Abstract
Transcriptional control in mammals and Drosophila is often mediated by regulatory sequences located far from gene promoters. Different classes of such elements – particularly enhancers, but also locus control regions and insulators -- have been defined by specific functional assays, although it is not always clear how these assays relate to the function of these elements within their native loci. Recent advances in genomics suggest, however, that such elements are highly abundant within the genome and may represent the primary mechanism by which cell- and developmental-specific gene expression is accomplished. In this review, we discuss the functional parameters of enhancers as defined by specific assays, along with the frequency with which they occur in the genome. In addition, we examine the available evidence for the mechanism by which such elements communicate or interact with the promoters they regulate.
Transcriptional regulation is accomplished via the association of trans-acting factors with regulatory DNA sequences. As might be expected, DNA sequences located close to gene promoters are universally involved in this regulation, and in fact in many organisms, including bacteria and unicellular eukaryotes, the vast majority of regulatory phenomena are limited to the few hundred base pairs upstream of the transcription start site. A wealth of studies of gene regulation in vertebrates and in Drosophila, however, indicate that at least in these organisms additional classes of regulatory elements – enhancers, locus control regions (LCRs) and insulators – can be located far from the transcription start site, and that at complex loci multiple elements that are arrayed over large regions can collaborate in regulation of expression of individual genes or gene clusters.
The existence of regulatory sequences located far from the promoters they regulate poses a number of questions, most notably how such elements bridge the distance to their cognate promoters to mediate whatever regulatory effects they may have, or alternatively what regulatory functions can be accomplished over such distances. Ongoing interest in these questions has led to the development of novel technologies to probe for interactions between distal sequence elements, which in turn have placed enhancer and LCR function squarely at the nexus of regulation of RNA polymerase II transcription, modulation of chromatin structure and organization of the genome within the nucleus.
Activities of Distal Regulatory Elements: Enhancers, LCRs and Boundary Elements
Although studies of the characteristics and function of distal regulatory sequences within their endogenous loci are becoming more frequent, most often such elements have been defined in artificial gain-of-function assays, which have revealed distinct classes of regulatory sequences (Maston et. al., 2006). Enhancers, for example, are defined by their activity in assays involving the transient transfection of reporter genes into cultured cells. Such elements are capable of activating transcription regardless of how far they are located from a promoter (within the spatial limitations of the plasmid DNA constructs used for the assay) or their position relative to it – i.e. upstream or downstream.
Another class of regulatory sequences is identified by the establishment of consistent levels of gene expression upon stable integration into the genome of either cultured cells or transgenic mice. Transgenes in general are subject to position effects, in which expression levels are highly dependent upon the genomic site into which a transgene is inserted. Gene promoters by themselves are capable of driving expression in only a small proportion of integration sites. Some sequence elements, however, are capable of conferring high-level gene expression to linked promoters in a position-independent fashion. This activity implies the ability to activate transcription at the majority of genomic locations, and is the functional definition of a locus control region (LCR).
Sequence elements that exhibit enhancer or LCR activity, as defined by these specific functional assays, have been derived from multiple gene loci and thus appear to represent a general phenomenon (Li et. al., 1999; Maston et. al., 2006). The distinct functional definitions of enhancers and LCRs, however, raise the question of how these elements differ intrinsically. At least some LCRs appear to subsume the function of enhancers. For example, the LCR within the human β-globin locus harbors at least one element, 5’ DNaseI-hypersensitive site 2 (5’HS2) that acts as an enhancer in transient assays (Hardison et. al., 1997). The immediate question would appear to be one of quantity vs. quality: does an LCR consist of the same activity as an enhancer, only more powerful, or does it possess a different class of activity?
Evidence exists to support both possibilities. HS2 of the human β-globin LCR can confer position-independent expression to transgenes in mice, but only when 5’HS2-containing transgenes are integrated with copy numbers greater than 3–4 (Ellis et. al., 1993). One interpretation of this behavior is that a sufficient number of HS2 enhancer elements can add up to an LCR, although this has never been investigated systematically.
On the other hand, the β-globin LCR also harbors at least two elements – 5’HSs 3 and 4 – that generally exhibit little activity in transient assays, but activate transcription of linked genes when stably integrated in the genome (Hardison et. al., 1997), and regulatory regions have been derived from other loci that behave similarly. This activity suggests that transient and stable assays of gene expression can reveal different mechanisms of gene activation, and that LCRs can function in ways that are distinct from enhancers.
Additional indications of a fundamental difference between LCR and enhancer activity are provided by studies of the immunoglobulin enhancer Eµ (Forrester et. al., 1994), the thymic enhancer of the adenosine deaminase gene (Aronow et. al., 1995) and the human CD2 LCR (Festenstein et. al. 1996). In each case, a core sequence possesses enhancer activity in transient assays but does not function as an LCR in transgenic assays. LCR activity is observed when additional flanking elements, which possess no measurable activity on their own, are included with the core enhancer. Such activity has been correlated with the propagation of a more accessible chromatin structure from the LCR to more distal sequences.
Another class of promoter-distal regulatory element consists of insulators (Gaszner and Felsenfeld, 2006). The term “insulator” in turn encompasses two distinct subclasses of activity. First, “barrier” elements are defined by their ability to insulate stably transfected transgenes from position effects. This is superficially similar to the activity that defines LCRs, but is different in mechanism: barrier elements are only effective when they flank the transgene to either side, while an LCR functions in any orientation with respect to a transgene. The most common interpretation of this behavior is that a barrier serves to block the encroachment of repressive chromatin structure from a given integration site, while an LCR actively functions to establish a chromatin structure that is more conducive to gene expression. The most well-characterized barrier element to date is 5’HS4 of the chicken β-globin locus; a specific sequence within this element is bound by USF transcription factors and has been shown in multiple studies to insulate transgenes from position effects (Huang et. al., 2007).
The second subclass of insulator consists of “enhancer-blocking” elements, which are defined by their ability to block the function of an enhancer on a linked promoter, but only when located between the enhancer and promoter. They do not appear to silence enhancers or promoters; an enhancer that is blocked from activating a promoter to one side of it by an enhancer-blocking element is still able to activate a promoter located on the other side (Dorsett, 1999). Prominent examples of such enhancer-blocking elements include (again) 5’HS4 of the chicken β-globin locus, specifically a binding site for the transcription factor CTCF (Bell et. al., 1999). In Drosophila, binding sites for the suppressor of Hairy wing [su(Hw)] factor can mediate the same effect (Gdula et. al., 1996).
It is not clear that sequences defined as enhancers, LCRs or enhancer-blocking elements in artificial assays necessarily mediate such functions within the loci from which they are derived. Endogenous regulatory elements normally have no need to ensure gene expression from more than one location within the genome, and so it is not obvious how the ability of an LCR to overcome transgene position effects relates to its function at its native location. For example, deletion of the immunoglobulin enhancer Eµ conforms to the model that transgenic studies would predict, in that expression from the mutant locus is variegated, involving complete silencing of the locus in the majority of cells (Ronai et. al., 1999). Deletion of the murine β-globin LCR, however, while leading to a severe reduction in β-globin expression levels in all cells, fails to affect any measurable feature of chromatin structure within the locus. The major function for this element, as revealed by the deletion, appears to be in transcriptional elongation (Bender et. al., 2000; Schubeler et. al., 2001; Sawado et. al., 2003).
Discrepancies like this could arise from the existence of additional, redundant activities at endogenous loci. Such studies do suggest, however, that the artificial assays used to characterize the function of distal regulatory elements should be interpreted with caution with respect to the role of such sequences within their native loci.
Distal Regulatory Elements in Mammalian Genomes
The development and generalized use of high-throughput and/or genome-wide methodologies for examining transcription factor binding, core histone modifications and RNA polymerase II association has drastically altered the perception of how regulatory sequences are distributed in mammalian genomes. In contrast to the budding yeast S. cerevisiae, for example, the human genome is only sparsely populated with protein-coding genes, and even when growing awareness of noncoding genes, such as small RNAs, is considered, it is readily apparent that the largest proportion of the genome consists of intergenic or intragenic (intronic) sequences for which a specific function is not obvious. Prior studies of selected gene loci have identified distal regulatory sequences such as enhancers and LCRs within these regions, but the gain-of-function assays used to characterize these elements have only served to delineate one or a few such elements for each locus, leaving the majority of noncoding DNA with no known function.
More recently, however, distal regulatory elements have been distinguished from gene promoters by a signature of histone modifications and trans-acting factor binding identified via genome-wide microarray and high-throughput sequencing (chIP-seq) (The ENCODE Project Consortium, 2007; Koch et. al., 2007; Heintzman et. al., 2007; Heintzman et. al., 2009; Visel et. al., 2009). Features of this signature include monomethylation of histone H3 lysine 4 (H3K4) and association of specific factors, such as the histone acetyltransferase and transcriptional coactivator p300. Levels of H3K4 monomethylation in particular peak at enhancers and not at transcription start sites. Conversely, H3K4 trimethylation appears to occur at promoters but not at enhancers. In addition, there is a strong correlation between these regulatory elements and the locations of DNaseI hypersensitive sites (DNaseI HSs), which are generally thought to mark regions where local chromatin structure is disrupted by transcription factor binding (Xi et. al., 2007)
Both H3K4 monomethylation and p300 binding have proven to be predictive for enhancer activity of genomic elements in functional assays (The ENCODE Project Consortium, 2007; Heintzman et. al., 2007; Visel et. al., 2009). This is perhaps not surprising – for example, any sequence that is bound by p300 might be expected to exhibit enhancer activity in a transient transfection assay when linked to a reporter gene, but this doesn’t necessarily indicate that such a sequence actually functions as an enhancer at its native location.
Still, current high-throughput studies are intriguing in several ways. First, they have revealed an unexpected abundance of putative enhancer sequences. A genome-wide study utilizing only two cell lines identified 55,000 sequences exhibiting the “chromatin signature” indicative of enhancers (Heintzman et. al., 2009), which is significantly larger than the number of genes expressed in these lines. The signature at most of these sequences was specific to one or the other cell type as well, and given the variety of cell types present in mammals, the authors extrapolated this figure to estimate that the human genome harbors 105–106 such elements in total. This would represent an average across the genome of one such element every 3,000–30,000 bp, with significantly higher densities in “gene-rich” regions. A pilot survey of 1% of the human genome by the ENCODE project revealed a similar frequency of occurrence of monomethyl H3K4 not associated with gene promoters (The ENCODE Project Consortium, 2007; Koch et. al., 2007).
Second, comparisons of patterns of histone modification and transcription factor association between putative enhancers and known transcription start sites have suggested that the greatest differences between cell types lie in the distal enhancers, not the promoters (Heintzman et. al., 2009). Similarly, mapping of DNaseI HSs across six different cell lines showed that the majority, which were common among all of the lines, were associated with promoters or putative insulator elements, while the remaining cell type-specific HSs were highly enriched for enhancer elements (Xi et. al. 2007). The implication is that development and differentiation of disparate cell types is accomplished for the most part via the differential activities of distal regulatory elements like enhancers.
Since the initial discovery of enhancers, it has been known that they are most often the dominant element in conferring tissue specificity to a linked gene. A hallmark of most enhancers is their ability to activate transcription from any linked promoter in reporter gene constructs, even if promoter and enhancer originate from gene loci with completely different expression patterns in vivo. Although there are exceptions to the general principle, expression of the reporter gene follows the pattern governed by the enhancer, not the promoter. This ability, in fact, has been used to identify enhancers (or, more correctly, regions of the genome) that drive specific expression patterns, via the “enhancer trap” – a transgene under the control of a weak promoter will only be expressed if it integrates into a genomic location that is under the influence of an enhancer that can activate the promoter. The importance of enhancers in determining patterns of eukaryotic gene expression is also illustrated by known examples of genes expressed in different tissues or locations in an organism, which in turn are regulated by multiple enhancers, each of which specifies part of the expression pattern.
On the other hand, the finding that differences in histone modification patterns and transcription factor binding between cell types localizes most often to enhancers and not promoters would seem to conflict with the known prevalence of genes with multiple promoters. Genome-wide analyses have shown that more than 50% of human genes (Kimura et. al., 2006; Carninci et. al., 2007), and ~14% of genes in Drosophila (Zhu and Halfon, 2009), are associated with multiple transcription start sites, and the literature is abundant with examples of genes that are expressed in different cell types via different promoters. It would appear, however, that expression from these alternate promoters is under the control of multiple, alternate enhancers, and that in the majority of cases tissue-, developmental- and/or differentiation stage-specific transcription is under the control of distal regulatory elements that are dominant over the promoter(s).
Third, genome-wide and otherwise high-throughput studies of putative enhancers have unexpectedly revealed that a substantial proportion of such elements are not evolutionarily constrained (The ENCODE Project Consortium, 2007; Margulies et. al., 2007). In the ENCODE pilot survey, roughly half of the sequences determined to have activity in functional assays did not appear to be subject to evolutionary constraint based on cross-species sequence comparisons. Previously, sequence conservation in regions of the genome not associated with gene-coding exons has been used to support other lines of evidence for function of distal regulatory elements, and in fact such conservation has been used as a predictive tool to identify potential regulatory regions, a technique termed “phylogenetic footprinting” (Hardison, 2000). The results of the ENCODE analysis indicate either that many distal regulatory elements cannot be identified on the basis of DNA sequence conservation, or that the conserved sequences within these elements are so small as to escape detection by commonly used computer-based algorithms.
A study of the embryonic enhancers of the even-skipped gene in Drosophila as compared to scavenger flies (Sepsidae) illustrates how this might occur (Hare et. al., 2008). Although the DNA sequences of the enhancer regions in either species are highly divergent, they function to accomplish embryonic patterns of even-skipped expression that are nearly identical. Conservation of small sequence motifs was also excluded. Thus, highly specific function of a set of distal regulatory elements can be conserved even when DNA sequence is not. Speculation for how this can occur has focused on the possibility of compensatory mutations – that is, pairs of mutations that together are not as deleterious as would be expected for single mutations (Veitia, 2008). In addition, an enhancer could conceivably recruit the same activating complex, but interact with different components of it. Over time, the entire sequence of an enhancer can be transformed while maintaining function.
Some caution in generalization of these studies is warranted, in that thus far they have exclusively utilized transformed cell lines, and so it is not yet clear that primary tissues follow the same pattern. Still, these genome-wide studies of histone modification patterns and transcription factor binding have provided a strong suggestion that in at least some metazoans the genome is rife with promoter-distal sequences that represent the dominant regulatory elements in gene expression.
Interactions of Distal Regulatory Elements and Nuclear Organization
Since the discovery of enhancers, one of the most important questions related to their function has been how they regulate promoters located far from them along a linear chromosome. The majority of speculation on this subject has involved the physical association of an enhancer with a promoter, although several mechanisms have been put forward to explain how such juxtapositions might take place. The development of the Chromosomal Conformation Capture (3C) technique and its variants has provided the means to map interactions between distal sequences within the nucleus, including those that may occur between enhancers and promoters (Dekker et. al., 2002). In this technique, cells are crosslinked with formaldehyde and subjected to digestion with restriction endonucleases, then incubated with DNA ligase. Genomic restriction fragments that colocalize in the nucleus are then able to ligate with each other, regardless of their positions on the linear genome, and at greater frequencies than sequences that are positioned in different regions of the nucleus. The technique was originally applied to verify the colocalization of telomeres in nuclei of the budding yeast S. cerevisiae (Dekker et. al., 2002), but was quickly extended to reveal interactions between distal regulatory sequences and promoters in mammalian cells, initially at the β-globin locus (Tolhuis et. al., 2002), and since then at a large and growing number of gene loci (Miele and Dekker, 2008).
The 3C assay has been used to demonstrate that within the murine β-globin locus, sequences dispersed throughout a region of 100–200 kb appear to colocalize within the nucleus in erythroid cells. These include the LCR, the active genes within the cluster, a sequence located ~20 kb downstream of the locus (3’HS1), and additional sequences that are located further upstream of the LCR (−60 HS) , to which erythroid-specific HSs have been mapped (Tolhuis et. al., 2002). This agglomeration of elements is termed the “active chromatin hub” (ACH), and the association of the β-globin gene promoters with the ACH has been precisely correlated with their activity. Thus, in erythroid cells that lack transcription factors crucial for β-globin gene expression, such as GATA-1 and EKLF, the β-globin genes do not appear to colocalize with the LCR or other sequences (Drissen et. al., 2004; Vakoc et. al., 2005). Furthermore, in erythroid progenitors that do not yet express the β-globin genes, a subset of the elements that make up the ACH – 3’HS1, −60 HS, and HS5 of the LCR – and which are bound by CTCF still colocalize. The presumed structure that results has been termed a “pre-ACH” or simply a “chromatin hub” (CH), and it has been speculated that the formation of the ACH occurs on this pre-formed core (Palstra et. al., 2003).
3C has also been used to elucidate association or colocalization of distal sequences within the Th2 cytokine locus in T-lymphocytes (Spilianakis and Flavell, 2004). In addition to a complex pattern of colocalization of various sequences within this region, however, selected promoters and other elements appear to colocalize with other elements within the Ifnγ locus, located on another chromosome (Spilianakis et. al., 2005). This apparent interchromosomal interaction is particularly interesting since genes within the Th2 cytokine locus and the Ifnγ locus are expressed specifically in lineages (Th2 and Th1 lymphocytes, respectively) that are both derived from naïve CD4+ T cells; the interchromosomal interaction appeared strongest in these cells and was diminished after differentiation into Th2 or Th1 cells.
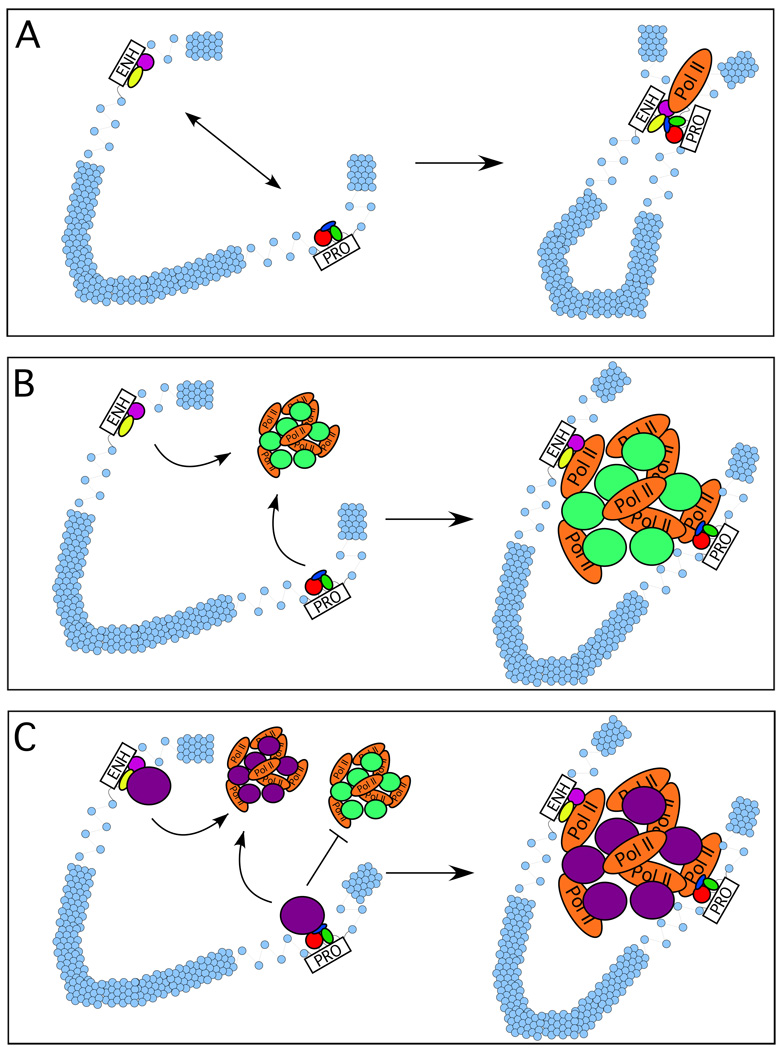
A similar picture has also emerged for the function of Polycomb group (PcG) proteins in gene repression (a comprehensive overview of PcG function is beyond the scope of this article, but for current reviews see Simon and Kingston, 2009 and Müller and Verrijzer, 2009). 3C-based studies, along with other approaches, have suggested that DNA sequences associated with PcG factors colocalize with silenced gene promoters, and that the PcG factors themselves are responsible for promoting long-range intra- and inter-chromosomal interactions to form repressive domains within the nucleus (Cleard et. al., 2006; Lanzuolo et. al., 2007; Tiwari et. al., 2008). These studies and a host of others suggest that distal regulatory elements function via colocalization with the promoters they regulate, resulting in the formation of specific three-dimensional conformations within the nucleus that facilitate the molecular steps required for gene activation (Figure 1A). Thus, enhancer function is linked to larger patterns of nuclear organization – most notably, the spatial segregation of the genome into compartments that harbor “open” and “closed” chromatin, respectively – that accompany gene expression patterns genome-wide (Lieberman-Aiden et. al., 2009).
Figure 1.
Potential mechanisms underlying distal enhancer-promoter colocalization. A. The “traditional” view of enhancer-promoter interactions, in which proteins bound to the enhancer (ENH) and other proteins bound to the promoter (PRO) directly interact with each other to facilitate transcriptional activation, whether by recruitment of RNA polymerase II (as shown), stimulation of RNA polymerase II elongation (as demonstrated for the β-globin LCR), or another mechanism. Blue circles represent nucleosomes; based on the best available evidence, we have depicted the majority of intervening sequence between the enhancer and promoter as condensed to the 30 nm fiber, although higher-order chromatin structure is not well understood and so compaction may be more extensive than this. B. Enhancer-promoter colocalization by association with RNA polymerase II transcription “factories”. In this model, factors bound to both enhancer and promoter independently recruit RNA polymerase II; in a nuclear environment consisting of transcription “factories”, however, this amounts to enhancer and promoter colocalizing to the same “factory”. The green circles represent as-yet undefined, non-Pol II proteins presumed to be present in transcription factories. C. Enhancer and promoter interactions with specific transcription factories. In this model, different genes associate with different kinds of transcription factories. Such specific association is presumably mediated by some common factor or complex recruited to both the enhancer and the promoter (denoted by violet or green circles, respectively), and would be expected to underlie specific interchromosomal interactions.
Thus far, however, while factor dependence for such associations has often been shown, the molecular interactions responsible for bringing about the observed juxtapositions between enhancers and promoter remain elusive. Factors required for function of enhancer-blocking elements [CTCF and, in Drosophila, su(Hw)] have been observed to form distinct foci within the nucleus, representing colocalization of multiple CTCF- and su(Hw)-bound sequences throughout the genome (Wallace and Felsenfeld, 2007; Gaszner and Felsenfeld, 2006). Evidence for a similar unifying principle for enhancer-promoter colocalization has been provided by studies suggesting that within eukaryotic nuclei, RNA polymerase II also forms distinct foci, implying the presence of so-called “transcription factories” (Iborra et. al., 1996). Notably, in addition to gene promoters, many enhancers are associated with RNA polymerase II, which would suggest that colocalization of enhancers and promoters involves association of both types of element with the same transcription factories (Figure 1B). A study of the α-globin locus that utilized 3C along with chromatin immunoprecipitation (ChIP) to reveal RNA polymerase II binding sites demonstrated a perfect correlation: all sequences found to colocalize by 3C were associated with RNA polymerase II and vice versa (Vernimmen et. al., 2007). In addition, in studies of the β-globin locus there is an excellent correlation between conditions under which promoter-LCR interactions are lost (for example, in cells lacking GATA-1 and EKLF) and the absence of RNA polymerase II at the promoter (Drissen et. al., 2004; Vakoc et. al., 2005). This raises an important question: is colocalization of enhancers and promoters, as revealed by 3C, an underlying mechanism of enhancer-promoter communication, or is it simply a consequence of the association of RNA polymerase II with both types of regulatory element?
Within the β-globin locus, simultaneous deletion of 3’HS1 and the −60 HS resulted in no measurable phenotype (Bender et. al., 2006); thus, loss of two of the three CTCF-bound elements known to comprise the “pre-ACH” had no effect on β-globin gene expression. In support of this, the absence of CTCF in murine erythroid progenitors similarly had no effect on β-globin gene expression or colocalization of other, non-CTCF-bound elements within the locus as determined by 3C (Splinter et. al., 2006), with the caveat that undifferentiated progenitors do not express high levels of β-globins normally. Thus, while some studies have shown that loss of gene expression correlates with loss of nuclear colocalization as revealed by 3C, other studies have shown that interactions revealed by 3C are not necessarily functional in gene regulation.
FISH analysis of erythroid cells has shown that active genes throughout mouse chromosome 7 colocalize within the nucleus, and at surprisingly high frequencies (Osborne et. al., 2004). For example, the active β-globin gene appears to colocalize with active genes at the other end of chromosome 7 in 40–60% of cells. In fact, combination of 3C with microarray technology – termed “4C” – has shown that in general, active genes appear to colocalize with other active genes on the same chromosome, with some interchromosomal interactions as well (Simonis et. al., 2006). Conversely, inactive genes tend to colocalize with other inactive genes on the same chromosome. Such studies have been interpreted to suggest an organization within the nucleus in which active gene loci form self-organized clusters, within which concentrations of activating factors are thus high enough to reinforce the active state. Transfection-based studies have suggested that different types of genes tend to cluster in the same RNA polymerase “factories”, implying that this self-organizing principle extends beyond active/inactive to include specific gene classifications within the nucleus (Xu and Cook, 2008) (Figure 1C).
The question that arises, however, is then to what degree specific enhancer-promoter interactions revealed by 3C actually contribute to gene activation. If the β-globin LCR colocalizes with other active genes on chromosome 7 nearly as frequently as it does with an active β-globin gene, then if this colocalization is important for function, the β-globin LCR should have a regulatory influence on the other genes as well. It is not yet known, however, if deletion of the β-globin LCR affects expression levels of other genes on mouse chromosome 7. Thus, the functional consequences of nuclear colocalization, or even of transcription factories, are still largely unknown, and most of the available evidence consists of correlations, not demonstrations of function. There is reason for skepticism. For example, the aggregates of the Su(Hw) protein that provided the initial evidence for the clustering of Su(Hw)-bound DNA sequences appear to be unrelated to the enhancer-blocking function of these sequences (Golovnin et. al. 2008).
Enhancer-Blocking and a Potential Role for Intervening DNA in Enhancer Function
Alternatively, LCR- or enhancer-promoter colocalization revealed by 3C or FISH may not reflect the major or only mechanism by which activation occurs. Several observations of enhancer function argue against a simple “looping” model for interactions with promoters, starting with the early finding that sequences and cognate binding factors that activate promoters over long distances in mammalian cells fail to do so when imported into the yeast S. cerevisiae (Dorsett, 1999). This has been interpreted as an indication that an activity required for long-range function is missing in yeast. In addition, even in mammals the ability of enhancers to activate transcription appears to be developmentally acquired, coinciding with the onset of transcriptional repression in a 2-cell embryo in mice (Majumder and DePamphilis, 1995).
The activity of enhancer-blocking elements suggests not only that the mechanism by which enhancers and promoters interact is more complex than colocalization alone, but that the intervening DNA between such elements is also important. This activity has long been recognized as a significant clue to how enhancer-promoter communication occurs, although its precise meaning has never been clear. Such elements do not appear to inactivate either the promoter or the enhancer, nor do they “trap” the enhancer such that it cannot interact with another promoter, or the promoter so that it cannot interact with another enhancer (Dorsett, 1999).
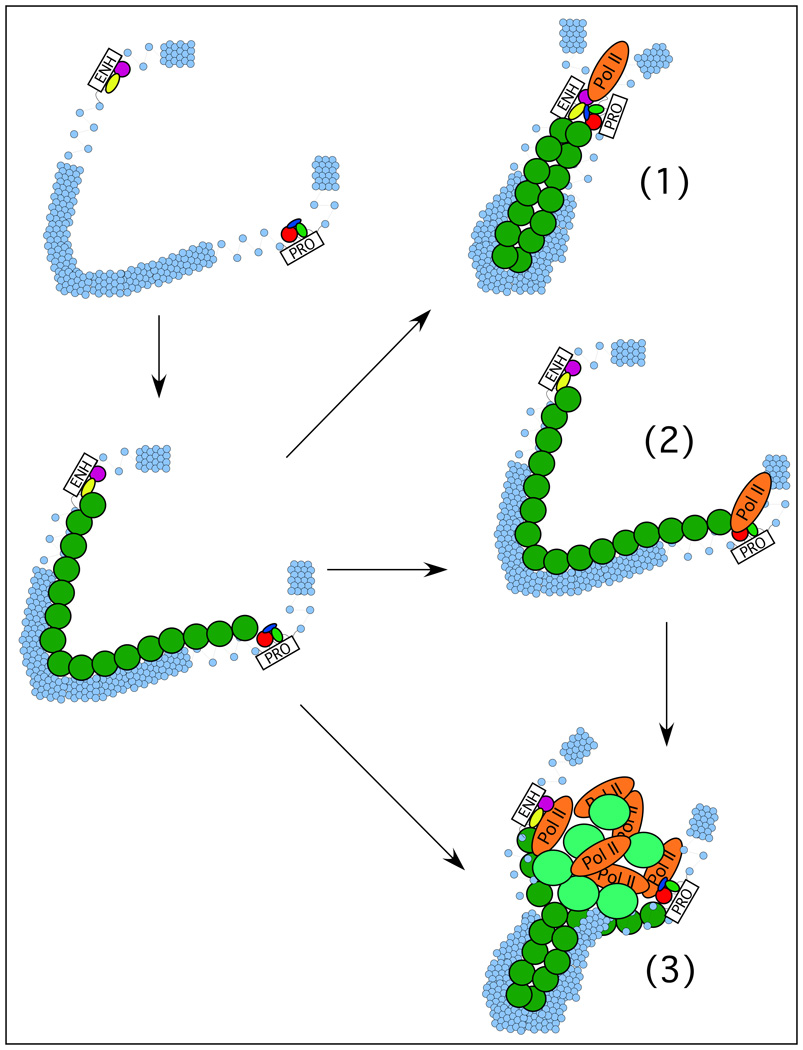
Perhaps the most popular model for the mechanism by which enhancer-blocking elements work is that they divide genomic DNA into “topologically distinct” domains across which sequence-specific trans-interactions cannot occur (Gaszner and Felsenfeld, 2006). It is not obvious, however, how two regions of the genome can be kept “topologically distinct” when the presumed mechanism for enhancer function involves direct interactions that can occur across different chromosomes. In addition, at least one process that is known to require direct interaction between DNA sequences that can be located far from each other within the genome – site-specific recombination by Flp recombinase – is not inhibited by interposing an enhancer-blocking element between recombinase sites (Dunaway et. al., 1997). Thus, although the best evidence suggests a correlation between gene activation and enhancer-promoter colocalization within the nucleus, such colocalization is either established by a process that involves the intervening DNA, or is accompanied by such a process (Figure 2). It is then this process that enhancer-blocking elements disrupt, and is in turn not present in S. cerevisiae.
Figure 2.
Potential role of intervening DNA between enhancers and promoters, and factors that associate with it, in enhancer function. An enhancer and a promoter within a gene locus (top left) are both bound by sequence-specific factors, which in turn serve to recruit a factor or complex that then associates with chromatin throughout the locus (bottom left). We depict several potential mechanisms that might then ensue: (1) factors bound between the enhancer and promoter serve to organize the intervening chromatin in order to bring the two elements together spatially; (2) factors extending from the enhancer to the promoter along the intervening chromatin serve as the primary signal for gene activation, with no necessary role for enhancer-promoter juxtaposition; (3) factors associating with the intervening chromatin organize the locus to bring both enhancer and promoter to a transcription factory and accomplish gene activation. Conceivably, enhancer-promoter juxtaposition at transcription factories may be an event secondary to gene activation, as in (2).
Some hints have been presented that suggest a mechanism by which enhancer-promoter interactions are mediated via intervening sequences. First, based on the activity of enhancer-blocking elements, a genetic screen was performed in Drosophila that identified a protein termed Chip (with homologues termed Ldb-1 and -2 in mammals) as a crucial cofactor in the activity of multiple enhancer elements (Morcillo et. al., 1997; Dorsett, 1999). Based on the known properties of Chip, it was suggested that this protein was responsible for forming multiple “mini-loops” between an enhancer and a promoter that effectively shortened the distance between them and facilitated their interaction. Notably, the other factor that emerged from the same genetic screen in Drosophila was Nipped-B, which is required for the loading of cohesins onto chromosomes during S phase, again implying a function in bridging the gap between enhancer and promoter by effectively shortening the distance between them (Dorsett, 2009).
A study of the β-globin locus in mammals has also shown that the histone methyltransferase MLL2 is associated with chromatin extending from the LCR to the active gene (Demers et. al., 2007). Loss of the LCR-binding transcription factor NF-E2 resulted in loss of transcription and also loss of association of MLL2 throughout the locus. Thus, the β-globin LCR, while apparently colocalizing with the active gene promoters, is also required for the association of a histone modifying enzyme throughout the locus, including regions with which it does not appear to colocalize. Notably, genomic mapping of Nipped-B binding across the Drosophila genome similarly revealed large blocks of sequence that were continuously associated with this factor (Dorsett, 2009). The data again suggest a role for the intervening DNA in enhancer function via the generalized association of cofactors.
Summary
The first demonstrations of gene activation by sequences located far from promoters are now nearly 30 years old (Banerji et. al. 1981;Moreau et. al. 1981); the first demonstration of LCR activity in transgenes was made more than 20 years ago (Grosveld et. al. 1987). In the time since these discoveries, major advances have been made in our understanding of gene regulation and the underlying processes – transcription by RNA polymerase II, modulation of chromatin structure and nuclear organization – accompanied by the development of new technologies that have suggested an unexpected breadth and complexity to the occurrence and activity of distal regulatory elements, and revealed enhancer-promoter interactions suggestive of the mechanism by which they function. Despite these advances, however, it is clear that answers to the most basic questions of enhancer function are as yet far from complete. Given the demonstrated importance of distal elements in the function of many gene loci, and the general importance implied by the potential for as many as a million enhancers in mammalian genomes, further studies of their function would appear to be a major requirement for a full understanding of gene regulation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aronow BJ, Ebert CA, Valerius MT, Potter SS, Wiginton DA, Witte DP, Hutton JJ. Dissecting a locus control region: facilitation of enhancer function by extended enhancer-flanking sequences. Mol. Cell. Biol. 1995;15:1123–1135. doi: 10.1128/mcb.15.2.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhancer by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- Bender MA, Bulger M, Close J, Groudine M. Beta-globin gene switching and DNaseI sensitivity of the endogenous beta-globin locus in mice do not require the locus control region. Mol. Cell. 2000;5:387–393. doi: 10.1016/s1097-2765(00)80433-5. [DOI] [PubMed] [Google Scholar]
- Bender MA, Byron R, Ragoczy T, Telling A, Bulger M, Groudine M. Flanking HS-62.5 and 3’ HS1, and regions upstream of the LCR, are not required for beta-globin transcription. Blood. 2006;108:1395–1401. doi: 10.1182/blood-2006-04-014431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engström PG, Frith MC, Forrest AR, Alkema WB, Tan SL, Plessy C, Kodzius R, Ravasi T, Kasukawa T, Fukuda S, Kanamori-Katayama M, Kitazume Y, Kawaji H, Kai C, Nakamura M, Konno H, Nakano K, Mottagui-Tabar S, Arner P, Chesi A, Gustincich S, Persichetti F, Suzuki H, Grimmond SM, Wells CA, Orlando V, Wahlestedt C, Liu ET, Cleard F, Moshkin Y, Karch F, Maeda RK. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nature Genet. 2006;38:931–935. doi: 10.1038/ng1833. [DOI] [PubMed] [Google Scholar]
- Harbers M, Kawai J, Bajic VB, Hume DA, Hayashizaki Y. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Demers C, Chaturvedi CP, Ranish JA, Juban G, Lai P, Morle F, Aebersold R, Dilworth FJ, Groudine M, Brand M. Activator-mediated recruitment of the MLL2 methyltransferase complex to the beta-globin locus. Mol. Cell. 27:573–584. doi: 10.1016/j.molcel.2007.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. Distant liaisons: long-range enhancer-promoter interactions in Drosophila. Curr. Opin. Genet. Dev. 1999;9:505–514. doi: 10.1016/s0959-437x(99)00002-7. [DOI] [PubMed] [Google Scholar]
- Dorsett D. Cohesin, gene expression and development: lessons from Drosophila. Chromosome Res. 2009;17:185–200. doi: 10.1007/s10577-009-9022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaway M, Hwang JY, Xiong M, Yuen HL. The activity of the scs and scs’ insulator elements is not dependent on chromosomal context. Mol. Cell. Biol. 1997;17:182–189. doi: 10.1128/mcb.17.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J, Talbot D, Dillon N, Grosveld F. Synthetic human beta-globin 5’HS2 constructs function as locus control regions only in multicopy transgene concatemers. EMBO J. 1993;12:127–134. doi: 10.1002/j.1460-2075.1993.tb05638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festenstein R, Tolaini M, Corbella P, Mamalaki C, Parrington J, Fox M, Miliou A, Jones M, Kioussis D. Locus control region function and heterochromatin-induced position effect variegation. Science. 1996;271:1123–1125. doi: 10.1126/science.271.5252.1123. [DOI] [PubMed] [Google Scholar]
- Forrester WC, van Genderen C, Jenuwein T, Grosschedl R. Dependence of enhancer-mediated transcription of the immunoglobulin mu gene on nuclear matrix attachment regions. Science. 1994;265:1221–1225. doi: 10.1126/science.8066460. [DOI] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Gdula DA, Gerasimova TI, Corces VG. Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc. Natl. Acad. Sci. USA. 1996;93:9378–9383. doi: 10.1073/pnas.93.18.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovnin A, Melnikova L, Volkov I, Kostuchenko M, Galkin AV, Georgiev P. ‘Insulator bodies’ are aggregates of proteins but not of insulators. EMBO Rep. 2008;9:440–445. doi: 10.1038/embor.2008.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosveld F, van Assendelft GB, Greaves DR, Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- Hardison R. Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 2000;16:369–372. doi: 10.1016/s0168-9525(00)02081-3. [DOI] [PubMed] [Google Scholar]
- Hardison R, Slightom JL, Gumucio DL, Goodman M, Stojanovic N, Miller W. Locus control regions of mammalian beta-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene. 1997;205:73–94. doi: 10.1016/s0378-1119(97)00474-5. [DOI] [PubMed] [Google Scholar]
- Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet. 2008;4:e1000106. doi: 10.1371/journal.pgen.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Huang S, Li X, Yusufzai TM, Qiu Y, Felsenfeld G. USF1 recruits histone modification complexes and is critical for maintenance of a chromatin barrier. Mol. Cell. Biol. 2007;27:7991–8002. doi: 10.1128/MCB.01326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra FJ, Pombo A, Jackson DA, Cook PR. Active RNA polymerases are localized within discrete transcription “factories” in human nuclei. J. Cell Sci. 1996;109:1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J, Sekine M, Tsuritani K, Wakaguri H, Ishii S, Sugiyama T, Saito K, Isono Y, Irie R, Kushida N, Yoneyama T, Otsuka R, Kanda K, Yokoi T, Kondo H, Wagatsuma M, Murakawa K, Ishida S, Ishibashi T, Takahashi-Fujii A, Tanase T, Nagai K, Kikuchi H, Nakai K, Isogai T, Sugano S. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch CM, Andrews RM, Flicek P, Dillon SC, Karaöz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, James KD, Lefebvre GC, Bruce AW, Dovey OM, Ellis PD, Dhami P, Langford CF, Weng Z, Birney E, Carter NP, Vetrie D, Dunham I. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nature Cell Biol. 2007;9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- Li Q, Harju S, Peterson KR. Locus control regions: coming of age at a decade plus. Trends Genet. 1999;15:403–408. doi: 10.1016/s0168-9525(99)01780-1. [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder S, DePamphilis ML. A unique role for enhancers is revealed during early mouse development. Bioessays. 1995;17:879–889. doi: 10.1002/bies.950171010. [DOI] [PubMed] [Google Scholar]
- Margulies EH, Cooper GM, Asimenos G, Thomas DJ, Dewey CN, Siepel A, Birney E, Keefe D, Schwartz AS, Hou M, Taylor J, Nikolaev S, Montoya-Burgos JI, Löytynoja A, Whelan S, Pardi F, Massingham T, Brown JB, Bickel P, Holmes I, Mullikin JC, Ureta-Vidal A, Paten B, Stone EA, Rosenbloom KR, Kent WJ, Bouffard GG, Guan X, Hansen NF, Idol JR, Maduro VV, Maskeri B, McDowell JC, Park M, Thomas PJ, Young AC, Blakesley RW, Muzny DM, Sodergren E, Wheeler DA, Worley KC, Jiang H, Weinstock GM, Gibbs RA, Graves T, Fulton R, Mardis ER, Wilson RK, Clamp M, Cuff J, Gnerre S, Jaffe DB, Chang JL, Lindblad-Toh K, Lander ES, Hinrichs A, Trumbower H, Clawson H, Zweig A, Kuhn RM, Barber G, Harte R, Karolchik D, Field MA, Moore RA, Matthewson CA, Schein JE, Marra MA, Antonarakis SE, Batzoglou S, Goldman N, Hardison R, Haussler D, Miller W, Pachter L, Green ED, Sidow A. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17:760–764. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Annu. Rev. Genomics Hum Genet. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- Miele A, Dekker J. Long-range chromosomal interactions and gene regulation. Mol. Biosyst. 2008;4:1046–1057. doi: 10.1039/b803580f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcillo P, Rosen C, Baylies MK, Dorsett D. Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer in Drosophila. Genes Dev. 1997;11:2729–2740. doi: 10.1101/gad.11.20.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau P, Hen R, Wasylyk B, Everett R, Gaub MP, Chambon P. The SV40 72 base pair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981;9:6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Verrijzer P. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr. Opin. Genet. Dev. 2009;19:150–158. doi: 10.1016/j.gde.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- Ronai D, Berru M, Shulman MJ. Variegated expression of the endogenous immunoglobulin heavy-chain gene in the absence of the intronic locus control region. Mol. Cell. Biol. 1999;19:7031–7040. doi: 10.1128/mcb.19.10.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawado T, Halow J, Bender MA, Groudine M. The beta-globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes Dev. 2003;17:1009–1018. doi: 10.1101/gad.1072303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D, Groudine M, Bender MA. The murine beta-globin locus control region regulates the rate of transcription but not the hyperacetylation of histones at the active genes. Proc. Natl. Acad. Sci. USA. 2001;98:11432–11437. doi: 10.1073/pnas.201394698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of Polycomb gene silencing: knowns and unknowns. Nature Rev. Mol. Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat. Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Flavell RA. Long-range chromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari VK, Cope L, McGarvey KM, Ohm JE, Baylin SB. A novel 6C assay uncovers Polycomb-mediated higher order chromatin conformations. Genome Res. 2008;18:1171–1179. doi: 10.1101/gr.073452.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol. Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Veitia RA. One thousand and one ways of making functionally similar transcriptional enhancers. Bioessays. 2008;30:1052–1057. doi: 10.1002/bies.20849. [DOI] [PubMed] [Google Scholar]
- Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 2007;26:2041–2051. doi: 10.1038/sj.emboj.7601654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H, Shulha HP, Lin JM, Vales TR, Fu Y, Bodine DM, McKay RD, Chenoweth JG, Tesar PJ, Furey TS, Ren B, Weng Z, Crawford GE. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3:e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Cook PR. Similar active genes cluster in specialized transcription factories. J. Cell Biol. 2008;181:615–623. doi: 10.1083/jcb.200710053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Halfon MS. Complex organizational structure of the genome revealed by genome-wide analysis of single and alternative promoters in Drosophila melanogaster. BMC Genomics. 2009;10:9. doi: 10.1186/1471-2164-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]