Abstract
Alternative pre-mRNA splicing is a central element of eukaryotic gene expression. Its deregulation can lead to disease, and methods to change splice site selection are developed as potential therapies. Spinal muscular atrophy is caused by the loss of the SMN1 (survival of motoneuron 1) gene. A therapeutic avenue for spinal muscular atrophy treatment is to promote exon 7 inclusion of the almost identical SMN2 (survival of motoneuron 2) gene. The splicing factor tra2-beta1 promotes inclusion of this exon and is antagonized by protein phosphatase (PP) 1. To identify new compounds that promote exon 7 inclusion, we synthesized analogs of cantharidin, an inhibitor of PP1, and PP2A. Three classes of compounds emerged from these studies. The first class blocks PP1 and PP2A activity, blocks constitutive splicing in vitro, and promotes exon 7 inclusion in vivo. The second class has no measurable effect on PP1 activity but activates PP2A. This class represents the first compounds described with these properties. These compounds cause a dephosphorylation of Thr-33 of tra2-beta1, which promotes exon 7 inclusion. The third class had no detectable effect on phosphatase activity and could promote exon 7 via allosteric effects. Our data show that subtle changes in similar compounds can turn a phosphatase inhibitor into an activator. These chemically related compounds influence alternative splicing by distinct mechanisms.
Keywords: Drug Action, Protein Phosphatase, RNA Processing, RNA Splicing, Spliceosome, Spinal Muscular Atrophy, Pseudocantharidin, Survival of Motoneuron Genes
Introduction
An estimated 95% of human multi-exon genes undergo alternative pre-mRNA splicing (1, 2). Unlike promoter activity that regulates the abundance of transcripts, alternative splicing influences the structure of the mRNAs and their encoded proteins (reviewed in Ref. 3). The high incidence of alternative splicing and its ability to increase the coding capacity of the genome makes it a central element in eukaryotic gene expression.
Despite its importance, we do not fully understand how splice sites, especially the alternative ones, are selected. The accurate recognition of splice sites in vivo is the result of a combinatorial mechanism (4–7) where protein complexes assemble on the nascent pre-mRNA, because of the combination of weak RNA/RNA, RNA/protein, and protein/protein interactions. Transient interaction between the spliceosome and regulatory proteins assembling on the pre-mRNA help in the recognition of splice sites (8). There is increasing evidence that the phosphorylation state of splicing regulatory proteins is a crucial element for this recognition process (reviewed in Ref. 9), because phosphorylation/dephosphorylation often changes the affinity between proteins in these complexes. The spliceosome acts in a stepwise fashion on the pre-mRNA, and four distinct splicing complexes, the H, A, B and C complexes, have been identified (7). The H and E complexes contain the pre-mRNA imbedded with various proteins, among them hnRNPs and SR proteins as well as the U1 snRNP bound to the 5′ splice site. The entry of U2 snRNP defines the branch point and leads to the formation of the A complex. The entry of the U4/5/6 snRNP marks the B complex that transitions to the C complex upon the exit of the U1 and U4 snRNPs. The formation of the lariat and the joining of exons mark steps 1 and 2 of splicing that occur in the catalytic C complex.
In contrast to the ribosome, where numerous substances have been identified that block protein synthesis at specific steps, relatively few compounds are known that uniquely affect splicing (10). Substances that change splice site selection or block splicing at well defined points are research tools that could provide a detailed understanding of the splicing reactions within the H, A, B, and C complexes. Because numerous human diseases are caused by the selection of incorrect splice sites (11, 12), substances that influence alternative splicing may also provide potential agents for the treatment of these diseases.
Spinal Muscular Atrophy
Spinal muscular atrophy (SMA)3 is a neurodegenerative disease that is currently the leading genetic cause of death in children (13). Because of an evolutionarily recent gene duplication, humans possess two related SMN genes, SMN1 and SMN2, that are prone to recombination. The loss of the SMN1 gene that encodes the survival-of-motoneuron (SMN) protein causes the death of neurons by an unknown mechanism, presumably involving numerous small changes in alternative splicing of pre-mRNAs and/or a defect in the neuronal transport system (14).
Because of the gene duplication, humans also possess one or more copies of the SMN2 gene that is almost identical to SMN1. One crucial difference is a silent C → T mutation in exon 7 of the pre-mRNA arising from SMN2 that causes predominant exon 7 skipping. The failure to include this exon leads to a truncated, unstable protein. A possible therapeutic approach for SMA involves altering the splicing pattern of the SMN2 pre-mRNA in favor of exon 7 inclusion. Exon 7 inclusion is regulated by several elements of the SMN2 pre-mRNA, including a central tra2-beta1 enhancer and an intronic silencer (13, 15, 16). Tra2-beta1 is a splicing regulator and a member of the SR protein family that binds to RNA using its central RNA recognition motif. We previously showed that tra2-beta1 contains a functional protein phosphatase 1 (PP1)-binding site in its RNA recognition motif. In all cases tested to date, PP1 promotes the skipping of tra2-beta1-dependent exons. Blocking PP1 activity promotes inclusion of tra2-beta1-dependent exons, including exon 7 in SMN2 (17). Inhibition of PP1 can be achieved by regulatory proteins, such as NIPP1 (nuclear inhibitor of PP1) or by chemical substances, such as tautomycin or cantharidin (17).
Synthesis of Pseudocantharidins
Cantharidin is a small molecule that has PP1 and PP2A as defined molecular targets and that changes alternative splicing (17). Both phosphatases are necessary for the splicing reaction (18, 19). Because of its defined cellular targets and simple chemical structure, we used cantharidin as a starting point to synthesize a series of phosphatase activity modulators, named “iso-” and “pseudocantharidins,” that were subsequently tested for their effect on alternative splicing.
Five novel compounds promote SMN2 exon 7 inclusion and lead to formation of SMN protein in patient fibroblasts. As expected for cantharidin-like structures, two of the compounds are PP1/PP2A inhibitors. Unexpectedly, two compounds activate PP2A but have no effect on PP1, which is the first time that a chemical PP2A agonist without an influence on PP1 has been described. PP1/PP2A-inhibiting pseudocantharidins block constitutive splicing. The PP2A activators dephosphorylate tra2-beta1 at position Thr-33, and mutating this position from threonine to alanine promotes exon inclusion, suggesting that the phosphorylation status of this residue is under cellular control and influences exon 7 usage.
EXPERIMENTAL PROCEDURES
Synthesis of Pseudocantharidins
(3aR,4S,7R,7aS)-4,7-Epoxy-2-(4-methoxyphenyl)-3a,7a-dimethyl-1H-isoindole-1,3(2H)-dione (Pseudocantharidin A)
A mixture of 80 mg (0.41 mmol) of cantharidin and 108 mg (0.88 mmol, 2.1 eq) of p-anisidine was heated at 150–155 °C in a 1-dram vial equipped with a rubber septum at reduced pressure (∼20 mm) for 30 min to afford, after repeatedly diluting the cooled product with 0.5 ml of methanol and decanting the dark-colored methanolic p-anisidine solution from the crude crystalline product and finally after chromatography using 1:5 ethyl acetate-hexane (two developments), 60 mg (48%) of pseudocantharidin A: mp 137–139 °C (recrystallized from ethyl acetate); IR (KBr) 1701 cm−1; 1H NMR (CDCl3) δ 1.24 (s, 6, CH3), 1.6–1.9 (m, 4, C-5 and C-6 CH2), 3.82 (two s, 3, OCH3), 4.68 (br s, 2, C-4 and C-7 CH), 6.9–7.25 (m, 4, ArH); 13C NMR (CDCl3) δ 13.1 (CH3), 24.0 (C-5 and C-6), 54.2 and 55.7 (C-4 and C-7), 84.3 (C-3a and C-7a), 114.6 (Ar C), 124.9 (Ar C), 127.9 (Ar C), 159.7 (Ar C), 181.1 (C = O); LRMS m/z (intensity): 108 (34), 232 (100), 301 (83); HRMS calculated for C17H19NO4: 301.1314, found 301.1323 (mean of five determinations, S.D. 1.8 ppm; error 3.1 ppm). Analysis calculated for C17H19NO4: C, 67.76; H, 6.36. Found: C, 67.93; H, 6.32.
(4-Bromophenyl) ((3aR,4R,7S,7aS)-rel-4,7-Epoxy-1H-isoindol-2(3H,3aH,4H,5H,6H,7H,7aH)-yl)methanone (Pseudocantharidin C)
To 14 ml of 2 m (28 mmol, 3 eq) lithium aluminum hydride in THF at 0 °C was added 1.5 g (9 mmol) of (3aR,4S,7R,7aS)-rel-octahydro-4,7-epoxy-1H-isoindole-1,3-dione (20) in 16 ml of anhydrous THF. The mixture was refluxed for 3 h. The reaction was quenched by the successive addition of 1 ml of water, 1 ml of 15% aqueous NaOH solution, and 2 ml of water. The product was filtered through Celite and concentrated. The product was diluted with ethyl acetate, washed with brine, and dried over anhydrous MgSO4. To 400 mg (2.9 mmol) of crude amine in 7.3 ml of anhydrous pyridine was added 834 mg (3.8 mmol, 1.3 eq) of 4-bromobenzoyl chloride. The mixture was stirred at 25 °C for 22 h. The mixture was quenched with ice, diluted with ethyl acetate, washed successively with saturated CuSO4 solution and brine, and dried over anhydrous MgSO4. The product was chromatographed in ethyl acetate to give 800 mg (86%) of pseudocantharidin C: mp 125–127° (recrystallized from 1:5 hexane-ethyl acetate); IR (KBr) 1638 cm−1; 1H NMR (CDCl3) δ 1.2–1.3 (m, 4, C-5 and C-6 H), 2.4–2.6 (m, 2, C-3a and C-7a H), 3.2–3.7 (m, 4, C-1 and C-3 H), 4.1–4.5 (m, 2, C-4 and C-7 H), 7.3–7.6 (m, 4, ArH); 13C NMR (CDCl3) δ 28.3 and 28.6 (C-5 and C-6), 47.3 and 49.3 (C-4 and C-7), 50.5 and 54.1 (C-1 and C-3), 80.8 and 81.2 (C-3a and C-7a), 124.2 (Ar C), 129.0 (Ar C), 131.7 (Ar C), 136.0 (Ar C), 168.0 (C = O); LRMS m/z (intensity) 138 (99), 183 (87), 321 (15) and 323 (15); HRMS C15H16O2N79Br: 321.0364, found 321.0361 (mean of five determinations, S.D. 0.6 ppm; error −0.9 ppm). Analysis calculated for C15H16NO2Br: C, 55.91; H, 5.01. Found: C, 55.76; H, 4.86.
(1R,2S,3S,4S)-rel-3-(Hydroxymethyl)-7-oxabicyclo[2.2.1]heptan-2-yl)diphenylmethanol (Pseudocantharidin B)
To a solution of 22.4 ml of a 0.4 m solution of phenylmagnesium bromide (9 mmol, 1.5 eq) in anhydrous THF was added 502 mg (3 mmol) of isocantharidin (21, 22) to afford the intermediate γ-lactone, (3aR,4R,7S,7aR)-rel-octahydro-3,3-diphenyl-4,7-epoxyisobenzofuran-1-one. The crude γ-lactone was reduced using 311 mg (8.2 mmol, 5.2 eq) of lithium aluminum hydride to afford, after chromatography on Merck silica F254 preparative layer plates using ethyl acetate, 406 mg (47%) of pseudocantharidin B: mp 174–175° (recrystallized from 1:1 CH2Cl2-hexane); IR (KBr) 3415 cm−1 (br OH); 1H NMR (CDCl3) δ 1.44–1.50 (m, 1, H-5β), 1.57–1.66 (m, 2, H-5α and H-6α), 1.77–1.86 (m, 1, H-6β), 2.25 (ddd, 1, H-3), 2.74 (dd, J = 3.6 and 7.2 Hz, 1, CH2OH), 3.20 (ddd, 1, CH2OH), 3.33 (d, J2,3 = 8.8 Hz, 1, H-2), 3.43 (ddd, 1, CH2OH), 4.43 (d, J = 4.8 Hz, 1, H-1), 4.59 (d, J = 5.2 Hz, 1, H-4), 5.14 (s, 1, C(OH)(C6H5)2), 7.07–7.17 (m, 2, para-ArH), 7.21–7.29 (m, 4, meta-ArH), 7.52–7.61 (m, 4, ortho-ArH); 13C NMR (CDCl3) δ 29.4 and 30.7 (C-5 and C-6), 50.1 and 55.3 (C-2 and C-3), 62.4 and 78.1 (CH2OH and C(OH)Ph2), 79.1 and 81.3 (C-1 and C-4), 125.1 (Ar C), 125.5 (Ar C), 126.4 (Ar C), 126.8 (Ar C), 128.5 (Ar C), 128.7 (Ar C), 148.2 (Ar C), 148.6 (Ar C); LRMS m/z (intensity) 105 (100), 183 (79), 215 (25), 292 (2); HRMS calculated for C20H20O2 (M+-H2O) 292.1463, found 292.1457 (mean of five determinations, S.D. 1.8 ppm; error −1.9 ppm). Analysis calculated for C20H22O3: C, 77.39; H, 7.14. Found: C, 77.32; H, 7.28.
(3aR,4S,7R,7aS)-4,7-Epoxy-3a,7a-dimethyl-2-phenyl-1H-isoindole-1,3(2H)-dione (Pseudocantharidin D)
A mixture of 72 mg (0.37 mmol) of cantharidin and 342 mg (3.7 mmol, 10 eq) of aniline was heated at 160–170 °C (760 mm) for 4 h to afford, after chromatography using 1:2 ethyl acetate-hexane, 94 mg (94%) of pseudocantharidin D: mp 114–116 °C (recrystallized from 1:5 ethyl acetate-hexane); IR (KBr) 1709 cm−1; 1H NMR (CDCl3) δ 1.26 (s, 6, CH3), 1.58–1.94 (m, 4, C-5 and C-6 CH2), 4.70 (dd, 2, J = 2.4 and 3.2 Hz, C-4 and C-7 CH), 7.25–7.55 (m, 5, ArH); 13C NMR (CDCl3) δ 13.1 (CH3), 24.0 (C-5 and C-6), 54.3 (C-4 and C-7), 84.3 (C-3a and C-7a), 126.7 (Ar C), 128.8 (Ar C), 129.3 (Ar C), 132.3 (Ar C), 180.9 (C = O); LRMS m/z (intensity): 203 (100), 271 (9); HRMS calculated for C16H17NO3: 271.1208, found 271.1206 (mean of five determinations, S.D. 1.8 ppm; error −0.7 ppm). Analysis calculated for C16H17NO3: C, 70.83; H, 6.32. Found: C, 70.93; H, 6.27.
Phosphatase Assays
PP1, PP2A, and glycogen phosphorylase were purified from rabbit skeletal muscle, and the phosphorylase phosphatase assays were performed as described previously (23).
Drug Treatment of Cells
Four hours after transfection with the reporter minigene, HEK293 cells were treated with the indicated concentration of compounds for an additional 12 h. At 16 h post-transfection, total RNA was extracted using Qiagen RNAeasy kit. To determine toxicity, 106 cells/cm2 SMAI cells were seeded into 24-well plates the day before treatment. The MTT assay was performed after adding different concentration of compounds to SMAI cells for 24 h. For protein level detection, 106 cells/cm2 SMAI cells were seeded into six-well plates. The cells were treated with compounds every other day for 15 days. The growth medium was changed prior to compound addition. The cells were lysed with radioimmune precipitation assay buffer after treatment with compounds.
RT-PCR
Reverse transcription was performed using SV40pA RT (TGGTTTGTCCAAACTCATCAA). PCR was performed using pCI For (GGTGTCCACTCCCAGTTCAA) and SMNex8rev (GCCTCACCACCGTGCTGG). For reverse transcription, 400 ng of total RNA (200 ng/μl), 5 pmol of reverse primer, and 40 units of SuperScript III reverse transcriptase were mixed in 5 μl of RT buffer. To reverse transcribe the RNA, the reaction was incubated at 55 °C for 50 min. One-third of the RT reaction was used for cDNA amplification. The reaction was performed in 25 μl and contained 10 pmol of specific forward and reverse primers, 200 μm dNTPs, 1× Taq polymerase buffer, and 1 unit of Taq DNA polymerase. The amplification was carried out in an Eppendorf PCR system thermocycler under the following conditions: initial denaturation for 4 min at 94 °C; 28 cycles of 30 s at 94 °C, 30 s at 60 °C; and extension of 45 s at 72 °C. After the last cycle, the reaction was held for 5 min at the extension temperature to complete the amplification of all products.
SMA Cells
Fibroblasts from a 7-month-old, male, Caucasian SMA type I patient was obtained from the Coriell Institute for Medical Research (clone number GM00232).
In Vitro Splicing and Spliceosomal Assembly
A transcription template for the MINX pre-mRNA was generated from the pMINX plasmid (24) by PCR. HeLa nuclear extract was prepared according to Dignam et al. (25). Splicing reactions contained 40% (v/v) HeLa nuclear extract in buffer D (20 mm HEPES-KOH, pH 7.9, 100 mm KCl, 1.5 mm MgCl2, 0.2 mm EDTA, 10% (v/v) glycerol, 0.5 mm DTT, 0.5 mm PMSF), 25 mm KCl, 3 mm MgCl2, 20 mm creatine phosphate, 2 mm ATP, 3 nm 32P-labeled pre-mRNA, and the indicated concentration of isocantharidin or pseudocantharidins. The nuclear extract was preincubated with the compounds (or just water as a control) for 10 min at 30 °C, and the reactions were then started by the addition of the other components. For the analysis of the splicing products, RNA was isolated by proteinase K treatment, phenol extraction, and ethanol precipitation, separated by denaturing polyacrylamide gel electrophoresis on an 8.3 m urea, 14% (w/v) polyacrylamide gel, and visualized by autoradiography. For the analysis of the spliceosomal complexes, 10 μl of the splicing reaction were added to 2.5 μl of load buffer (1× Tris base/boric acid/EDTA, 30% (v/v) glycerol, 1.25 μg/μl heparin) at the time points indicated and then placed on ice. The complexes were separated on 1.8% (w/v) agarose gels (26).
Cell Viability
Viability was tested by metabolizing MTT by mitochondrial dehydrogenases (Sigma): 1.000,000 cells were treated with the compounds overnight, and the cells were subsequently analyzed with MTT. The MTT staining obtained from cells receiving just Me2SO control was set as 100% signal.
Antisera
Anti-SAP130 and anti-actin antisera were from Abcam. The anti-Thr-33 phospho-specific antiserum was made by immunizing rabbits with CKSARH-pT-PARSR peptide, followed by subsequent affinity purification (Biogenes, Germany).
RESULTS
Synthesis of Pseudocantaridins, a New Series of Cantharidin-like Compounds
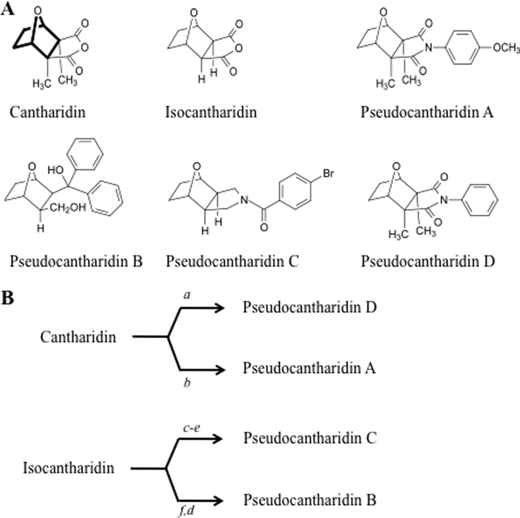
We previously showed that PP1-inhibiting substances, like cantharidin or tautomycin, influence alternative splicing (17). PP1 dephosphorylates several splicing factors after binding to an evolutionarily conserved RVxF-type docking motif in the beta 4 strand of their RNA recognition motif. One of these proteins is tra2-beta1, a splicing factor that binds to SMN2 exon 7 and promotes its inclusion (27). Because of its simple chemical structure, we used cantharidin as a starting point to generate new substances, called pseudocantharidins, which were tested for their effect on splice site selection. The pseudocantharidins A, B, C, and D (Fig. 1A) were synthesized either from commercially available cantharidin or the previously reported isocantharidin (21, 22) according to the procedures summarized in Fig. 1B. For purposes of this paper, pseudocantharidin refers to the cantharidin-like structures in Fig. 1A. Pseudocantharidins A, C, and D were synthesized as achiral meso-compounds, and pseudocantharidin B was synthesized as a racemic mixture.
FIGURE 1.
Structure and synthesis of pseudocantharidins. A, structures of pseudocantharidins used in this study. The 7-oxabicyclo[2.2.1]heptane skeleton common to all compounds is shown in bold. The formal names for the pseudocantharidins are: A, (3aR,4S,7R,7aS)-4,7-epoxy-2-(4-methoxyphenyl)-3a,7a-dimethyl-1H-isoindole-1,3(2H)-dione; B, (1R,2S,3S,4S)-rel-3-(hydroxymethyl)-7-oxabicyclo[2.2.1]heptan-2-yl)diphenylmethanol; C, (4-bromophenyl) (3aR,4R,7S,7aS)-rel-4,7-epoxy-1H-isoindol-2(3H,3aH,4H,5H,6H,7H, 7aH)-yl)methanone; D, (3aR,4S,7R,7aS)-4,7-epoxy-3a,7a-dimethyl-2-phenyl-1H-isoindole-1,3(2H)-dione; B, schematic overview of the synthesis of pseudocantharidins. a, aniline, 160–170 °C; b, p-anisidine, 150–155 °C (20 mm); c, urea, 150 °C (20 mm); d, LiA1H4, THF; e, 5-bromobenzoyl chloride, Py; f, C6H5MgBr.
Pseudocantharidins A and D were synthesized from cantharidin using the procedure of Bockstahler (28) in which cantharidin was melted with aniline or p-anisidine at 150–170 °C in the absence of solvent. Other reported procedures for the synthesis of similar derivatives (29–34) of cantharidin or isocantharidin involved heating amines in solution or in sealed glass tubes under pressure. Pseudocantharidin C was synthesized using a three-step procedure: (i) fusion of isocantharidin with urea to afford (3aR,4S,7R,7aS)-rel-octahydro-4,7-epoxy-1H-isoindole-1,3-dione (20); (ii) reduction of this intermediate imide with lithium aluminum hydride to furnish the intermediate pyrrolidine; and (iii) acylation of the pyrrolidine with 4-bromobenzoyl chloride. Pseudocantharidin B was synthesized by the addition of phenylmagnesium bromide to isocantharidin (21, 22) and the subsequent reduction of the intermediate γ-lactone with lithium aluminum hydride. Complete characterization of these products through a combination of infrared and NMR spectroscopy, mass spectrometry, and combustion analyses unequivocally established the assigned structures and purity of these compounds.
Pseudocantharidins Change Alternative Splicing of the SMN2 pre-mRNA and Exhibit Different Cellular Toxicity
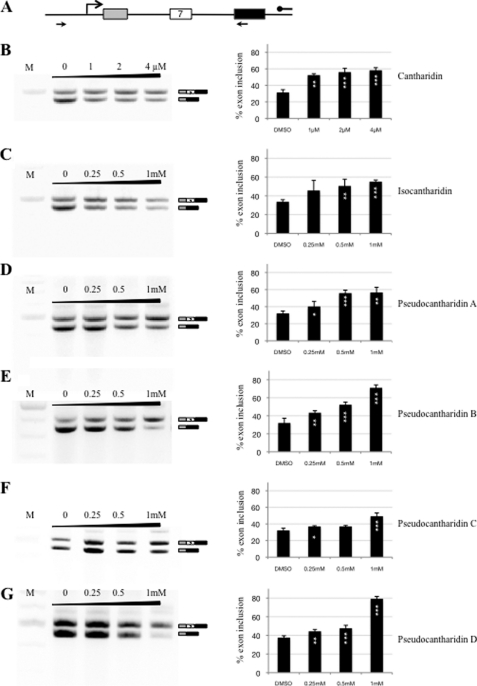
Using cantharidin or isocantharidin (21, 22) as starting materials, we synthesized over 50 different pseudocantharidins and tested them for their ability to promote inclusion of exon 7 of the SMN2 gene. HEK293 cells were transfected with 1 μg of pCl-SMN2 reporter gene (27, 35) (Fig. 2A) and treated with different pseudocantharidins at a concentration between 0.25 and 1 mm. Similar to cantharidin (Fig. 2B), isocantharidin promotes exon 7 inclusion (Fig. 2C). However, the effective concentration range (0.25–1 mm) for isocantharidin was ∼250 times higher than that of cantharidin that already exhibited a pronounced effect at 4 μm. Among the pseudocantharidins that were synthesized and tested for their ability to promote exon 7 inclusion in vivo, the most active pseudocantharidins A–D are those shown in Fig. 1. These pseudocantharidins increased the inclusion of exon 7 by more than 40%, a result that is comparable with the action of cantharidin (Fig. 2B) or tautomycin (17).
FIGURE 2.
Pseudocantharidins promote inclusion of SMN2 exon 7. A, schematic overview of the reporter gene construct. B–G, HEK293 cells were transfected with an SMN2 splicing reporter, consisting of exons 6–8, as well as the included introns that is schematically shown in A. After 4 h, the cells were treated with the compounds at the concentrations indicated. All of the compounds were dissolved in Me2SO, 0 refers to pure Me2SO. RNA was isolated after 16 h, and the SMN2 mRNA was amplified using the primers shown in A. The graphs on the right show the percentage of exon 7 inclusion, determined by RT-PCR. B, cantharidin; C, isocantharidin; D, pseudocantharidin A; E, pseudocantharidin B; F, pseudocantharidin C; G, pseudocantharidin D.
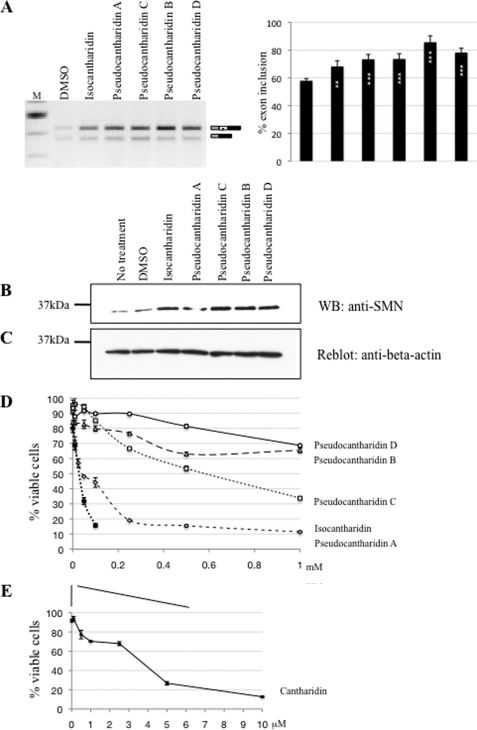
To determine whether the effect of pseudocantharidins on exon 7 inclusion affects the endogenous genes and is reflected in protein levels, we treated SMA patient fibroblasts with isocantharidin or the pseudocantharidins. These fibroblasts lack the SMN1 gene, and all SMN protein expression is derived from the fraction of SMN2 mRNA that includes exon 7.
We treated SMA patient fibroblasts with a final 1 mm concentration of the compounds and performed RT-PCR after 16 h. As shown in Fig. 3A, the compounds promoted inclusion of exon 7 from the endogenous SMN2 gene in SMA patient fibroblasts. To determine the effect on protein expression, we treated these cells with 50 μm compounds for 2 weeks, a concentration where we observed little toxic effect. As shown in Fig. 3B, untreated patient fibroblasts express residual levels of SMN protein. When compared with β-actin, this expression is increased 2–3.8-fold when pseudocantharidins are added (Fig. 3, B and C). The data show that isocantharidin and pseudocantharidins A–D promote the formation of the SMN protein from the endogenous, physiological relevant gene.
FIGURE 3.
Pseudocantharidins promote SMN2 protein formation and are less toxic than cantharidin. A, RT-PCR analysis of the endogenous SMN2 RNA after treatment with substances. SMN1 type 1 (−/− for SMN2) fibroblasts were treated for 16 h with 1 mm of the substances indicated. The usage of exon 7 was analyzed by RT-PCR. B, the SMN1 type 1 fibroblasts were treated with 50 μm of the compounds every other day for 2 weeks, and an anti-SMN antiserum was used to detect SMN accumulation. C, reblot of the Western blot (WB) filter with an anti-actin serum to show the loading of the gel. D and E, MTT toxicity assay of pseudocantharidins: SMN1 type 1 (−/− for SMN2) fibroblasts were incubated with the compounds indicated. The survival of cells treated with the solvent (DMSO, dimethyl sulfoxide) was set to be 100%. The cell viability was determined with the MTT assay (36) that measures mitochondrial dehydrogenase activity.
We next tested the toxicity of isocantharidin and pseudocantharidins A–D using the MTT test (36), in which MTT was added to cells. The mitochondrial dehydrogenases of live cells metabolize MTT, which is measured spectroscopically. Again, we used human fibroblast from SMA type I patients for the experiments. As shown in Fig. 3 (D and E), cantharidin causes the death of more than 80% of the cells at 10 μm concentration. Isocantharidin had a similar toxicity at 500 μm concentration. Surprisingly, pseudocantharidin A was more toxic than isocantharidin, causing the death of ∼90% of the cells at 100 μm concentration. Pseudocantharidin C reduced the number of viable cells by 60% when used at a concentration of 1 mm. In contrast, pseudocantharidins B and D were considerably less toxic and had 70% survival at 1 mm concentration. The toxicity of pseudocantharidins was more severe in SMA type I fibroblast than in HEK293 cells used for the splicing assays. We did not observe significant death of cells in HEK293 cells using pseudocantharidin treatment in the transfection assays in Fig. 2. In summary, we generated novel compounds that are less toxic than cantharidin at concentrations where they promote exon 7 inclusion.
We next determined how many other alternative exons are influenced by pseudocantharidins. As shown in supplemental Fig. S1, some but not all of the exons analyzed are changed by pseudocantharidins. This indicates that pseudocantharidins will affect multiple exons. The individual composition of exonic regulatory elements determines how these exons are influenced by tra2-beta1 and PP1 activity.
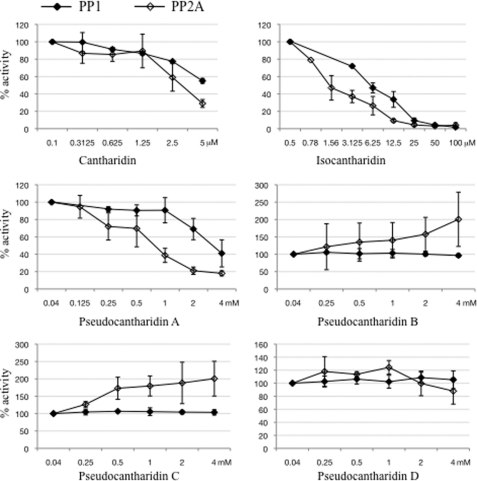
Pseudocantharidins Differ in Their Ability to Modulate the Activity on PP1 and PP2A
We next wanted to understand what determines the difference in toxicity of the pseudocantharidins. Because cantharidin inhibits PP1 and PP2A, we tested whether pseudocantharidins change the activities of these phosphatases. We determined the dephosphorylation of glycogen phosphorylase by purified PP1 and PP2A in the presence of pseudocantharidins. As shown in Fig. 4, cantharidin, isocantharidin, and pseudocantharidin A inhibited the activity of both phosphatases. Interestingly, pseudocantharidin B–D did not inhibit PP1. Surprisingly, pseudocantharidins B and C stimulated PP2A activity up to 2-fold. The only compounds known to activate PP2A are certain ceramides (38) and substances that act together with ceramides on PP2A (39). These results indicate that modification of the simple rigid 7-oxabicyclo[2.2.1]heptane structure that forms the core of cantharidin can turn a phosphatase inhibitor into an phosphatase activator. Furthermore, the substances that are most toxic are the ones that inhibit PP1.
FIGURE 4.
Modulation of PP1 and PP2A activity by pseudocantharidins. Radioactively labeled glycogen phosphorylase was incubated with PP1 or PP2A, and the phosphate release was measured in the presence of the indicated concentration of pseudocantharidins. The activity without inhibitor was set to 100%. The results are expressed as the means ± S.E. of three assays.
PP1 and PP2A Inhibition by Isocantharidin and Pseudocantaridin A Blocks Constitutive Splicing
To gain insight into the molecular mechanism that leads to exon 7 inclusion, we performed in vitro splicing assays. Because the SMN2 exon 7 splices poorly in vitro, we used the well established adenovirus-derived MINX reporter gene system to determine the effect of pseudocantharidins on the splicing reaction. As shown in Fig. 5A, isocantharidin and pseudocantharidin A blocked in vitro splicing at 0.15–5 mm concentration. In contrast, the other pseudocantharidins did not decrease the amount of product at the 1.5 mm concentration where they influenced alternative splicing. It is noteworthy that the only compounds that had a strong effect on constitutive splicing in vitro were substances that inhibit PP1. The inhibition of splicing by isocantharidin and pseudocantharidin A further correlates well with the higher toxicity of the two compounds when compared with other pseudocantharidins (Figs. 3C and 4).
FIGURE 5.
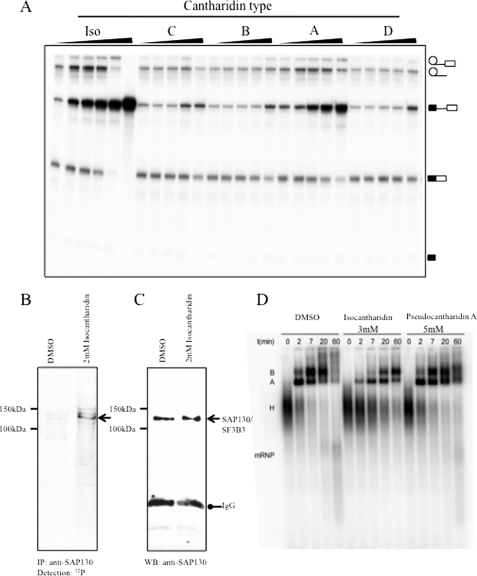
PP1-inhibiting substances block constitutive splicing and increase phosphorylation of the spliceosomal component SAP130. A, in vitro splicing assay of a MINX substrate in HeLa nuclear extract. The compounds were added at 0.05, 0.15, 0.5, 1.5, and 5 mm concentrations. B, in vitro spliceosomal assembly assay using the MINX substrate. C and D, isocantharidin promotes phosphorylation of SAP130 in nuclear extract. HeLa nuclear extract was incubated with 4 μCi of [γ-32P]ATP for 30 min at 37 °C in the presence of isocantharidin or its solvent, dimethyl sulfoxide (DMSO). SAP130 was immunoprecipitated (IP), and the immunoprecipitates were analyzed by autoradiography (C). The same filter was then reblotted with anti-SAP130 antiserum (D) to show loading of the protein. WB, Western blot.
We next asked whether isocantharidin that showed the strongest effect on blocking constitutive splicing could change the phosphorylation of spliceosomal components. To obtain candidates, we labeled nuclear extract by incubating it with [γ-32P]ATP and tested the effect of isocantharidin on the phosphorylation status of nuclear proteins. After gel electrophoresis, bands that showed an increase in [32P]ATP incorporation were analyzed by mass spectrometry. One of the identified proteins was SAP130, which we further analyzed by immunoprecipitation. We incubated nuclear extract with [γ-32P]ATP and immunoprecipitated SAP130. As shown in Fig. 5 (B and C), we observed an increase of phosphorylation signal for SAP130 in the presence of isocantharidin when compared with the Me2SO control. This indicates that isocantharidin most likely blocks the dephosphorylation of this factor. Experiments with other identified splicesomal proteins remained inconclusive, because the antisera did not work in the immunoprecipitations. The data indicate that a simultaneous inhibition of PP1 and PP2A activity evoked by isocantharidin has a strong influence on the phosphorylation of the U2 snRNP component SAP130.
It is well established that PP1 and PP2A activity is necessary for the constitutive splicing reaction (19). The phosphatases act on proteins in the second step of the splicing reaction (18). We therefore examined which stage in the spliceosomal reaction was blocked by isocantharidin and pseudocantharidin A and performed assembly assays using the MINX substrate. As shown in Fig. 5D, isocantharidin slowed down the transition from the H to A complex and blocks C complex formation. Pseudocantharidin A has no discernable effect on H complex formation but again blocks C complex formation. These data are in agreement with an inhibition of phosphatase activity needed for the B to C transition. It is likely that a partial inhibition of constitutive splicing is a reason for the toxicity of isocantharidin and pseudocantharidin A.
PP2A Activation by Pseudocantharidin B and C Changes Phosphorylation of tra2-beta1 at Position Thr-33 and Promotes Exon 7 Inclusion
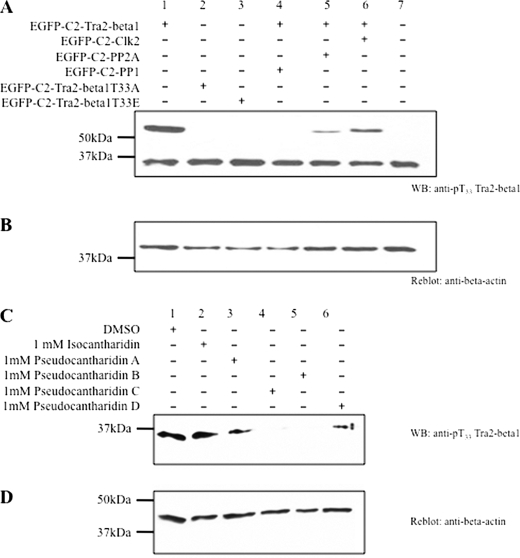
We next asked why pseudocantharidins B and C, which activate PP2A but have no measurable effect on PP1, promote exon 7 inclusion. Previously, we used mass spectrometry to characterize the phosphorylation status of splicing regulatory proteins and found that the phosphorylation of tra2-beta1 at position Thr-33 is changed during the splicing reaction in vitro (40). Phosphorylated Thr-33 could be detected in the A complex assembling on the MINX substrate but not in other complexes. Tra2-beta1 is an SR-like protein that binds to the central enhancer of exon 7 (27). Its binding is most likely stabilized by interactions with hnRNPG and SRp30c (41). Together, this enhancer complex causes recognition of the exon by the spliceosome and its subsequent inclusion. To investigate whether pseudocantharidins interfere with the phosphorylation at Thr-33, we created a phosphorylation-selective polyclonal antisera. As shown in Fig. 6A, this antiserum recognizes EGFP-tra2-beta1 that is transfected into HEK293 cells. The antiserum did not detect tra2-beta1 mutants where we changed the threonine 33 to either alanine (T33A) or glutamic acid (T33E), which demonstrates its selectivity. Because tra2-beta1 binds directly to PP1, we tested the influence of PP1 on the phosphorylation on this site and found that transfection of PP1 expression constructs completely abolishes the phosphorylation signal. Similarly, because PP2A activity is necessary for the splicing reaction, we tested the influence of PP2A overexpression on this site and found a significant reduction (Fig. 6A). The SR protein kinase CLK2 was included as a positive control. We next tested the effect of pseudocantharidins on the phosphorylation of tra2-beta1 on this site. We treated HEK293 cells with 1 mm pseudocantharidins and tested the phosphorylation of endogenous tra2-beta1. As shown in Fig. 6 (C and D), pseudocantharidins B and C cause a dephosphorylation at this site, which is in agreement with their ability to promote PP2A activity when assayed on the glycogen phosphorylate substrate (Fig. 4). The data suggest that PP2A activation, caused by pseudocantharidin C and B, causes a dephosphorylation of tra2-beta1 at a specific site.
FIGURE 6.
PP2A activation causes dephosphorylation of tra2-beta1 residue Thr-33. A, characterization of the antiserum. The tra2-beta1expressing cDNAs were transfected with cDNAs expressing the proteins indicated and analyzed with an affinity-purified antiserum that detects the phosphorylated form of Thr-33. B, reblot of the proteins in A with β-actin, which shows loading. C, HEK293 cells were treated with the pseudocantharidins indicated, and the effect of the phosphorylation on Thr-33 was determined by Western blot (WB). D, reblot of the proteins in C with β-actin.
Phosphorylation of tra2-beta1 Thr-33 Influences Exon 7 Inclusion
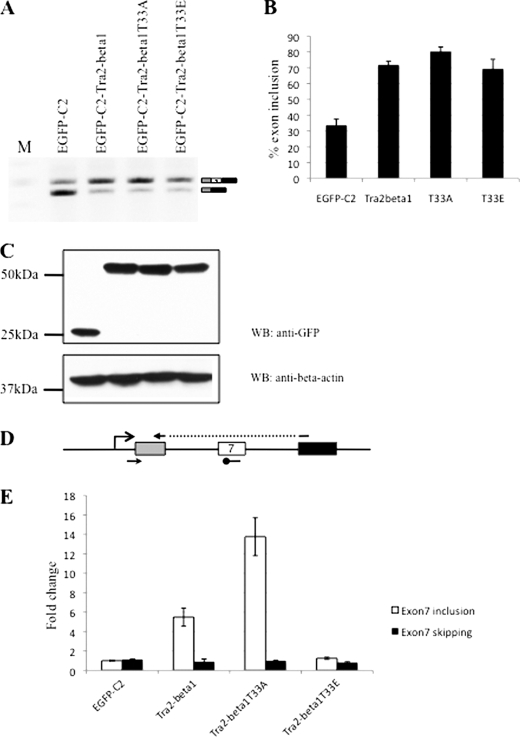
To test the role of Thr-33 in SMN2 exon 7 inclusion, we analyzed Thr-33 mutants in cotransfection experiments, using the SMN2 reporter minigene. We used cDNA constructs expressing Tra2-beta1 wild type, as well as mutants where Thr-33 was changed to alanine (T33A) or glutamic acid (T33E). As shown in Fig. 7 (A and B), the T33A mutant that mimics the dephosphorylated form of tra2-beta1 induced exon 7 inclusion stronger than the wild type. The T33E mutant that mimics the phosphorylated form had an effect similar to the wild type. To test the effect of these mutants quantitatively, we performed real time PCR experiments and found a 2-fold stronger effect of the T33A compared with wild type (Fig. 7E).
FIGURE 7.
Mutation of tra2-beta1 residue Thr-33 to alanine promotes exon 7 inclusion. A, the SMN2 reporter minigene, schematically shown in Fig. 2A, was transfected with cDNAs expressing tra2-beta1 wild type and the mutants indicated. The RNAs were analyzed by end point RT-PCR. B, quantification of three independent experiments from A. C, quantification of the tra2-beta1 expression constructs and in the experiment in A by Western blot (WB) of cellular lysates. β-Actin was used as a loading control. D and E, real time PCR analysis of RNAs from three independent experiments from A. The location of the primers is shown in D, and the quantification is in E.
These experiments support the idea that dephosphorylation of tra2-beta1 at residue 33 promotes exon 7 inclusion. In vivo, this dephosphorylation can be caused by pseudocantharidins B and C acting on the endogenous gene.
DISCUSSION
Modification of Natural Products Allows Generation of New Compounds That Influence Alternative Splicing
We used the established PP1 and PP2A inhibitor cantharidin as a starting point to generate new compounds that change alternative splicing and promote inclusion of SMN2 exon 7. Previous efforts to develop broad spectrum, PP1/PP2A inhibitors as antineoplastic agents led to published chemical modifications of isocantharidin (21, 34, 42–46) principally in two ways: (i) acyl substitution reactions of the anhydride and (ii) substitution-elimination reactions of the anhydride with amines to give imide products. We used both routes to generate approximately 50 pseudocantharidins and tested their effect on SMN2 exon 7 inclusion. We selected the compounds that had the strongest effect on exon 7 inclusion and that are shown in Fig. 1A. These compounds activate exon 7 when used in reporter gene assays and promote production of full-length SMN protein in patient fibroblasts, demonstrating their effect in a physiological system. Most importantly, some pseudocantharidins exhibit a lower toxicity than cantharidin at the concentration where they promote exon 7 inclusion.
Because PP1 and PP2A are structurally similar in their active center, it was not surprising that inhibitors related to cantharidin, such as isocantharidin and pseudocantharidin A, inhibit both phosphatases. However, further chemical modifications resulted in pseudocantharidin B and C that activate PP2A while having no effect on PP1. Currently, several ceramides are the only substances known to activate PP2A, but they also activate PP1 (47).
The mechanism of PP2A activation is not clear, and there are no chemical similarities between pseudocantharidin B or C and ceramides. Cantharidin most likely binds to the active center of PP2A (21), and given their chemical resemblance, pseudocantharidins B and C could stabilize a more open and accessible form of the active center of the phosphatase, leading to its activation.
Mechanism of Isocantharidin and Pseudocantharidin A on Splice Site Selection
Because all pseudocantharidins are derived from the PP1/PP2A inhibitor cantharidin, we first determined their effect on these phosphatases, using glycogen phosphorylase as the traditional substrate. Based on these studies, the compounds fall into three groups: (i) isocantharidin and pseudocantharidin A that inhibit both PP1 and PP2A, (ii) pseudocantharidin B and C that activate PP2A and have no effect on PP1, and (iii) pseudocantharidin D that does not show any phosphatase inhibition.
The phosphatase inhibitors isocantharidin and pseudocantharidin A block constitutive splicing, which is in agreement with previous studies indicating that PP1 and PP2A activity is necessary for the splicing reaction (18, 19). We next used these compounds in a cell-free system to determine what phosphorylation events they regulate and identified SAP130 (SF3b3), a U2 component as a target. SAP130 is part of the SF3a/3b complex that binds to introns upstream of the branch point and helps to position U2. It has been reported that PP1 and PP2A act on the U2 and U5 components SAP155 and U5–116kd during the second step of splicing (18). Using assembly assays, we found that both phosphatase inhibitors act earlier, in the transition from B to C complex. Furthermore, Sap130 is destabilized and partially dissociates from the C complex prior to step 2 (40). Therefore, our data support an earlier finding that the phosphatases are required for both splicing steps (19) and furthermore identify SAP130 as another protein that is subject to reversible phosphorylation during the splicing reaction.
Our in vitro assays indicate that the splicing of the MINX substrate is not completely blocked but severely slowed down. The reaction starts slowing down in the 0.5–1.5 mm range, comparable with the concentrations where we see a change in alternative splicing in vivo. It is possible that the lack of a necessary dephosphorylation of SAP130 slows down the spliceosomal assembly or necessary rearrangements within the splicing complexes. It is not clear how this favors exon 7 inclusion. One possibility is that the commitment complex for exon 7 inclusion is less dependent on an efficient dephosphorylation of some of its components.
Pseudocantharidins B and C Activate PP2A and Change the Phosphorylation of tra2-beta1, a Protein Promoting Exon 7 Inclusion
The second group of compounds, pseudocantharidin B and C, activates PP2A and has no effect on PP1. This is a highly surprising result, because currently only ceramides are known to be PP2A activators (47, 48). Ceramides activate both PP1 and PP2A. They influence alternative splicing of caspase-9 and bcl-x by activating PP1 (49). Therefore pseudocantharidin B and C appear to be the first substances that are activating PP2A without influencing PP1. Because they show low toxicity and, compared with ceramides, improved water solubility, the compounds are the most interesting ones that promote exon 7 inclusion.
We used a phospho-selective antisera against the residue Thr-33 of tra2-beta1 to show that pseudocantharidin B and C promote dephosphorylation of this amino acid. Tra2-beta1 binds to the central enhancer of exon 7 and strongly promotes exon inclusion (27). The role of its Thr-33 residue in exon 7 inclusion was supported by mutational analysis, because the alanine mutant had a stronger effect than the wild type and the glutamic acid mutant. The exact molecular role of this phosphorylation remains to be determined. It is likely that Thr-33 indicates the dephosphorylation of other, as yet unidentified residues. Collectively, these changes could influence splice site selection. Although the composition of the complexes forming on exon 7 has not been determined, it is likely that similar to other SR proteins (7), tra2-beta1 is present in spliceosomal A, B, and C complexes. A dephosphorylation on this and possibly other sites could increase the affinity of the spliceosome to exon 7, resulting in improved exon recognition.
Action of Pseudocantharidin D
Pseudocantharidin D has no influence on PP1 or PP2 activity and does not block splicing in vitro. However, it strongly promotes inclusion of exon 7, indicating that it has an effect on alternative splice site selection. It is possible that this effect is due to an allosteric effect on PP1. Crystallographic studies of PP1 bound to a peptide representing its targeting unit GM (50) indicated that the conformation of GM changes upon PP1 binding. This suggests that PP1 acting on the spliceosome and pre-mRNP not only causes dephosphorylation but also causes conformational changes. It is notable that PP1-binding sites of several splicing proteins, including tra2-beta1, are located in the beta 4 strand of the RRM. This part of the RRM contributes to RNA binding, and it is possible that binding of PP1 and RNA to an RRM are mutually exclusive. One possible mechanism of action for pseudocantharidin D is that the compound interferes with this regulation.
Do Pseudocantharidins Recapitulate a Physiological Process?
It is likely that reversible phosphorylation achieved by a tightly regulated interplay of kinases and phosphatases plays an important role in alternative splice site selection (9, 51). Phosphatases are typically present in complexes with inhibiting proteins and are activated by cellular signals. The example of ceramides shows that this activation can also be achieved by low molecular weight substances (52). We showed that pseudocantharidins that are derived from a natural compound have a similar ability to alter alternative splicing, although they are not chemically related to ceramides. This suggests the existence of more natural metabolites that influence alternative splicing. Such metabolites could contribute to tissue-specific alternative splicing patterns and could also be useful lead compounds to combat diseases caused by mis-splicing.
Acknowledgments
We thank Dr. H. Peter Spielmann for discussions. The mass spectrometry analysis was performed at the University of Kentucky Center for Structural Biology Protein Core Facility.
This work was supported, in whole or in part, by National Institutes of Health Research Resources Grant P20 RR020171 (to the University of Kentucky Center for Structural Biology Protein Core Facility) and Center of Biomedical Research Excellence in the Molecular Basis of Human Disease Grant P20 RR020171 (to the Organic Synthesis Core Facility). This work was also supported by the Muscular Dystrophy Association and EURASNET.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- SMA
- spinal muscular atrophy
- PP
- protein phosphatase
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- THF
- tetrahydrofuran.
REFERENCES
- 1. Pan Q., Shai O., Lee L. J., Frey B. J., Blencowe B. J. (2008) Nat. Genet. 40, 1413–1415 [DOI] [PubMed] [Google Scholar]
- 2. Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S. F., Schroth G. P., Burge C. B. (2008) Nature 456, 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stamm S., Ben-Ari S., Rafalska I., Tang Y., Zhang Z., Toiber D., Thanaraj T. A., Soreq H. (2005) Gene 344, 1–20 [DOI] [PubMed] [Google Scholar]
- 4. Black D. L. (2003) Annu. Rev. Biochem. 72, 291–336 [DOI] [PubMed] [Google Scholar]
- 5. Maniatis T., Tasic B. (2002) Nature 418, 236–243 [DOI] [PubMed] [Google Scholar]
- 6. Maniatis T., Reed R. (2002) Nature 416, 499–506 [DOI] [PubMed] [Google Scholar]
- 7. Wahl M. C., Will C. L., Lührmann R. (2009) Cell 136, 701–718 [DOI] [PubMed] [Google Scholar]
- 8. Berget S. M. (1995) J. Biol. Chem. 270, 2411–2414 [DOI] [PubMed] [Google Scholar]
- 9. Stamm S. (2008) J. Biol. Chem. 283, 1223–1227 [DOI] [PubMed] [Google Scholar]
- 10. Sumanasekera C., Watt D. S., Stamm S. (2008) Biochem. Soc. Trans. 36, 483–490 [DOI] [PubMed] [Google Scholar]
- 11. Tazi J., Bakkour N., Stamm S. (2009) Biochim. Biophys. Acta 1792, 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cooper T. A., Wan L., Dreyfuss G. (2009) Cell 136, 777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lunn M. R., Wang C. H. (2008) Lancet 371, 2120–2133 [DOI] [PubMed] [Google Scholar]
- 14. Burghes A. H., Beattie C. E. (2009) Nat. Rev. Neurosci. 10, 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Singh N. N., Shishimorova M., Cao L. C., Gangwani L., Singh R. N. (2009) RNA Biol. 6, 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Singh R. N. (2007) RNA Biol. 4, 7–10 [DOI] [PubMed] [Google Scholar]
- 17. Novoyatleva T., Heinrich B., Tang Y., Benderska N., Butchbach M. E., Lorson C. L., Lorson M. A., Ben-Dov C., Fehlbaum P., Bracco L., Burghes A. H., Bollen M., Stamm S. (2008) Hum Mol. Genet. 17, 52–70 [DOI] [PubMed] [Google Scholar]
- 18. Shi Y., Reddy B., Manley J. L. (2006) Mol. Cell 23, 819–829 [DOI] [PubMed] [Google Scholar]
- 19. Mermoud J. E., Cohen P., Lamond A. I. (1992) Nucleic Acids Res. 20, 5263–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jolivet J. (1960) Ann. Chim. 5, 1165–1213 [Google Scholar]
- 21. Baba Y., Hirukawa N., Tanohira N., Sodeoka M. (2003) J. Am. Chem. Soc. 125, 9740–9749 [DOI] [PubMed] [Google Scholar]
- 22. Politis J. K., Nemes J. C., Curtis M. D. (2001) J. Am. Chem. Soc. 123, 2537–2547 [DOI] [PubMed] [Google Scholar]
- 23. Beullens M., Van Eynde A., Stalmans W., Bollen M. (1992) J. Biol. Chem. 267, 16538–16544 [PubMed] [Google Scholar]
- 24. Zillmann M., Zapp M. L., Berget S. M. (1988) Mol. Cell. Biol. 8, 814–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Das R., Reed R. (1999) RNA 5, 1504–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hofmann Y., Lorson C. L., Stamm S., Androphy E. J., Wirth B. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9618–9623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bockstahler E. R. (July 19, 1966) U. S. Patent 3,261,845
- 29. Nakatani T., Konishi T., Miyahara K., Noda N. (2004) Chem. Pharm. Bull 52, 807–809 [DOI] [PubMed] [Google Scholar]
- 30. Kok S. H., Chui C. H., Lam W. S., Chen J., Tang J. C., Lau F. Y., Cheng G. Y., Wong R. S., Chan A. S. (2006) Int. J. Mol. Med. 17, 151–157 [PubMed] [Google Scholar]
- 31. Kok S. H., Chui C. H., Lam W. S., Chen J., Lau F. Y., Cheng G. Y., Wong R. S., Lai P. P., Leung T. W., Tang J. C., Chan A. S. (2006) Int. J. Mol. Med. 17, 945–949 [PubMed] [Google Scholar]
- 32. Kok S. H., Chui C. H., Lam W. S., Chen J., Lau F. Y., Wong R. S., Cheng G. Y., Tang W. K., Teo I. T., Cheung F., Cheng C. H., Chan A. S., Tang J. C. (2006) Int. J. Mol. Med. 18, 1217–1221 [PubMed] [Google Scholar]
- 33. Kok S. H., Chui C. H., Lam W. S., Chen J., Lau F. Y., Wong R. S., Cheng G. Y., Lai P. B., Leung T. W., Yu M. W., Tang J. C., Chan A. S. (2007) Bioorg. Med. Chem. Lett. 17, 1155–1159 [DOI] [PubMed] [Google Scholar]
- 34. Hill T. A., Stewart S. G., Ackland S. P., Gilbert J., Sauer B., Sakoff J. A., McCluskey A. (2007) Bioorg. Med. Chem. 15, 6126–6134 [DOI] [PubMed] [Google Scholar]
- 35. Lorson C. L., Hahnen E., Androphy E. J., Wirth B. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 6307–6311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mosmann T. (1983) J. Immunol. Methods 65, 55–63 [DOI] [PubMed] [Google Scholar]
- 37. Deleted in proof.
- 38. Galadari S., Kishikawa K., Kamibayashi C., Mumby M. C., Hannun Y. A. (1998) Biochemistry 37, 11232–11238 [DOI] [PubMed] [Google Scholar]
- 39. Leoni L. M., Shih H. C., Deng L., Tuey C., Walter G., Carson D. A., Cottam H. B. (1998) Biochem. Pharmacol. 55, 1105–1111 [DOI] [PubMed] [Google Scholar]
- 40. Bessonov S., Anokhina M., Will C. L., Urlaub H., Lührmann R. (2008) Nature 452, 846–850 [DOI] [PubMed] [Google Scholar]
- 41. Singh N. N., Androphy E. J., Singh R. N. (2004) Crit. Rev. Eukaryot. Gene Expr. 14, 271–285 [DOI] [PubMed] [Google Scholar]
- 42. Hill T. A., Stewart S. G., Sauer B., Gilbert J., Ackland S. P., Sakoff J. A., McCluskey A. (2007) Bioorg. Med. Chem. Lett. 17, 3392–3397 [DOI] [PubMed] [Google Scholar]
- 43. Stewart S. G., Hill T. A., Gilbert J., Ackland S. P., Sakoff J. A., McCluskey A. (2007) Bioorg. Med. Chem. 15, 7301–7310 [DOI] [PubMed] [Google Scholar]
- 44. McCluskey A., Ackland S. P., Bowyer M. C., Baldwin M. L., Garner J., Walkom C. C., Sakoff J. A. (2003) Bioorg. Chem. 31, 68–79 [DOI] [PubMed] [Google Scholar]
- 45. Hart M. E., Chamberlin A. R., Walkom C., Sakoff J. A., McCluskey A. (2004) Bioorg. Med. Chem. Lett. 14, 1969–1973 [DOI] [PubMed] [Google Scholar]
- 46. Pang S. K., Yu C. W., Au-Yeung S. C., Ho Y. P. (2007) Biochem. Biophys. Res. Commun. 363, 235–240 [DOI] [PubMed] [Google Scholar]
- 47. Chalfant C. E., Szulc Z., Roddy P., Bielawska A., Hannun Y. A. (2004) J. Lipid Res. 45, 496–506 [DOI] [PubMed] [Google Scholar]
- 48. Chalfant C. E., Kishikawa K., Mumby M. C., Kamibayashi C., Bielawska A., Hannun Y. A. (1999) J. Biol. Chem. 274, 20313–20317 [DOI] [PubMed] [Google Scholar]
- 49. Chalfant C. E., Rathman K., Pinkerman R. L., Wood R. E., Obeid L. M., Ogretmen B., Hannun Y. A. (2002) J. Biol. Chem. 277, 12587–12595 [DOI] [PubMed] [Google Scholar]
- 50. Egloff M. P., Johnson D. F., Moorhead G., Cohen P. T., Cohen P., Barford D. (1997) EMBO J. 16, 1876–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shin C., Manley J. L. (2004) Nat. Rev. Mol. Cell Biol. 5, 727–738 [DOI] [PubMed] [Google Scholar]
- 52. Pettus B. J., Chalfant C. E., Hannun Y. A. (2002) Biochim. Biophys. Acta 1585, 114–125 [DOI] [PubMed] [Google Scholar]