Abstract
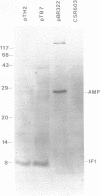
The gene for translation initiation factor IF1, infA, has been identified by using two synthetic oligonucleotides to screen a Charon 30 library of Escherichia coli DNA. A recombinant lambda phage, C1921, was purified from a plaque positive for both probes. A 2.8 kb BglII fragment and a 2.0 kb HindIII fragment isolated from C1921 were subcloned into the BamHI and HindIII sites of pBR322 to yield pTB7 and pTH2 respectively. Synthesis of IF1 in maxicells transformed with pTB7 or pTH2 indicates the presence of inf A in both inserts. This was confirmed by DNA sequencing: a region was found that codes for a 8,119 dalton protein with an amino acid sequence corresponding to IF1. The chromosomal location of inf A was determined by mapping the closely linked beta-lactamase gene (Ampr) in pTB7 and pTH2. pTB7 and pTH2 were transformed into polA Hfr hosts, and integration of the plasmid by homologous recombination near inf A was selected on the basis of ampicillin resistance. The site of integration was confirmed by Southern blot analysis of restriction nuclease digested wild type and transformed genomic DNA. The Ampr marker (and therefore inf A) was mapped to about 20 minutes by Hfr interrupted matings and P1 transduction experiments. The structure and regulation of the inf A operon currently are being investigated.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Howe J. G., Hershey J. W. Initiation factor and ribosome levels are coordinately controlled in Escherichia coli growing at different rates. J Biol Chem. 1983 Feb 10;258(3):1954–1959. [PubMed] [Google Scholar]
- Ikemura T., Ozeki H. Codon usage and transfer RNA contents: organism-specific codon-choice patterns in reference to the isoacceptor contents. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1087–1097. doi: 10.1101/sqb.1983.047.01.123. [DOI] [PubMed] [Google Scholar]
- Ishii S., Kuroki K., Imamoto F. tRNAMetf2 gene in the leader region of the nusA operon in Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jan;81(2):409–413. doi: 10.1073/pnas.81.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Plumbridge J. A., Deville F., Sacerdot C., Petersen H. U., Cenatiempo Y., Cozzone A., Grunberg-Manago M., Hershey J. W. Two translational initiation sites in the infB gene are used to express initiation factor IF2 alpha and IF2 beta in Escherichia coli. EMBO J. 1985 Jan;4(1):223–229. doi: 10.1002/j.1460-2075.1985.tb02339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumbridge J. A., Howe J. G., Springer M., Touati-Schwartz D., Hershey J. W., Grunberg-Manago M. Cloning and mapping of a gene for translational initiation factor IF2 in Escherichia coli. Proc Natl Acad Sci U S A. 1982 Aug;79(16):5033–5037. doi: 10.1073/pnas.79.16.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumbridge J. A., Springer M. Organization of the Escherichia coli chromosome around the genes for translation initiation factor IF2 (infB) and a transcription termination factor (nusA). J Mol Biol. 1983 Jun 25;167(2):227–243. doi: 10.1016/s0022-2836(83)80333-7. [DOI] [PubMed] [Google Scholar]
- Pon C. L., Wittmann-Liebold B., Gualerzi C. Structure--function relationships in Escherichia coli initiation factors. II. Elucidation of the primary structure of initiation factor IF-1. FEBS Lett. 1979 May 1;101(1):157–160. doi: 10.1016/0014-5793(79)81316-2. [DOI] [PubMed] [Google Scholar]
- Rimm D. L., Horness D., Kucera J., Blattner F. R. Construction of coliphage lambda Charon vectors with BamHI cloning sites. Gene. 1980 Dec;12(3-4):301–309. doi: 10.1016/0378-1119(80)90113-4. [DOI] [PubMed] [Google Scholar]
- Sacerdot C., Dessen P., Hershey J. W., Plumbridge J. A., Grunberg-Manago M. Sequence of the initiation factor IF2 gene: unusual protein features and homologies with elongation factors. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7787–7791. doi: 10.1073/pnas.81.24.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacerdot C., Fayat G., Dessen P., Springer M., Plumbridge J. A., Grunberg-Manago M., Blanquet S. Sequence of a 1.26-kb DNA fragment containing the structural gene for E.coli initiation factor IF3: presence of an AUU initiator codon. EMBO J. 1982;1(3):311–315. doi: 10.1002/j.1460-2075.1982.tb01166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Yura T. Chromosomal location and expression of the structural gene for major outer membrane protein Ia of Escherichia coli K-12 and of the homologous gene of Salmonella typhimurium. J Bacteriol. 1979 Aug;139(2):468–477. doi: 10.1128/jb.139.2.468-477.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver P., Wickner W. Genetic mapping of the Escherichia coli leader (signal) peptidase gene (lep): a new approach for determining the map position of a cloned gene. J Bacteriol. 1983 May;154(2):569–572. doi: 10.1128/jb.154.2.569-572.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M., Graffe M., Grunberg-Manago M. Characterization of an E. coli mutant with a thermolabile initiation factor IF3 activity. Mol Gen Genet. 1977 Feb 28;151(1):17–26. doi: 10.1007/BF00446908. [DOI] [PubMed] [Google Scholar]
- Springer M., Graffe M., Grunberg-Manago M. Genetic organization of the E. coli chromosome around the structural gene for initiation factor IF3 (infC). Mol Gen Genet. 1979 Feb 1;169(3):337–343. doi: 10.1007/BF00382279. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Shaffer J., Murphy R. F., Bonner J., Hirose T., Itakura K. Hybridization of synthetic oligodeoxyribonucleotides to phi chi 174 DNA: the effect of single base pair mismatch. Nucleic Acids Res. 1979 Aug 10;6(11):3543–3557. doi: 10.1093/nar/6.11.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Yeast RNA polymerase II genes: isolation with antibody probes. Science. 1983 Nov 18;222(4625):778–782. doi: 10.1126/science.6356359. [DOI] [PubMed] [Google Scholar]