Abstract
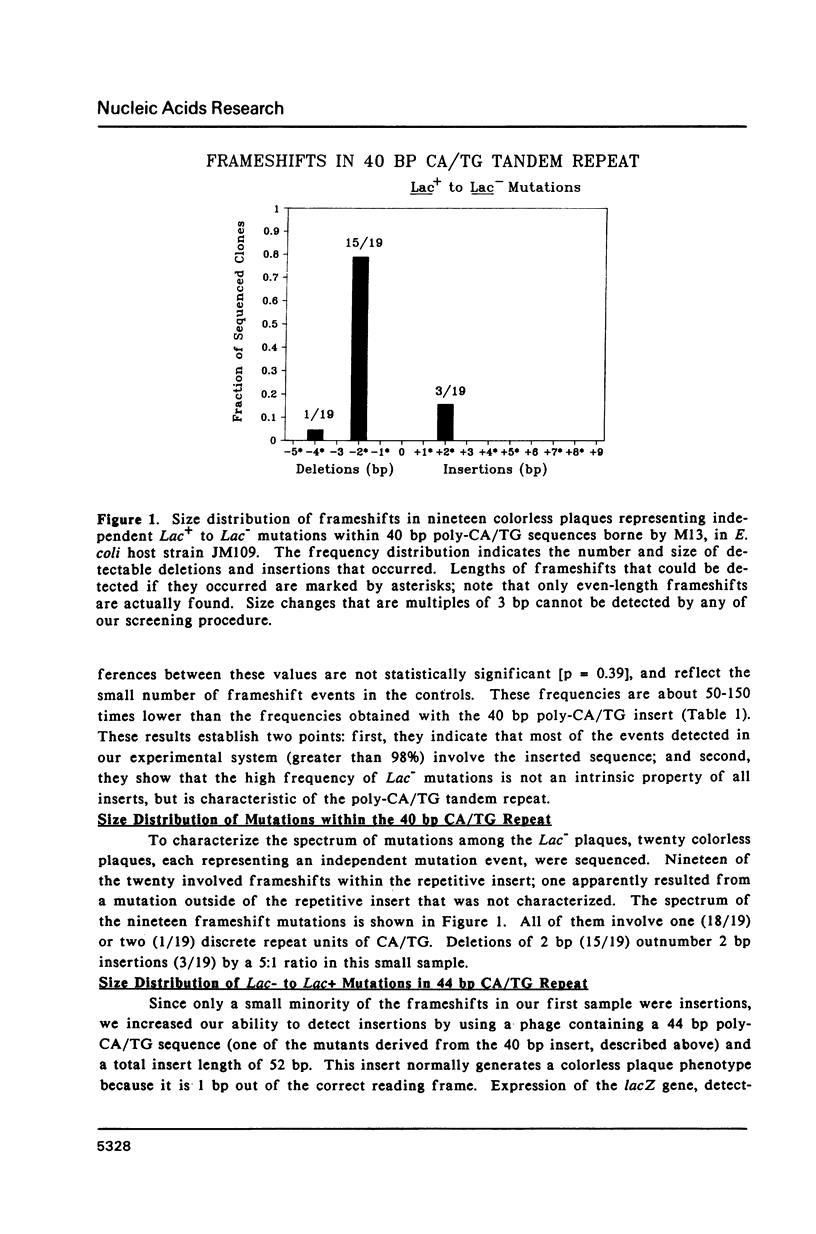
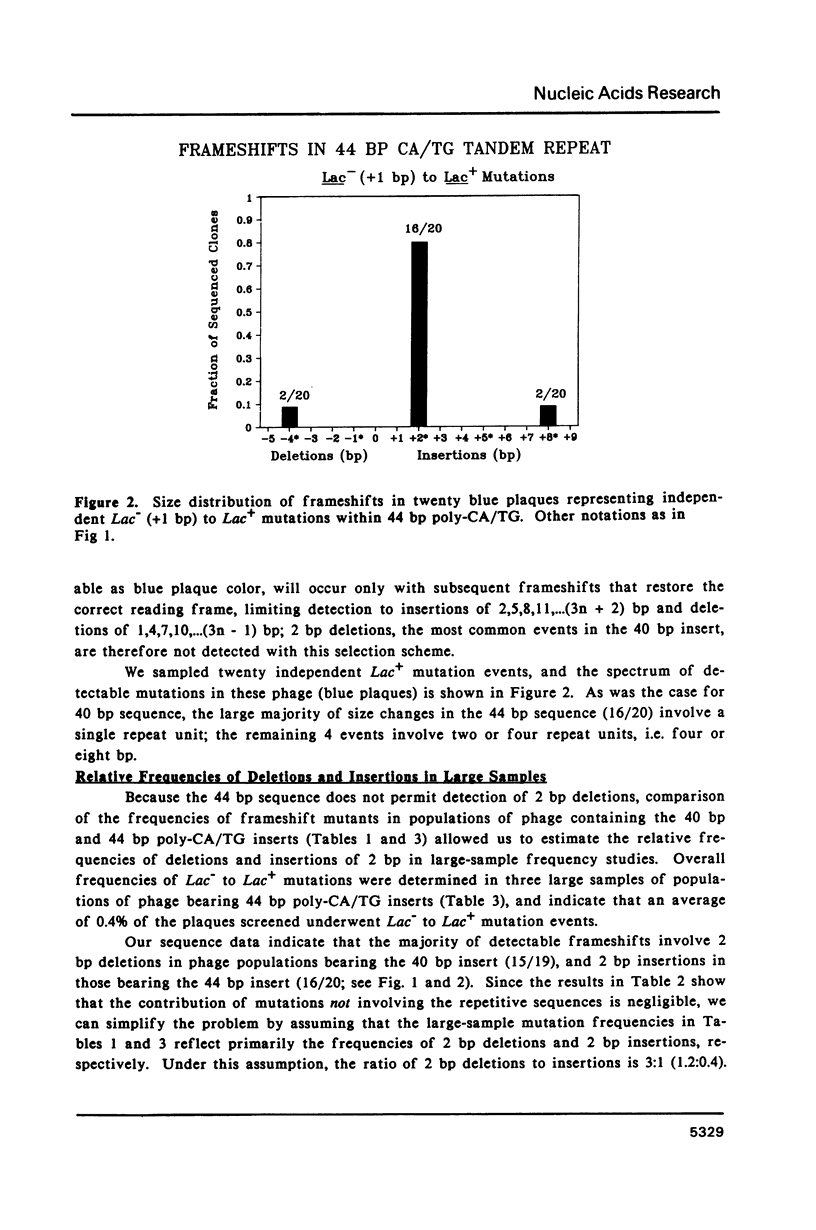
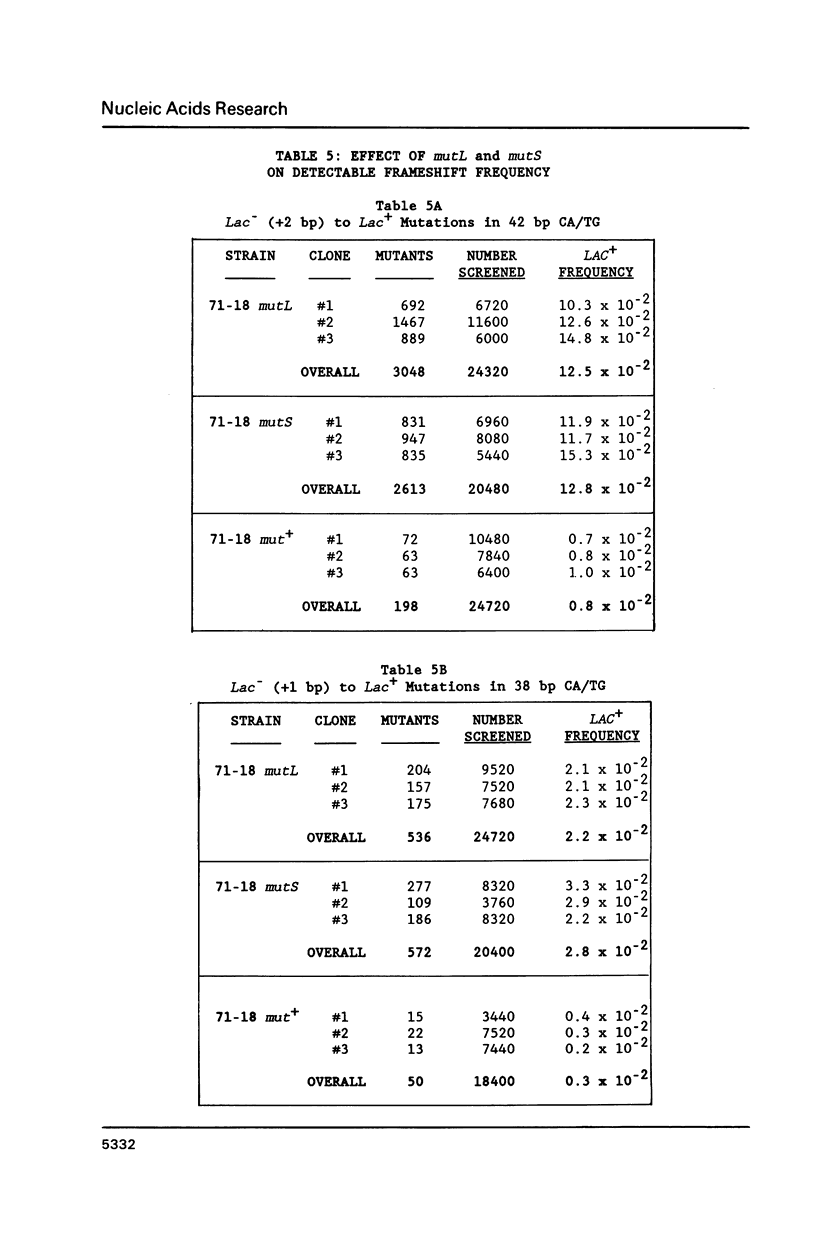
Slipped-strand mispairing (SSM) may play an major role in repetitive DNA sequence evolution by generating large numbers of short frameshift mutations within simple tandem repeats. Here we examine the frequency and size spectrum of frameshifts generated within poly-CA/TG sequences inserted into bacteriophage M13 in Escherichia coli hosts. The frequency of detectable frameshifts within a 40 bp tract of poly-CA/TG is greater than one percent and increases more than linearly with length, being lower by a factor of four in a 22 bp target sequence. The frequency increases more than 13-fold in mutL and mutS host cells, suggesting that a high proportion of frameshift events are normally repaired by methyl-directed mismatch repair. Of the 87 sequenced frameshifts in this study, 96% result from deletion or insertion of only or two 2 bp repeat units. The most frequent events are 2 bp deletions, 2 bp insertions, and 4 bp deletions, the relative frequencies of these events being about 18:6:1.
Full text
PDF












Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson R. P., Roth J. R. Tandem genetic duplications in phage and bacteria. Annu Rev Microbiol. 1977;31:473–505. doi: 10.1146/annurev.mi.31.100177.002353. [DOI] [PubMed] [Google Scholar]
- Clark A. J. Recombination deficient mutants of E. coli and other bacteria. Annu Rev Genet. 1973;7:67–86. doi: 10.1146/annurev.ge.07.120173.000435. [DOI] [PubMed] [Google Scholar]
- Clarke C. H., Johnston A. W. Intragenic mutational spectra and hot spots. Mutat Res. 1976 Aug;36(2):147–164. doi: 10.1016/0027-5107(76)90003-8. [DOI] [PubMed] [Google Scholar]
- Cox E. C. Bacterial mutator genes and the control of spontaneous mutation. Annu Rev Genet. 1976;10:135–156. doi: 10.1146/annurev.ge.10.120176.001031. [DOI] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Genes involved in transitory recombination between phage M13 and plasmid pHV33. EMBO J. 1984 Jan;3(1):87–89. doi: 10.1002/j.1460-2075.1984.tb01765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Transitory recombination between plasmid pHV33 and phage M13. EMBO J. 1983;2(12):2117–2122. doi: 10.1002/j.1460-2075.1983.tb01711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohet C., Wagner R., Radman M. Methyl-directed repair of frameshift mutations in heteroduplex DNA. Proc Natl Acad Sci U S A. 1986 May;83(10):3395–3397. doi: 10.1073/pnas.83.10.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Flanagan J. G., Lefranc M. P., Rabbitts T. H. Mechanisms of divergence and convergence of the human immunoglobulin alpha 1 and alpha 2 constant region gene sequences. Cell. 1984 Mar;36(3):681–688. doi: 10.1016/0092-8674(84)90348-9. [DOI] [PubMed] [Google Scholar]
- Fresco J. R., Alberts B. M. THE ACCOMMODATION OF NONCOMPLEMENTARY BASES IN HELICAL POLYRIBONUCLEOTIDES AND DEOXYRIBONUCLEIC ACIDS. Proc Natl Acad Sci U S A. 1960 Mar;46(3):311–321. doi: 10.1073/pnas.46.3.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman L. Enzymes involved in the repair of damaged DNA. Arch Biochem Biophys. 1981 Oct 15;211(2):511–522. doi: 10.1016/0003-9861(81)90485-9. [DOI] [PubMed] [Google Scholar]
- Hamada H., Kakunaga T. Potential Z-DNA forming sequences are highly dispersed in the human genome. Nature. 1982 Jul 22;298(5872):396–398. doi: 10.1038/298396a0. [DOI] [PubMed] [Google Scholar]
- Hamada H., Petrino M. G., Kakunaga T. A novel repeated element with Z-DNA-forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6465–6469. doi: 10.1073/pnas.79.21.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Petrino M. G., Kakunaga T., Seidman M., Stollar B. D. Characterization of genomic poly(dT-dG).poly(dC-dA) sequences: structure, organization, and conformation. Mol Cell Biol. 1984 Dec;4(12):2610–2621. doi: 10.1128/mcb.4.12.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Seidman M., Howard B. H., Gorman C. M. Enhanced gene expression by the poly(dT-dG).poly(dC-dA) sequence. Mol Cell Biol. 1984 Dec;4(12):2622–2630. doi: 10.1128/mcb.4.12.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hasson J. F., Mougneau E., Cuzin F., Yaniv M. Simian virus 40 illegitimate recombination occurs near short direct repeats. J Mol Biol. 1984 Jul 25;177(1):53–68. doi: 10.1016/0022-2836(84)90057-3. [DOI] [PubMed] [Google Scholar]
- Hentschel C. C. Homocopolymer sequences in the spacer of a sea urchin histone gene repeat are sensitive to S1 nuclease. Nature. 1982 Feb 25;295(5851):714–716. doi: 10.1038/295714a0. [DOI] [PubMed] [Google Scholar]
- Jeang K. T., Hayward G. S. A cytomegalovirus DNA sequence containing tracts of tandemly repeated CA dinucleotides hybridizes to highly repetitive dispersed elements in mammalian cell genomes. Mol Cell Biol. 1983 Aug;3(8):1389–1402. doi: 10.1128/mcb.3.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Thein S. L. Individual-specific 'fingerprints' of human DNA. Nature. 1985 Jul 4;316(6023):76–79. doi: 10.1038/316076a0. [DOI] [PubMed] [Google Scholar]
- Jones C. W., Kafatos F. C. Accepted mutations in a gene family: evolutionary diversification of duplicated DNA. J Mol Evol. 1982;19(1):87–103. doi: 10.1007/BF02100227. [DOI] [PubMed] [Google Scholar]
- KORNBERG A., BERTSCH L. L., JACKSON J. F., KHORANA H. G. ENZYMATIC SYNTHESIS OF DEOXYRIBONUCLEIC ACID, XVI. OLIGONUCLEOTIDES AS TEMPLATES AND THE MECHANISM OF THEIR REPLICATION. Proc Natl Acad Sci U S A. 1964 Feb;51:315–323. doi: 10.1073/pnas.51.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Kramer W., Schughart K., Fritz H. J. Directed mutagenesis of DNA cloned in filamentous phage: influence of hemimethylated GATC sites on marker recovery from restriction fragments. Nucleic Acids Res. 1982 Oct 25;10(20):6475–6485. doi: 10.1093/nar/10.20.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebkowski J. S., Clancy S., Miller J. H., Calos M. P. The lacI shuttle: rapid analysis of the mutagenic specificity of ultraviolet light in human cells. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8606–8610. doi: 10.1073/pnas.82.24.8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. E., Yamasaki E., Ames B. N. A new Salmonella tester strain, TA97, for the detection of frameshift mutagens. A run of cytosines as a mutational hot-spot. Mutat Res. 1982 Jun;94(2):315–330. doi: 10.1016/0027-5107(82)90294-9. [DOI] [PubMed] [Google Scholar]
- Levinson G., Marsh J. L., Epplen J. T., Gutman G. A. Cross-hybridizing snake satellite, Drosophila, and mouse DNA sequences may have arisen independently. Mol Biol Evol. 1985 Nov;2(6):494–504. doi: 10.1093/oxfordjournals.molbev.a040374. [DOI] [PubMed] [Google Scholar]
- Lorenzetti R., Cesareni G., Cortese R. Frameshift mutations induced by an Escherichia coli strain carrying a mutator gene, mutD5. Mol Gen Genet. 1983;192(3):515–516. doi: 10.1007/BF00392200. [DOI] [PubMed] [Google Scholar]
- Lu A. L., Clark S., Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace H. A., Pelham H. R., Travers A. A. Association of an S1 nuclease-sensitive structure with short direct repeats 5' of Drosophila heat shock genes. Nature. 1983 Aug 11;304(5926):555–557. doi: 10.1038/304555a0. [DOI] [PubMed] [Google Scholar]
- Matfield M., Badawi R., Brammar W. J. Rec-dependent and Rec-independent recombination of plasmid-borne duplications in Escherichia coli K12. Mol Gen Genet. 1985;199(3):518–523. doi: 10.1007/BF00330768. [DOI] [PubMed] [Google Scholar]
- Michel B., Ehrlich S. D. Illegitimate recombination at the replication origin of bacteriophage M13. Proc Natl Acad Sci U S A. 1986 May;83(10):3386–3390. doi: 10.1073/pnas.83.10.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. M., Weinger R. S. Pseudo-drumsticks in granulocytes of a male with a Yqh+ polymorphism. Am J Hematol. 1980;8(4):411–414. doi: 10.1002/ajh.2830080410. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Leppert M., O'Connell P., Wolff R., Holm T., Culver M., Martin C., Fujimoto E., Hoff M., Kumlin E. Variable number of tandem repeat (VNTR) markers for human gene mapping. Science. 1987 Mar 27;235(4796):1616–1622. doi: 10.1126/science.3029872. [DOI] [PubMed] [Google Scholar]
- Nickol J. M., Felsenfeld G. DNA conformation at the 5' end of the chicken adult beta-globin gene. Cell. 1983 Dec;35(2 Pt 1):467–477. doi: 10.1016/0092-8674(83)90180-0. [DOI] [PubMed] [Google Scholar]
- Owen J. E., Schultz D. W., Taylor A., Smith G. R. Nucleotide sequence of the lysozyme gene of bacteriophage T4. Analysis of mutations involving repeated sequences. J Mol Biol. 1983 Apr 5;165(2):229–248. doi: 10.1016/s0022-2836(83)80255-1. [DOI] [PubMed] [Google Scholar]
- Pribnow D., Sigurdson D. C., Gold L., Singer B. S., Napoli C., Brosius J., Dull T. J., Noller H. F. rII cistrons of bacteriophage T4. DNA sequence around the intercistronic divide and positions of genetic landmarks. J Mol Biol. 1981 Jul 5;149(3):337–376. doi: 10.1016/0022-2836(81)90477-0. [DOI] [PubMed] [Google Scholar]
- RADDING C. M., JOSSE J., KORNBERG A. Enzymatic synthesis of deoxyribonucleic acid. XII. A polymer of deoxyguanylate and deoxycytidylate. J Biol Chem. 1962 Sep;237:2869–2876. [PubMed] [Google Scholar]
- Radding C. M. Homologous pairing and strand exchange in genetic recombination. Annu Rev Genet. 1982;16:405–437. doi: 10.1146/annurev.ge.16.120182.002201. [DOI] [PubMed] [Google Scholar]
- Rodakis G. C., Lecanidou R., Eickbush T. H. Diversity in a chorion multigene family created by tandem duplications and a putative gene-conversion event. J Mol Evol. 1984;20(3-4):265–273. doi: 10.1007/BF02104732. [DOI] [PubMed] [Google Scholar]
- Rogers J. Molecular biology. CACA sequences - the ends and the means? Nature. 1983 Sep 8;305(5930):101–102. doi: 10.1038/305101a0. [DOI] [PubMed] [Google Scholar]
- Roth J. R. Frameshift mutations. Annu Rev Genet. 1974;8:319–346. doi: 10.1146/annurev.ge.08.120174.001535. [DOI] [PubMed] [Google Scholar]
- SCHACHMAN H. K., ADLER J., RADDING C. M., LEHMAN I. R., KORNBERG A. Enzymatic synthesis of deoxyribonucleic acid. VII. Synthesis of a polymer of deoxyadenylate and deoxythymidylate. J Biol Chem. 1960 Nov;235:3242–3249. [PubMed] [Google Scholar]
- SWARTZ M. N., TRAUTNER T. A., KORNBERG A. Enzymatic synthesis of deoxyribonucleic acid. XI. Further studies on nearest neighbor base sequences in deoxyribonucleic acids. J Biol Chem. 1962 Jun;237:1961–1967. [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sheppard H. W., Gutman G. A. Rat kappa-chain J-segment genes: two recent gene duplications separate rat and mouse. Cell. 1982 May;29(1):121–127. doi: 10.1016/0092-8674(82)90096-4. [DOI] [PubMed] [Google Scholar]
- Slightom J. L., Blechl A. E., Smithies O. Human fetal G gamma- and A gamma-globin genes: complete nucleotide sequences suggest that DNA can be exchanged between these duplicated genes. Cell. 1980 Oct;21(3):627–638. doi: 10.1016/0092-8674(80)90426-2. [DOI] [PubMed] [Google Scholar]
- Smith G. P. Evolution of repeated DNA sequences by unequal crossover. Science. 1976 Feb 13;191(4227):528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Owen J. Mechanisms of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics. 1985 Apr;109(4):633–659. doi: 10.1093/genetics/109.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D., Renz M. Simple DNA sequences of Drosophila virilis isolated by screening with RNA. J Mol Biol. 1984 Jan 15;172(2):229–235. doi: 10.1016/s0022-2836(84)80041-8. [DOI] [PubMed] [Google Scholar]
- Tautz D., Renz M. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res. 1984 May 25;12(10):4127–4138. doi: 10.1093/nar/12.10.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. Inducible DNA repair systems. Annu Rev Biochem. 1985;54:425–457. doi: 10.1146/annurev.bi.54.070185.002233. [DOI] [PubMed] [Google Scholar]
- Weintraub H. A dominant role for DNA secondary structure in forming hypersensitive structures in chromatin. Cell. 1983 Apr;32(4):1191–1203. doi: 10.1016/0092-8674(83)90302-1. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Blair J. E. Studies on polynucleotides. LXXI. Sedimentation and buoyant density studies of some DNA-like polymers with repeating nucleotide sequences. J Mol Biol. 1967 Jul 28;27(2):273–288. doi: 10.1016/0022-2836(67)90020-4. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Büchi H., Kössel H., Ohtsuka E., Khorana H. G. Studies on polynucleotides. LXX. Synthetic deoxyribopolynucleotides as templates for the DNA polymerase of Escherichia coli: DNA-like polymers containing repeating tetranucleotide sequences. J Mol Biol. 1967 Jul 28;27(2):265–272. doi: 10.1016/0022-2836(67)90019-8. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Jacob T. M., Narang S. A., Khorana H. G. Studies on polynucleotides. LXIX. Synthetic deoxyribopolynucleotides as templates for the DNA polymerase of Escherichia coli: DNA-like polymers containing repeating trinucleotide sequences. J Mol Biol. 1967 Jul 28;27(2):237–263. doi: 10.1016/0022-2836(67)90018-6. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yourno J., Ino I., Kono T. A hotspot for spontaneous frameshift mutations in the histidinol dehydrogenase gene of Salmonella typhimurium. J Mol Biol. 1971 Nov 28;62(1):233–240. doi: 10.1016/0022-2836(71)90142-2. [DOI] [PubMed] [Google Scholar]


