Abstract
Most plant glycoproteins contain substantial amounts of paucimannosidic N-glycans instead of their direct biosynthetic precursors, complex N-glycans with terminal N-acetylglucosamine residues. We now demonstrate that two β-N-acetylhexosaminidases (HEXO1 and HEXO3) residing in different subcellular compartments jointly account for the formation of paucimannosidic N-glycans in Arabidopsis thaliana. Total N-glycan analysis of hexo knock-out plants revealed that HEXO1 and HEXO3 contribute equally to the production of paucimannosidic N-glycans in roots, whereas N-glycan processing in leaves depends more heavily on HEXO3 than on HEXO1. Because hexo1 hexo3 double mutants do not display any obvious phenotype even upon exposure to different forms of abiotic or biotic stress, it should be feasible to improve the quality of glycoprotein therapeutics produced in plants by down-regulation of endogenous β-N-acetylhexosaminidase activities.
Keywords: Arabidopsis, Glycoprotein Biosynthesis, Glycosylation, Plant, Protein Targeting, Hexosaminidase
Introduction
Plant-based expression systems are an attractive alternative technology for the production of recombinant glycoproteins. Potential drawbacks, however, are variations in the final steps of N-glycan processing, which differ significantly between mammals and plants (1–3). Plant glycoproteins contain two major types of processed oligosaccharides: paucimannosidic and complex N-glycans. Complex N-glycans carry β1,2-linked xylose and α1,3-linked fucose, as well as one or two terminal N-acetylglucosamine (GlcNAc) residues attached to the core Man3GlcNAc2 structure. In contrast, paucimannosidic N-glycans lack the two terminal GlcNAc residues at their nonreducing ends (Fig. 1). These truncated N-glycan structures are generated in late stages of the N-glycosylation pathway due to the action of β-N-acetylhexosaminidases. Paucimannosidic N-glycans constitute the majority of glycans present on vacuolar glycoproteins and occur in smaller amounts on extracellular plant glycoproteins (4–7). Because these truncated N-glycan structures are not found on mammalian glycoproteins, it is possible that these glycoforms negatively affect the biological activity of a recombinant glycoprotein therapeutic produced in plants. Consequently, strategies need to be developed to overcome this limitation of plant-based expression systems.
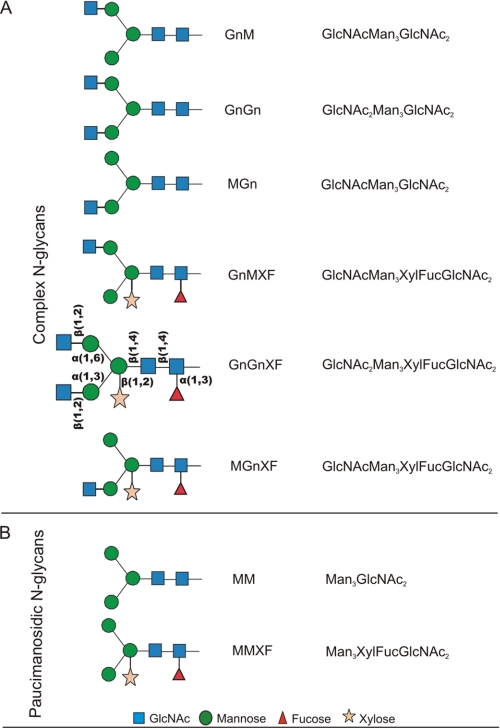
FIGURE 1.
N-Glycan structures relevant for this study. A, complex N-glycans carrying one or two GlcNAc residues linked to the α1,3- or/and α1,6-mannosyl branch. B, paucimannosidic N-glycans, lacking the two terminal GlcNAc residues at the nonreducing end. For a detailed description of the nomenclature see also the Proglycan Web site.
Recently, three putative β-N-acetylhexosaminidases encoded by the Arabidopsis thaliana genome were cloned, heterologously expressed in insect cells, and analyzed with respect to their enzymatic properties in vitro (8). All three enzymes (termed HEXO1–3) were capable of removing GlcNAc residues from synthetic substrates and from the α1,3- and α1,6-mannosyl branches of different N-glycan substrates, without displaying any strict branch preference. Fluorescently tagged forms of HEXO1–3 were found to be located in different subcellular compartments when transiently expressed in Nicotiana benthamiana leaf epidermal cells, indicating that the enzymes are involved in different cellular processes.
Here, we investigated the role of HEXO1, HEXO2, and HEXO3 in the generation of paucimannosidic N-glycans in Arabidopsis. Single, double, and triple knock-out mutants were generated and assessed for changes in β-N-acetylhexosaminidase activity and protein N-glycosylation. These studies revealed that both HEXO1 and HEXO3 contribute to the conversion of complex into paucimannosidic N-glycans in planta. The subcellular localization of HEXO1 and HEXO3 differs also in Arabidopsis, indicating that each enzyme acts in situ on a separate subset of glycoproteins. Intriguingly, the functional expression of HEXO2 was not detected in any Arabidopsis tissue analyzed, questioning the physiological relevance of this protein.
EXPERIMENTAL PROCEDURES
Plant Material
Arabidopsis ecotype Columbia (Col-0) and mutant plants were grown in a growth chamber at 22 °C under long day conditions (16-h light/8-h dark photoperiod) on soil or on 1 × Murashige and Skoog medium (1 × MS medium2; M5519; Sigma-Aldrich) containing 2% (w/v) sucrose and 1% (w/v) agar. All seeds were cold-treated for 2 days in the dark before incubation at 22 °C. For root growth analysis, plants were grown on 1 × MS medium containing 2% (w/v) sucrose and 2% (w/v) agar, stratified for 2 days, and then incubated for 5 days at 22 °C. Seedlings were transferred to medium supplemented with various concentrations of NaCl or sucrose and grown for another 7–9 days, and root length was scored. To test heat and cold tolerance, seedlings were sown on soil or 1 × MS medium containing 2% (w/v) sucrose and 1% (w/v) agar. After incubation at 22 °C for 7 days, seedlings were subjected to 30 °C for 14 days or to 4 °C for 40 days. Seedling growth inhibition and oxidative burst assays were performed as described earlier (9, 10).
Isolation of Hexo Mutants
Arabidopsis T-DNA insertion lines hexo1-1 (SALK_026094), hexo2-1 (SALK_052154), hexo3-1 (SALK_022485), and hexo3-2 (SAIL_325_B01) were obtained from the European Arabidopsis Stock Center. The hexo1-1 T-DNA insertion was analyzed by sequencing of PCR-amplified products using left border primer LBa1 and gene-specific primer At3g55260-2R and At3g55260-1F. Homozygous lines were identified using primers At3g55260-1F and At3g55260-2R. For the hexo2-1 T-DNA insertion primers LBb1 and At1g05590-1F were used, for hexo3-1 LBa1 and At1g65590-4F, and for hexo3-2 LBsail1/At1g65590-6F and At1g65590-5R/RBsail2. Homozygous lines were identified using primers At1g05590-1F/-2R (hexo2-1), At1g65590-4F/-7R (hexo3-1), and At1g65590-6F/-5R (hexo3-2). For hexo2-1 and hexo3-1 the second T-DNA-genome junction could not be identified. Double and triple mutants were obtained by crossing and genotyping by PCR. Sequences of primers are listed in supplemental Table S1.
Protein Extraction for Activity Assays with Synthetic and Pyridylaminated Substrates
Root or leaf material from Col-0 wild-type, single, double, and triple knock-out plants was ground in liquid nitrogen, resuspended in 50 mm sodium citrate buffer (pH 4.6) and 1% (v/v) plant protease inhibitor mixture (P9599; Sigma) for activity tests with synthetic substrates or in 0.1 m sodium citrate/phosphate buffer (pH 5.0) and 1% (v/v) plant protease inhibitor mixture (Sigma) for activity tests with N-glycan substrates. Aliquots of the crude extracts were subjected to a centrifugation step for 1 h at 129,400 × g and 4 °C using a SW 41 Ti rotor (Beckman). The supernatant (soluble protein fraction) was saved, and the pellet was resuspended in the respective buffer and centrifuged as above. After discarding the resulting supernatant, the pellet was again resuspended in buffer (insoluble protein fraction).
Assays with Synthetic Substrates
Soluble and insoluble protein fractions from roots and leaves were incubated with 5 mm pNP-GlcNAc (4-nitrophenyl N-acetyl-β-d-glucosaminide; Sigma N9376) or pNP-Man (p-nitrophenyl α-d-mannopyranoside; Sigma N2127) in a total volume of 40 μl of assay buffer (50 mm sodium citrate buffer (pH 4.6), 1 mg/ml bovine serum albumin, 0.02% (w/v) NaN3) for 1h at 22 °C. The reactions were stopped by the addition of 80 μl of 0.4 m glycine buffer (pH 10.4), boiled for 5 min at 95 °C, and centrifuged at 16,000 × g prior to spectrophotometric analysis at 405 nm. Assays of sucrose gradient fractions were performed in a total volume of 40 μl of assay buffer containing 1 mm MU-GlcNAc (4-methylumbelliferyl-N-acetyl-β-d-glucosaminide; Sigma M2133). After incubation at 22 °C for an appropriate time, the reactions were stopped by the addition of 80 μl of 0.4 m glycine buffer (pH 10.4), boiled for 5 min at 95 °C, and centrifuged at 16,000 × g prior to spectrofluorometry using excitation and emission wavelengths of 365 and 460 nm, respectively. Protein extracts heat-inactivated prior to incubation with the substrate were used as controls.
Activity Assay with GnGn-PA
The pyridylaminated oligosaccharide substrate GnGn-PA (GlcNAc2Man3GlcNAc2-PA; Fig. 1) was prepared as described previously (11). Assays with PA-labeled oligosaccharides were performed in a total volume of 20 μl of 0.1 m sodium citrate buffer (pH 5.0) containing 5 μm GnGn-PA and were incubated at 22 °C for an appropriate time. After the addition of 80 μl of water, the reactions were stopped by heating for 5 min at 95 °C. Aliquots of 25 μl were analyzed by reverse-phase HPLC as described previously (8). Protein extracts heat-inactivated prior to incubation with the substrate were used as controls.
Subcellular Localization and Imaging
The construct pPFH3-HEXO1 (8) was introduced into Col-0 wild type, and p20-HEXO3 (8) was introduced into Col-0 and sgs2 mutant plants (12) by the floral dip procedure. Seeds were selected on 1 × MS agar containing 50 μg/ml kanamycin. Arabidopsis lines stably expressing the fluorescent proteins were grown for 5–10 days on agar growth medium at 22 °C and examined by confocal laser scanning microscopy. Imaging was performed using a Leica TCS SP2 confocal microscope as described in detail recently (13).
For plasmolysis experiments, transgenic seedlings were incubated in 1 m KNO3 for 10–15 min. The seedlings were mounted in 1 m KNO3 prior to confocal laser scanning microscopy. Plasma membrane intrinsic protein 2a (PIP2a) transgenic seedlings were obtained from the European Arabidopsis Stock Center.
Extraction of Apoplastic Proteins
The protocols for vacuum infiltration of rosette leaves and the extraction of apoplastic proteins were modified from Ref. 14 as follows. 1 g of rosette leaves of 4–5-week-old wild-type and hexo knock-out plants were harvested, immersed in 0.3 m mannitol in a desiccator connected to a vacuum pump, and vacuum-infiltrated for 4 min at 4 °C. The infiltrated leaves were then wrapped in Miracloth (Calbiochem) and centrifuged at 900 × g for 20 min at 4 °C. The supernatants were analyzed by SDS-PAGE and silver staining or lectin blots using biotinylated Griffonia simplicifolia lectin II (see below).
Complementation of hexo1 and hexo3 Plants
For complementation of hexo1 and hexo3 mutants, the single knock-out mutants were crossed with transgenic Arabidopsis lines carrying the pPFH3-HEXO1 or p20-HEXO3 constructs. F2 seedlings were transferred to soil and genotyped by PCR using the following primer combinations: At3g55260-1F/-2R and At3g55260-1F/mRFP-4R, At1g65590-6F/GFP-5R, and At1g65590-4F/-10R. Activity assays with protein extracts from root and leaf material of stable transgenic lines were performed with pNP-GlcNAc as described above.
Immunoblotting Analysis of HEXO Proteins
Protein extracts were subjected to SDS-PAGE (12%) under reducing conditions and blotted onto Hybond ECL nitrocellulose membranes (GE Healthcare). The blots were blocked in PBS containing 0.1% (v/v) Tween 20 and 5% milk powder and were then incubated with the respective primary antibodies. The detection was performed using horseradish peroxidase-conjugated goat anti-rabbit IgG antibodies (Sigma) with Super Signal West Femto Chemiluminescent substrate (Thermo Scientific). The generation of HEXO-specific antibodies is described in detail in supplemental Methods.
Subcellular Fractionation
Fresh root material of 14-day-old seedlings was ground and resuspended in extraction buffer containing 50 mm Tris-HCl (pH 7.5), 2.5 mm KCl, 10% sucrose (w/w), 10 mm EDTA, 7.5 mm EGTA, 10 mm sodium β-glycerophosphate, 2 mm dithioerythritol, 2 mm phenylmethylsulfonylfluoride (PMSF), and 1% protease inhibitor mixture (Sigma). The homogenate was centrifuged several times (500–3,000 × g), and the supernatant was layered onto a 20–52% (w/w) discontinuous sucrose density gradient that contained 5 mm EDTA. The gradient was centrifuged at 134,400 × gav for 5 h at 4 °C in a SW 41 Ti rotor. Samples were fractionated into 300-μl (10–25%) and 600-μl aliquots (30–52%). 10 μl of each fraction was used to perform activity assays (in triplicate) with MU-GlcNAc as described above. For immunoblot analysis, the fractions were precipitated with methanol/chloroform/water (4:1:3) (15). The pellets were resuspended in SDS-PAGE loading buffer, subjected to SDS-PAGE (12%) under reducing conditions, and blotted onto Hybond ECL nitrocellulose membranes (GE Healthcare). The blots were blocked in PBS containing 0.1% (v/v) Tween 20 and 3% BSA or 5% milk powder and were then incubated with rabbit antisera to PIP2;1 (1:6000) (16) and TIP1;1 (1:6,000) (17). Anti-HEXO1 antibodies and anti-HEXO3 antibodies were used as described in supplemental Methods.
N-Glycan Analysis
Protein gel blot analysis of crude protein extracts was performed using anti-horseradish peroxidase antibodies (Sigma-Aldrich) as described (18, 19). Lectin blotting with biotinylated G. simplicifolia lectin II (GSL II; Vector Laboratories) was done according to the instructions of the manufacturer. Total N-glycan analysis was performed using 500 mg of plant material by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) as described previously (18).
RESULTS
Generation and Characterization of HEXO1-, HEXO2-, and HEXO3-deficient Plants
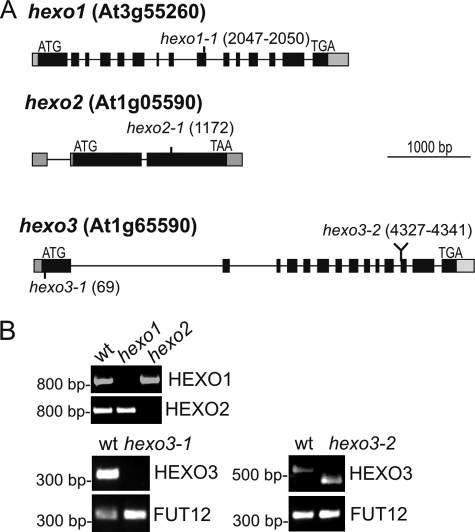
The enzymatic properties of recombinantly expressed HEXO1–3 indicate that each of these enzymes could be involved in N-glycan trimming in planta (8). To explore the physiological function of HEXO1–3 in vivo, we characterized Arabidopsis mutants with disrupted HEXO expression (Fig. 2A). T-DNA insertion lines for HEXO1, HEXO2, and HEXO3 were obtained from the SALK and SAIL collections, and homozygous T-DNA insertion lines were identified by PCR. Semiquantitative RT-PCR was performed on RNA isolated from rosette leaves to show that the single knockouts hexo1-1, hexo2-1, and hexo3-1 lack full-length transcripts (Fig. 2B). In the case of HEXO3, a second T-DNA insertion line was also analyzed. The T-DNA insertion of hexo3-2 was located at an intron-exon junction (Fig. 2A), which entails an incorrect splicing event leading to a smaller aberrant transcript (Fig. 2B). For hexo2-1 and hexo3-1 only, the left border T-DNA junction could be identified (Fig. 2A). For those two lines, the second junction could neither be obtained with right border nor with left border primers, indicating the presence of a deletion or a complex insertion of the T-DNA. Single knock-out lines were then crossed to obtain the double knock-out mutants hexo1 hexo2, hexo1 hexo3-1, and hexo2 hexo3-2. To generate a hexo1 hexo2 hexo3 triple mutant, the homozygous hexo1 hexo2 and hexo1 hexo3-1 double knock-out mutants were crossed.
FIGURE 2.
Characterization of hexo knock-out mutants. A, schematic overview of hexo alleles. Boxes represent exons (black area represents the coding region), and vertical lines indicate the position of T-DNA insertions. T-DNA insertions are found at positions 2047–2050 (hexo1-1), 1172 (hexo2-1), and 69 (hexo3-1). The T-DNA insertion in hexo3-2 results in a small deletion (4327–4341) at an intron-exon junction. B, RT-PCR analysis of hexo mutants. RT-PCR (two independent experiments) was performed on RNA isolated from rosette leaves of the indicated lines. Primers specific for the indicated transcripts were then used for amplification. Core α1,3-fucosyltransferase (FUT12) served as a positive control.
Endogenous HEXO Proteins Are Active on Synthetic and N-Glycan Substrates
The availability of single, double, and triple knock-out mutants allowed us to determine the contribution of the HEXO1–3 proteins to the total β-N-acetylhexosaminidase activity present in Arabidopsis tissues. Assays with pNP-GlcNAc and GnGn-PA were performed on leaf and root extracts, using either crude tissue homogenates or supernatant and pellet fractions to detect not only soluble β-N-acetylhexosaminidases but also membrane-bound or insoluble enzyme activity.
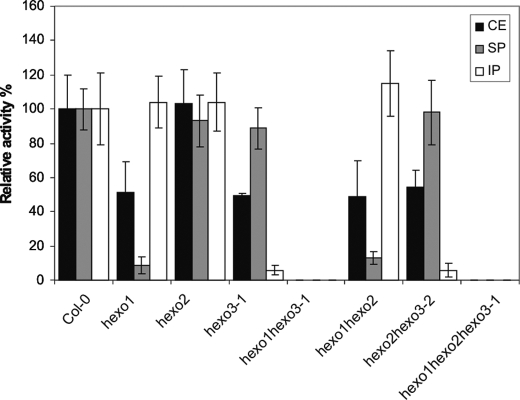
With the synthetic substrate pNP-GlcNAc, Col-0 and hexo2 show very similar β-N-acetylhexosaminidase activity in total leaf extracts. In contrast, hexo1, hexo3-1, and hexo3-2 knock-out mutants as well as hexo1 hexo2, hexo2 hexo3-2 double mutants and the F1 hybrid hexo3-1 hexo3-2 displayed reduced activity. No difference was observed between hexo1 and hexo1 hexo2 as well as hexo3-1 and hexo2 hexo3-2, respectively, indicating that HEXO2 does not contribute to the β-N-acetylhexosaminidase activity present in leaves (Fig. 3 and supplemental Fig. S1). When analyzing the soluble protein fraction, the β-N-acetylhexosaminidase activity of hexo1 and hexo1 hexo2 extracts was only about 10% of that of wild-type plants (Fig. 3). The residual activity seen in hexo1 and hexo1 hexo2 samples is very likely due to a small portion of soluble HEXO3 because this activity is absent in soluble fractions of hexo1 hexo3-1 and hexo1 hexo2 hexo3-1 leaves. In the pellet fraction, significant activity was found in the hexo1 and hexo1 hexo2 mutants compared with wild-type plants (Fig. 3). Only little insoluble enzyme activity was observed in the hexo3-1 and hexo2 hexo3-2 mutants, which is most likely derived from residual soluble HEXO1 contaminating the pellet fraction. Similar results were obtained when roots were analyzed (supplemental Fig. S2). In control α-mannosidase assays, all tested lines showed similar specific activities (data not shown).
FIGURE 3.
Endogenous HEXO1 and HEXO3 hydrolyze the synthetic substrate pNP-GlcNAc. Crude (CE, black bars), soluble (SP, gray bars), and insoluble protein extracts (IP, white bars) from leaves of wild-type and knock-out plants were incubated with 5 mm pNP-GlcNAc for 1 h at 22 °C. Data are expressed as means ± S.D. (error bars) (n = 8). In wild-type plants, specific HEXO activity was found to be 14.5 milliunits/mg in soluble protein extracts and 8 milliunits/mg in insoluble protein extracts.
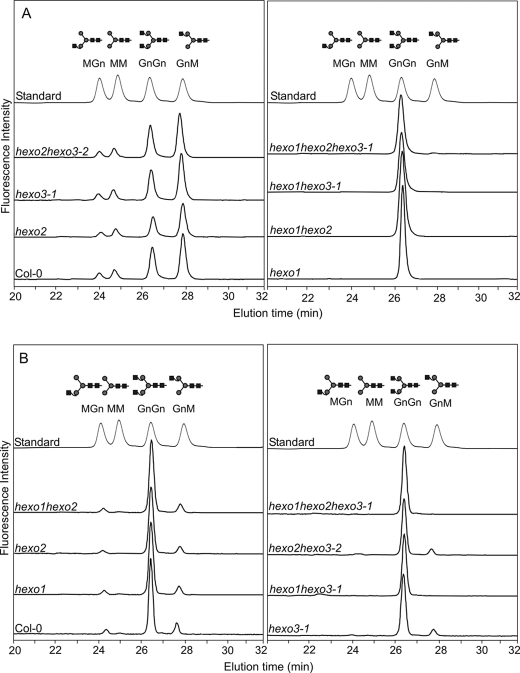
To address the question of whether complex N-glycans are substrates for the endogenous β-N-acetylhexosaminidases, activity assays were performed using GnGn-PA (GlcNAc2Man3GlcNAc2-PA; Fig. 1) as substrate. The removal of either one or two GlcNAc residues results in a characteristic shift of the retention times when analyzed by reverse-phase HPLC. Release of GlcNAc residues was clearly detected for Col-0, hexo2, hexo3-1, and hexo2 hexo3-2 leaf extracts. In each case, incubation with crude homogenates (data not shown) and soluble protein fractions (Fig. 4A) led to the formation of GnM-PA, MGn-PA, and MM-PA (GlcNAcMan3GlcNAc2-PA; GlcNAcMan3GlcNAc2-PA and Man3GlcNAc2-PA; Fig. 1). In contrast, soluble extracts of hexo1 mutants did not hydrolyze GnGn-PA to a detectable extent. When we analyzed the insoluble protein fractions of wild-type and hexo mutants, only little substrate turnover was obtained even upon extended incubation periods. Most of this activity was found to be due to HEXO3 (Fig. 4B). The residual insoluble enzyme activity in hexo3-1 and hexo2 hexo3-2 extracts is presumably derived from HEXO1 trapped in the pellet fraction. Similar results were obtained when roots were analyzed (supplemental Fig. S3).
FIGURE 4.
Activity assays with leaf protein extracts from wild-type and knock-out plants with GnGn-PA as substrate. Soluble protein extracts were incubated for 1.5 h (A), and insoluble protein extracts were incubated for 16 h with 5 μm GnGn-PA at 22 °C (B). The samples were then analyzed by reverse-phase HPLC and fluorescence detection. The elution positions of standards are shown (MGn-, MM-, GnGn-, and GnM-PA).
HEXO1 and HEXO3 Are Responsible for the Formation of Paucimannosidic N-Glycans in A. thaliana
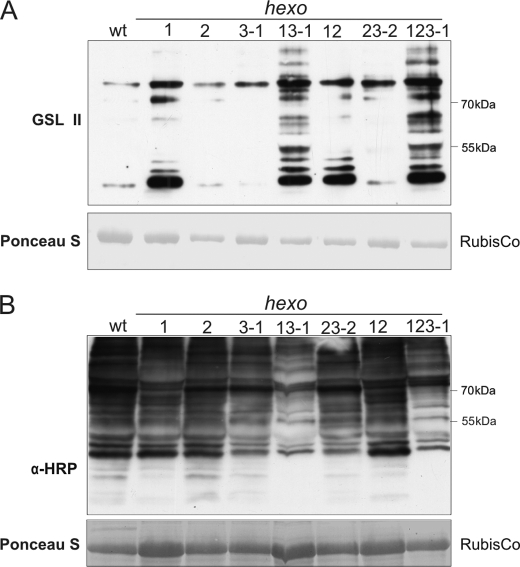
To analyze whether HEXO1 and HEXO3 could indeed act on N-glycans in planta, we performed total N-glycan analysis of wild-type and knock-out plants. First, N-glycosylation changes in hexo1, hexo2, hexo3-1, hexo1 hexo2, hexo2 hexo3-2, hexo1 hexo3-1, and hexo1 hexo2 hexo3-1 plants were analyzed by immunoblotting and lectin overlays. A lectin blot with GSL II, which binds preferentially to terminal GlcNAc residues (20), displayed a stronger signal in hexo1 knockouts compared with wild type, which could be further enhanced by the concomitant absence of functional HEXO3 (Fig. 5A). Immunoblot analysis of protein extracts from leaves of wild-type and hexo knock-outs with anti-horseradish peroxidase (HRP) antibodies which recognize β1,2-xylose and core α1,3-fucose revealed that the staining intensities of protein extracts of the single hexo knock-outs and hexo1 hexo2 and hexo2 hexo3-2 double mutants were similar to that of wild type, but hexo1 hexo3-1 and hexo1 hexo2 hexo3-1 displayed a less pronounced signal (Fig. 5B). It has been shown previously that anti-HRP binds more efficiently to MMXF than to GnGnXF structures (21). We therefore hypothesized that the hexo1 hexo3-1 double and the hexo1 hexo2 hexo3-1 triple mutant contain lower levels of paucimannosidic N-glycan structures.
FIGURE 5.
hexo1 hexo3-1 and hexo1 hexo2 hexo3-1 plants display overall changes in N-glycosylation. Total protein extracts from leaves were subjected to SDS-PAGE under reducing conditions. Blots were incubated with GSL II, which binds to terminal GlcNAc residues (A) and with anti-HRP (α-HRP) antibodies, which recognize β1,2-xylose and α1,3-fucose residues on N-glycans (B).
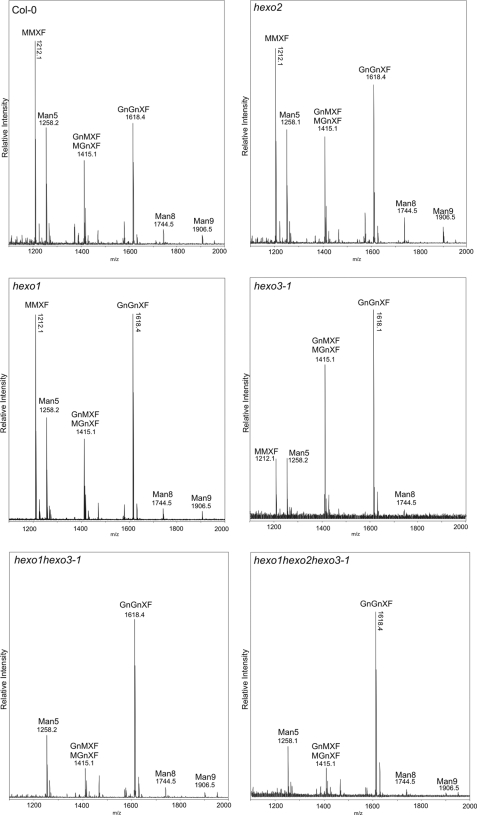
A detailed N-glycan analysis by MALDI-TOF-MS was carried out on leaves (Fig. 6 and supplemental Fig. S4) and roots (supplemental Fig. S5) of wild-type and mutant plants to assess relative changes in their N-glycan profiles. The hexo2 single knock-out mutant did not show any obvious alterations in the N-glycan pattern compared with wild type. Small peaks corresponding to the masses of complex N-glycans (GnGnXF: m/z = 1618.4; GnMXF/MGnXF: m/z = 1415.1) were detectable, but the predominant N-glycan species found was paucimannosidic MMXF (m/z = 1212.1). In the hexo1 and hexo3-1 mutants, GnGnXF was far more prominent (Fig. 6), as also observed for hexo1 hexo2 and hexo2 hexo3-2 plants (supplemental Fig. S4). Mass spectrometric N-glycan profiling of hexo1 hexo3-1 and hexo1 hexo2 hexo3-1 revealed a complete absence of MMXF, with GnGnXF being the major N-glycan species present in leaf and root extracts of these mutants (Fig. 6 and supplemental Fig. S5).
FIGURE 6.
Increased levels of complex N-glycans in leaves in the absence of HEXO proteins. MALDI-TOF-MS spectra of total N-glycans extracted from leaves of wild type, (Col-0) hexo1, hexo2, hexo3-1, hexo1 hexo3-1, and hexo1 hexo2 hexo3-1 knock-out plants.
Despite undetectable endogenous β-N-acetylhexosaminidase activity, a small GnMXF/MGnXF peak was found in N-glycan spectra of hexo1 hexo3-1 and hexo1 hexo2 hexo3-1 plants. To distinguish between these two isobaric structures a detailed glycan analysis was performed using porous graphitized carbon chromatography coupled to electrospray ionization-TOF-MS (22, 23). These experiments demonstrated the complete absence of the GnMXF isomer (supplemental Fig. S6). In contrast to GnMXF, the occurrence of the other isomer (MGnXF) in plant tissues does not necessarily require the action of β-N-acetylhexosaminidases. MGnXF could be the product of incomplete N-glycan processing by N-acetylglucosaminyltransferase II (24).
HEXO1-mRFP and HEXO3-GFP Are Located in Different Compartments
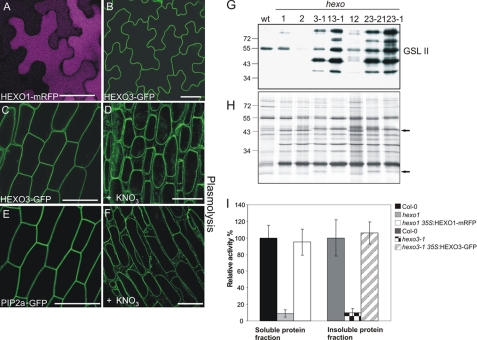
HEXO1-mRFP and HEXO3-GFP transgenes were introduced into Col-0 and the gene silencing mutant sgs2, which has been shown to provide more stable, high level transgene expression (12). Arabidopsis lines stably expressing the fusion constructs were then examined by confocal laser scanning microscopy. Analysis of transgenic HEXO1-mRFP plants revealed a fluorescence pattern reminiscent of a vacuolar protein (Fig. 7A). In contrast, HEXO3-GFP clearly labeled the outline of the cell, indicating targeting to the plasma membrane or the extracellular space (Fig. 7B). To distinguish plasma membrane targeting from cell wall/apoplast localization, plasmolysis experiments were performed. Transgenic plants expressing PIP2a fused to GFP, a well known plasma membrane marker (25), were used as a control. After incubation of the HEXO3-GFP and PIP2a-GFP seedlings in a hypertonic salt solution for an appropriate time, shrinking of the protoplasts was observed in HEXO3-GFP as well as in PIP2a-GFP expressing Arabidopsis lines indicating mainly plasma membrane localization (Fig. 7, C–F). Although we could not detect significant amounts of secreted HEXO3 by confocal laser scanning microscopy, activity assays indicate that a minor amount of HEXO3 is soluble and therefore secreted into the cell wall or apoplast. A subsequent analysis of the apoplastic fluid of wild-type plants and knock-out mutants by performing lectin overlays with GSL II clearly showed an increase in the signal intensity in hexo3 mutants (Fig. 7G). In addition, SDS-PAGE analysis of apoplastic exudates revealed several proteins in hexo3 mutants with altered migration (Fig. 7H, arrows). These results suggest changes in the GlcNAc levels of apoplastic glycoproteins in the absence of a functional HEXO3 and therefore imply a role of HEXO3 in the removal of GlcNAc residues from secreted glycoproteins.
FIGURE 7.
HEXO1 and HEXO3 are localized in different subcellular compartments. A and B, leaf epidermal cells of 5–10-day-old seedlings, stably expressing HEXO fluorescent fusion proteins, were analyzed by confocal laser scanning microscopy. A, HEXO1-mRFP displays uniform fluorescence across the whole cell. Scale bar, 50 μm. B, HEXO3-GFP labels the outline of the cell. Scale bar, 25 μm. C–F, hypocotyl cells of 5–10-day-old seedlings show that (C) HEXO3 localizes to the plasma membrane. E, PIP2a-GFP was used as a plasma membrane marker. Scale bar, 25 μm. HEXO3-GFP (D) and PIP2a-GFP (F) localization in plasmolysed hypocotyl cells is shown. Scale bar, 25 μm. G and H, analysis of apoplastic fluid isolated from leaves of wild-type and hexo knock-out plants by lectin overlays with GSL II (G) and by silver staining (H) is shown. Arrows indicate proteins that migrate slower when HEXO3 is knocked out. I, HEXO1-mRFP and HEXO3-GFP are functional enzymes. Activity assays with soluble and insoluble protein extracts were performed with 5 mm pNP-GlcNAc as substrate for 1 h at 22 °C. The mean values ± S.D. (error bars) of two independent experiments are shown.
We also tested whether the fluorescent fusion proteins are catalytically functional and thus can complement the individual mutants. Transgenic expression of HEXO1-mRFP and HEXO3-GFP restored the β-N-acetylhexosaminidase deficiencies in hexo1 and hexo3-1 mutants. hexo1 35S:HEXO1-mRFP displayed around 95% of wild-type activity, and 35S:HEXO3-GFP fully restored β-N-acetylhexosaminidase activity in the hexo3-1 mutant when activity assays with pNP-GlcNAc were performed (Fig. 7I).
A comparison of the N-glycan patterns of the transgenic lines with that of wild-type plants revealed that the complemented hexo1 and hexo3-1 single knock-out plants contain an even higher percentage of paucimannosidic N-glycan structures than Col-0, probably due to overexpression of the transgenes. Accordingly, the relative levels of complex N-glycans (MGnXF/GnMXF and GnGnXF) were found to be reduced (supplemental Fig. S7).
Distinct Subcellular Distribution of HEXO1 and HEXO3
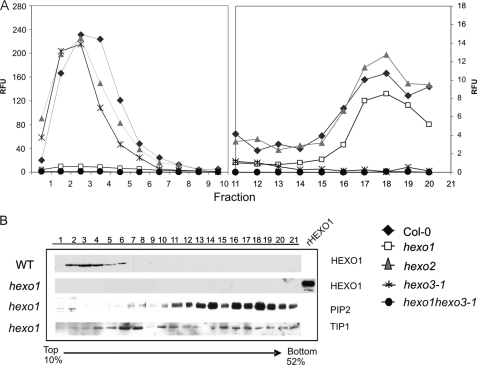
To extend our analysis of the subcellular localization of HEXO1 and HEXO3 to the endogenous forms of these proteins, we decided to perform sucrose density gradient centrifugation with root extracts of Col-0 and different knock-out lines. The distribution of HEXO1 and HEXO3 in each fraction was examined by activity assays with the synthetic substrate MU-GlcNAc. In Col-0 wild-type roots, β-N-acetylhexosaminidase activity was separated into two fractions. The major peak was found in the low density fractions, whereas a minor peak was detected in high density fractions (Fig. 8A). With a HEXO1-specific antibody (supplemental Fig. S8), immunoreactivity was detected only in the low density fractions as typical for vacuolar proteins (Fig. 8B). In hexo1 mutants, much less β-N-acetylhexosaminidase activity was found in the low density fractions, most probably resulting from membrane-shed HEXO3. About half of the HEXO3 activity was detected in the low density fractions, the other half in the high density fractions (Fig. 8A, fractions 16–20). The latter fractions displayed strong signals with anti-PIP2;1 (Fig. 8B), indicating the presence of plasma membrane proteins (16). The same results were obtained with hexo1 hexo2 and hexo2 hexo3-2 double mutants (supplemental Fig. S9). No activity was detectable in gradients performed with root extracts of hexo1 hexo3-1 double and hexo1 hexo2 hexo3-1 triple mutants (Fig. 8A and supplemental Fig. S9). Anti-TIP1;1 was used to identify the tonoplast fractions (17), which were enriched in fractions 6 and 7 (Fig. 8B). These data strongly indicate that endogenous HEXO3 is located mainly in the plasma membrane.
FIGURE 8.
Subcellular distribution of HEXO1 and HEXO3 as assessed by sucrose gradient centrifugation. Sucrose density gradient centrifugation was performed with extracts of roots from wild-type (WT), hexo1, hexo2, hexo3-1, and hexo1 hexo3-1 plants. A, β-N-acetylhexosaminidase activity was analyzed with 1 mm MU-GlcNAc as substrate. RFU, relative fluorescence units. B, aliquots of the fractions were subjected to SDS-PAGE followed by immunoblotting with anti-HEXO1, anti-PIP2;1, and anti-TIP1;1 antibodies. Anti-PIP2;1 was used as plasma membrane marker, and anti-TIP1;1 was used as tonoplast marker. rHEXO1, recombinant HEXO1 (50 ng).
hexo Mutant Plants Do Not Display Any Obvious Phenotype
Despite the lack of functional HEXO transcripts, the single, double, and triple hexo mutants did not display any obvious growth, morphological, or developmental differences compared with wild-type plants when grown on MS medium or on soil. Recently it was shown that the ability to synthesize complex N-glycans seems to be necessary for plant abiotic stress tolerance (26). Therefore, we investigated whether the altered N-glycan patterns of hexo knock-out plants have an influence on root growth when exposed to osmotic stress conditions. Root growth was not sensitive to NaCl or high amounts of sucrose, and the root morphology did not differ significantly between wild-type and mutant plants (supplemental Fig. S10). In addition, we tested whether hexo mutants are affected by cold or heat stress. Seedlings and plants grown at 4 °C or 30 °C were indistinguishable from wild-type Col-0 (data not shown).
Furthermore, it has been shown that proper N-glycosylation of pattern recognition receptors is essential for microbe/pathogen-associated molecular pattern (MAMP/PAMP)-triggered immune reactions by affecting cell surface accumulation and ligand binding properties of these proteins (27). The receptor kinases FLS2 and EFR, which are heavily glycosylated, recognize flagellin and EF-Tu, respectively, through their elicitor active PAMPs flg22 and elf18 (9, 28). They induce a broad array of plant defense responses, including seedling growth inhibition and the production of reactive oxygen species. Because it was shown that several Arabidopsis N-glycosylation mutants display altered immune reactions (27), we investigated whether hexo1 hexo3-1 and hexo1 hexo2 hexo3-1 are impaired in flg22- and elf18-triggered seedling growth inhibition. Five-day-old seedlings were treated with different concentrations of the two peptides (supplemental Fig. S11). Over the whole range of concentrations tested, flg22 and elf18 sensitivity of hexo1 hexo3-1 and hexo1 hexo2 hexo3-1 was comparable with wild-type Col-0 and xylt fucta fuctb, which also contains increased amounts of complex N-glycans (18).
DISCUSSION
Occurrence of paucimannosidic N-glycans is a unique feature of invertebrates and plants and thus, presents a problem for using plant-based expression platforms for the production of recombinant glycoproteins intended for therapeutic use in humans. Major efforts are currently undertaken for remodeling the N-glycosylation pathway of plants to obtain more mammalian-like glycoforms. This includes (i) the elimination of the endogenous plant-specific β1,2-xylosyltransferase and core α1,3-fucosyltransferase activities (18, 29–31); (ii) elongation of N-glycans with β1,4-galactose residues (32–34); (iii) attachment of a bisecting GlcNAc (35–37); (iv) the generation of core α1,6-fucosylated N-glycans (38); and (v) finally the generation of sialylated glycoforms (39). In this study, we established that two β-N-acetylhexosaminidases, HEXO1 and HEXO3, account for the conversion of complex into paucimannosidic N-glycans in Arabidopsis. Interestingly, the two enzymes were found to reside in different cellular compartments. Although HEXO1 is a soluble vacuolar protein, HEXO3 is largely insoluble and located in the plasma membrane. Total N-glycan analysis of mutant plants revealed that HEXO1 and HEXO3 contribute equally to the formation of paucimannosidic N-glycans in roots. In leaves, this process seems to depend more strongly on HEXO3 than on HEXO1. This is most likely due to qualitative and/or quantitative variations between the vacuolar and secretory glycoprotein subsets of leaves and roots because there were no significant differences between the relative HEXO1 and HEXO3 activities of these proteins when tested in vitro. Taken together, our data indicate that HEXO1 and HEXO3 display complementary activities in planta despite their similar catalytic properties (8). The A. thaliana genome encodes a third β-N-acetylhexosaminidase, HEXO2. Although recombinant HEXO2 is capable of cleaving synthetic and N-glycan substrates (8), this enzyme is not involved in complex N-glycan processing in Arabidopsis. Even though HEXO2 transcripts could be detected by RT-PCR in different plant organs, we failed to detect HEXO2 in tissue extracts by enzymatic and immunological means. Hence, it appears unlikely that A. thaliana HEXO2 plays a prominent role in any cellular process requiring β-N-acetylhexosaminidases, including the removal of O-GlcNAc residues from cytoplasmic proteins (40, 41). Interestingly, recombinant HEXO2 was found to be far less stable than HEXO1 and HEXO3, which could also apply to the endogenous protein.
Because the different hexo mutants did not display any detectable phenotype under standard growth conditions, we subjected the mutants to different stress treatments. A. thaliana cgl1 plants lacking complex N-glycans or xylt fucta fuctb mutants show increased salt sensitivity (26). In contrast, none of our hexo mutants displayed an altered salt tolerance. It has been also shown that proper N-glycosylation is indispensable for the function of the pattern recognition receptors EFR and FLS2 (10, 27, 42), which are two heavily glycosylated leucine-rich repeat receptor kinases. Our findings of wild-type like elf18 and fls22-triggered immune reactions in hexo mutants are in full agreement with data from others (27), who found that late N-glycan processing events are not necessary for EFR and FLS2 function. Although the N-glycan structures present on these two receptors have not been determined yet, these findings indicate that paucimannosidic N-glycans are not involved in signal perception by EFR and FLS2.
In summary, we have shown that HEXO1 and HEXO3 are together responsible for the formation of paucimannosidic N-glycans in Arabidopsis. Although HEXO1 represents a classical vacuolar β-N-acetylhexosaminidase, HEXO3 accounts for the processing of secreted glycoproteins. Because A. thaliana plants lacking HEXO1 and HEXO3 lack an overt phenotype, interference with functional β-N-acetylhexosaminidase expression bears promise for the improvement of plant-based expression systems for the production of glycoprotein therapeutics.
Acknowledgments
We thank Barbara Svoboda, Ulrike Vavra, and Karin Polacsek for expert technical assistance and Lindy Abas for sharing reagents and advice. We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants and the European Arabidopsis Stock Centre (United Kingdom) for seed stocks. We thank Hervé Vaucheret for the kind gift of sgs2 seeds and Masayoshi Maeshima for anti-PIP2;1 and anti-TIP1;1 antibodies.
This work was supported by the Austrian Science Fund Grants P19092 and P20817. Research in the Zipfel laboratory is supported by the Gatsby Charitable Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S11, Table S1, and Methods.
- MS medium
- Murashige and Skoog medium
- GSL II
- G. simplicifolia lectin II
- MU-GlcNAc
- 4-methylumbelliferyl-N-acetyl-β-d-glucosaminide
- PIP
- plasma membrane intrinsic protein
- pNP-GlcNAc
- 4-nitrophenyl N-acetyl-β-d-glucosaminide
- pNP-Man
- p-nitrophenyl α-d-mannopyranoside
- MAMP/PAMP
- microbe/pathogen-associated molecular pattern.
REFERENCES
- 1. Lerouge P., Cabanes-Macheteau M., Rayon C., Fischette-Lainé A. C., Gomord V., Faye L. (1998) Plant Mol. Biol. 38, 31–48 [PubMed] [Google Scholar]
- 2. Wilson I. B., Zeleny R., Kolarich D., Staudacher E., Stroop C. J., Kamerling J. P., Altmann F. (2001) Glycobiology 11, 261–274 [DOI] [PubMed] [Google Scholar]
- 3. Zeleny R., Kolarich D., Strasser R., Altmann F. (2006) Planta 224, 222–227 [DOI] [PubMed] [Google Scholar]
- 4. Takahashi N., Hotta T., Ishihara H., Mori M., Tejima S., Bligny R., Akazawa T., Endo S., Arata Y. (1986) Biochemistry 25, 388–395 [Google Scholar]
- 5. Sturm A. (1991) Eur. J. Biochem. 199, 169–179 [DOI] [PubMed] [Google Scholar]
- 6. Fitchette-Lainé A. C., Gomord V., Cabanes M., Michalski J. C., Saint Macary M., Foucher B., Cavelier B., Hawes C., Lerouge P., Faye L. (1997) Plant J. 12, 1411–1417 [DOI] [PubMed] [Google Scholar]
- 7. Dirnberger D., Steinkellner H., Abdennebi L., Remy J. J., van de Wiel D. (2001) Eur. J. Biochem. 268, 4570–4579 [DOI] [PubMed] [Google Scholar]
- 8. Strasser R., Bondili J. S., Schoberer J., Svoboda B., Liebminger E., Glössl J., Altmann F., Steinkellner H., Mach L. (2007) Plant Physiol. 145, 5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J. D., Boller T., Felix G. (2006) Cell 125, 749–760 [DOI] [PubMed] [Google Scholar]
- 10. Nekrasov V., Li J., Batoux M., Roux M., Chu Z. H., Lacombe S., Rougon A., Bittel P., Kiss-Papp M., Chinchilla D., van Esse H. P., Jorda L., Schwessinger B., Nicaise V., Thomma B. P., Molina A., Jones J. D., Zipfel C. (2009) EMBO J. 28, 3428–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Altmann F., Schwihla H., Staudacher E., Glössl J., März L. (1995) J. Biol. Chem. 270, 17344–17349 [DOI] [PubMed] [Google Scholar]
- 12. Butaye K. M., Goderis I. J., Wouters P. F., Pues J. M., Delauré S. L., Broekaert W. F., Depicker A., Cammue B. P., De Bolle M. F. (2004) Plant J. 39, 440–449 [DOI] [PubMed] [Google Scholar]
- 13. Schoberer J., Vavra U., Stadlmann J., Hawes C., Mach L., Steinkellner H., Strasser R. (2009) Traffic 10, 101–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boudart G., Jamet E., Rossignol M., Lafitte C., Borderies G., Jauneau A., Esquerré-Tugayé M. T., Pont-Lezica R. (2005) Proteomics 5, 212–221 [DOI] [PubMed] [Google Scholar]
- 15. Wessel D., Flügge U. I. (1984) Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 16. Suga S., Imagawa S., Maeshima M. (2001) Planta 212, 294–304 [DOI] [PubMed] [Google Scholar]
- 17. Kobae Y., Mizutani M., Segami S., Maeshima M. (2006) Biosci. Biotechnol. Biochem. 70, 980–987 [DOI] [PubMed] [Google Scholar]
- 18. Strasser R., Altmann F., Mach L., Glössl J., Steinkellner H. (2004) FEBS Lett. 561, 132–136 [DOI] [PubMed] [Google Scholar]
- 19. Liebminger E., Hüttner S., Vavra U., Fischl R., Schoberer J., Grass J., Blaukopf C., Seifert G. J., Altmann F., Mach L., Strasser R. (2009) Plant Cell 21, 3850–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu K., Huesing J. E., Shade R. E., Bressan R. A., Hasegawa P. M., Murdock L. L. (1996) Plant Physiol. 110, 195–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jin C., Bencúrová M., Borth N., Ferko B., Jensen-Jarolim E., Altmann F., Hantusch B. (2006) Glycobiology 16, 349–357 [DOI] [PubMed] [Google Scholar]
- 22. Pabst M., Bondili J. S., Stadlmann J., Mach L., Altmann F. (2007) Anal. Chem. 79, 5051–5057 [DOI] [PubMed] [Google Scholar]
- 23. Stadlmann J., Pabst M., Kolarich D., Kunert R., Altmann F. (2008) Proteomics 8, 2858–2871 [DOI] [PubMed] [Google Scholar]
- 24. Strasser R., Steinkellner H., Borén M., Altmann F., Mach L., Glössl J., Mucha J. (1999) Glycoconj. J. 16, 787–791 [DOI] [PubMed] [Google Scholar]
- 25. Cutler S. R., Ehrhardt D. W., Griffitts J. S., Somerville C. R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kang J. S., Frank J., Kang C. H., Kajiura H., Vikram M., Ueda A., Kim S., Bahk J. D., Triplett B., Fujiyama K., Lee S. Y., von Schaewen A., Koiwa H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5933–5938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Häweker H., Rips S., Koiwa H., Salomon S., Saijo Y., Chinchilla D., Robatzek S., von Schaewen A. (2010) J. Biol. Chem. 285, 4629–4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chinchilla D., Bauer Z., Regenass M., Boller T., Felix G. (2006) Plant Cell 18, 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koprivova A., Stemmer C., Altmann F., Hoffmann A., Kopriva S., Gorr G., Reski R., Decker E. L. (2004) Plant Biotechnol. J. 2, 517–523 [DOI] [PubMed] [Google Scholar]
- 30. Cox K. M., Sterling J. D., Regan J. T., Gasdaska J. R., Frantz K. K., Peele C. G., Black A., Passmore D., Moldovan-Loomis C., Srinivasan M., Cuison S., Cardarelli P. M., Dickey L. F. (2006) Nat. Biotechnol. 24, 1591–1597 [DOI] [PubMed] [Google Scholar]
- 31. Strasser R., Stadlmann J., Schähs M., Stiegler G., Quendler H., Mach L., Glössl J., Weterings K., Pabst M., Steinkellner H. (2008) Plant Biotechnol. J. 6, 392–402 [DOI] [PubMed] [Google Scholar]
- 32. Palacpac N. Q., Yoshida S., Sakai H., Kimura Y., Fujiyama K., Yoshida T., Seki T. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4692–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bakker H., Bardor M., Molthoff J. W., Gomord V., Elbers I., Stevens L. H., Jordi W., Lommen A., Faye L., Lerouge P., Bosch D. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 2899–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Strasser R., Castilho A., Stadlmann J., Kunert R., Quendler H., Gattinger P., Jez J., Rademacher T., Altmann F., Mach L., Steinkellner H. (2009) J. Biol. Chem. 284, 20479–20485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rouwendal G. J., Wuhrer M., Florack D. E., Koeleman C. A., Deelder A. M., Bakker H., Stoopen G. M., van Die I., Helsper J. P., Hokke C. H., Bosch D. (2007) Glycobiology 17, 334–344 [DOI] [PubMed] [Google Scholar]
- 36. Karg S. R., Frey A. D., Kallio P. T. (2010) J. Biotechnol. 146, 54–65 [DOI] [PubMed] [Google Scholar]
- 37. Frey A. D., Karg S. R., Kallio P. T. (2009) Plant Biotechnol. J. 7, 33–48 [DOI] [PubMed] [Google Scholar]
- 38. Forthal D. N., Gach J. S., Landucci G., Jez J., Strasser R., Kunert R., Steinkellner H. (2010) J. Immunol. 185, 6876–6882 [DOI] [PubMed] [Google Scholar]
- 39. Castilho A., Strasser R., Stadlmann J., Grass J., Jez J., Gattinger P., Kunert R., Quendler H., Pabst M., Leonard R., Altmann F., Steinkellner H. (2010) J. Biol. Chem. 285, 15923–15930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hartweck L. M., Scott C. L., Olszewski N. E. (2002) Genetics 161, 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thornton T. M., Swain S. M., Olszewski N. E. (1999) Trends Plant Sci. 4, 424–428 [DOI] [PubMed] [Google Scholar]
- 42. Lu X., Tintor N., Mentzel T., Kombrink E., Boller T., Robatzek S., Schulze-Lefert P., Saijo Y. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 22522–22527 [DOI] [PMC free article] [PubMed] [Google Scholar]