Abstract
High-throughput tools for nucleic acid characterization now provide the means to conduct comprehensive analyses of all somatic alterations in the cancer genomes. Both large-scale and focused efforts have identified new targets of translational potential. The deluge of information that emerges from these genome-scale investigations has stimulated a parallel development of new analytical frameworks and tools. The complexity of somatic genomic alterations in cancer genomes also requires the development of robust methods for the interrogation of the function of genes identified by these genomics efforts. Here we provide an overview of the current state of cancer genomics, appraise the current portals and tools for accessing and analyzing cancer genomic data, and discuss emerging approaches to exploring the functions of somatically altered genes in cancer.
Keywords: functional validation, genomic data portal, integrative analysis, large-scale cancer genomics
The development of powerful and scalable methods to analyze nucleic acids has transformed biological inquiry and has the potential to alter the practice of medicine (Lander 1996; Collins et al. 2003). The application of such technologies, together with powerful computational methods in human disease and animal model systems, has facilitated the study of both normal and disease-affected tissues in a manner previously not possible. Indeed, the connection between basic inquiry and potential clinical translation has never been more intimate.
This convergence is particularly evident in cancer, a complex multigenic disease characterized by a diversity of genetic and epigenetic alterations (Vogelstein and Kinzler 1993; Weir et al. 2004; Jones and Baylin 2007; Stratton et al. 2009). Early cancer genome analysis has already led to new targets for cancer therapy and new insights into the relationship of specific genetic mutations and clinical response, as well as new approaches useful for diagnosis and prognosis. These initial efforts have motivated large-scale coordinated cancer genomic efforts to obtain complete catalogs of the genomic alterations in specific cancer types (The Cancer Genome Atlas [TCGA], http://cancergenome.nih.gov; Hudson et al. 2010). Moreover, the current pace of technological advances make it increasingly clear that the ability to perform prospective and comprehensive molecular profiling of tumors will become commonplace and enable genome-informed personalized cancer medicine.
However, the bottlenecks along this path are formidable and numerous. For one, these large-scale genome characterization efforts involve the generation and interpretation of data at an unprecedented scale, which has brought into sharp focus the need for improved information technology infrastructure and new computational tools to render the data suitable for meaningful analyses. Moreover, exploiting this information to develop new therapeutic strategies depends on further biological insights derived from understanding the functional consequences of such genomic alterations. Thus, it is also clear that new approaches that permit the efficient validation of genomic data are required as a first step in distinguishing mutations responsible for disease pathogenesis from other mutations that are the consequence of genomic instability; defining genes involved in cancer initiation, progression, or maintenance; and identifying the optimal ways to exploit this information therapeutically.
In this review, we provide an overview of the current state of cancer genomics, describe the types of data being generated and where they can be accessed, and discuss recent progress in developing tools, models, and methods for analysis of gene functions in cancer, which is the requisite next step in the translation of cancer genome information.
State of cancer genomics
Nearly all cancer genomes contain many nucleotide sequence changes compared with the germline of the cancer patient (Vogelstein and Kinzler 1993; Stratton et al. 2009). These variations include the genomic alterations that cause or promote cancer, often referred to colloquially as “drivers,” as well as alterations present in the cancer genome but without obvious advantage to the cancerous cells when they occurred, referred to as “passengers” (Davies et al. 2005). The major known somatic alterations in the cancer genome include nucleotide substitution mutations and small insertion/deletions (indels), copy number gains and losses, chromosomal rearrangements, and nucleic acids of foreign origin (e.g., oncogenic viruses) (Weir et al. 2004; Chin and Gray 2008; Stratton et al. 2009). In addition, alterations in the epigenetic mechanisms that regulate gene expression occur in most cancers; this subject has been covered elsewhere (Jones and Baylin 2002, 2007) and is not treated in detail here. All of these acquired changes occur in the setting of germline variations of copy number and nucleotide sequence, which may influence the rate of occurrence and/or the effects of somatic genetic alterations (Balmain et al. 2003). Moreover, although somatic mutations occur in tumor cells, it is increasingly clear that the tumor microenvironment mediates important heterotypic signals between the tumor and stromal cells for growth and survival (Hanahan and Weinberg 2000). In this respect, global gene expression encompassing the full transcriptome—including coding messenger RNAs (mRNAs) (Schena et al. 1995) and noncoding microRNAs (miRNAs) (Lu et al. 2005)—of the complex tumor tissue reflects the panoply of somatic and epigenomic alterations, together with the state of cell differentiation of the tumor and the admixture of noncancerous cells. Hence, transcriptional profiling can define a unique gene expression signature for each tumor that may prove useful for classification and prognosis (Golub et al. 1999; Alizadeh et al. 2000; van't Veer et al. 2002).
Evolution of cancer genomics
Over the past decade, technologies for detection of each of these types of alterations have been developed and applied to analyses of the cancer genomes. The initial studies focused on a single technology platform and/or type of genetic alteration. For example, high-resolution copy number profiling has led to the discovery of novel oncogenes in ovarian cancer (Nanjundan et al. 2007), melanoma (Garraway et al. 2005; Kim et al. 2006; Scott et al. 2009), lung carcinoma (Weir et al. 2007; Bass et al. 2009), and colon carcinoma (Firestein et al. 2008), and tumor suppressor genes in leukemias (Mullighan et al. 2007, 2008). Similarly, the application of directed sequencing of specific classes of genes has identified novel genes involved in specific types of cancer (Davies et al. 2002; Lynch et al. 2004; Paez et al. 2004; Pao et al. 2004; Samuels et al. 2004; Stephens et al. 2004; Baxter et al. 2005; James et al. 2005; Kralovics et al. 2005; Levine et al. 2005; Zhao et al. 2005; Pollock et al. 2007; Chen et al. 2008; Dutt et al. 2008; George et al. 2008; Janoueix-Lerosey et al. 2008; Mosse et al. 2008). These observations underscore the contribution of different types of somatic genome alterations in different subsets of cancer, and that comprehensive profiling of the cancer genome will require interrogation of different types of genome alterations in diverse cancer types and subtypes.
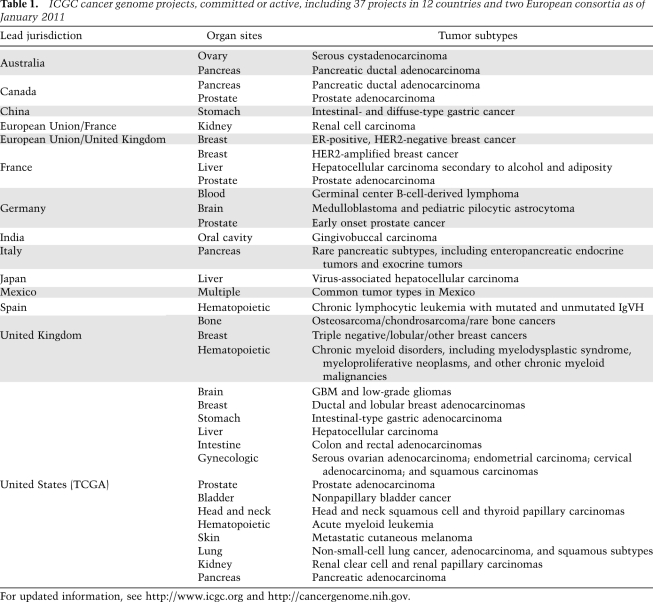
As technologies to perform comprehensive profiling of the cancer genome progressed, different technology platforms from microarrays to capillary sequencing were brought together on unified sample sets (The Cancer Genome Atlas Network 2008; S Jones et al. 2008; Parsons et al. 2008). For example, Velculescu and colleagues (S Jones et al. 2008) integrated sequencing with expression and copy number profiling to identify IDH1 mutations in glioblastomas (GBM). The Cancer Genome Atlas pilot project applied targeted sequencing, copy number, and expression profiling, in addition to epigenetic assessment to a large number of stringently qualified tumor samples to define core pathways of deregulation in GBM (The Cancer Genome Atlas Network 2008) and discover genomic and epigenomic definition of molecular subtypes (Noushmehr et al. 2010; Verhaak et al. 2010). Indeed, global comprehensive analysis with complementary genome annotation tools in statistically powered high-quality sample cohorts is a key aspect of the current consensus standard for large-scale cancer genomics efforts under the International Cancer Genome Consortium (ICGC) (Hudson et al. 2010), now encompassing >20 projects from 14 countries (Table 1).
Table 1.
ICGC cancer genome projects, committed or active, including 37 projects in 12 countries and two European consortia as of January 2011
For updated information, see http://www.icgc.org and http://cancergenome.nih.gov.
Second-generation sequencing technologies
During the past several decades, continuous improvements in genomic technology have led to a series of breakthroughs in our understanding of cancer genetics. The advent of second-generation sequencing technologies and their applications to cancer have already accelerated the pace of genome discovery, as summarized in recent reviews (Shendure and Ji 2008; Meyerson et al. 2010). During the last 5 years, a variety of array-based methods have been developed, including picotiter plate pyrosequencing (Margulies et al. 2005; Wheeler et al. 2008), single-nucleotide fluorescent base extension with reversible terminators (Bentley et al. 2008), and ligation-based sequencing (Shendure et al. 2005; Drmanac et al. 2010). All of these second-generation methods involve the amplification of individual DNA molecules on arrays or beads prior to massively parallel sequence generation.
The throughput limitation and cost of first-generation Sanger-based capillary sequencing technology had, until now, dictated two predominant study designs for cancer genome discovery: one in which large numbers of samples were analyzed but only a small number of genes were interrogated (Greenman et al. 2007; The Cancer Genome Atlas Network 2008; Dalgliesh et al. 2010; Kan et al. 2010), versus a second in which all coding genes were sequenced but in only a handful of discovery samples, followed by targeted sequencing of candidates in an extension cohort comprised of an independent set of samples (Sjoblom et al. 2006; Wood et al. 2007; S Jones et al. 2008; Parsons et al. 2008). Second-generation sequencing technology enables the complete sequencing of entire genomes in a time- and cost-efficient manner. Today, a single sequencing run of an Illumina HiSeq 2000 sequencer can generate ∼200 gigabases of sequence data in 8 d—an output that easily exceeds the annual sequencing production of a genome sequencing center a few years ago (http://www.genome.gov/10001691). This astronomical increase in sequencing capacity, along with the rapid reduction in sequencing cost (which is faster than the doubling of semiconductor/computer capacity every 18 mo, known as Moore's law) (Pettersson et al. 2009), has completely transformed cancer genome discovery science.
Second-generation sequencing offers several advantages over previous technologies. It has the power to identify mutations in highly admixed samples by virtue of deep coverage (Thomas et al. 2006), overcoming a major limitation of Sanger-based capillary sequencing technology. Whereas previous technologies could query one modality of cancer genome alteration (mutation, copy number, or expression) at a time, second-generation sequencing analyses permit the identification of all such alterations simultaneously. For example, one can obtain high-resolution and accurate measurements of somatic copy number alterations (SCNAs) from whole-genome sequencing (Campbell et al. 2008; Chiang et al. 2009), and the same data can identify nucleotide substitutions. Furthermore, second-generation sequencing offers structural information never before available from other genomic platforms, thus enabling for the first time global assessment of chromosomal rearrangements in cancer (Campbell et al. 2008; Mardis et al. 2009; Stephens et al. 2009; Ding et al. 2010a; Pleasance et al. 2010a,b). Similar approaches can be applied to cDNA, also known as RNA-seq, which permits accurate digital measurements of gene expression across the whole transcriptome. Importantly, this latter approach provides the means to measure known, and discover novel, splice variants as well as aberrant transcripts generated by somatic structural genome rearrangements (Maher et al. 2009a,b; Berger et al. 2010; Palanisamy et al. 2010). This type of data will undoubtedly reveal new insights into the regulation of gene transcription and RNA processing.
In the near future, sequencing-based approaches will be applied to nearly all aspects of cancer genome characterization. For example, current TCGA projects involve the comprehensive sequencing of all protein-coding genes and transcripts by hybrid capture/whole-exome sequencing in hundreds of tumor- and germline-matched pairs, complemented by deep sequencing of the whole genomes (WGS) (at >30-fold coverage) in 10% of the samples. Given the rate at which sequencing capacity is increasing and cost is shrinking, it is highly likely that deep-coverage WGS will soon be applied to the majority of the discovery samples. In parallel, efforts to use low-coverage sequencing to conduct structural and copy number analyses are likely to replace array-based technologies in the near future. Similar study designs are being adopted by ICGC projects.
Accessing cancer genomics data
Although the data generated from these large-scale cancer genome characterization efforts have been and will continue to be made publicly available, accessing and using these cancer genome data remains a major challenge. In this section, we attempt to provide a framework for nongenomic noncomputational cancer biologists to become familiar with what data are available and where to download or query each of the major genomic data types. Specifically, we describe briefly the technology platform(s) used to generate each data type, followed by common public sites where cancer genome data can be downloaded and summarized results can be queried. We also point out basic open source analytical tools or computational algorithms for manipulation and analysis of cancer genome data. However, it should be noted that the tools described here are illustrative examples, rather than a complete survey of sites, data sources, or analytical tools, as these are rapidly evolving.
Data structure and data access policies
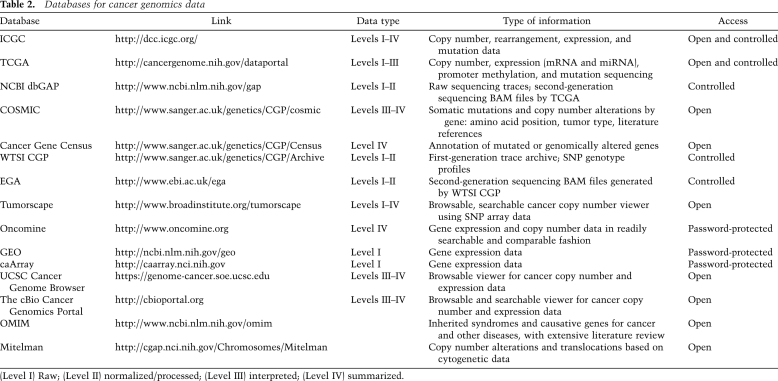
Generally speaking, cancer genomic data can be divided into (1) raw, (2) processed or normalized, (3) interpreted, and (4) summarized categories based on the degree of computational modification and integration applied to the data. These categories are sometimes referred to as Level I–Level IV data. Raw, processed, and interpreted (Level I–III) data apply to individual samples, while summarized (Level IV) data refer to analyses across sample sets. For example, normalized or processed data represent data that have been assigned to a genome reference, such as alignment of sequences to reference genome or mapping of probes to chromosomal positions. For microarray-based platforms, normalization refers to combining multiple probes measuring a single genomic locus to a single value and transforming the measured intensities such that the values can be compared between experiments; examples of normalization steps include correction for background noise and total brightness. Interpreted data represent meaningful biological results extracted from each specimen, such as genome-wide copy number profile, where copy number breakpoints have been statistically defined, or gene expression profiles, where individual gene expression levels have been collated from multiple loci across the gene. Summarized (Level IV) data represent analysis of interpreted data across a cohort of samples, where statistical methodologies can be applied to define significant events or molecular subtypes. This category of analyzed data is often presented as the findings of a genomic study in a publication. Major sites where these data sets can be accessed are listed in Table 2 and are described in some detail below.
Table 2.
Databases for cancer genomics data
(Level I) Raw; (Level II) normalized/processed; (Level III) interpreted; (Level IV) summarized.
Although cancer genomic data from various large-scale projects—including the Cancer Genome Project (CGP) at Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/genetics/CGP), TCGA (http://cancergenome.nih.gov/dataportal), and ICGC (http://dcc.icgc.org)—are publically available, for protection of patient privacy, access is either open or controlled. Prior to second-generation sequencing, most raw data and some type of normalized data (e.g., single nucleotide polymorphism [SNP] profiles) are subjected to controlled-access restriction, while interpreted and summarized data are openly accessible. With the transition to second-generation sequencing data, it is likely that raw and processed data, and possibly some interpreted data, will fall under the “controlled-access” category, since the level of resolution may provide the means to identify specific patients. Controlled-access data are restricted to qualified researchers (with certification by host institution) with a specific proposal of data use that is deemed compliant with the project's data access policy, typically requiring preapproval by the institutional review board of the requesting investigator. For TCGA, access to controlled data is obtained through dbGAP (http://www.ncbi.nlm.nih.gov/gap); for ICGC projects, access is obtained through its Data Access Compliance Office (http://www.icgc.org/daco). In the case of the Sanger Institute's Cancer Genome Project, genotyping and first-generation sequencing traces can be requested at its data archive (http://www.sanger.ac.uk/genetics/CGP/Archive), and its second-generation sequencing data must be obtained through the European Genome-Phenome Archive (EGA, http://www.ebi.ac.uk/ega). At present, downloading raw data from these sources is a technically and logistically challenging task that requires significant network infrastructure to handle the size of the data files.
Nucleotide sequence mutations
Nucleotide substitutions and small insertions/deletions are common mechanisms for activating oncogenes and inactivating tumor suppressor genes. The initial development of methods to determine the nucleotide sequence of DNA in 1975 (Sanger and Coulson 1975) led to the discovery of cancer-specific somatic mutations in the RAS gene family in the early 1980s (Parada et al. 1982; Shimizu et al. 1983; Santos et al. 1984; Bos et al. 1985) and, later, mutations in human tumor suppressor genes (Friend et al. 1986; Hahn et al. 1996). Subsequently, the invention of automated sequencing instruments (Hunkapiller et al. 1991) led to the initial sequencing of the human genome (Lander et al. 2001; Venter et al. 2001), and then to systematic efforts to sequence gene families (Davies et al. 2005; Stephens et al. 2005; Greenman et al. 2007). These latter efforts identified several new oncogene mutations that are targets for cancer therapy—most notably the BRAF and EGFR protein kinase genes and the PIK3CA phosphatidylinositol kinase gene (Davies et al. 2002; Lynch et al. 2004; Paez et al. 2004; Pao et al. 2004; Samuels et al. 2004)—leading to approved and in-development targeted therapeutics for cancers.
For sequencing data, the normalized data category represents sequencing reads that have been aligned to a specific version of the human reference genome. As the reference genome is refined and filled in with each new version, mapping data may change; thus, researchers should pay attention to and always note the specific reference genome build used in an analysis. Raw sequencing reads from both Sanger-based capillary sequencing and second-generation platforms are stored at NCBI Sequence Read Archive and dbGAP (see Table 2). Access to these sequencing data is restricted and requires data use approval by the appropriate data access committee. The interpreted category for mutation data includes the sequence variant calls, the types of sequence variants (such as synonymous versus nonsynonymous or missense versus indel), and the location of the nucleotide change in relation to annotated gene structure; e.g., intron versus exon and consequent amino acid changes. One aspect of this analysis includes an annotation of whether such sequence variants are reported in dbSNP, a database representing likely common SNPs. If not found in dbSNP and not observed in matched germline-derived sequences, such variants are generally considered somatic in nature. Putative variants should be verified (e.g., result reproduced in an independent assay using the same technology platform) or validated (e.g., result observed by an orthogonal method) by methods including genotyping. For TCGA projects, all verified, validated, or putative somatic mutations and associated descriptions discovered in a sample or a cohort of samples can be found in the .MAF file, which is available on the TCGA data portal (http://cancergenome.nih.gov/dataportal). Similar data files can be found at the ICGC Data Coordination Center (http://dcc.icgc.org) under the Download Data page. New file formats will likely emerge in the near future to support the increasing applications of next-generation sequencing platforms for data generation.
A key downstream (Level IV) analysis of verified or validated mutations is the determination of significance, accounting for the background mutation rate and size as well as composition of a gene. Several methodologies have been developed for this purpose (Getz et al. 2007; Greenman et al. 2007). For example, in the MutSig algorithm, a P-value is calculated for each gene, testing the hypothesis that all of the observed mutations in that gene are a consequence of random background mutation processes, taking into account the list of bases that are successfully interrogated by sequencing (i.e., “covered”) and the list of observed somatic mutations, as well as the length and composition of the gene in addition to the background mutation rates in different sequence contexts. As in analyses of other genomic data, such calculations must then be corrected for multiple hypothesis testing (see below). Using these types of significance analyses, the majority of the somatic mutations found in cancer genomes is likely to represent passenger events and only a minority is likely drivers. For example, of the 453 validated nonsilent mutations in GBM scattered across 223 genes, only eight genes were considered having higher than background mutation frequency, suggestive of positive selection pressure (The Cancer Genome Atlas Network 2008). However, it is worth noting that, as for any statistical test, the lack of statistical significance by MutSig or similar analyses does not preclude true cancer relevance, as, relatively speaking, the number of samples having been adequately sequenced is still low. Moreover, computational algorithms for mutation calling and statistical frameworks for significance calculation are still being developed and refined.
Beyond statistical analyses, there are various theoretical and computational models designed to predict the likely functional consequences of specific nucleotide mutations, particularly for mutations in coding genes. These models are often based on the impact of specific amino acid substitutions on protein structure or known functional domains or evolutionarily conserved regions. For example, the PolyPhen (for polymorphism phenotyping) tool predicts the possible impact of an amino acid substitution on the structure and function of a protein using a variety of structural and chemical parameters in addition to evolutionary conservation (Sunyaev et al. 2001; Ramensky et al. 2002). Indeed, a recurring theme in analysis of genomic data is the leverage of evolutionary information. MutationAssessor (Reva et al. 2007) is a recently published algorithm for predicting potential functional impact of a sequence mutation based heavily on the assumption that if a highly conserved residue is changed to a different residue type, the change is presumed to have high functional impact on the function of the affected protein. By analyzing aligned sequence families of paralogous and orthologous proteins within the human genome and across many other species, this algorithm calculates the functional impact (FI) score for a mutation. Both of these tools are Web accessible (http://genetics.bwh.harvard.edu/pph; http://mutationassessor.org) and offer an intuitive and easy-to-use query interface as well as a batch processing feature. With MutationAssessor, in addition to the calculated FI score for each variance, users can inspect the placement of mutations in a multiple sequence alignment relative to amino acid residues that are conserved globally or in a specific subfamily, as well as observe the consequences of the residue change in an interactive three-dimensional protein structure. Other examples of prediction algorithms include SIFT, CanPredict, and CHASM (Ng and Henikoff 2001; Kaminker et al. 2007; Carter et al. 2009; Adzhubei et al. 2010). In the end, beyond statistical analysis or the prediction of functional impact, the relevance of any mutational event to human cancer will require functional validation (see below).
The COSMIC (Catalog of Somatic Mutations in Cancer) site is the single most comprehensive source of curated analyzed somatic mutation data in cancers developed and maintained by the Cancer Genome Project at the Wellcome Trust Sanger Institute (Futreal et al. 2004; Forbes et al. 2008). COSMIC is an open source, easily accessible, and searchable database containing >140,000 somatic mutations of 18,000+ genes in >550,000 tumor samples curated from 2.8 million experiments (COSMIC version 49, http://www.sanger.ac.uk/genetics/CGP/cosmic; Forbes et al. 2011). While it clearly provides a valuable source of collated mutation data in human cancers, users should also be cognizant of the fact that there is likely curational bias, as in any database of this type.
For germline cancer-associated mutations, no similar effort exists to collect such mutations, but the Online Mendelian Inheritance in Man (OMIM, http://www.ncbi.nlm.nih.gov/omim; McCusick 1998) provides an excellent literature summary regarding each major familial cancer gene and susceptibility loci.
SCNAs and structural rearrangements
Cancer genomes are highly disordered compared with normal genomes, with extensive changes in chromosome structure and copy number. The SCNAs found in cancer include whole-chromosome or regional alterations spanning part to whole arms of a chromosomes, as well as focal events involving one or a few genes. The development of array-based comparative genomic hybridization using bacterial artificial chromosomes (Hodgson et al. 2001; Cai et al. 2002), cDNA (Pollack et al. 1999), oligomers (O'Hagan et al. 2003; Brennan et al. 2004), and SNP arrays (Lindblad-Toh et al. 2000; Mei et al. 2000; Bignell et al. 2004; Zhao et al. 2004) has enabled systematic analyses of the cancer genome and defined many new recurrent SCNAs in cancer (Beroukhim et al. 2010). To date, the major CNAs linked to cancer are somatic changes, although many germline copy number variations are found in human populations (Sebat et al. 2004; Redon et al. 2006). Further work is needed to determine the role of germline copy number aberrations in cancer.
Most of the available genome-wide high-resolution copy number profiling data were generated on either Agilent or Affymetrix microarray platforms. Performance of these various platforms has been compared (Brennan et al. 2004; Lai et al. 2005, 2008; Willenbrock and Fridlyand 2005) and used comparatively on the same sample cohort by TCGA during its pilot phase (The Cancer Genome Atlas Network 2008). Raw data for copy number are probe-level signals, whereas processed data are the results of normalization, calculation of tumor-to-normal copy number ratio, and mapping to chromosomal positions. Interpreted data generally contain segmented copy number profiles where breakpoints along the chromosomes have been defined in each individual tumor using segmentation methods such as circular binary segmentation (CBS) (Venkatraman and Olshen 2007),GLAD (Hupe et al. 2004), and others (Picard et al. 2005; Wang et al. 2005; Day et al. 2007; Ben-Yaacov and Eldar 2008). Recent methods for measuring and determining segments of absolute allele-specific copy number from SNP arrays provide a more accurate description of allelic gains and losses and loss of heterozygosity in cancer genomes (LaFramboise et al. 2005; Bengtsson et al. 2010; Greenman et al. 2010; Van Loo et al. 2010).
When a cohort of segmented copy number profiles is analyzed together, several methodological tools are available to define the Level IV summarized data, where significantly altered regions are defined (by specifying the boundaries of “peaks” and significance). GISTIC is a popular algorithm based on statistical considerations (Beroukhim et al. 2007), allowing users to define the most significant regions and peaks; GISTIC works most robustly in large sample cohorts. RAE uses a similar methodology (Taylor et al. 2008). GTS (Wiedemeyer et al. 2008) provides independent measures of recurrence frequency and focality; the latter can be a strong indicator for relevance when sample size is small. For those with basic R programming skills, cghMCR and CNTools are two Bioconductor packages (http://www.r-project.org; Gentleman et al. 2004) available for copy number data analyses. The former provides a fast and platform-independent approach to identifying and visualizing altered regions, while the latter enables the conversion of segmented copy number profiles into a matrix structure to allow further downstream analyses. It should be noted that most of these algorithms are built using the prevalence and shape (e.g., focal and high amplitude vs. flat and broad) of a numerical copy number aberration to discriminate likely target(s) at the peaks from passengers. Although this framework has led to identification of new cancer genes that have been experimentally validated, next-generation sequencing is beginning to provide information on sequence-level structures underlying these numerical copy number aberrations. These emerging data will provide much finer details of structural rearrangements in addition to simple numerical changes such as duplication or amplification or deletion. Such new insights will likely lead to different or improved algorithms to identify candidate targets of these genomic alterations.
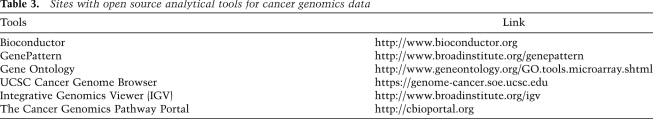
The major repositories for somatic copy number data in cancer include Gene Expression Omnibus (GEO) (Edgar et al. 2002; Barrett et al. 2009), Tumorscape (Beroukhim et al. 2010), TCGA (http://cancergenome.nih.gov), COSMIC (Forbes et al. 2008), and Oncomine (Rhodes et al. 2004, 2007) (Table 3). The GEO site contains predominantly raw data, while COSMIC and Oncomine emphasize normalized and interpreted data. All three categories of data are available from the TCGA DCC as well as at Tumorscape. For systematic queries of analyzed summary data on CNAs in human cancers, Tumorscape is particularly useful, as it provides segmented data for >3000 high-resolution copy number profiles from SNP arrays in a format that can be visualized with the interactive Integrative Genome Viewer (http://www.broadinstitute.org/igv). COSMIC also contains copy number data annotated on the gene level in addition to raw data download at the CGP archive, while Oncomine has recently begun to curate copy number profile data sets in addition to transcriptome data sets.
Table 3.
Sites with open source analytical tools for cancer genomics data
Before the advent of second-generation sequencing (Maher et al. 2009a,b; Berger et al. 2010; Palanisamy et al. 2010), conventional cytogenetic methodologies such as FISH were the primary means for identification of chromosomal translocations in cancers, predominantly in leukemia and lymphoma (Rabbitts 1994). One of the most useful compilations of cytogenetic alterations was established by Mitelman et al. (2007), which includes both translocations and CNAs in >50,000 samples. The recent identification of translocations in epithelial cancers suggests that the application of high-throughput sequencing will expand the number of such translocations (Tomlins et al. 2005), although the number and significance of such recurrent translocations in solid tumors remains to be delineated.
Expression analysis of cancer
Global gene expression profiles offer a global view to the transcriptome of a tumor, the signature of which has the potential to provide diagnostic (Golub et al. 1999; Alizadeh et al. 2000) or prognostic (van de Vijver et al. 2002; Paik et al. 2004) information. This approach has also been used to molecularly subclassify tumors that are currently assigned to one histopathological class. Gene expression is also one measure of functional consequence of a genomic alteration, thus potentially enabling the interpretation of inactivating genetic changes on the DNA level or epigenomic (methylation) on DNA promoters. Interpreted expression data typically represent gene-level data where multiple probes reporting on one annotated gene are collapsed (by taking either the median or mean values or best probe).
A major summarized category for expression data type is definition of molecular subtypes. The discovery of novel molecular subclasses is based primarily on differences in gene expression between groups within a cohort using various unsupervised classification methodologies, such as hierarchical clustering (Eisen et al. 1998), self-organizing maps (Tamayo et al. 1999), and nonnegative matrix factorization (Kim and Tidor 2003; Brunet et al. 2004). Visual verification or identification of clusters requires transforming the high-dimensional expression data (each sample is represented by the expression values of all genes) to two or three dimensions. This is often performed using principal component analysis (PCA) (Raychaudhuri et al. 2000) or multidimensional scaling (MDS) (Khan et al. 1998). However, it should be noted that, given the large number of available gene expression profiles and the number of independent measurements contained within each profile, one can produce class discriminations that may or may not reflect underlying biological differences. For example, batch effects, introduced by profiling samples on different days using different lots of reagents or at different sites, can introduce variations and confound such analyses. These considerations require reproducing the classification results in independent test cohorts of samples and using multiple hypothesis testing correction methods.
Once subtypes are defined, there are several user-friendly analytical algorithms that can be employed to interrogate these data sets. For example, one of the most commonly asked questions with gene expression data is: What gene expression difference exists between two biologically, clinically, or molecularly defined subgroups? Significance analysis of microarrays (SAM, http://www-stat.stanford.edu/∼tibs/SAM; Tusher et al. 2001) is a commonly used tool to discern genes that are significantly different between two groups of samples. In GenePattern, ComparativeMarkerSelection (Gould et al. 2006) is another tool that can be used to define genes that are characteristic of a subgroup. Finally, one of the most commonly used sites for querying analyzed cancer gene expression data is Oncomine, which allows comparison of any two data sets from different cancers or normal tissues to determine the genes that are specifically expressed in the data set of interest (Rhodes et al. 2004, 2007).
Once differential expression gene lists or signatures for each subtype are generated, these lists can be interrogated for pathway activation using knowledge-based pathway analysis tools such as Ontologizer (Bauer et al. 2008), which looks for statistical enrichment of Gene Ontology terms, or Gorilla, a tool to visualize Gene Ontology terms that are enriched in gene lists ranked by a user-defined criterion (Eden et al. 2009). Gene set enrichment analysis (GSEA) is an algorithm that allows one to assess whether the differentially expressed genes are enriched for particular gene sets even though each member of the gene set individually is not necessarily strongly differentially expressed. Gene sets can be defined in various ways, such as manual curation of pathways, based on genomic position or shared motifs, expression correlation, or by experimentally identifying signatures that represent a molecular event or phenotype (Subramanian et al. 2005), such as KRAS activation (Sweet-Cordero et al. 2005). Gene signatures that can be used as input for GSEA can be downloaded from the Molecular Signatures Database (MSigDB, http://www.broadinstitute.org/gsea/msigdb/index.jsp). Other gene set and pathway repositories include Pathway Commons (http://www.pathwaycommons.org), KEGG (Kanehisa et al. 2006), and others. Another commonly used tool for pathway and Gene Ontology enrichment is DAVID (http://david.abcc.ncifcrf.gov/home.jsp; Huang et al. 2009). These types of analyses provide a framework for generating hypotheses for further testing. However, users should be reminded that annotation of pathways or definition of gene sets is biased by what is known and published. Genes that are widely studied will be linked to many different processes, while ones that have not been explored in depth would have few connections to others.
Second-generation sequencing data
It is clear that, in the very near future, all genome characterization data will be generated by second-generation sequencing technologies. Output from these sequencing platforms is in the form of short-sequence reads, ranging from 35 base pairs (bp) to 100 bp or longer. These raw sequencing reads and their mapped reads after alignments (to reference genome) are captured in a BAM file. Therefore, one can consider the BAM file as containing both raw and normalized (Level I–II) data; hence, BAM files are controlled access data. For TCGA, these data are stored at the Sequence Read Archive of NCBI (http://www.ncbi.nlm.nih.gov/Traces/sra) and accessed through dbGAP. Similar second-generation sequencing data from the Sanger CGP are accessed through EGA (http://www.ebi.ac.uk/ega). The size of these data files (typically in tens to hundreds of gigabytes) and the complexity of manipulating such data make it challenging to access and analyze these data by noncomputational groups. A detailed discussion of the computational challenges, algorithms, and software for analyzing second-generation sequencing for cancer is provided in recent reviews (Ding et al. 2010b; Meyerson et al. 2010).
Viewers and tools
Some of the open source software tools and viewers are summarized in Table 3. Several user-friendly Web-based viewers provide intuitive environments with which to visualize and explore cancer genome data, although most do not allow fully interactive queries at the present time. The University of California at Santa Cruz (UCSC) Cancer Genome Browser (Zhu et al. 2009) is perhaps the most commonly accessed site, which allows users to view and query a variety of cancer data types in the context of the widely used UCSC Genome Browser (https://genome-cancer.soe.ucsc.edu). The Cancer Genomics Pathway Portal (http://www.cbioportal.org), hosted at Memorial Sloan-Kettering Cancer Center (MSKCC), is a recently launched site aiming to provide direct visualization and queries of summarized results by cancer biologists with little to no bioinformatic expertise as well as download of large-scale cancer genomics data sets for bioinformatic power users. For example, its major feature, Oncoprints, provides an easy way to visualize distinct genomic alterations (e.g., somatic mutations, CNAs, and mRNA expression changes) of a gene or genes of interest across a set of tumor samples. Another tool is the Integrative Genome Viewer (IGV, http://www.broadinstitute.org/igv), a scalable and readily browsable interface for accessing any type of cancer genome data, including mutations, expression, and copy number, particularly suitable for second-generation sequencing data. IGV also allows browsing clinical annotations alongside the genomic data. In addition to local data, a user can load data from a server that contains many data sets, including open access TCGA data. IGV can be downloaded and launched from a desktop computer. Similarly, for viewing and manipulating cancer copy number data and gene expression data to generate heat map figures, the dChip software system (Li and Wong 2001) can be installed on a standard desktop or laptop computer.
To perform customized analyses beyond querying summarized results, many open source analytical tools requiring only basic programming and bioinformatics expertise are now available. For example, Bioconductor (Gentleman et al. 2004) is an open source site with many useful analytical packages, written in the R language (http://www.r-project.org), for the analysis and interpretation of cancer genomic data, including second-generation sequencing data. Another site is GenePattern (Reich et al. 2006) (http://www.broadinstitute.org/cancer/software/genepattern), which is a user-friendly Web-based interface for >125 different genomics analysis tools and pipelines for various types of data, including gene expression, copy number data, proteomic data, and others. GenePattern can also keep track of parameters and versions of tools, and thus achieves an important goal of enabling reproducible research. The Gene Ontology site (http://www.geneontology.org/GO.tools.microarray.shtml) also maintains a collection of open source analytical tools contributed by its consortial members for analyses of microarray-based expression data.
Computational considerations in cancer genome analysis
In addition to specific challenges inherent in analysis of each type of cancer genome data, several general considerations should be kept in mind when one analyzes, interprets, and uses cancer genomics data. These include (1) quality control (QC) of data, (2) the accurate estimation of signal and noise in large data sets, (3) reproducible approaches to complex genomic analyses, and (4) achieving sufficient power in the face of multiple hypothesis testing. We briefly touch on each of these issues, but recommend several books for a broader introduction to bioinformatics; these include ones that are focused on principles and applications (Xiong 2006; Pevsner 2009), as well as on computational methodologies (Jones and Pevzner 2004).
Before analyzing genomic data, either publicly available or locally produced, and generating hypotheses for further experimental follow-up, one has to ensure the data are of sufficient quality. Two key aspects contributing to raw data quality are biospecimen and technical execution. Criteria used for biospecimen inclusion and exclusion can influence data quality; for example, a tumor specimen with a high proportion of stromal contamination will reduce one's ability to detect somatic alterations in the tumor cells. On the data generation front, standardization of technical methods (i.e., standard operating procedures [SOPs]) and execution by highly trained individuals can minimize experimental variation and ensure reproducibility and high data quality. Once data are generated, the normalization step (Level II) attempts to remove any experimental artifacts that can negatively impact the data quality; however, some level of batch effects often remains and, if not controlled, may lead to spurious findings. A simple approach for detecting batch effects and estimating their extent is to use standard supervised methods to search for differences between batches; e.g., use SAM, ComparativeMarkerSelection, or ANOVA to look for genes that are differentially expressed between batches.
Another potential problem with large genomic data sets beyond data generation is sample mismatching; hence, it is recommended that one double-checks that the data originated from the intended sample. This is particularly important when analyzing multiple data types, since a mix-up could happen in some but not other data types. A useful approach to screening for sample mismatching is to leverage our understanding of the data types. For example, one can (1) correlate copy number and expression data, expecting overall positive correlation; (2) correlate expression and promoter methylation data with the expectation of finding a negative correlation; and (3) detect a drop in expression levels of specific genes in samples harboring truncating mutations (nonsense and frameshift insertions or deletions). With second-generation sequencing for somatic alterations, a common QC check is to compare the SNP genotype profiles between array-based and sequencing data to ensure that there is a match between tumor and germline DNAs, as a mismatch will result in erroneous calls of somatic mutations.
Like any other type of experimental data acquisition and analysis, maximizing signal and minimizing noise is essential in cancer genomics. In particular, using noise filters to remove poor-quality samples helps to achieve the most accurate analysis (Kauffmann and Huber 2010). Platform-specific noise measures can be used to optimize data quality for each type of genomic measurement. With such measures, the analysis of independent genomic data sets has proven to be reproducible across multiple independent laboratories, for example, for microarray-based gene expression analysis of human cancers (Dobbin et al. 2005).
Another key element in any computational analysis is reproducibility, a concept that is familiar to experimentalists, but the framework to ensure reproducibility in computational analysis is still in development. Naturally, the reproducibility standards expected from in silico experiments are beyond what is possible from bench experiments, since the initial raw data are available and computational tools (at least deterministic ones) should always reproduce the same values for all measurements. For example, if 200 genes are reported to be differentially expressed between two tumor subtypes, one would expect to obtain the exact same list of 200 genes when reproducing the analysis using the same tool, parameters, and input data. To aid in reproducibility, one must have a clear record of all inputs, parameters, and data manipulations for tracking and debugging. Such information will permit one to identify, for example, an input error that results in mislabeling of subsequent samples. Therefore, some of the key elements in reproducible bioinformatics include associating each analysis with a freeze of not just the data, but also the analytic software (codes) and parameters (Mesirov 2010). For example, recording the specific reference genome build used for a particular analysis is critical to one's ability to reproduce a result using the same input data. Increasing use of tools that have automated version tracking capability will greatly enhance reproducibility.
Unlike traditional molecular biology experiments, where only relatively few measurements are generally made (<10) in any single experiment, the number of individual measurements in a cancer genomics study is in the thousands to millions. This scale requires not only a large number of samples to achieve statistical power, but also addressing the issue of multiple hypothesis testing. Statistical power is a concept discussed in general publications on biostatistics, but it is also specifically illustrated in the case of whole-exome sequencing (Getz et al. 2007; Hudson et al. 2010), where it was calculated that 500 tumor samples are needed to detect, with ∼80% power, genes that are mutated in 3% of patients (assuming a typical background mutation rate). In other words, a small discovery cohort will have very little power to detect infrequently mutated genes. Therefore, a lack of somatic mutation in a gene of interest in a study of a small number of samples should be considered noninformative, rather than interpreted as the gene is not mutated in cancers.
The concept of correcting for multiple hypothesis testing is very important in rigorous data interpretation of cancer genomics data, as it addresses the issue of false discovery. When thousands to millions of measurements are queried in each sample, significant differences will be observed, but the likelihood that such observed differences would occur by chance is extremely high. For example, when searching for differentially expressed genes by comparing two transcriptomes of 20,000 coding genes, a typical P-value cutoff of 0.05 will mean that, regardless of biological relevance, 5% of 20,000 genes (1000 genes) will be identified as differentially expressed by chance alone. To compensate for this, a q-value or false discovery rate (FDR) (Benjamini and Hochberg 1995) is calculated to bound the expected fraction of false discoveries in the results. In the above example of comparing two transcriptomes, if a set of genes in the differentially expressed list has a calculated FDR value of 0.2 (a commonly used cutoff) or less, it implies that the expected fraction of false discoveries among the list of differentially expressed genes is, at most, 20%. The FDR approach has become widely used in cancer genome analyses, as controlling the FDR is a more liberal approach than the standard Bonferroni correction, which bounds the chance of having even a single false discovery.
In summary, when using publically available cancer genomic data, one should pay attention to the design of the study that generated the data; when expertise is available, it is recommended that one downloads the raw data to repeat normalization and other QC checks prior to analyses. Analysts performing analyses, whether using off-the-shelf tools or developing new algorithms, must be mindful of reproducibility. With respect to interpretation of results, one must keep in mind the statistical power of a particular study and correct for multiple hypothesis testing. Finally, the most stringent test to assess the significance of these conclusions is the ability to reproduce the findings in independent data sets.
Integrative analyses of the cancer genomes
Cancer genomic data remains incomplete due to limitations of the current experimental methods as well as the inherent complexity of its biology. One method to identify genes of particular interest involves integrating the outputs from different types of experiments. For example, finding that a gene is targeted for genomic deletion, inactivating mutations, promoter hypermethylation, alterations of miRNA expression, and/or transcriptional down-regulation in different tumor samples would collectively suggest that this gene is a candidate tumor suppressor gene, even if each type of genomic alteration may be infrequent.
By extension, one can interrogate several types of data derived from the same set of tumors for evidence of dysregulation in a functional complex or pathway based on known constituents. The recent work in clear cell renal cancer is an excellent example of integrative analysis of a functional complex (Dalgliesh et al. 2010). Here, the finding of low-frequency (3%) mutations in several enzyme-coding genes (SETD2, UTX, and JARID1C), all implicated in modifying regulatory lysine residues on histone H3, was interpreted collectively as evidence pinpointing the deregulation of H3 histone modification as a likely cancer-promoting process in a significant proportion of clear cell RCC.
An example of pathway integration is the analysis of GBM by TCGA. In this study, hundreds of clinically annotated GBM with matched blood normal (white blood cells) as a reference were characterized for (1) somatic mutation in 600 known or candidate cancer genes, (2) global SCNA patterns, (3) mRNA and miRNA expression, and (4) DNA promoter methylation status. By analyzing mutations and SCNAs in the same samples, TCGA showed that nearly all GBM harbored alterations in some components of the receptor tyrosine kinase/PI3K/RAS, p53, and RB/cell cycle pathways (The Cancer Genome Atlas Network 2008), providing definitive genomic evidence of the deregulation of these core pathways as obligate events for this cancer. In addition, by integrating sequencing and copy number data with expression profile analyses, TCGA linked major molecular subtypes of GBM defined by transcriptional profiles to specific genotypes (Verhaak et al. 2010). In addition, analysis of DNA promoter methylation defined a further subclass of GBM exhibiting a CpG island Methylator Phenotype linked to IDH1 mutation and better survival (Noushmehr et al. 2010).
Cross-species comparative oncogenomics
The structural complexity of human cancer genomes has motivated efforts to leverage different data sets involving complementary information to identify somatically altered genes that likely contribute to tumorigenesis. In addition to integrating information derived from the same samples, the comparison of genetic alterations found in murine cancer models with those found in human cancers provides another method to identify cancer drivers. The rationale for the use of model organisms rests on the view that truly important driver genes and their linked mechanisms will be evolutionarily conserved, while bystander events not linked to biological processes are less likely to be shared in these cross-species comparisons.
Several studies have successfully leveraged the mouse cancer genomics along with human cancer genome data to identify novel cancer genes (Kim et al. 2006; Zender et al. 2006). For example, Zender et al. (2006) compared regions that were amplified in both murine and human liver cancer genomes and found that cIAP1 and Yap are coamplified. Similarly, Kim et al. (2006) investigated a focal amplification in a murine model of melanoma that acquired new metastatic capability and found NEDD9 as a metastasis gene that is the target of the large 6p23 regional gain observed in ∼30% of human metastatic melanomas. In a more recent example using second-generation sequencing technology, a mouse mammary tumor was found to carry an internal deletion of exons of the Lrp1b gene, an event similar to internal deletions found in the corresponding human ortholog in ∼4% of human cancer cell lines (Varela et al. 2010). Unlike what is usually observed in human cancer genomes, most mouse cancer genomes in genetically engineered mouse (GEM) strains harbor relatively few structural and copy number aberrations. Although this species difference limits the number of events available for comparison across the species, it also means that the presence of a focal amplification or deletion in a mouse cancer genome reflects strong selective pressure, thus providing strong evolutionary evidence that implicates the syntenic event in humans as a likely driver event. Indeed, when the mouse genome is engineered to experience telomere dysfunction and consequent genome instability like most human cells, the resultant tumors acquired a human-like genome harboring complex rearrangement and alterations that are syntenic to loci altered in human cancers (Maser et al. 2007). These examples provide a rationale for cross-species comparative oncogenomics as an efficient strategy for the annotation of human cancer genes.
A complementary way to use mouse cancer models to identify cancer genes involves the use of forward genetic screens. For example, Berns and colleagues (Uren et al. 2008) have created cohorts of mice containing retroviral insertions for identification of genes that cooperate in a specific genetic background to drive tumorigenesis. In the setting of Ink4a/Arf or p53 deletion, they have identified many candidate oncogenes and tumor suppressor genes that corresponded to known human cancer-associated mutations. Similarly, Copeland and Jenkins (2010) have deployed the Sleeping Beauty transposon system to generate both activation and inactivation of genes in the murine genome, and have used this system to discover genes involved in colorectal and other cancers (Dupuy et al. 2009; Starr et al. 2009). In another recent study, genome-wide copy number profiles of hundreds of human cancer cell lines were compared with hundreds of common insertion sites isolated from >1000 mouse tumors induced with the murine leukemia virus (MuLV), showing a significant enrichment of human ortholog genes with known mutations in COSMIC and overrepresentation of human orthologs that are annotated as oncogenes in the Cancer Gene Census (http://www.sanger.ac.uk/genetics/CGP/Census; Futreal et al. 2004). Taken together, these efforts thus far have demonstrated the power of cross-species comparisons as a powerful approach to identify cancer genes, and argue for systematic and comprehensive genomic characterization of appropriately engineered mouse models of human cancers.
Converting genomic data into biological knowledge
The current large-scale cancer genomics efforts will identify and enumerate the frequency of every genetic element of interest that is structurally altered in the cancer genome, including those impacting annotated genes, noncoding miRNA, or other conserved elements. While statistical significance based on frequency will be an important filter to cull bystanders from likely driver events, it is likely that mutations that occur at a lower frequency may also contribute to specific types of cancer. For example, mutations or translocations involving ALK occur in 4%–5% of non-small-cell lung cancers (NSCLCs) (Soda et al. 2007), but confer sensitivity to small-molecule ALK inhibitors (McDermott et al. 2008). In such cases, additional experimental evidence is necessary to identify such low-frequency events as contributors to cancer initiation or progression (Chin and Gray 2008). Indeed, since it is likely that multiple genetic alterations are required to program the behavior of any specific cancer, functional analyses will be required to complement genome annotation. For example, the recent demonstration that the mutation status of KRAS dictates the response of tumors that harbor mutant EGFR to treatment with EGFR inhibitors and the complex interplay between BRAF, CRAF, and RAS in response to selective BRAF inhibitor (Heidorn et al. 2010; Joseph et al. 2010; Poulikakos et al. 2010; Whittaker et al. 2010) confirm that further functional studies will be required to exploit knowledge of somatic mutations. In addition, genes that are not mutated in cancers may also contribute to the survival of cancers harboring other genetic alterations, such as the PARP1 gene, whose inactivation is synthetically lethal in breast and ovarian cancers that lack BRCA1 or BRCA2 (Fong et al. 2009, 2010). Thus, functional analyses of cancer genomes are needed to complement structural analyses of cancer genomes.
High-throughput evaluation of gene function in cancer
In addition to laying the foundation for analysis of the cancer genomes, the information provided by the human genome project has facilitated the development of reagents to perform comprehensive somatic cell genetics in mammalian cells through the systematic manipulation of gene expression. Specifically, libraries that permit the expression or suppression of the majority of human or murine genes now exist in several formats. Although these tools can be used to study nearly any aspect of biology, early studies focused on cancer phenotypes have not only provided a proof-of-principle demonstration of the utility of such systematic functional approaches, but have also begun to provide insights into novel aspects of cancer biology.
Expression-based studies
Although cDNA libraries have been used for many years to identify genes whose overexpression confers specific phenotypes (Seed and Aruffo 1987; Lin et al. 1991; Wang et al. 1991), and played a key role in the discovery of some of the first oncogenes (Shih and Weinberg 1982), such screens have often been limited by the efficiency of gene transduction as well as the representation of genes present in such libraries. As such, most successful screens involved positive selection strategies. However, several vector systems now exist that permit high-efficiency transduction of cDNAs into a wide range of mammalian cells (Koh et al. 2002), and increasingly complete collections of human cDNAs or ORFs now permit more comprehensive gain-of-function screens. Probably the most dramatic recent example of such a gain-of-function screen is the discovery of the EML4-ALK translocation in NSCLC by a retroviral expression system (Soda et al. 2007).
Using a positive selection screening strategy, several groups have identified genes that bypass specific anti-proliferative signals. For example, building on work that identified the retinoblastoma and p53 signaling networks as key regulators of cell proliferation and survival, TBX2 was found as a gene amplified in breast cancer that permits cells to proliferate in Bmi1-deficient fibroblasts (Jacobs et al. 2000), DRIL1 was discovered as a gene that bypassed RAS-induced senescence (Peeper et al. 2002), and BCL6 was found to permit cell proliferation in the presence of active p19ARF/p53 signaling (Shvarts et al. 2002).
This approach has also been used to identify genes involved in other cancer-related phenotypes. For example, the prostate-derived ETS factor gene SPDEF was identified as a gene that permits immortalized mammary epithelial cells to invade and migrate (Gunawardane et al. 2005). SPDEF was subsequently found to be overexpressed in both breast and prostate cancers, and to cooperate with receptor tyrosine kinase genes such as ERBB2 and CSF1R to drive cell transformation.
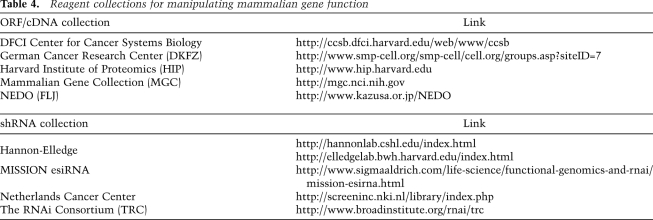
In each of these examples, the cDNA libraries used by these investigators were derived from cell lines by reverse transcription of mRNA. Although this approach has been used successfully, several limitations of this methodology are that each gene is not represented at equal frequency in the library, longer cDNAs are underrepresented, and the full repertoire of splice variants is not present. With the development of large collections of ORFs (Table 4; Lamesch et al. 2007; Rolfs et al. 2008; Varjosalo et al. 2008), it is now possible to create expression libraries in which at least one splice version of every gene is present at equal numbers. For example, analysis of a relatively small cDNA library targeting 353 kinases identified IKBKE as a breast cancer oncogene that substitutes for AKT to permit cell transformation (Boehm et al. 2007). In addition to this example of intersecting hits from a genetic screen with genomic profiles of human cancers, another approach is to create customized libraries of candidate genes—e.g., a library of amplified genes or mutated genes—to assess which ones have oncogenic activities. This type of approach can now be expanded to hundreds, rather than tens, of genes that emerge from genomic efforts, so one can enlist many candidates into a gain-of-function genetic screen for functional activity.
Table 4.
Reagent collections for manipulating mammalian gene function
Loss-of-function approaches
Similar to the cDNA or ORF libraries used for gain-of-function approaches, libraries of RNAi reagents can be introduced into cells either stably or transiently. In mammalian cells, RNAi-mediated gene suppression can be induced by the introduction of chemically synthesized siRNAs or plasmids expressing RNA hairpins, known as shRNAs, which are processed to siRNAs by Dicer (Bernstein et al. 2001). Chemically synthesized siRNAs are available from many different commercial sources as individual reagents, as pools targeting specific genes, or as genome-scale libraries. In general, siRNAs are easily synthesized and highly effective in inducing gene knockdown. However, such oligonucleotide reagents are relatively expensive and can be used only for transient loss-of-function experiments.
Vector-based systems to express RNAi provide several advantages compared with siRNA. By creating viruses, these vectors permit long-term, stable expression of the RNAi construct and expand the range and type of cells into which such constructs can be introduced. Both academic and commercial groups have produced large libraries of shRNAs in a variety of expression vectors (Table 4; Brummelkamp and Bernards 2003; Paddison et al. 2004; Silva et al. 2005, 2008; Buchholz et al. 2006; Moffat et al. 2006; Luo et al. 2009). Each of these systems has unique features, including high-efficiency infections, ease of recombination-based cloning, and inducible expression. In addition, these vector-based systems are useful for both arrayed and pooled screening approaches.
Both siRNA and shRNA libraries have been used successfully in arrayed screens (Aza-Blanc et al. 2003; Kittler and Buchholz 2005; MacKeigan et al. 2005; Whitehurst et al. 2007). For many other cancer-related phenotypic assays—such as anchorage-independent colony formation, bypass of senescence, or tumor xenografts—long-term gene suppression is essential, requiring stable integration and expression of the RNAi vector. Recent work from several laboratories has shown that these approaches are tractable in human cells. For example, PITX1 was found as a negative regulator of RAS signaling (Kolfschoten et al. 2005), REST1 has been identified a negative regulator of PI3K signaling (Westbrook et al. 2005), CDK8 has been identified as a regulator of β-catenin signaling in colon cancer (Firestein et al. 2008), SIK1 was found to be a negative regulator of aniokis and metastasis (Cheng et al. 2009), and CDK6 has been shown to be an oncogene in GBM (Wiedemeyer et al. 2010). Although such arrayed format screens require assays that are amenable to well-based miniaturization, this experimental design permits the use of high-content imaging to identify subtle or complex phenotypes such as changes in cell morphology (Moffat et al. 2006; TR Jones et al. 2008).
In addition, vector-based shRNA libraries can be used to interrogate gene function in a massively parallel manner by creating pools of shRNAs. The advantages of this approach are that such pooled screens permit the study of a larger number of genes with decreased cost and provide the possibility of using loss-of-function genetics in assays that cannot be performed in vitro. Several large-scale screens using pooled libraries have been performed (Brummelkamp et al. 2006; Ngo et al. 2006; Luo et al. 2008; Schlabach et al. 2008), demonstrating that both positive and negative selection screens are possible using these formats. To facilitate the deconvolution of genes targeted by shRNAs in these screens, each of these groups has developed strategies to quantify the abundance of each shRNA at the beginning and end of each screen by using the sequence of the shRNA or another unique sequence in the shRNA vector. Indeed, the use of a pooled format screen, together with microarrays to identify the abundance of shRNA sequences, has been used to identify the NF-κB pathway and CARD11 in particular as essential in the activated B-cell-like subtype of diffuse large B-cell lymphoma (Ngo et al. 2006), TTI1 (Tel two-interacting protein 1) and TTI2 as members of a complex with TEL2 and ATM that mediate resistance to ionizing radiation (Hurov et al. 2010), 53BP1 as an essential mediator of nutlin-3-induced cytotoxicity (Brummelkamp et al. 2006), and CRKL as a NSCLC oncogene (Luo et al. 2008). As deep-sequencing technologies become widely available, this technology will increasingly be used for deconvolution of both focused and genome-scale screens.
In addition to the identification of genes that are oncogenes or that act in specific pathways, the use of loss-of-function screens has also facilitated the identification of genes whose expression is essential in a particular context. For example, several groups have identified genes that, when suppressed, lead to cell death only in the context of cells that are dependent on oncogenic KRAS. Specifically, TBK1, STK33, PLK1, WT1, and SNAIL2 have been identified as genes that are required for the survival of cells dependent on KRAS (Barbie et al. 2009; Luo et al. 2009; Scholl et al. 2009; Wang et al. 2010). Although not yet reaching saturation, these studies already provide a path toward defining enhancers and suppressors that may also serve as therapeutic targets. Indeed, a similar strategy was used to identify genes that enhance PARP inhibitor sensitivity (Turner et al. 2008). Taken together, the systematic manipulation of gene function promises to provide information complementary to that derived from characterizing mutations in cancer genomes.
Manipulating miRNA expression
miRNAs are endogenous small noncoding RNAs that function by down-regulating expression of their target genes, either primarily through induction of transcript degradation or through translational inhibition. Approximately 500 annotated human miRNAs have been described to date, although the targets of most of these miRNAs remain undefined (Griffiths-Jones 2006). Several lines of evidence have established that dysregulation of miRNAs contributes to malignant transformation. Indeed, mice lacking Dicer, the endo-ribonuclease that is required for miRNA processing, show an increased susceptibility to cancer (Sekine et al. 2009). In addition, specific miRNAs have been implicated in particular tumors. For example, let-7, a negative regulator of RAS, is up-regulated in a subset of lung cancers (Johnson et al. 2005); the miR-17-92 cluster is amplified and up-regulated in lymphomas and promotes lymphomagenesis (He et al. 2005), and miR-15 and miR-16, negative regulators of BCL2, are down-regulated in chronic lymphocytic leukemia (Cimmino et al. 2005). Recent work has confirmed that, just as has been reported for protein-coding oncogenes, such miRNAs are also essential for tumor maintenance (Medina et al. 2010).
Several groups have now created expression libraries composed of miRNAs. For example, using a retroviral expression library of miRNAs, Voorhoeve et al. (2006) identified miR-372 and miR-373 in a Ras-induced senescence bypass screen. Since a relatively small number of miRNAs exist, one advantage of screening with current miRNA expression libraries is that it is possible to comprehensively query each of the miRNAs. However, with the development of large-scale libraries of cDNAs or ORFs for the majority of human genes, similar experiments at the genome scale will be increasingly possible in the near future.
Model systems
A wide spectrum of model systems exists that permits the investigation of the context necessary for cell transformation and progression. The commonly used model systems include large panels of genome-annotated human cancer cells, early passage primary cancer cells from patients, genetically engineered immortal primary human cells, and GEM models and their derivative primary or transformed cells. The optimal use of these models requires an understanding of their ideal applications and experimental limitations, and the most predictive results will come from complementary uses of multiple models.
Established cancer cell model systems
Established human cancer cell lines and primary human cancer cell cultures have been used extensively in functional validation assays due to their ease of manipulation and versatility. Although such systems only partially model the more complex biological features of cancers in vivo, they have proven powerful in advancing the validation of novel cancer genes, defining signaling pathways, and establishing pharmacogenomic relationships. A major challenge to the optimal use of these established cancer cell line panels has been the lack of a complete atlas of the genetic alterations in these cells, as it is appreciated that the specific genotypes in these cell models will dictate or modify the response to any molecular (RNAi or overexpression) or pharmacological perturbation. Using the same tools described above to characterize tumor genomes, several groups are engaged in characterizing large panels of cell lines, such as the Cancer Cell Line Project (http://www.sanger.ac.uk/genetics/CGP/CellLines), or the Cancer Cell Line Encyclopedia Project (http://www.broadinstitute.org/ccle). Additionally, when multiple independent cell lines with molecular diversity are used, the risk that the observed phenotypes are idiosyncratic to a particular cell line can be minimized. Although these cell line models are powerful in throughput, they do not capture all molecular subtypes of a particular tumor type, nor do they retain any interaction with stromal microenvironment, thus raising the possibility that certain gene functions will be audited or represent artifacts of in vitro biology. Therefore, re-enforcing data from other model systems will continue to be important to complement information derived from established cell lines.
Nontransformed genetically engineered cells
Primary human cells engineered with initiating events that are insufficient to achieve full transformation represent a powerful system to validate novel genes. In particular, such experimental models permit the creation of isogenic cells with specific mutations found in specific tumor subtypes, enabling the investigation of the role of a candidate cancer driver in a stringently defined genetic context. Such models can complement the use of established human cancer cell lines to interrogate functions of a candidate gene through both suppression by RNAi in established cancer cell lines and overexpression by expression in engineered primary human cells.
GEM models
GEM models of cancer have proven invaluable in cancer gene validation and in revealing mechanistic insights of a novel gene's role in the cancer process. While tumors from these GEM models are tremendously useful for comparative oncogenomics (see above), the practical challenges of time and expense involved in the creation, characterization, and uses of such GEM tumor models limit their utility as a high-throughput system.
The recent advances in nongermline GEM (nGEM) models offered different approaches that mitigate some of these limitations (for review, see Heyer et al. 2010)). Inspired by the use of nontransformed stem/progenitor cells from GEM models, termed stem transgenesis (Bachoo et al. 2002), these nGEM systems make use of tissue-restricted stem and progenitor cells that are engineered with signature mutations encountered in specific human cancer subtypes for transplantation into a primed syngeneic recipient, in which the engineered primary cells hone in to the appropriate tissue for tumor development. When these primary cells are engineered in such a way that they are poised for (but not capable of on their own) transformation, transduction with a library of vectors encoding candidate oncogene ORFs or shRNAs targeting a candidate tumor suppressor gene will permit selection for cooperating event(s) to achieve tumorigenesis. For example, Rad17 was shown to be a haploinsufficient tumor suppressor in lymphoma in a study in which a library of shRNA targeting a curated list of 1000 cancer genes was introduced into hematopoietic progenitor cells derived from Eμ-myc transgenic mice and screened for lymphomagenesis following engraftment into syngeneic recipients. Although this is often used in hematopoietic systems as hematopoietic stem and progenitor cells are readily isolated from bone marrow or fetal livers, conceptually similar approaches can be applied to other organ systems, such as liver or brain, as stem or progenitor cells have been identified, isolated, and transplanted in these systems (Bachoo et al. 2002; Zender et al. 2006; Zindy et al. 2007).
Approaches to functional validation
To fully credential a candidate gene as a therapeutic target or a diagnostic biomarker requires extensive functional assays and downstream biological studies that are time- and labor-intensive (Chin and Gray 2008). One approach to validation is to begin with assays that offer throughput rather than depth, and move only the validated ones onto lower-throughput labor-intensive assays that provide insight into specific biological aspects. However, as noted, every assay—whether it is low or high throughput—will yield biological false positives and false negatives due to the context-specific nature of gene function.
Context can relate to cellular, genetic, and microenvironmental factors. One well-known example is the opposing roles of TGFβ signaling in initiation versus progression (Massague 2008); hence, it is possible that the role of a candidate cancer gene may be oncogenic or tumor-suppressive, depending on the specific cellular or developmental contexts. Alternatively, a candidate may require cooperation of another genetic event to manifest its oncogenicity. An example is NEDD9, a metastasis gene that resides on chromosome 6 that is frequently gained in melanoma (Kim et al. 2006). NEDD9 exhibited robust invasion activity in a Modified Boyden Chamber assay only in melanocytes harboring RAS mutations, consistent with its focal amplification in RAS mutant melanoma cells that have acquired metastatic capacity in vivo (Kim et al. 2006). Thus, the specific genetic and cellular background is necessary to understand the context in which a candidate oncogene or tumor suppressor gene operates.
Biological false positives can also emerge as a direct consequence of the artificial nature of experimental models. For example, overexpression may induce phenotypes due to supraphysiologic levels of expression, or suppression of a gene may have a different phenotype in vivo. The combination of both functional studies and information derived from the analysis of cancer genomes can help mitigate these concerns. In addition, clinicopathological validation using tumor tissue microarrays can be highly informative, offering added evidentiary support for cancer relevance by demonstrating the prevalence of dysregulation on DNA (by FISH) and protein (by immunohistochemistry or immunofluorescence) levels in independent large cohorts of specific tumor types and of broad tumor spectrums.
Although we have at our disposal a series of assays that permit the assessment of a large number of candidates with high throughput, as well as ones that enable deeper interrogation with lower throughput, the existing repertoire of cancer-related assays remains incomplete. Even in assays that are well developed, cellular as well as genetic contexts are important in interpretation of the results. Hence, it is important that conclusions drawn from these functional assays performed in experimental model systems are validated in human cancer specimens.
Conclusions and challenges
Comprehensive characterization of human cancer genomes will soon generate comprehensive lists of genomic and epigenomic alterations present in diverse human cancers. The integration of these genome characterization efforts—both structural and functional—promises to provide the foundation for a complete understanding of the somatic alterations that program cancer initiation, maintenance, and progression. Although the majority of early efforts have focused on known cancer pathways as a means to validate these methodologies, the full application of these approaches will identify new pathways and networks necessary for establishment and maintenance of the malignant state. Moreover, such efforts will provide a framework on which to develop new therapeutics and rational combination regimens.
Although the throughput and reproducibility of methods for analyzing cancer genomes has improved at a rapid rate, many challenges remain. For example, collecting accurate clinical information on tumor samples remains an important and difficult task, but one that is necessary to afford the interpretation of genomic findings in a larger context. However, most available samples are associated with incomplete annotation or lack appropriate consent to permit the linkage of clinical information. Further progress will require the development and expansion of an infrastructure to collect samples and associated data with appropriate consent. The challenges—scientific, operational, and legal—of this process should not be underestimated.
In addition, it is clear that future efforts will need to account for genetic heterogeneity within tumors. As technologies improve in the near future, one will be able to explore patterns of molecular heterogeneity on the single-cell level to determine how such heterogeneity affects tumor biology and the response to treatment.
Finally, as emphasized above, integrative analyses of comprehensive cancer genomics data will generate hypotheses (such as candidate targets) that require experimental testing and validation. Such experimental validation results will guide further refinements of these analytical tools and approaches. The release of data sets prior to publication will aid in these efforts, but it is clear that more work is necessary to make these data sets available in useful formats.
In summary, cancer is the consequence of accumulated somatic genomic and epigenomic alterations within the tumor cells, influenced by their heterotypic interactions with a tumor microenvironment. The ability to catalog the somatic genomic alterations in large numbers of cancers, together with systematic functional analyses, will provide a foundation that will facilitate efforts to understand how these alterations induce malignant transformation. Moreover, as these genome characterization efforts are applied prospectively in patients, these efforts will provide a framework for relating studies in experimental models to the corresponding human tumors.
Acknowledgments
We apologize to colleagues whose studies, algorithms, or portals are not cited in this review due to limitation in space. L.C., G.G., and M.M. are TCGA-funded investigators (NIH U24CA143845, U24CA144025, and U24CA143867). L.C. and W.C.H. are CTD2 (Cancer Target Discovery and Development) network investigators (NIH RC2CA148268).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2017311.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR 2010. A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403: 503–511 [DOI] [PubMed] [Google Scholar]
- Aza-Blanc P, Cooper CL, Wagner K, Batalov S, Deveraux QL, Cooke MP 2003. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol Cell 12: 627–637 [DOI] [PubMed] [Google Scholar]
- Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ, Tang Y, DeFrances J, Stover E, Weissleder R, et al. 2002. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell 1: 269–277 [DOI] [PubMed] [Google Scholar]
- Balmain A, Gray J, Ponder B 2003. The genetics and genomics of cancer. Nat Genet 33: 238–244 [DOI] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. 2009. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. 2009. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res 37: D885–D890 doi: 10.1093/nar/gkn764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, et al. 2009. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 41: 1238–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S, Grossmann S, Vingron M, Robinson PN 2008. Ontologizer 2.0—a multifunctional tool for GO term enrichment analysis and data exploration. Bioinformatics 24: 1650–1651 [DOI] [PubMed] [Google Scholar]
- Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, et al. 2005. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 365: 1054–1061 [DOI] [PubMed] [Google Scholar]
- Bengtsson H, Neuvial P, Speed TP 2010. TumorBoost: normalization of allele-specific tumor copy numbers from a single pair of tumor-normal genotyping microarrays. BMC Bioinformatics 11: 245 doi: 10.1186/1471-2105-11-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300 [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yaacov E, Eldar YC 2008. A fast and flexible method for the segmentation of aCGH data. Bioinformatics 24: i139–i145 doi: 10.1093/bioinformatics/btn272 [DOI] [PubMed] [Google Scholar]
- Berger MF, Levin JZ, Vijayendran K, Sivachenko A, Adiconis X, Maguire J, Johnson LA, Robinson J, Verhaak RG, Sougnez C, et al. 2010. Integrative analysis of the melanoma transcriptome. Genome Res. 20: 413–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, et al. 2007. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci 104: 20007–20012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell GR, Huang J, Greshock J, Watt S, Butler A, West S, Grigorova M, Jones KW, Wei W, Stratton MR, et al. 2004. High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome Res 14: 287–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, Sjostrom SK, Garraway LA, Weremowicz S, Richardson A, et al. 2007. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell 129: 1065–1079 [DOI] [PubMed] [Google Scholar]
- Bos JL, Toksoz D, Marshall CJ, Verlaan-de Vries M, Veeneman GH, van der Eb AJ, van Boom JH, Janssen JW, Steenvoorden AC 1985. Amino-acid substitutions at codon 13 of the N-ras oncogene in human acute myeloid leukaemia. Nature 315: 726–730 [DOI] [PubMed] [Google Scholar]
- Brennan C, Zhang Y, Leo C, Feng B, Cauwels C, Aguirre AJ, Kim M, Protopopov A Chin L 2004. High-resolution global profiling of genomic alterations with long oligonucleotide microarray. Cancer Res 64: 4744–4748 [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R 2003. New tools for functional mammalian cancer genetics. Nat Rev Cancer 3: 781–789 [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Fabius AW, Mullenders J, Madiredjo M, Velds A, Kerkhoven RM, Bernards R, Beijersbergen RL 2006. An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat Chem Biol 2: 202–206 [DOI] [PubMed] [Google Scholar]
- Brunet JP, Tamayo P, Golub TR, Mesirov JP 2004. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci 101: 4164–4169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz F, Kittler R, Slabicki M, Theis M 2006. Enzymatically prepared RNAi libraries. Nat Methods 3: 696–700 [DOI] [PubMed] [Google Scholar]
- Cai WW, Mao JH, Chow CW, Damani S, Balmain A Bradley A 2002. Genome-wide detection of chromosomal imbalances in tumors using BAC microarrays. Nat Biotechnol 20: 393–396 [DOI] [PubMed] [Google Scholar]
- Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. 2008. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet 40: 722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter H, Chen S, Isik L, Tyekucheva S, Velculescu VE, Kinzler KW, Vogelstein B, Karchin R 2009. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res 69: 6660–6667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M, Wang L, Soda M, Kikuchi A, Igarashi T, et al. 2008. Oncogenic mutations of ALK kinase in neuroblastoma. Nature 455: 971–974 [DOI] [PubMed] [Google Scholar]
- Cheng H, Liu P, Wang ZC, Zou L, Santiago S, Garbitt V, Gjoerup OV, Iglehart JD, Miron A, Richardson AL, et al. 2009. SIK1 couples LKB1 to p53-dependent anoikis and suppresses metastasis. Sci Signal 2: ra35 doi: 10.1126/scisignal.2000369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang DY, Getz G, Jaffe DB, O'Kelly MJ, Zhao X, Carter SL, Russ C, Nusbaum C, Meyerson M, Lander ES 2009. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods 6: 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Gray JW 2008. Translating insights from the cancer genome into clinical practice. Nature 452: 553–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. 2005. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci 102: 13944–13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Green ED, Guttmacher AE, Guyer MS 2003. A vision for the future of genomics research. Nature 422: 835–847 [DOI] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA 2010. Harnessing transposons for cancer gene discovery. Nat Rev Cancer 10: 696–706 [DOI] [PubMed] [Google Scholar]
- Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, et al. 2010. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463: 360–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. 2002. Mutations of the BRAF gene in human cancer. Nature 417: 949–954 [DOI] [PubMed] [Google Scholar]
- Davies H, Hunter C, Smith R, Stephens P, Greenman C, Bignell G, Teague J, Butler A, Edkins S, Stevens C, et al. 2005. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res 65: 7591–7595 [DOI] [PubMed] [Google Scholar]
- Day N, Hemmaplardh A, Thurman RE, Stamatoyannopoulos JA, Noble WS 2007. Unsupervised segmentation of continuous genomic data. Bioinformatics 23: 1424–1426 [DOI] [PubMed] [Google Scholar]
- Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, et al. 2010a. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464: 999–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Wendl MC, Koboldt DC, Mardis ER 2010b. Analysis of next-generation genomic data in cancer: accomplishments and challenges. Hum Mol Genet 19: R188–R196 doi: 10.1093/hmg/ddq391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbin KK, Beer DG, Meyerson M, Yeatman TJ, Gerald WL, Jacobson JW, Conley B, Buetow KH, Heiskanen M, Simon RM, et al. 2005. Interlaboratory comparability study of cancer gene expression analysis using oligonucleotide microarrays. Clin Cancer Res 11: 565–572 [PubMed] [Google Scholar]
- Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, et al. 2010. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327: 78–81 [DOI] [PubMed] [Google Scholar]
- Dupuy AJ, Rogers LM, Kim J, Nannapaneni K, Starr TK, Liu P, Largaespada DA, Scheetz TE, Jenkins NA, Copeland NG 2009. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res 69: 8150–8156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutt A, Salvesen HB, Chen TH, Ramos AH, Onofrio RC, Hatton C, Nicoletti R, Winckler W, Grewal R, Hanna M, et al. 2008. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc Natl Acad Sci 105: 8713–8717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48 doi: 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, Freed E, Ligon AH, Vena N, Ogino S, et al. 2008. CDK8 is a colorectal cancer oncogene that regulates β-catenin activity. Nature 455: 547–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, et al. 2009. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 361: 123–134 [DOI] [PubMed] [Google Scholar]
- Fong PC, Yap TA, Boss DS, Carden CP, Mergui-Roelvink M, Gourley C, De Greve J, Lubinski J, Shanley S, Messiou C, et al. 2010. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol 28: 2512–2519 [DOI] [PubMed] [Google Scholar]
- Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR 2008. The catalogue of somatic mutations in cancer (COSMIC). Curr Protoc Hum Genet 57: 10.11.1–10.11.26 doi: 10.1002/0471142905.hg1011s57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al. 2011. COSMIC: mining complete cancer genomes in the catalogue of somatic mutations in cancer. Nucleic Acids Res 39: D945–D950 doi: 10.1093/nar/gkq929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend SH, Bernards R, Rogelj S, Weinberg RA, Rapaport JM, Albert DM, Dryja TP 1986. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 323: 643–646 [DOI] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR 2004. A census of human cancer genes. Nat Rev Cancer 4: 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. 2005. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 436: 117–122 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80 doi: 11.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George RE, Sanda T, Hanna M, Frohling S, Luther W II, Zhang J, Ahn Y, Zhou W, London WB, McGrady P, et al. 2008. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature 455: 975–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getz G, Hofling H, Mesirov JP, Golub TR, Meyerson M, Tibshirani R, Lander ES 2007. Comment on ‘The consensus coding sequences of human breast and colorectal cancers.’ Science 317: 1500. [DOI] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. 1999. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286: 531–537 [DOI] [PubMed] [Google Scholar]
- Gould J, Getz G, Monti S, Reich M, Mesirov JP 2006. Comparative gene marker selection suite. Bioinformatics 22: 1924–1925 [DOI] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. 2007. Patterns of somatic mutation in human cancer genomes. Nature 446: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman CD, Bignell G, Butler A, Edkins S, Hinton J, Beare D, Swamy S, Santarius T, Chen L, Widaa S, et al. 2010. PICNIC: an algorithm to predict absolute allelic copy number variation with microarray cancer data. Biostatistics 11: 164–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S 2006. miRBase: the microRNA sequence database. Methods Mol Biol 342: 129–138 [DOI] [PubMed] [Google Scholar]
- Gunawardane RN, Sgroi DC, Wrobel CN, Koh E, Daley GQ, Brugge JS 2005. Novel role for PDEF in epithelial cell migration and invasion. Cancer Res 65: 11572–11580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, et al. 1996. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 271: 350–353 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2000. The hallmarks of cancer. Cell 100: 57–70 [DOI] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. 2005. A microRNA polycistron as a potential human oncogene. Nature 435: 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer CJ, Pritchard C, et al. 2010. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell 140: 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer J, Kwong LN, Lowe SW, Chin L 2010. Non-germline genetically engineered mouse models for translational cancer research. Nat Rev Cancer 10: 470–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson G, Hager JH, Volik S, Hariono S, Wernick M, Moore D, Nowak N, Albertson DG, Pinkel D, Collins C, et al. 2001. Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nat Genet 29: 459–464 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, Bernabe RR, Bhan MK, Calvo F, Eerola I, Gerhard DS, et al. 2010. International network of cancer genome projects. Nature 464: 993–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunkapiller T, Kaiser RJ, Koop BF, Hood L 1991. Large-scale and automated DNA sequence determination. Science 254: 59–67 [DOI] [PubMed] [Google Scholar]
- Hupe P, Stransky N, Thiery JP, Radvanyi F, Barillot E 2004. Analysis of array CGH data: from signal ratio to gain and loss of DNA regions. Bioinformatics 20: 3413–3422 [DOI] [PubMed] [Google Scholar]
- Hurov KE, Cotta-Ramusino C, Elledge SJ 2010. A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev 24: 1939–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JJ, Keblusek P, Robanus-Maandag E, Kristel P, Lingbeek M, Nederlof PM, van Welsem T, van de Vijver MJ, Koh EY, Daley GQ, et al. 2000. Senescence bypass screen identifies TBX2, which represses Cdkn2a (p19(ARF)) and is amplified in a subset of human breast cancers. Nat Genet 26: 291–299 [DOI] [PubMed] [Google Scholar]
- James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C, Garcon L, Raslova H, Berger R, Bennaceur-Griscelli A, et al. 2005. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 434: 1144–1148 [DOI] [PubMed] [Google Scholar]
- Janoueix-Lerosey I, Lequin D, Brugieres L, Ribeiro A, de Pontual L, Combaret V, Raynal V, Puisieux A, Schleiermacher G, Pierron G, et al. 2008. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature 455: 967–970 [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ 2005. RAS is regulated by the let-7 microRNA family. Cell 120: 635–647 [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB 2002. The fundamental role of epigenetic events in cancer. Nat Rev Genet 3: 415–428 [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB 2007. The epigenomics of cancer. Cell 128: 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones NC, Pevzner PA 2004. An introduction to bioinformatics algorithms. MIT Press, Cambridge, MA [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. 2008. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321: 1801–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TR, Kang IH, Wheeler DB, Lindquist RA, Papallo A, Sabatini DM, Golland P, Carpenter AE 2008. CellProfiler analyst: data exploration and analysis software for complex image-based screens. BMC Bioinformatics 9: 482 doi: 10.1186/1471-2105-482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph EW, Pratilas CA, Poulikakos PI, Tadi M, Wang W, Taylor BS, Halilovic E, Persaud Y, Xing F, Viale A, et al. 2010. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc Natl Acad Sci 107: 14903–14908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker JS, Zhang Y, Watanabe C, Zhang Z 2007. CanPredict: a computational tool for predicting cancer-associated missense mutations. Nucleic Acids Res 35: W595-W598 doi: 10.1093/nar/gkm405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J, et al. 2010. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466: 869–873 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M 2006. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34: D354–D357 doi: 10.1093/nar/gkj102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffmann A, Huber W 2010. Microarray data quality control improves the detection of differentially expressed genes. Genomics 95: 138–142 [DOI] [PubMed] [Google Scholar]
- Khan J, Simon R, Bittner M, Chen Y, Leighton SB, Pohida T, Smith PD, Jiang Y, Gooden GC, Trent JM, et al. 1998. Gene expression profiling of alveolar rhabdomyosarcoma with cDNA microarrays. Cancer Res 58: 5009–5013 [PubMed] [Google Scholar]
- Kim PM, Tidor B 2003. Subsystem identification through dimensionality reduction of large-scale gene expression data. Genome Res 13: 1706–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Gans JD, Nogueira C, Wang A, Paik JH, Feng B, Brennan C, Hahn WC, Cordon-Cardo C, Wagner SN, et al. 2006. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell 125: 1269–1281 [DOI] [PubMed] [Google Scholar]
- Kittler R, Buchholz F 2005. Functional genomic analysis of cell division by endoribonuclease-prepared siRNAs. Cell Cycle 4: 564–567 [PubMed] [Google Scholar]
- Koh EY, Chen T, Daley GQ 2002. Novel retroviral vectors to facilitate expression screens in mammalian cells. Nucleic Acids Res 30: e142 doi: 10.1093/nar/gnf142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolfschoten IG, van Leeuwen B, Berns K, Mullenders J, Beijersbergen RL, Bernards R, Voorhoeve PM, Agami R 2005. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell 121: 849–858 [DOI] [PubMed] [Google Scholar]
- Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M, Skoda RC 2005. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med 352: 1779–1790 [DOI] [PubMed] [Google Scholar]
- LaFramboise T, Weir BA, Zhao X, Beroukhim R, Li C, Harrington D, Sellers WR, Meyerson M 2005. Allele-specific amplification in cancer revealed by SNP array analysis. PLoS Comput Biol 1: e65 doi: 10.1371/journal.pcbi.0010065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WR, Johnson MD, Kucherlapati R, Park PJ 2005. Comparative analysis of algorithms for identifying amplifications and deletions in array CGH data. Bioinformatics 21: 3763–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W, Choudhary V, Park PJ 2008. CGHweb: a tool for comparing DNA copy number segmentations from multiple algorithms. Bioinformatics 24: 1014–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Li N, Milstein S, Fan C, Hao T, Szabo G, Hu Z, Venkatesan K, Bethel G, Martin P, et al. 2007. Human ORFeome 3.1: a resource of human open reading frames covering over 10,000 human genes. Genomics 89: 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES 1996. The new genomics: global views of biology. Science 274: 536–539 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, et al. 2005. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 7: 387–397 [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci 98: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HY, Harris TL, Flannery MS, Aruffo A, Kaji EH, Gorn A, Kolakowski LF Jr, Lodish HF, Goldring SR 1991. Expression cloning of an adenylate cyclase-coupled calcitonin receptor. Science 254: 1022–1024 [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Tanenbaum DM, Daly MJ, Winchester E, Lui WO, Villapakkam A, Stanton SE, Larsson C, Hudson TJ, Johnson BE, et al. 2000. Loss-of-heterozygosity analysis of small-cell lung carcinomas using single-nucleotide polymorphism arrays. Nat Biotechnol 18: 1001–1005 [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. 2005. MicroRNA expression profiles classify human cancers. Nature 435: 834–838 [DOI] [PubMed] [Google Scholar]
- Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, et al. 2008. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci 105: 20380–20385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong KK, Elledge SJ 2009. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 137: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. 2004. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350: 2129–2139 [DOI] [PubMed] [Google Scholar]
- MacKeigan JP, Murphy LO, Blenis J 2005. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat Cell Biol 7: 591–600 [DOI] [PubMed] [Google Scholar]
- Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM 2009a. Transcriptome sequencing to detect gene fusions in cancer. Nature 458: 97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher CA, Palanisamy N, Brenner JC, Cao X, Kalyana-Sundaram S, Luo S, Khrebtukova I, Barrette TR, Grasso C, Yu J, et al. 2009b. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci 106: 12353–12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. 2009. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 361: 1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser RS, Choudhury B, Campbell PJ, Feng B, Wong KK, Protopopov A, O'Neil J, Gutierrez A, Ivanova E, Perna I, et al. 2007. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature 447: 966–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J 2008. TGFβ in cancer. Cell 134: 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusick VA 1998. Mendelian inheritance in man: a catalog of human genes and genetic disorders. Johns Hopkins University Press, Baltimore, MD [Google Scholar]
- McDermott U, Iafrate AJ, Gray NS, Shioda T, Classon M, Maheswaran S, Zhou W, Choi HG, Smith SL, Dowell L, et al. 2008. Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res 68: 3389–3395 [DOI] [PubMed] [Google Scholar]
- Medina PP, Nolde M, Slack FJ 2010. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 467: 86–90 [DOI] [PubMed] [Google Scholar]
- Mei R, Galipeau PC, Prass C, Berno A, Ghandour G, Patil N, Wolff RK, Chee MS, Reid BJ, Lockhart DJ 2000. Genome-wide detection of allelic imbalance using human SNPs and high-density DNA arrays. Genome Res 10: 1126–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesirov JP 2010. Computer science. Accessible reproducible research. Science 327: 415–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M, Gabriel S, Getz G 2010. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet 11: 685–696 [DOI] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F 2007. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer 7: 233–245 [DOI] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124: 1283–1298 [DOI] [PubMed] [Google Scholar]
- Mosse YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, Laquaglia MJ, Sennett R, Lynch JE, Perri P, et al. 2008. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 455: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, et al. 2007. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446: 758–764 [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, White D, Hughes TP, Le Beau MM, Pui CH, et al. 2008. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 453: 110–114 [DOI] [PubMed] [Google Scholar]
- Nanjundan M, Nakayama Y, Cheng KW, Lahad J, Liu J, Lu K, Kuo WL, Smith-McCune K, Fishman D, Gray JW, et al. 2007. Amplification of MDS1/EVI1 and EVI1, located in the 3q26.2 amplicon, is associated with favorable patient prognosis in ovarian cancer. Cancer Res 67: 3074–3084 [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S 2001. Predicting deleterious amino acid substitutions. Genome Res 11: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, et al. 2006. A loss-of-function RNA interference screen for molecular targets in cancer. Nature 441: 106–110 [DOI] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. 2010. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17: 510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan RC, Brennan CW, Strahs A, Zhang X, Kannan K, Donovan M, Cauwels C, Sharpless NE, Wong WH, Chin L 2003. Array comparative genome hybridization for tumor classification and gene discovery in mouse models of malignant melanoma. Cancer Res 63: 5352–5356 [PubMed] [Google Scholar]
- Paddison PJ, Silva JM, Conklin DS, Schlabach M, Li M, Aruleba S, Balija V, O'Shaughnessy A, Gnoj L, Scobie K, et al. 2004. A resource for large-scale RNA-interference-based screens in mammals. Nature 428: 427–431 [DOI] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. 2004. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304: 1497–1500 [DOI] [PubMed] [Google Scholar]
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, et al. 2004. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351: 2817–2826 [DOI] [PubMed] [Google Scholar]
- Palanisamy N, Ateeq B, Kalyana-Sundaram S, Pflueger D, Ramnarayanan K, Shankar S, Han B, Cao Q, Cao X, Suleman K, et al. 2010. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med 16: 793–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, et al. 2004. EGF receptor gene mutations are common in lung cancers from ‘never smokers' and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci 101: 13306–13311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada LF, Tabin CJ, Shih C, Weinberg RA 1982. Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus ras gene. Nature 297: 474–478 [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. 2008. An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeper DS, Shvarts A, Brummelkamp T, Douma S, Koh EY, Daley GQ, Bernards R 2002. A functional screen identifies hDRIL1 as an oncogene that rescues RAS-induced senescence. Nat Cell Biol 4: 148–153 [DOI] [PubMed] [Google Scholar]
- Pettersson E, Lundeberg J, Ahmadian A 2009. Generations of sequencing technologies. Genomics 93: 105–111 [DOI] [PubMed] [Google Scholar]
- Pevsner J 2009. Bioinformatics and functional genomics. Wiley-Blackwell, New York [Google Scholar]
- Picard F, Robin S, Lavielle M, Vaisse C, Daudin JJ 2005. A statistical approach for array CGH data analysis. BMC Bioinformatics 6: 27 doi: 10.1186/1471-2105-6-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, et al. 2010a. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463: 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. 2010b. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO 1999. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet 23: 41–46 [DOI] [PubMed] [Google Scholar]
- Pollock PM, Gartside MG, Dejeza LC, Powell MA, Mallon MA, Davies H, Mohammadi M, Futreal PA, Stratton MR, Trent JM, et al. 2007. Frequent activating FGFR2 mutations in endometrial carcinomas parallel germline mutations associated with craniosynostosis and skeletal dysplasia syndromes. Oncogene 26: 7158–7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N 2010. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 464: 427–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbitts TH 1994. Chromosomal translocations in human cancer. Nature 372: 143–149 [DOI] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S 2002. Human non-synonymous SNPs: server and survey. Nucleic Acids Res 30: 3894–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Stuart JM, Altman RB 2000. Principal components analysis to summarize microarray experiments: application to sporulation time series. Pac Symp Biocomput 2000: 455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. 2006. Global variation in copy number in the human genome. Nature 444: 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP 2006. GenePattern 2.0. Nat Genet 38: 500–501 [DOI] [PubMed] [Google Scholar]
- Reva B, Antipin Y, Sander C 2007. Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol 8: R232 doi: 10.1186/gb-2007-8-11-r232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A Chinnaiyan AM 2004. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. 2007. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9: 166–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfs A, Montor WR, Yoon SS, Hu Y, Bhullar B, Kelley F, McCarron S, Jepson DA, Shen B, Taycher E, et al. 2008. Production and sequence validation of a complete full length ORF collection for the pathogenic bacterium Vibrio cholerae. Proc Natl Acad Sci 105: 4364–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. 2004. High frequency of mutations of the PIK3CA gene in human cancers. Science 304: 554. [DOI] [PubMed] [Google Scholar]
- Sanger F, Coulson AR 1975. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol 94: 441–448 [DOI] [PubMed] [Google Scholar]
- Santos E, Martin-Zanca D, Reddy EP, Pierotti MA, Della Porta G, Barbacid M 1984. Malignant activation of a K-ras oncogene in lung carcinoma but not in normal tissue of the same patient. Science 223: 661–664 [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270: 467–470 [DOI] [PubMed] [Google Scholar]
- Schlabach MR, Luo J, Solimini NL, Hu G, Xu Q, Li MZ, Zhao Z, Smogorzewska A, Sowa ME, Ang XL, et al. 2008. Cancer proliferation gene discovery through functional genomics. Science 319: 620–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl C, Frohling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY, Silver SJ, Tamayo P, Wadlow RC, Ramaswamy S, et al. 2009. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell 137: 821–834 [DOI] [PubMed] [Google Scholar]
- Scott KL, Kabbarah O, Liang MC, Ivanova E, Anagnostou V, Wu J, Dhakal S, Wu M, Chen S, Feinberg T, et al. 2009. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature 459: 1085–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, et al. 2004. Large-scale copy number polymorphism in the human genome. Science 305: 525–528 [DOI] [PubMed] [Google Scholar]
- Seed B, Aruffo A 1987. Molecular cloning of the CD2 antigen, the T-cell erythrocyte receptor, by a rapid immunoselection procedure. Proc Natl Acad Sci 84: 3365–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine S, Ogawa R, Ito R, Hiraoka N, McManus MT, Kanai Y, Hebrok M 2009. Disruption of Dicer1 induces dysregulated fetal gene expression and promotes hepatocarcinogenesis. Gastroenterology 136:2304–2315e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J, Ji H 2008. Next-generation DNA sequencing. Nat Biotechnol 26: 1135–1145 [DOI] [PubMed] [Google Scholar]
- Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM 2005. Accurate multiplex polony sequencing of an evolved bacterial genome. Science 309: 1728–1732 [DOI] [PubMed] [Google Scholar]
- Shih C, Weinberg RA 1982. Isolation of a transforming sequence from a human bladder carcinoma cell line. Cell 29: 161–169 [DOI] [PubMed] [Google Scholar]
- Shimizu K, Goldfarb M, Suard Y, Perucho M, Li Y, Kamata T, Feramisco J, Stavnezer E, Fogh J, Wigler MH 1983. Three human transforming genes are related to the viral ras oncogenes. Proc Natl Acad Sci 80: 2112–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvarts A, Brummelkamp TR, Scheeren F, Koh E, Daley GQ, Spits H, Bernards R 2002. A senescence rescue screen identifies BCL6 as an inhibitor of anti-proliferative p19(ARF)-p53 signaling. Genes Dev 16: 681–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. 2005. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet 37: 1281–1288 [DOI] [PubMed] [Google Scholar]
- Silva JM, Marran K, Parker JS, Silva J, Golding M, Schlabach MR, Elledge SJ, Hannon GJ, Chang K 2008. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science 319: 617–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. 2006. The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274 [DOI] [PubMed] [Google Scholar]
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al. 2007. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448: 561–566 [DOI] [PubMed] [Google Scholar]
- Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL, Gupta M, O'Sullivan MG, Matise I, Dupuy AJ, et al. 2009. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science 323: 1747–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens P, Hunter C, Bignell G, Edkins S, Davies H, Teague J, Stevens C, O'Meara S, Smith R, Parker A, et al. 2004. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature 431: 525–526 [DOI] [PubMed] [Google Scholar]
- Stephens P, Edkins S, Davies H, Greenman C, Cox C, Hunter C, Bignell G, Teague J, Smith R, Stevens C, et al. 2005. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet 37: 590–592 [DOI] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. 2009. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462: 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA 2009. The cancer genome. Nature 458: 719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Koch I, Lathe W III, Kondrashov AS, Bork P 2001. Prediction of deleterious human alleles. Hum Mol Genet 10: 591–597 [DOI] [PubMed] [Google Scholar]
- Sweet-Cordero A, Mukherjee S, Subramanian A, You H, Roix JJ, Ladd-Acosta C, Mesirov J, Golub TR, Jacks T 2005. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet 37: 48–55 [DOI] [PubMed] [Google Scholar]
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR 1999. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci 96: 2907–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BS, Barretina J, Socci ND, Decarolis P, Ladanyi M, Meyerson M, Singer S, Sander C 2008. Functional copy-number alterations in cancer. PLoS ONE 3: e3179 doi: 10.1371/journal.pone.0003179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RK, Nickerson E, Simons JF, Janne PA, Tengs T, Yuza Y, Garraway LA, LaFramboise T, Lee JC, Shah K, et al. 2006. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat Med 12: 852–855 [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. 2005. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310: 644–648 [DOI] [PubMed] [Google Scholar]
- Turner NC, Lord CJ, Iorns E, Brough R, Swift S, Elliott R, Rayter S, Tutt AN, Ashworth A 2008. A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. EMBO J 27: 1368–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci 98: 5116–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren AG, Kool J, Matentzoglu K, de Ridder J, Mattison J, van Uitert M, Lagcher W, Sie D, Tanger E, Cox T, et al. 2008. Large-scale mutagenesis in p19(ARF)- and p53-deficient mice identifies cancer genes and their collaborative networks. Cell 133: 727–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. 2002. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347: 1999–2009 [DOI] [PubMed] [Google Scholar]
- Van Loo P, Nordgard SH, Lingjaerde OC, Russnes HG, Rye IH, Sun W, Weigman VJ, Marynen P, Zetterberg A, Naume B, et al. 2010. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci 107: 16910–16915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415: 530–536 [DOI] [PubMed] [Google Scholar]
- Varela I, Klijn C, Stephens PJ, Mudie LJ, Stebbings L, Galappaththige D, van der Gulden H, Schut E, Klarenbeek S, Campbell PJ, et al. 2010. Somatic structural rearrangements in genetically engineered mouse mammary tumors. Genome Biol 11: R100 doi: 10.1186/gb-2010-11-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varjosalo M, Bjorklund M, Cheng F, Syvanen H, Kivioja T, Kilpinen S, Sun Z, Kallioniemi O, Stunnenberg HG, He WW, et al. 2008. Application of active and kinase-deficient kinome collection for identification of kinases regulating hedgehog signaling. Cell 133: 537–548 [DOI] [PubMed] [Google Scholar]
- Venkatraman ES, Olshen AB 2007. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics 23: 657–663 [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. 2001. The sequence of the human genome. Science 291: 1304–1351 [DOI] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. 2010. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17: 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW 1993. The multistep nature of cancer. Trends Genet 9: 138–141 [DOI] [PubMed] [Google Scholar]
- Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, et al. 2006. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 124: 1169–1181 [DOI] [PubMed] [Google Scholar]
- Wang XF, Lin HY, Ng-Eaton E, Downward J, Lodish HF, Weinberg RA 1991. Expression cloning and characterization of the TGF-β type III receptor. Cell 67: 797–805 [DOI] [PubMed] [Google Scholar]
- Wang P, Kim Y, Pollack J, Narasimhan B, Tibshirani R 2005. A method for calling gains and losses in array CGH data. Biostatistics 6: 45–58 [DOI] [PubMed] [Google Scholar]
- Wang Y, Ngo VN, Marani M, Yang Y, Wright G, Staudt LM, Downward J 2010. Critical role for transcriptional repressor Snail2 in transformation by oncogenic RAS in colorectal carcinoma cells. Oncogene 29: 4658–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B, Zhao X, Meyerson M 2004. Somatic alterations in the human cancer genome. Cancer Cell 6: 433–438 [DOI] [PubMed] [Google Scholar]
- Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. 2007. Characterizing the cancer genome in lung adenocarcinoma. Nature 450: 893–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B, Zhao JJ, Roberts TM, Mandel G, Hannon GJ, et al. 2005. A genetic screen for candidate tumor suppressors identifies REST. Cell 121: 837–848 [DOI] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, et al. 2008. The complete genome of an individual by massively parallel DNA sequencing. Nature 452: 872–876 [DOI] [PubMed] [Google Scholar]
- Whitehurst AW, Bodemann BO, Cardenas J, Ferguson D, Girard L, Peyton M, Minna JD, Michnoff C, Hao W, Roth MG, et al. 2007. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature 446: 815–819 [DOI] [PubMed] [Google Scholar]
- Whittaker S, Kirk R, Hayward R, Zambon A, Viros A, Cantarino N, Affolter A, Nourry A, Niculescu-Duvaz D, Springer C, et al. 2010. Gatekeeper mutations mediate resistance to BRAF-targeted therapies. Sci Transl Med 2: 35ra41 doi: 10.1126/scitranslmed.3000758 [DOI] [PubMed] [Google Scholar]
- Wiedemeyer R, Brennan C, Heffernan TP, Xiao Y, Mahoney J, Protopopov A, Zheng H, Bignell G, Furnari F, Cavenee WK, et al. 2008. Feedback circuit among INK4 tumor suppressors constrains human glioblastoma development. Cancer Cell 13: 355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemeyer WR, Dunn IF, Quayle SN, Zhang J, Chheda MG, Dunn GP, Zhuang L, Rosenbluh J, Chen S, Xiao Y, et al. 2010. Pattern of retinoblastoma pathway inactivation dictates response to CDK4/6 inhibition in GBM. Proc Natl Acad Sci 107: 11501–11506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willenbrock H, Fridlyand J 2005. A comparison study: applying segmentation to array CGH data for downstream analyses. Bioinformatics 21: 4084–4091 [DOI] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. 2007. The genomic landscapes of human breast and colorectal cancers. Science 318: 1108–1113 [DOI] [PubMed] [Google Scholar]
- Xiong J 2006. Essential bioinformatics. Cambridge University Press, New York [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et al. 2006. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell 125: 1253–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Li C, Paez JG, Chin K, Janne PA, Chen TH, Girard L, Minna J, Christiani D, Leo C, et al. 2004. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res 64: 3060–3071 [DOI] [PubMed] [Google Scholar]
- Zhao R, Xing S, Li Z, Fu X, Li Q, Krantz SB, Zhao ZJ 2005. Identification of an acquired JAK2 mutation in polycythemia vera. J Biol Chem 280: 22788–22792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Sanborn JZ, Benz S, Szeto C, Hsu F, Kuhn RM, Karolchik D, Archie J, Lenburg ME, Esserman LJ, et al. 2009. The UCSC Cancer Genomics Browser. Nat Methods 6: 239–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F, Uziel T, Ayrault O, Calabrese C, Valentine M, Rehg JE, Gilbertson RJ, Sherr CJ, Roussel MF 2007. Genetic alterations in mouse medulloblastomas and generation of tumors de novo from primary cerebellar granule neuron precursors. Cancer Res 67: 2676–2684 [DOI] [PubMed] [Google Scholar]