Abstract
Although local regulation of T-cell responses by epithelial cells is increasingly viewed as important, few molecules mediating such regulation have been identified. Skint1, a recently identified member of the Ig-supergene family expressed by thymic epithelial cells and keratinocytes, specifies the murine epidermal intraepithelial lymphocyte (IEL) repertoire. Investigating whether Skint1-related molecules might regulate IEL in other compartments, this study focuses on buytrophilin-like 1 (Btnl1), which is conspicuously similar to Skint1 and primarily restricted to small intestinal epithelium. Btnl1 protein is mostly cytoplasmic, but surface expression can be induced, and in vivo Btnl1 can be detected adjacent to the IEL. In a newly developed culture system, enforced epithelial cell expression of Btnl1 attenuated the cells’ response to activated IEL, as evidenced by suppression of IL-6 and other inflammatory mediators. These findings offer a unique perspective on emerging genetic data that Btnl genes may comprise novel and important local regulators of gut inflammation.
Keywords: epithelial immunity, intraepithelial lymphocyte-culture, B7 family, inflammation, immunosuppression
Lymph nodes have been traditionally viewed as the sites for priming lymphocyte responses and epithelial tissues have been viewed as the sites where those responses are made manifest. Recently, however, intense interest has developed as to how initial events within epithelia influence lymphocyte priming, with significant impact on the functional complexion of resulting effector responses. Thus, particular attention has been paid to how microbial products affect the capacity of tissue-associated dendritic cells (DC) to induce specific differentiation programs in the T cells that they subsequently activate (1). In contrast, much less attention has been paid to the regulation of immune responses by epithelial cells, despite their being primary targets of infection and dysregulaton, and despite their potential to produce myriad innate immune effectors (2, 3). Furthermore, many epithelia are constitutively populated by T-cell receptor (TCR)αβ+ and TCRγδ+ intraepithelial lymphocytes (IEL) that compose a large T-cell subset whose localization offers obvious opportunities for interactions with epithelial cells (4). Indeed, mice lacking TCRγδ+ IEL show dyregulated intestinal epithelial cell turnover and differentiation (5). Hence, there is much interest in identifying epithelial molecules that might regulate interactions of epithelial cells and IELs.
One recently identified axis involves Skint1, the founding member of a hitherto undescribed Ig (Ig)-supergene family, all of the members of which are expressed by thymic epithelial cells and keratinocytes (6). Skint1 regulates epidermal γδ IEL development, with such cells displaying an atypical TCR repertoire in mice with dysfunctional Skint1 (6, 7). In seeking further epithelial regulators of local immune interactions, we sought molecules strikingly similar to Skint1. Prominent among these are members of the butyrophilin-like (Btnl) gene family (8, 9) that exhibit various criteria of putative immune regulators. Among the criteria are: they are conserved in mice and humans; several are encoded within the MHC-locus; they share strong homologies with the B7-family of costimulators; and certain murine and human Btnl/Btn gene products reportedly reduce the proliferative responses of costimulated T cells, with human BTN2A1 reportedly modulating DCs by engaging the lectin DC-SIGN (9–15). The potential importance of such interactions is further implied by genetic associations of human Btnl2 and Btn2A1 polymorphisms with sarcoidosis, myositis, and inflammatory bowel disease (16–18). Indeed, it has been proposed that Btnl proteins might be particularly important in mediating immune regulation in tissues, notably the gut (12), although the specific regulation of IEL–epithelial cell interactions by any Btnl/Btn gene has not hitherto been experimentally investigated.
Results
Btnl1, -4, and -6 Genes and Their Expression.
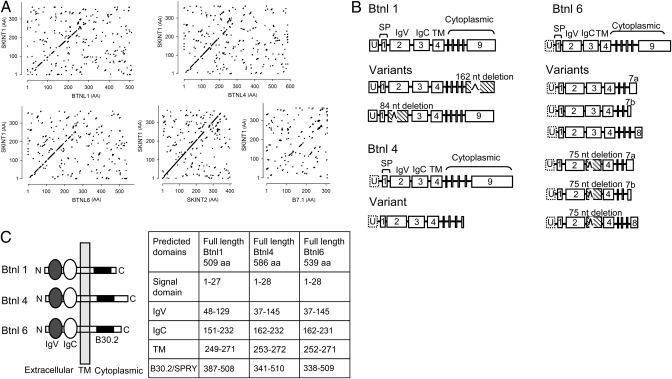
Beyond the Skint family, the genes most homologous to Skint1 are the murine butyrophilin-like Btnl1, -4, and -6 genes that are almost equivalently similar, closely followed by the butyrophilin Btn1a1 gene (Table S1). Whereas Skint1and Skint2 are homologous across their lengths, the similarity to Btnl genes is concentrated in the predicted Ig (V and C) ectodomains (Fig. 1A). Skint1 shows slightly less similarity to Btnl7, and conspicuously less to Btnl2 (Table S1), the focus of most previous investigations of murine Btnl genes (9, 11, 12). Similarity to Skint1 declines further for prototypic B7-family genes (Fig. 1A and Table S1). Hence, a subfamily of Btnl genes is highly and specifically similar to Skint1, meriting their biological comparison.
Fig. 1.
Structural analysis of Btnl1, -4, and -6. (A) Dot matrix comparison of Skint1 (y axis) with Btnl1, -4, and -6, and Skint2, and B7.1. (B) Full-length Btnl1, -4, and -6 cDNA and minor splice variants. Hatched boxes depict exons with internal in-frame deletions. The 5′ untranslated exon, U, identified for Btnl1 remains putative for Btnl4 and -6. SP, signal peptide; TM, transmembrane domain. (C) Inferred protein organization of Btnl1, -4, and -6, with predicted lengths of each domain as predicted by Pfam, SMART, TMHMM, and SignalP databases.
To better understand murine Btnl genes most closely related to Skint1, intron-exon boundaries of predominant transcripts and of rare splice variants were redefined by RACE-analysis and cloning. A 5′ untranslated exon (U) initiates Btnl1 and possibly Btnl4 and -6 mRNAs (Fig. 1B). The nine residual exons predict Btnl1, -4, and -6 protein structures (Fig. 1C) featuring a cytoplasmic B30.2 domain: this combines a SPRY domain with a PRY domain (19), and is to date unique to Btn/Btnl proteins and to tripartite motif (TRIM) proteins, in which it facilitates multimerisation (20, 21). In contrast, Btnl2 is predicted to comprise three or four Ig ectodomains and to lack B30.2 (Fig. S1).
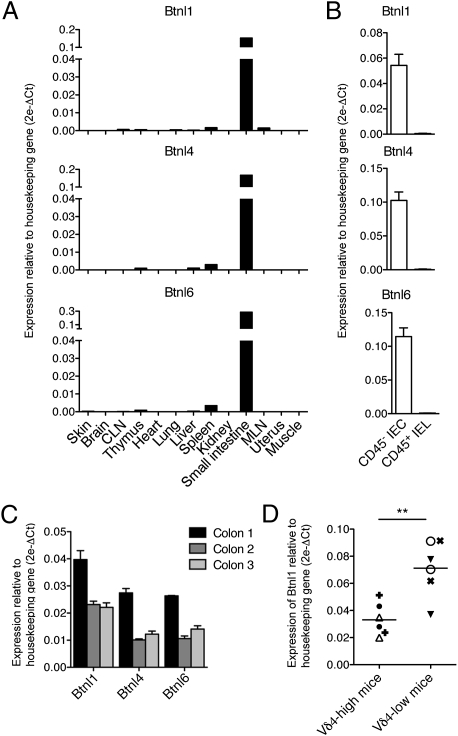
Btnl1, -4, and -6 transcripts were almost exclusively expressed in the gut, with high levels in epithelial, CD45(−) cells of the small intestine, and with lower and variable levels in the colon (Fig. 2 A–C). Thus, Btnl1, -4, and -6 share with Skint1 high structural similarity, and predominant expression in epithelia constitutively rich in T cells. Interestingly, mouse strains in which intestinal IELs include large numbers of Vδ4(+) cells usually expressed lower levels of Btnl1 RNA than did strains with few Vδ4(+) IELs (Fig. 2D). Intriguingly, such IEL repertoire variation maps to the MHC Class II, I-Eα region (22, 23), which also contains the Btnl1 gene. No consistent variation in expression was shown by Btnl4, -6, or -2 (Fig. S2).
Fig. 2.
Quantitative RT-PCR analysis of Btnl1, -4, and -6 mRNA (A) in normal mouse tissues; (B) in CD45(−) small intestinal epithelial cells (IECs) versus CD45(+) small intestinal IEL; (C) in large intestines harvested from three mice; and (D) in the small intestines of two mice of each of three strains having low (Balb/CJ, DBA/2J, C57L/J) or high numbers of Vδ4(+) IEL (B10.BR, CBA/J, C58/J) (different symbols depict different strains). **P < 0.005 for comparison by unpaired two-tailed t test of means and data-points distribution. Error bars in B and C indicate SEM.
Btnl1 Protein and Its Expression.
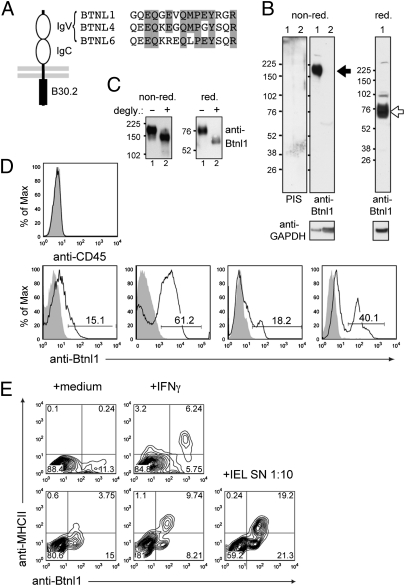
To detect Btnl1 protein, a rabbit polyclonal antibody was generated to a peptide from the putative IgV region that was <54% conserved with other Btnl proteins (Fig. 3A). Antibody fidelity was confirmed by stringent criteria. Thus, peptide-purified antibody immunoprecipitated specific proteins from human 293T cells transfected with a Btnl1 cDNA that included a FLAG-epitope C terminal to the putative signal cleavage site (Fig. S3A). The precipitated proteins migrated on reducing and nonreducing gels at ~53 kD (the predicted molecular mass for FLAG-tagged Btnl1) and >150 kD, respectively; they were not detected by preimmune serum and they comigrated with proteins detected by anti-FLAG, albeit that anti-Btnl1 preferentially detected the 150 kD unreduced complex, perhaps reflecting better reactivity toward conformational determinants (Fig. S3B). Both anti-Btnl1 and anti-FLAG detected surface expression of FLAG-tagged Btnl1 following transfection of the murine enterocyte cell line, MODE-K (24) or HEK293T cells (neither ordinarily express surface Btnl1), with anti-Btnl1 reactivity largely confined to GFP+ cells. Anti-Btnl1 did not detect Btnl4 or Btnl6-transfectants but they were detected with anti-FLAG (Fig. S3 C–E).
Fig. 3.
Btnl1 Protein. (A) A Btnl1-reactive antibody was raised against a synthetic peptide (residues 82–96); peptide alignment across Btnl1, -4, and -6 is shown. (B) Cleared lysates of enterocytes (lane 1) and liver (lane 2) resolved on nonreducing or reducing SDS/PAGE were Western-blotted with preimmune rabbit serum or peptide-purified anti-Btnl1 antibody. The GAPDH immunoblot acts as a loading control; size markers are in kilodaltons; specific antibody-reactive species are shown by arrows. (C) Btnl1 immunopurified from enterocyte lysates, was treated with N-glycosidase F (degly) as indicated, and the protein resolved as in B. (D) IECs derived from different mice at different times from different colonies, gated on CD45(−) cells (Upper), were stained with peptide-purified Btnl1-specific antibody (solid black line) or preimmune serum (shaded histogram). (E) Cultured enterocytes were treated with IFN-γ or activated IEL supernatant diluted 1:10 in enterocyte medium, or left untreated for 24 h, and then stained for Btnl1 and MHCII. Two independent experiments are shown. Differences in Btnl1 staining levels are partly because of different secondary reagents used, labeled with either AlexaFluor 488 or APC.
In lysates of primary enterocytes, anti-Btnl1 (but not preimmmune sera) detected a diffuse nonreduced complex of >150 kD, that upon reduction migrated at ~75 kD (Fig. 3B). The Btnl1 sequence predicts N-glycosylation, consistent with which the 75 kD form resolved to ~54 kD upon N-glycosidase-F treatment (Fig. 3C). Moreover, if as expected, the B30.2 motif acts as a multimerisation domain, then the nonreduced complex may include dimers of the 75-kD form. Finally, antibody reactivity was consistent with the RNA analyses (Fig. 2) in detecting no protein in liver or keratinocyte lysates (Fig. 3B and Fig. S3F).
Directly ex vivo, viable 7AAD(−) CD45(−) small intestinal cells with side- and forward-scatter typical of enterocytes showed variable levels of surface Btnl1 (~15% to ~60%) seemingly reflecting variations in animal husbandry and environment (Fig. 3D). IFN-γ treatment of enterocytes up-regulates several immunological properties, including surface MHC Class II (25, 26). Conspicuously, MHCII was almost exclusively induced on enterocytes expressing surface Btnl1 (Fig. 3E). Whereas IFN-γ did not markedly increase surface Btnl1, activated IEL supernatants strongly up-regulated Btnl1 and MHCII, likely reflecting the combined action of IFN-γ and other regulators, such as TNF (4, 27). Indeed, human BTN3 was reportedly up-regulated by IFN-γ + TNF, but not IFN-γ alone (28). Hence, the variable surface expression of Btnl1 can be regulated by immunological mediators.
Btnl1 in Situ.
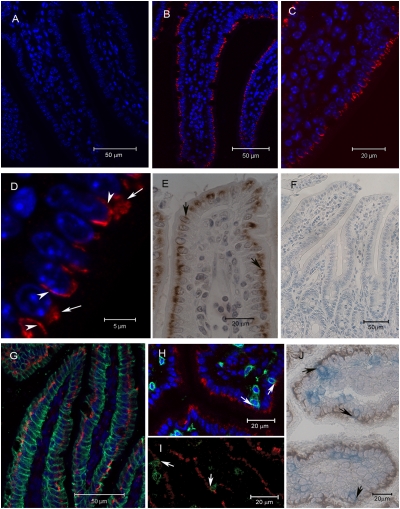
In 5-μm sections of murine small intestine, Btnl1 was detected in essentially all enterocytes (Fig. 4 A–C) but, consistent with its variable surface expression on explanted enterocytes, it was detected in either of two forms: a “lobular/reticular” pattern (Fig. 4D, long arrows) or, less commonly, a uniform decoration, evoking intercellular surface expression (Fig. 4D, short arrows). Consistent data were obtained by immunochemistry (Fig. 4 E and F), with further instances of lateral expression (Fig. 4 E, arrows). The lobular pattern is not easily attributed to apoptotic cells, because it was not enriched in villus tip enterocytes, where apoptosis is prevalent, and because enterocytes with this pattern stained with fidelity for the surface protein A33 (29) (Fig. 4G). Triple-staining for A33 (green), Btnl1 (red), and nuclei (blue; DAPI) suggested that Btnl1 was most commonly perinuclear (Fig. 4G), evocative of staining data presented for Btnl2 (12). Nonetheless, double-staining for Btnl1 and the TCR (anti-CD3) showed incidences of close juxtaposition when examined by either of two techniques (Fig. 4 H–J). Enumeration of 20 sections found ~10% of IEL juxtaposed to Btnl1.
Fig. 4.
Localization of Btnl1 in murine small intestine. Small intestinal sections were immunostained with peptide-purified anti-Btnl1 antibody (red in B–D and G–I; brown in E and J), with antibody (anti-A33) to a well-established enterocyte surface marker (green in G) or with anti-CD3 (green in H and I; blue in J). Btnl1+ epithelial cells were detected in most villi, equally distributed from crypts to tips (B and C). Btnl1 adopted either a lobular/reticular pattern, exemplified by long arrows in D, or a lateral pattern, exemplified by arrowheads in D and arrows in E. Triple-staining for A33, Btnl1, and nuclei indicated that Btnl1 most commonly localized to a perinuclear region (G). Sections stained for Btnl1 and CD3 indicated prominent incidences of close juxtaposition, as depicted by arrows in H to J. No staining was detected using preimmune serum (A and F). Sections A to D and G and H were counterstained with DAPI (blue) to visualize nuclei.
Epithelial–T-Cell Communication.
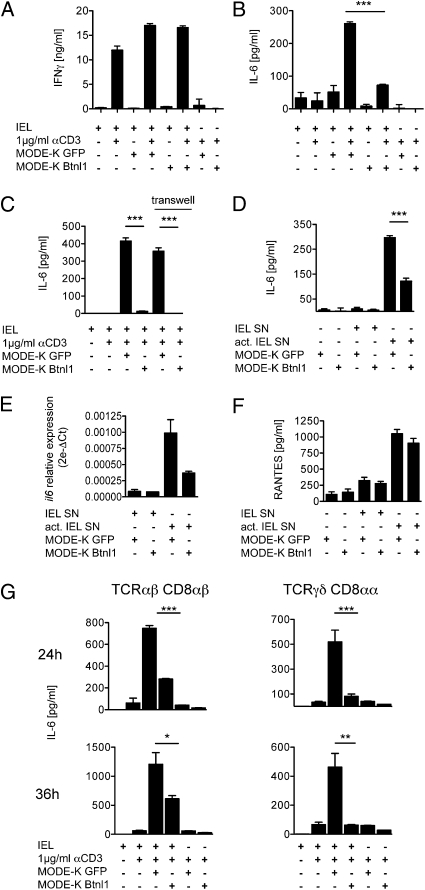
Given claims that Btnl genes suppress T-cell activation (11–13, 15), we sought an analogous effect of Btnl1 on IELs by exploiting a culture system (SI Materials and Methods) that overcomes the rapid apoptosis of IELs ex vivo (30, 31). This system permits IELs to be rested as viable cells that rapidly reactivate IFN-γ when stimulated via the TCR (Fig. 5A). The induction of IFN-γ and other major IEL cytokines (e.g., TNF, IL-3) was largely unaffected when IELs were activated in the presence of MODE-K enterocytes expressing either transfected gfp or gfp + Btnl1 (compare columns 2, 4, and 6 in Fig. 5A and Fig. S4A). Other cytokines (e.g., IL-4, IL-5, IL-10, IL-13, and IL-17) were produced by activated IELs at negligible levels and not reproducibly affected by coculture with MODE-K.gfp or MODE-K.Btnl1.
Fig. 5.
Btnl1 selectively modulates cytokine expression. Culture of total IEL (A–C) or TCRγδ and TCRαβ IEL subsets (G) with (+) or without (−) anti-CD3, and with or without MODE-K cells transfected with gfp, or Btnl1-IRES-gfp, respectively, as indicated. In C, transwells separated IELs and MODE-K. Alternatively, MODE-K.gfp or MODE-K.gfp.Btnl1 cells were cultured with supernatants (SN) from activated or unactivated IEL (D–F). IFN-γ, IL-6, and RANTES were assessed by ELISA (A–D, F) and Luminex (G). Relative expression of il6 mRNA was determined by quantitative RT-PCR (E). *P < 0.05, **P ≤ 0.005, and ***P ≤ 0.0001 as calculated for means and SD by unpaired two-tailed t test.
Despite recording no clear suppression of IEL activation, a clear and unanticipated effect of Btnl1 expression on IEL-enterocyte cocultures was observed as follows: cocultures of activated IEL with primary enterocytes or MODE-K cells substantially up-regulate IL-6 (Fig. 5B), the key source of which is the enterocytes, because induction occurs when transwells separate the IEL and enterocytes; when activated IEL supernatants are added to enterocytes; and when enterocyte RNA is examined (Fig. 5 C–E). The same was true for RANTES (regulated upon activation, normal T cell expressed and secreted) (CCL5) (Fig. 5F). The nature of the activated IEL cytokine that strongly up-regulates enterocyte cytokine expression is being investigated. Germane to this study, however, IL-6 but not RANTES induction was significantly inhibited (>50%) when independent lines of MODE-K enterocytes expressed Btnl1 introduced using different vectors (Fig. 5 B–G). Btnl1 likewise impaired IL-6 up-regulation when MODE-K cells were cocultured with activated CD8αβ+ TCRαβ+ IEL [often regarded as conventional, effector-memory T cells (4)], with activated CD8αα+ γδ IEL [prototypic unconventional T cells (4)] (Fig. 5G), or with activated systemic T cells (Fig. S4B).
IL-6 is a uniquely important regulator of intestinal biology (32). Nonetheless, the effects of dysregulated Btnl1 expression were not unique to IL-6, as the up-regulation by MODE-K cells of proteins and RNA for KC/CXCL1, MIP1β/CCL4, and IL-15 was also inhibited (from ~33% to >90%), whereas MCP1/CCL2 (like CCL5) was not (Fig. S5). Hence, Btnl1 can selectively regulate how enterocytes respond to activated T cells.
Discussion
Epithelia are common sites of infection and dysregulation, where immuno-protection must be rapid, effective, and durable, but not develop into inflammatory disease and associated carcinoma. Hence, there is intense interest in how epithelial cells may regulate immune responses (2, 3). A recently elucidated member of the Ig-superfamily, Skint1, expressed exclusively by thymic and skin epithelium, regulates intraepidermal γδ T-cell development (6, 7). Investigating whether Skint-related genes might share a similar biology, we found that Btnl1, -4, and -6 show striking sequence similarity to Skint1; are also largely restricted to an epithelial tissue (the small intestine) replete with T cells; and that at least one, Btnl1, can act as a novel immune regulator. In this regard, it may be appropriate to distinguish those Btnl genes that are essentially restricted to specific epithelia from others that may be more broadly expressed and that may have properties more akin to inhibitory B-7 family members.
This unique study of the effect of a Btnl gene on IEL–enterocyte interactions has revealed a previously unidentified immuno-regulatory mechanism: namely suppression of proinflammatory mediators, such as IL-6, CXCL1, IL-15, and CCL4, that are ordinarily up-regulated by epithelial cells in response to activated T-cell cytokines. Because all such mediators are regulated by NFκB (33–36), the possibility exists that Btnl1 inhibits this pathway. Nonetheless, any effects must be selective, because NFκB also contributes to the up-regulation of genes (e.g., CCL5 and CCL2) unaffected by Btnl1 (37, 38).
There may be additional targets of Btnl1 regulation, but those so far identified are provocative in their major contributions to intestinal immuno-pathobiology. CXCL1, IL-6, and CCL4 promote the influx of neutrophils, monocytes, and CD8+ memory T cells, respectively (39–41), and IL-15 promotes the growth and cytolytic potential of CD8+ memory cells and of intestinal IEL. Although each of these functions may be host-protective, attenuation by Btnl1 may limit progression to chronic inflammation (40). IL-6 also directs intestinal CD4 T-cell differentiation toward IL-17 production, particularly in response to specific bacterial colonization (42–44). Although such IL-6 is commonly attributed to intestinal DCs (45, 46), its production by enterocytes may reinforce IL-17 production by infiltrating systemic T cells. Hence, attenuation of IL-6 expression may limit immunopathologies involving IL17-dependent neutrophilic infiltrates, consistent with the genetic associations of Btnl genes with colitis and sarcoidosis (16, 18).
IL-6 and IL-15 also contribute to epithelial growth (32), in part by rescuing cells from apoptosis. In the steady state, this may promote wound healing and barrier integrity, and it is noteworthy that intestinal epithelial turnover and differentiation is dysregulated in mice lacking γδ IELs that may up-regulate IL-6 production by enterocytes (Fig. 5) (5). However, the same actions of IL-6 in the chronic phase can contribute to colitis-associated colorectal cancer (47): hence, attenuation by Btnl genes may pose an important barrier to dysplasia and neoplasia. Each of these scenarios starkly highlights the power of the epithelial cell to regulate local immune responses and their consequences. By regulating how epithelial cells respond to neighboring activated IEL, Btnl1 may attenuate local responses to day-to-day infection, whereas by attenuating responsiveness to systemic T cells, Btnl1 may also antagonize the pathologic consequences of inflammatory T-cell infiltrates.
The mode of action of Btnl1 examined in this study departs from the cited capacity of Btnl2, Btn1A1, Btn2A2, B7S3, and, very recently, Btnl1 to directly suppress protagonist T cells by counter receptor engagement (11–13, 15, 48). Indeed, despite initially investigating such a capacity, we found no reproducible significant suppression of T-cell cytokine production by Btnl1, just as we could not confirm the broad expression pattern for Btnl1 cited in the same recent report. Likewise, we could not confirm the same group's claims for a broad expression pattern of B7S3 (48) that corresponds to Skint2, a gene largely restricted to thymic and skin epithelium. Hence, although evidence for immunoregulation by Btnl genes is clearly growing, it seems premature to attempt to capture this with a single mechanism of action.
This finding is certainly not to preclude that some functions of Btnl1 are effected by engaging a counter receptor, on T cells, and/or neighboring epithelial cells. However, given the largely intracellular distribution of Btnl1 in vivo, this aspect of the protein's properties seems likely to be important. Upon T-cell activation, there is a regulated movement of CTLA4 from a perinuclear pool to the cell surface, facilitating T-cell down-regulation (49, 50). Surface Btnl1 expression can also be up-regulated following T-cell activation, possibly to attenuate the sensitivity of local cells to activated T-cell effectors. It is also provocative that Btnl1 expression shows interstrain variation that correlates with IEL repertoire variation. Clearly, the regulation of Btnl1 protein and the elucidation of interacting proteins promise to shed new light on local immune regulation in the gut. This form of regulation resonates with the emerging perspective that tissues can intrinsically regulate their sensitivity to immune effectors (51). The genetic data attest to the importance of the Btnl genes, and the potential clinical utility of tissue-associated regulators of inflammation.
Materials and Methods
Methods related to animals, cell lines, expression vectors, PCR, cytokine measurements, and flow cytometry are in SI Materials and Methods.
Cloning.
Murine Btnl1, -4, and -6 (accession nos. NM_001111094, NM_030746, and NM_030747) were identified from the National Centre for Biotechnology Information database. GeneRacer Kit (Invitrogen) was used for 3′ RACE PCR of RNA from C57BL/6 small intestine. Protein domains were predicted with TMHMM (www.cbs.dtu.dk/services/TMHMM), SignalP (www.cbs.dtu.dk/services/SignalP-2.0), SMART (http://smart.embl-heidelberg.de/), and Pfam (pfam.sanger.ac.uk).
Generation of Stably-Transfected N-FLAG-Btnl1-pMX-IRES-GFP MODE-K Cells.
HEK293 cells were transfected using polyethylenimine (Polysciences) by standard procedures. MODE-K cells were transduced with viral supernatants and sorted for GFPhi cells (SI Materials and Methods).
Generation of Btnl1 Polyclonal Antibody.
A KLH-conjugated synthetic peptide from the extracellular murine Btnl1 protein sequence was injected into New Zealand White rabbits. Preimmune sera was collected from each rabbit as a negative control. Antisera were collected postimmunization, and reactivity tested by ELISA against the original peptide. The antibody was purified on a peptide column, and preimmune sera were purified over protein A.
Isolation and Culture of Intestinal Cells.
IECs and IEL from murine small bowel were isolated according to previously described procedures (52–54), and either analyzed directly or cultured as described in SI Materials and Methods.
Protein Purifications, SDS/PAGE and Immunoblotting.
Purified mouse IECs or MODE-K cells were lysed in buffer [20 mM TrisHCl, pH 8, 137 mM NaCl, 2 mM EDTA, 10% glycerol, 0.1% Triton-X 100 with protease-inhibitor mixture (Roche)], and clarified by centrifugation. Immunopurification was performed using the anti-Btnl1 antibody covalently coupled to beads using the AminoLink plus Immobilization kit (Pierce). Purified Btnl1 was treated with 1 U of N-glycosidase F (Roche) according to manufacturer's instructions. SDS/PAGE and blotting onto PVDF membranes were performed according to standard protocols, and the membranes developed with either Rabbit preimmune Ig, or the anti-Btnl1 antibody, as indicated. As loading and transfer controls, membranes were blotted against GAPDH (6C5; Millipore); the secondary antibodies used were HRPO-labeled anti-rabbit and anti-mouse antisera from Thermo Scientific.
Immunohistochemistry.
Acetone-fixed frozen sections or Methanol-Carnoy's fixed and paraffin-embedded sections were stained with anti-Btnl1 only or double stained for expression of Btnl1 and CD3. The alkaline phosphatase conjugated anti-CD3 staining was developed with Vector Blue Alkaline Phosphatase Substrate Kit III (Vector Laboratories) and peroxidase stained Btnl1 sections developed using the 3, 3′-diaminobenzidine substrate kit (Vector Laboratories). Staining is described in SI Materials and Methods.
Confocal Microscopy.
Frozen sections or Methanol-Carnoy's fixed and paraffin embedded sections were double stained for Btnl1 and CD3 or Btnl1 and A33. Staining is described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. D. Kaiserlian for the gift of MODE-K cells, and Professor J. Spencer for guidance in immunohistology. This work is supported in part by the Wellcome Trust (A.C.H.); a European Molecular Biology Organization long-term fellowship and a Marie-Curie Intra-European fellowship (to M.S.); a National Institutes of Health Research Biomedical Research Centre Fellowship (to L.A.-D.); and the Swedish Foundation for Strategic Research, through its support of the Mucosal Immunobiology and Vaccine Center (A.B.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010647108/-/DCSupplemental.
References
- 1.Kapsenberg ML. Dendritic-cell control of pathogen-driven T-cell polarization. Nat Rev Immunol. 2003;3:984–993. doi: 10.1038/nri1246. [DOI] [PubMed] [Google Scholar]
- 2.Zaph C, et al. Epithelial-cell-intrinsic IKK-beta expression regulates intestinal immune homeostasis. Nature. 2007;446:552–556. doi: 10.1038/nature05590. [DOI] [PubMed] [Google Scholar]
- 3.Swamy M, Jamora C, Havran W, Hayday A. Epithelial decision makers: In search of the “epimmunome”. Nat Immunol. 2010;11:656–665. doi: 10.1038/ni.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayday A, Theodoridis E, Ramsburg E, Shires J. Intraepithelial lymphocytes: Exploring the Third Way in immunology. Nat Immunol. 2001;2:997–1003. doi: 10.1038/ni1101-997. [DOI] [PubMed] [Google Scholar]
- 5.Komano H, et al. Homeostatic regulation of intestinal epithelia by intraepithelial gamma delta T cells. Proc Natl Acad Sci USA. 1995;92:6147–6151. doi: 10.1073/pnas.92.13.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyden LM, et al. Skint1, the prototype of a newly identified immunoglobulin superfamily gene cluster, positively selects epidermal gammadelta T cells. Nat Genet. 2008;40:656–662. doi: 10.1038/ng.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis JM, et al. Selection of the cutaneous intraepithelial gammadelta+ T cell repertoire by a thymic stromal determinant. Nat Immunol. 2006;7:843–850. doi: 10.1038/ni1363. [DOI] [PubMed] [Google Scholar]
- 8.Rhodes DA, Stammers M, Malcherek G, Beck S, Trowsdale J. The cluster of BTN genes in the extended major histocompatibility complex. Genomics. 2001;71:351–362. doi: 10.1006/geno.2000.6406. [DOI] [PubMed] [Google Scholar]
- 9.Stammers M, Rowen L, Rhodes D, Trowsdale J, Beck S. BTL-II: A polymorphic locus with homology to the butyrophilin gene family, located at the border of the major histocompatibility complex class II and class III regions in human and mouse. Immunogenetics. 2000;51:373–382. doi: 10.1007/s002510050633. [DOI] [PubMed] [Google Scholar]
- 10.Linsley PS, Peach R, Gladstone P, Bajorath J. Extending the B7 (CD80) gene family. Protein Sci. 1994;3:1341–1343. doi: 10.1002/pro.5560030820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen T, Liu XK, Zhang Y, Dong C. BTNL2, a butyrophilin-like molecule that functions to inhibit T cell activation. J Immunol. 2006;176:7354–7360. doi: 10.4049/jimmunol.176.12.7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnett HA, et al. BTNL2, a butyrophilin/B7-like molecule, is a negative costimulatory molecule modulated in intestinal inflammation. J Immunol. 2007;178:1523–1533. doi: 10.4049/jimmunol.178.3.1523. [DOI] [PubMed] [Google Scholar]
- 13.Smith IA, et al. BTN1A1, the mammary gland butyrophilin, and BTN2A2 are both inhibitors of T cell activation. J Immunol. 2010;184:3514–3525. doi: 10.4049/jimmunol.0900416. [DOI] [PubMed] [Google Scholar]
- 14.Malcherek G, et al. The B7 homolog butyrophilin BTN2A1 is a novel ligand for DC-SIGN. J Immunol. 2007;179:3804–3811. doi: 10.4049/jimmunol.179.6.3804. [DOI] [PubMed] [Google Scholar]
- 15.Yamazaki T, et al. A butyrophilin family member critically inhibits T cell activation. J Immunol. 2010;185:5907–5914. doi: 10.4049/jimmunol.1000835. [DOI] [PubMed] [Google Scholar]
- 16.Valentonyte R, et al. Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nat Genet. 2005;37:357–364. doi: 10.1038/ng1519. [DOI] [PubMed] [Google Scholar]
- 17.Price P, et al. Two major histocompatibility complex haplotypes influence susceptibility to sporadic inclusion body myositis: Critical evaluation of an association with HLA-DR3. Tissue Antigens. 2004;64:575–580. doi: 10.1111/j.1399-0039.2004.00310.x. [DOI] [PubMed] [Google Scholar]
- 18.Mochida A, et al. Butyrophilin-like 2 gene is associated with ulcerative colitis in the Japanese under strong linkage disequilibrium with HLA-DRB1*1502. Tissue Antigens. 2007;70:128–135. doi: 10.1111/j.1399-0039.2007.00866.x. [DOI] [PubMed] [Google Scholar]
- 19.Rhodes DA, de Bono B, Trowsdale J. Relationship between SPRY and B30.2 protein domains. Evolution of a component of immune defence? Immunology. 2005;116:411–417. doi: 10.1111/j.1365-2567.2005.02248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mische CC, et al. Retroviral restriction factor TRIM5alpha is a trimer. J Virol. 2005;79:14446–14450. doi: 10.1128/JVI.79.22.14446-14450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhodes DA, Trowsdale J. TRIM21 is a trimeric protein that binds IgG Fc via the B30.2 domain. Mol Immunol. 2007;44:2406–2414. doi: 10.1016/j.molimm.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 22.Lefrancois L, LeCorre R, Mayo J, Bluestone JA, Goodman T. Extrathymic selection of TCR gamma delta + T cells by class II major histocompatibility complex molecules. Cell. 1990;63:333–340. doi: 10.1016/0092-8674(90)90166-c. [DOI] [PubMed] [Google Scholar]
- 23.Pereira P, Lafaille JJ, Gerber D, Tonegawa S. The T cell receptor repertoire of intestinal intraepithelial gammadelta T lymphocytes is influenced by genes linked to the major histocompatibility complex and to the T cell receptor loci. Proc Natl Acad Sci USA. 1997;94:5761–5766. doi: 10.1073/pnas.94.11.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vidal K, Grosjean I, Evillard JP, Gespach C, Kaiserlian D. Immortalization of mouse intestinal epithelial cells by the SV40-large T gene. Phenotypic and immune characterization of the MODE-K cell line. J Immunol Methods. 1993;166:63–73. doi: 10.1016/0022-1759(93)90329-6. [DOI] [PubMed] [Google Scholar]
- 25.Sturgess RP, et al. Effects of interferon-gamma and tumour necrosis factor-alpha on epithelial HLA class-II expression on jejunal mucosal biopsy specimens cultured in vitro. Scand J Gastroenterol. 1992;27:907–911. doi: 10.3109/00365529209000161. [DOI] [PubMed] [Google Scholar]
- 26.Hughes A, et al. Expression of MHC class II (Ia) antigen by the neonatal enterocyte: The effect of treatment with interferon-gamma. Immunology. 1991;72:491–496. [PMC free article] [PubMed] [Google Scholar]
- 27.Ebert EC. Intra-epithelial lymphocytes: Interferon-gamma production and suppressor/cytotoxic activities. Clin Exp Immunol. 1990;82:81–85. doi: 10.1111/j.1365-2249.1990.tb05407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Compte E, Pontarotti P, Collette Y, Lopez M, Olive D. Frontline: Characterization of BT3 molecules belonging to the B7 family expressed on immune cells. Eur J Immunol. 2004;34:2089–2099. doi: 10.1002/eji.200425227. [DOI] [PubMed] [Google Scholar]
- 29.Johnstone CN, et al. Characterization of mouse A33 antigen, a definitive marker for basolateral surfaces of intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2000;279:G500–G510. doi: 10.1152/ajpgi.2000.279.3.G500. [DOI] [PubMed] [Google Scholar]
- 30.Viney JL, MacDonald TT. Selective death of T cell receptor gamma/delta+ intraepithelial lymphocytes by apoptosis. Eur J Immunol. 1990;20:2809–2812. doi: 10.1002/eji.1830201242. [DOI] [PubMed] [Google Scholar]
- 31.Brunner T, Arnold D, Wasem C, Herren S, Frutschi C. Regulation of cell death and survival in intestinal intraepithelial lymphocytes. Cell Death Differ. 2001;8:706–714. doi: 10.1038/sj.cdd.4400854. [DOI] [PubMed] [Google Scholar]
- 32.Naugler WE, Karin M. The wolf in sheep's clothing: The role of interleukin-6 in immunity, inflammation and cancer. Trends Mol Med. 2008;14:109–119. doi: 10.1016/j.molmed.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 33.Libermann TA, Baltimore D. Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol Cell Biol. 1990;10:2327–2334. doi: 10.1128/mcb.10.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JM, et al. Nuclear factor-kappa B activation pathway in intestinal epithelial cells is a major regulator of chemokine gene expression and neutrophil migration induced by Bacteroides fragilis enterotoxin. Clin Exp Immunol. 2002;130:59–66. doi: 10.1046/j.1365-2249.2002.01921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirose K, et al. Interleukin-15 may be responsible for early activation of intestinal intraepithelial lymphocytes after oral infection with Listeria monocytogenes in rats. Infect Immun. 1998;66:5677–5683. doi: 10.1128/iai.66.12.5677-5683.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z, et al. CCAAT/Enhancer-binding protein beta and NF-kappaB mediate high level expression of chemokine genes CCL3 and CCL4 by human chondrocytes in response to IL-1beta. J Biol Chem. 2010;285:33092–33103. doi: 10.1074/jbc.M110.130377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Page K, et al. Regulation of airway epithelial cell NF-kappa B-dependent gene expression by protein kinase C delta. J Immunol. 2003;170:5681–5689. doi: 10.4049/jimmunol.170.11.5681. [DOI] [PubMed] [Google Scholar]
- 38.Ramana CV, et al. Lung epithelial NF-kappaB and Stat1 signaling in response to CD8+ T cell antigen recognition. J Interferon Cytokine Res. 2006;26:318–327. doi: 10.1089/jir.2006.26.318. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe K, Kinoshita S, Nakagawa H. Purification and characterization of cytokine-induced neutrophil chemoattractant produced by epithelioid cell line of normal rat kidney (NRK-52E cell) Biochem Biophys Res Commun. 1989;161:1093–1099. doi: 10.1016/0006-291x(89)91355-7. [DOI] [PubMed] [Google Scholar]
- 40.Hurst SM, et al. Il-6 and its soluble receptor orchestrate a temporal switch in the pattern of leukocyte recruitment seen during acute inflammation. Immunity. 2001;14:705–714. doi: 10.1016/s1074-7613(01)00151-0. [DOI] [PubMed] [Google Scholar]
- 41.Taub DD, Conlon K, Lloyd AR, Oppenheim JJ, Kelvin DJ. Preferential migration of activated CD4+ and CD8+ T cells in response to MIP-1 alpha and MIP-1 beta. Science. 1993;260:355–358. doi: 10.1126/science.7682337. [DOI] [PubMed] [Google Scholar]
- 42.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 43.Bettelli E, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 44.Ivanov II, et al. Specific microbiota direct the differentiation of IL-17–producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lombardi V, Van Overtvelt L, Horiot S, Moingeon P. Human dendritic cells stimulated via TLR7 and/or TLR8 induce the sequential production of Il-10, IFN-gamma, and IL-17A by naive CD4+ T cells. J Immunol. 2009;182:3372–3379. doi: 10.4049/jimmunol.0801969. [DOI] [PubMed] [Google Scholar]
- 47.Greten FR, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 48.Yang Y, et al. Characterization of B7S3 as a novel negative regulator of T cells. J Immunol. 2007;178:3661–3667. doi: 10.4049/jimmunol.178.6.3661. [DOI] [PubMed] [Google Scholar]
- 49.Linsley PS, et al. Intracellular trafficking of CTLA-4 and focal localization towards sites of TCR engagement. Immunity. 1996;4:535–543. doi: 10.1016/s1074-7613(00)80480-x. [DOI] [PubMed] [Google Scholar]
- 50.Iida T, et al. Regulation of cell surface expression of CTLA-4 by secretion of CTLA-4-containing lysosomes upon activation of CD4+ T cells. J Immunol. 2000;165:5062–5068. doi: 10.4049/jimmunol.165.9.5062. [DOI] [PubMed] [Google Scholar]
- 51.Jamieson AM, Yu S, Annicelli CH, Medzhitov R. Influenza virus-induced glucocorticoids compromise innate host defense against a secondary bacterial infection. Cell Host Microbe. 2010;7:103–114. doi: 10.1016/j.chom.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lundqvist C, Hammarström ML, Athlin L, Hammarström S. Isolation of functionally active intraepithelial lymphocytes and enterocytes from human small and large intestine. J Immunol Methods. 1992;152:253–263. doi: 10.1016/0022-1759(92)90147-l. [DOI] [PubMed] [Google Scholar]
- 53.Booth C, O'Shea JA. Isolation and culture of intestinal epithelial cells. In: Freshney RI, Freshney MG, editors. Culture of Epithelial Cells, Second Edition. New York: John Wiley and Sons, Inc.; 2002. [Google Scholar]
- 54.Guy-Grand D, Griscelli C, Vassalli P. The mouse gut T lymphocyte, a novel type of T cell. Nature, origin, and traffic in mice in normal and graft-versus-host conditions. J Exp Med. 1978;148:1661–1677. doi: 10.1084/jem.148.6.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.