms prior to nucleotide flipping is consistent with a similar initial recognition complex in which a partially flipped εA lesion interacts directly with the tryptophan side chains at the active site. However, we cannot exclude the possibility that other conformational changes in the protein or in the DNA (perhaps accompanying DNA bending) are responsible for the concerted changes in εA and tryptophan fluorescence.
Substitution of tyrosine with tryptophan is expected to be a relatively conservative mutation, since both side chains occupy similar volumes and can make similar stacking interactions. However, positions 127 and 159 in homologs of human AAG are invariably tyrosine. Extensive mutagenesis of AAG and selection for active site variants that conferred resistance to DNA alkylating agents has identified hundreds of allowed mutations, but no active variants at position 127 and only two active variants at position 159 were identified (5, 22). The finding that Y159N and Y159C had detectable activity is consistent with the finding that Y159W has less severe defects on nucleotide flipping than Y127W. Another study sought to identify AAG mutants that increased the frequency of mutations and identified Y127I as conferring a strong mutator phenotype (11).
We were surprised to find that the Y127W mutation has such severe defects in flipping, as a tryptophan side chain would be well-accommodated in the crystal structure of the εA-DNA complex (see Supporting Information). Nevertheless, our results and the failure to observe active variants at this position point toward a very precise role in positioning the substrate in the flipped out, active conformation. It is interesting that the Y127I mutation has been reported to exhibit only a 3-fold decrease in the single turnover excision of εA, but a much greater decrease in the single turnover excision of Hx (11). This is similar to the 4-fold defect in excision of εA that we measured for the Y127W mutant. It has been suggested that the mutator phenotype of Y127I is due to tighter binding to undamaged, bulged nucleotides that can occur from polymerase errors in repetitive sequences (11). It will be interesting to test whether the Y127I mutation changes the rate constants for flipping of normal bases, but this will require the development of new fluorescence-based assays.
Supplementary Material
ACKNOWLEDGMENT
We thank Dave Ballou and Bruce Palfey for help with the stopped-flow experiments and members of the O'Brien laboratory for helpful discussions and comments on the manuscript.
Abbreviations
- AAG
Alkyladenine DNA glycosylase, also known as methylpurine DNA glycosylase (MPG) and 3-methyladenine DNA glycosylase
- BER
base excision repair
- DTT
Dithiothreitol
- EDTA
Ethylenediaminetetraacetic acid
- εA
1,N6-ethenoadenine
- Hx
hypoxanthine (the base moiety of inosine)
- I
inosine
- NaHEPES
sodium N-(2-hydroxyethyl)piperazine-N′-(2-ethanesulfonate)
- NaMES
sodium 2-(N-morpholino)ethanesulfonate
- PAGE
polyacrylamide gel electrophoresis
Footnotes
This work was supported in part by a fellowship from the Chemical Biology Interface training grant to A.E.W., a Faculty Research Grant from the Horace H. Rackham School of Graduate Studies to P. O., and by a grant from the National Institutes of Health to P.O. (CA122254).
SUPPORTING INFORMATION AVAILABLE
Nine supporting figures are provided that include the sequences of the oligonucleotide substrates, views of the native and introduced tryptophan residues from the published crystal structure, additional analysis of the stopped-flow fluorescence experiments, and evidence that saturating concentration of enzyme was used for the determination of the single turnover rate constants. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Engelward BP, Weeda G, Wyatt MD, Broekhof JL, de Wit J, Donker I, Allan JM, Gold B, Hoeijmakers JH, Samson LD. Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13087–13092. doi: 10.1073/pnas.94.24.13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larsen E, Meza TJ, Kleppa L, Klungland A. Organ and cell specificity of base excision repair mutants in mice. Mutat. Res. 2007;614:56–68. doi: 10.1016/j.mrfmmm.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 3.O'Brien PJ, Ellenberger T. Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. J. Biol. Chem. 2004;279:9750–9757. doi: 10.1074/jbc.M312232200. [DOI] [PubMed] [Google Scholar]
- 4.Hitchcock TM, Dong L, Connor EE, Meira LB, Samson LD, Wyatt MD, Cao W. Oxanine DNA glycosylase activity from mammalian alkyladenine glycosylase. J. Biol. Chem. 2004;279:38177–38183. doi: 10.1074/jbc.M405882200. [DOI] [PubMed] [Google Scholar]
- 5.Guo HH, Choe J, Loeb LA. Protein tolerance to random amino acid change. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9205–9210. doi: 10.1073/pnas.0403255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glassner BJ, Rasmussen LJ, Najarian MT, Posnick LM, Samson LD. Generation of a strong mutator phenotype in yeast by imbalanced base excision repair. Proc. Natl. Acad. Sci. U. S. A. 1998;95:9997–10002. doi: 10.1073/pnas.95.17.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meira LB, Moroski-Erkul CA, Green SL, Calvo JA, Bronson RT, Shah D, Samson LD. Aag-initiated base excision repair drives alkylation-induced retinal degeneration in mice. Proc. Natl. Acad. Sci. U. S. A. 2009;106:888–893. doi: 10.1073/pnas.0807030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rinne ML, He Y, Pachkowski BF, Nakamura J, Kelley MR. N-methylpurine DNA glycosylase overexpression increases alkylation sensitivity by rapidly removing non-toxic 7-methylguanine adducts. Nucleic Acids Res. 2005;33:2859–2867. doi: 10.1093/nar/gki601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trivedi RN, Wang XH, Jelezcova E, Goellner EM, Tang JB, Sobol RW. Human methyl purine DNA glycosylase and DNA polymerase beta expression collectively predict sensitivity to temozolomide. Mol. Pharmacol. 2008;74:505–516. doi: 10.1124/mol.108.045112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofseth LJ, Khan MA, Ambrose M, Nikolayeva O, Xu-Welliver M, Kartalou M, Hussain SP, Roth RB, Zhou X, Mechanic LE, Zurer I, Rotter V, Samson LD, Harris CC. The adaptive imbalance in base excision,repair enzymes generates microsatellite instability in chronic inflammation. J. Clin. Invest. 2003;112:1887–1894. doi: 10.1172/JCI19757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klapacz J, Lingaraju GM, Guo HH, Shah D, Moar,Shoshani A, Loeb LA, Samson LD. Frameshift mutagenesis and microsatellite instability induced by human alkyladenine DNA glycosylase. Mol. Cell. 2010;37:843–853. doi: 10.1016/j.molcel.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau AY, Scharer OD, Samson L, Verdine GL, Ellenberger T. Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision. Cell. 1998;95:249–258. doi: 10.1016/s0092-8674(00)81755-9. [DOI] [PubMed] [Google Scholar]
- 13.Lau AY, Wyatt MD, Glassner BJ, Samson LD, Ellenberger T. Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13573–13578. doi: 10.1073/pnas.97.25.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolfe AE, O'Brien PJ. Kinetic mechanism for the flipping and excision of 1,N(6)-ethenoadenine by human alkyladenine DNA glycosylase. Biochemistry. 2009;48:11357–11369. doi: 10.1021/bi9015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hedglin M, O'Brien PJ. Human alkyladenine DNA glycosylase employs a processive search for DNA damage. Biochemistry. 2008;47:11434–11445. doi: 10.1021/bi801046y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuznetsov NA, Koval VV, Zharkov DO, Nevinsky GA, Douglas KT, Fedorova OS. Kinetics of substrate recognition and cleavage by human 8-oxoguanine-DNA glycosylase. Nucleic Acids Res. 2005;33:3919–3931. doi: 10.1093/nar/gki694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuznetsov NA, Koval VV, Zharkov DO, Vorobjev YN, Nevinsky GA, Douglas KT, Fedorova OS. Pre-steady-state kinetic study of substrate specificity of Escherichia coli formamidopyrimidine--DNA glycosylase. Biochemistry. 2007;46:424–435. doi: 10.1021/bi060787r. [DOI] [PubMed] [Google Scholar]
- 18.Stivers JT, Pankiewicz KW, Watanabe KA. Kinetic mechanism of damage site recognition and uracil flipping by Escherichia coli uracil DNA glycosylase. Biochemistry. 1999;38:952–963. doi: 10.1021/bi9818669. [DOI] [PubMed] [Google Scholar]
- 19.Wong I, Lundquist AJ, Bernards AS, Mosbaugh DW. Presteady-state analysis of a single catalytic turnover by Escherichia coli uracil-DNA glycosylase reveals a “pinch-pull-push” mechanism. J. Biol. Chem. 2002;277:19424–19432. doi: 10.1074/jbc.M201198200. [DOI] [PubMed] [Google Scholar]
- 20.Chelli R, Gervasio FL, Procacci P, Schettino V. Stacking and T-shape competition in aromatic-aromatic amino acid interactions. J. Am. Chem. Soc. 2002;124:6133–6143. doi: 10.1021/ja0121639. [DOI] [PubMed] [Google Scholar]
- 21.Bordo D, Argos P. Suggestions for “safe” residue substitutions in site-directed mutagenesis. J. Mol. Biol. 1991;217:721–729. doi: 10.1016/0022-2836(91)90528-e. [DOI] [PubMed] [Google Scholar]
- 22.Chen CY, Guo HH, Shah D, Blank A, Samson LD, Loeb LA. Substrate binding pocket residues of human alkyladenine-DNA glycosylase critical for methylating agent survival. DNA Repair (Amst) 2008;7:1731–1745. doi: 10.1016/j.dnarep.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Brien PJ, Ellenberger T. Human alkyladenine DNA glycosylase uses acid-base catalysis for selective excision of damaged purines. Biochemistry. 2003;42:12418–12429. doi: 10.1021/bi035177v. [DOI] [PubMed] [Google Scholar]
- 24.Secrist JA, 3rd, Barrio JR, Leonard NJ, Weber G. Fluorescent modification of adenosine,containing coenzymes. Biological activities and spectroscopic properties. Biochemistry. 1972;11:3499–3506. doi: 10.1021/bi00769a001. [DOI] [PubMed] [Google Scholar]
- 25.Lyons DM, O'Brien PJ. Efficient recognition of an unpaired lesion by a DNA repair glycosylase. J. Am. Chem. Soc. 2009;131:17742–17743. doi: 10.1021/ja908378y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsieh J, Walker SC, Fierke CA, Engelke DR. Pre-tRNA turnover catalyzed by the yeast nuclear RNase P holoenzyme is limited by product release. RNA. 2009;15:224–234. doi: 10.1261/rna.1309409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Connor TR. Purification and characterization of human 3-methyladenine-DNA glycosylase. Nucleic Acids Res. 1993;21:5561–5569. doi: 10.1093/nar/21.24.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baldwin MR, O'Brien PJ. Human AP endonuclease I stimulates multiple, turnover base excision by alkyladenine DNA glycosylase. Biochemistry. 2009;48:6022–6033. doi: 10.1021/bi900517y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scharer OD, Nash HM, Jiricny J, Laval J, Verdine GL. Specific binding of a designed pyrrolidine abasic site analog to multiple DNA glycosylases. J. Biol. Chem. 1998;273:8592–8597. doi: 10.1074/jbc.273.15.8592. [DOI] [PubMed] [Google Scholar]
- 30.Vallur AC, Feller JA, Abner CW, Tran RK, Bloom LB. Effects of hydrogen bonding within a damaged base pair on the activity of wild type and DNA,intercalating mutants of human alkyladenine DNA glycosylase. J. Biol. Chem. 2002;277:31673–31678. doi: 10.1074/jbc.M204475200. [DOI] [PubMed] [Google Scholar]
- 31.Parker JB, Bianchet MA, Krosky DJ, Friedman JI, Amzel LM, Stivers JT. Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature. 2007;449:433–437. doi: 10.1038/nature06131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 33.Banerjee A, Santos WL, Verdine GL. Structure of a DNA glycosylase searching for lesions. Science. 2006;311:1153–1157. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- 34.Friedman JI, Stivers JT. Detection of damaged DNA bases by DNA glycosylase enzymes. Biochemistry. 49:4957–4967. doi: 10.1021/bi100593a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.