Abstract
Eukaryotic cold shock domain proteins are nucleic acid-binding proteins that are involved in transcription, translation via RNA chaperone activity, RNA editing, and DNA repair during tissue developmental processes and stress responses. Cold shock domain proteins have been functionally implicated in important developmental transitions, including embryogenesis, in both animals and plants. Arabidopsis thaliana cold shock domain protein 4 (AtCSP4) contains a well conserved cold shock domain (CSD) and glycine-rich motifs interspersed by two retroviral-like CCHC zinc fingers. AtCSP4 is expressed in all tissues but accumulates in reproductive tissues and those undergoing cell divisions. Overexpression of AtCSP4 reduces silique length and induces embryo lethality. Interestingly, a T-DNA insertion atcsp4 mutant does not exhibit any morphological abnormalities, suggesting that the related AtCSP2 gene is functionally redundant with AtCSP4. During silique development, AtCSP4 overexpression induced early browning and shrunken seed formation beginning with the late heart embryo stage. A 50% segregation ratio of the defective seed phenotype was consistent with the phenotype of endosperm development gene mutants. Transcripts of FUS3 and LEC1 genes, which regulate early embryo formation, were not altered in the AtCSP4 overexpression lines. On the other hand, MEA and FIS2 transcripts, which are involved in endosperm development, were affected by AtCSP4 overexpression. Additionally, AtCSP4 overexpression resulted in up-regulation of several MADS-box genes (AP1, CAL, AG, and SHP2) during early stages of silique development. Collectively, these data suggest that AtCSP4 plays an important role during the late stages of silique development by affecting the expression of several development-related genes.
Keywords: Cold shock domain, embryo development, gene expression, silique development
Introduction
Cold shock domain proteins (CSPs) are among the most conserved nucleic acid-binding proteins consisting of a small gene family in bacteria, animals, and plants. CSPs participate in various cellular functions, which are mediated by their characteristic ability to bind nucleic acids (Braun, 2000; Evdokimova et al., 2001; Kohno et al., 2003). CSPs are mainly implicated in growth and development in eukaryotes and they are well studied in relation to cold stress responses in bacteria. In humans, the Y-box binding protein 1 (YB-1) is transferred from the cytosol into the nucleus in response to specific physiological, environmental, and growth stimuli. YB-1 functions in transcription regulation, DNA repair, and mRNA splicing, thereby affecting the growth and development of organisms (Wilkinson and Shyu, 2001; Faustino and Cooper, 2003; Kohno et al., 2003; Raffetseder et al., 2003). Under normal conditions, YB-1 is localized in the cytoplasm and functions as an RNA chaperone in cytoplasmic ribonucleoproteins (RNPs) and mediates translational repression (Evdokimova et al., 2001, 2006; Bader and Vogt, 2005). Mouse YB-1 is expressed during embryogenesis, and a YB-1-deficient mutant exhibits an embryo-lethal phenotype (Lu et al., 2005). MSY4, an additional mouse CSP, is also highly expressed during embryogenesis and can rescue a YB-1 loss-of-function mutant during early embryogenesis (Lu et al., 2006). Similar to animal CSPs, CSPs from plants, such as Arabidopsis and rice, have also been implicated in development (Nakaminami et al., 2006; Fusaro et al., 2007; Sasaki et al., 2007; Chaikam and Karlson, 2008; Park et al., 2009). Among the four Arabidopsis CSPs, AtCSP2 (AtGRP2/CSDP2; At4g38680) is characterized for its function in affecting flowering time and reproductive tissue development, including seed development (Fusaro et al., 2007). Further studies are necessary to understand fully the functions of AtCSPs in relation to plant reproductive development.
The physiology and genetics of the embryogenesis process are well understood (Goldberg et al., 1994; Jurgens, 1995, 2001; Laux et al., 2004; Park and Harada, 2008). After fertilization, the fertilized zygote divides to establish a position-dependent root and shoot meristem, which determines the fate of the plant body post-germination (Mayer and Jurgens, 1998). The mature seed consists of the embryo and endosperm, whose pattern formation and future developmental plan are decided by a specific gene regulatory network (for a review, see Laux et al., 2004). The transcriptional regulation of embryo formation is initiated specifically from the first asymmetric division of the fertilized single precursor cell, resulting in a primary apical–basal embryo. Apical and basal embryo development is genetically regulated by WUSCHEL-RELATED HOMEOBOX 2 (WOX2) and WOX8 genes, respectively (Haecker et al., 2004). GNOM/EMB30, PINFORMED 7 (PIN7), PIN1, GURKE, FACKEL (FK), and MONOPTEROS (MP) genes also regulate apical–basal embryo formation (Torres-Ruiz et al., 1996; Galweiler et al., 1998; Schrick et al., 2000; Geldner et al., 2001, 2003; Aida et al., 2002; Friml et al., 2003).
Post-embryonic development is controlled by the LEAFY COTYLEDON (LEC) class of genes including LEC1, LEC2, and FUSCA3 genes (Parcy et al., 1997; Lotan et al., 1998; Luerssen et al., 1998; Stone et al., 2001). Loss-of-function mutants for each of these genes exhibit defects in the storage of nutrients that are required for germination and desiccation tolerance during seed dormancy. During late embryogenesis in Arabidopsis, endosperm development is a pivotal process for seed maturation. Four genes belonging to the fertilization-independent seed (FIS) family repress endosperm maturation in non-pollinated and non-fertilized embryos (Kohler and Makarevich, 2006). The FIS family Polycomb group (PcG) proteins include: MEDEA (MEA/FIS1) (Grossniklaus et al., 1998; Kiyosue et al., 1999), Fertilization Independent Seed 2 (FIS2) (Luo et al., 1999, 2000), Fertilization Independent Endosperm 3 (FIE/FIS3) (Ohad et al., 1999), and MULTI-COPY SUPPRESSOR OF IRA1 (MSI1) (Kohler et al., 2003a; Guitton et al., 2004). Loss-of-function mutants of FIS genes result in two distinct phenotypes depending on pollination and fertilization. In the absence of pollination, loss-of-function mutants can initiate seed development. In contrast, in the presence of pollination and fertilization, embryogenesis is arrested at the heart stage of embryo development, resulting in embryo lethality (Ohad et al., 1996; Chaudhury et al., 1997, 2001; Grossniklaus et al., 1998; Chaudhury and Berger, 2001).
MADS-box proteins have been extensively studied in relation to their functional role in floral organ identity determination and transition of the floral meristem. MADS-box genes included in the ABC model are highly expressed during silique development and embryogenesis (Alvarez-Buylla et al., 2000; Parenicova et al., 2003; de Folter et al., 2004; Lehti-Shiu et al., 2005). Gene expression of AG, which is a carpel development gene, increases particularly again during fruit development and ripening (Seymour et al., 2008). Also, SHATTERPROOF (SHP) and FRUITFULL (FUL) are highly expressed and are functionally important during silique development (Gu et al., 1998; Ferrandiz et al., 2000; Liljegren et al., 2000; de Folter et al., 2004). CAULIFLOWER (CAL) also functions in floral meristem identity determination along with AP1. Many AGAMOUS-like proteins are also highly expressed during fruit development (Lehti-Shiu et al., 2005), especially AGAMOUS like-15 (AGL15) and AGL18 which are both expressed in endosperm and embryo tissue (Heck et al., 1995; Alvarez-Buylla et al., 2000). Constitutive overexpression of AGL15 in Arabidopsis delays leaf and flower senescence, as well as fruit maturation (Fernandez et al., 2000). AGL15 was recently shown to bind AtCSP2 and AtCSP4 promoters using a chromatin immunoprecipitation (ChIP) assay. These data support the hypothesis that AtCSP genes are regulated by MADS-box proteins and suggest that AtCSPs may function during silique development (Nakaminami et al., 2009).
In the present study, morphological analyses were performed in T-DNA insertion and gain-of-function AtCSP4 (AtGRP2b; At2g21060) overexpression mutants. Overexpression of AtCSP4 impairs normal silique size determination and embryo development. In addition, the expression of several MADS-box genes and endosperm development genes is altered in AtCSP4 overexpression lines. These data suggest that AtCSP4 alters silique development and embryogenesis by affecting genes involved in silique and seed development.
Materials and methods
Plant materials and growth conditions
An AtCSP4 T-DNA insertion mutant was obtained from GABI-Kat (Stock number: GK 623B08.01, www.gabi-kat.de), and Col-0 wild-type seeds were purchased from Lehle Seeds (Round Rock, TX, USA). Seeds were stratified at 4 °C for 4 d under dark conditions. All plants were grown in Metromix 360 soil (Scott Co., Marysville, OH, USA) under long-day conditions at 23 °C (16 h/8 h for light/dark cycle). To generate AtCSP4 overexpression lines, the coding region of AtCSP4 was amplified with gene-specific primers from a Col-0 wild-type plant with KOD Hot Start high fidelity Taq polymerase (Novagen, Gibbstown, NJ, USA) with the following primers: forward primer 5′-CAC CAT GAG CGG AGG AGG AGA CGT GAA C-3′; reverse primer 5′-ACG AGC ACC ACC GCT AGT GCA ATC CCT TGC-3′. The amplified coding sequence of AtCSP4 was cloned into the pENTR entry vector plasmid (Invitrogen, Carlsbad, CA, USA) according to standard procedures. The pN-TAP binary vector (NTAPa) was obtained from ABRC (http://www.arabidopsis.org, stock number:CD3 696). The AtCSP4 gene sequence was then transferred from pENTR into the NTAPa vector using the LR reaction according to standard procedures (Invitrogen) and the resultant plasmid was designated as 35S:NTAP:AtCSP4. DNA sequencing was performed to confirm sequence integrity (Macrogen, Rockville, MD, USA) and the construct was subsequently transformed into the GV3103 Agrobacterium strain using a MicroPulser electoporator with an electronic pulse of 2.4 kV, 25 μF for 5 ms according to standard procedures (BioRad, Hercules, CA, USA). The 35S:NTAP:AtCSP4 gene construct was transformed into Arabidopsis by the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on 1× Murashinge and Skoog (MS) plates including Gamborg's vitamins, 1% phytoagar, 1% sucrose, and 25 μg ml−1 gentamycin (Caisson Labs, North Logan, UT, USA).
Gene expression analysis
For characterizing gene expression in the atcsp4 T-DNA insertion mutant and 35S:NTAP:AtCSP4 overexpression lines, total RNA was extracted from leaf tissue using TRIzol® reagent (Invitrogen). For studying the expression of MADS-box and embryogenesis-related genes, total RNA was isolated from siliques using the Plant RNA extraction Reagent (Invitrogen). cDNA was synthesized using 500 ng of total RNA with the QuantiTect Reverse Transcription kit (Qiagen, Valencia, CA, USA). Semi-quantitative RT-PCR analysis was performed with the Go-Taq Flexi PCR kit (Promega, Madison, WI, USA) with the following thermocycling conditions: 95 °C for a 2 min initial denaturation followed by 95 °C for 30 s, 58 °C for 30 s, 72 °C for 30 s, for a specified number of cycles (as described within individual figures) with a final extension step for 7 min at 72 °C. Primers are listed in Supplementary Tables S1 and S2 available at JXB online.
For AtCSP4 quantification of transcripts within different tissues, Taq-Man probe quantitative real-time PCR (qRT-PCR) analysis with a TaqMan® Universal PCR Master mix (Applied Biosystems, Foster City, CA, USA) was utilized. Primers and TaqMan® probes for AtCSP4 and ACT2 genes were purchased from Applied Biosystems, and primer sequences are described in Supplementary Table S3 at JXB online. Thermocycling conditions were as follows: 95 °C for 10 min followed by 50 cycles of 95 °C for 15 s and 60 °C for 1 min. With the exception of leaves, total RNA from different tissues was extracted with the Plant RNA extraction Reagent (Invitrogen) and cDNA was prepared as described above.
β-Glucuronidase (GUS) expression plant and histological analysis
To generate a GUS expression gene construct fused to the AtCSP4 promoter, a 2 kb upstream fragment of the AtCSP4 gene was amplified with KOD Hot Start DNA polymerase (Applied Biosystems). The following primers were designed, and incorporated KpnI and NcoI restriction sites: forward 5′-TCT GGT ACC GGG AAA AAC CCA CGC TTG-3′and reverse 5′-TCT CCA TGG TCA CCG TTC CCT TGC GTC-3′. The amplified DNA fragment was digested by restriction enzymes and subsequently ligated into a pre-digested pCAMBIA1303 binary vector in-frame with the GUS gene. This vector was transformed into the LBA4404 Agrobacterium strain with a MicroPulser electoporator as previously described. Transformation of this vector into Col-0 wild-type Arabidopsis was performed by the floral dip method (Clough and Bent, 1998). Transgenic Arabidopsis were screened on 1× MS containing 1% sucrose, 1% phytoagar, and 25 μg ml−1 hygromycin.
GUS staining was performed on non-fixed tissues harvested from various developmental stages. Collected tissues were incubated overnight at 37 °C in GUS staining buffer {50 mM Na2HPO4, 30 mM NaH2PO4, 0.4 mM K3[Fe(CN6)], 0.4 mM K4[Fe(CN6)], 8 mM EDTA, 0.1% Triton X-100, 10% methanol, and 1 μg ml−1 5-bromo-4-chloro-3-indolyl-β-D-glucuronide cyclohexylamine salt (X-Gluc)}. Stained tissue was washed several times in 70% ethanol and samples were subsequently observed under a Nikon SMZ-U dissecting microscope equipped with a Nikon DXM 1200 CCD camera (Nikon, Melville, NY, USA).
Microscopic analysis of seeds
Seeds were dissected from different silique developmental stages and cleared in modified Hoyer's solution (80% chloral hydrate, 20% glycerol, 10% water, v/v/v) for 2 h. Cleared seeds were mounted on glass slides and images were acquired with a Nikon ECLIPSE E600 differential interference contrast (DIC) microscope equipped with Nomarski optics and a Nikon DXM 1200 CCD camera system (Nikon).
Subcellular localization of AtCSP4
The coding region of the AtCSP4 gene was amplified with the following primers: 5′-TCTGTCGACATGAGCGGAGGAGGAGACGT-3′, and 5′-AGACCATGGTACGAGCACCACCGCTAGTGC-3′ containing the NcoI and SalI restriction enzyme digestion sites, respectively. Amplified PCR products were digested with these restriction enzymes and ligated into a pre-digested synthetic GFP(s65T) [sGFPs65T)] plasmid. Sequence integrity and maintenance of the reading frame were confirmed by DNA sequencing. A 5 μg aliquot of sGFP empty plasmid or sGFP-AtCSP4 plasmids was coated onto DM-10 Tungsten particles according to the manufacturer's instructions (BioRad). A Biolistic® PDS-1000 particle bombardment system was used for transient transformation of onion cells by employing 25 inches of a Hg vacuum, a 1000 psi rupture disc, and a 12 cm target distance (BioRad). Bombarded onion epidermal cell layers were incubated at 22 °C overnight under dark conditions. DIC and GFP images were obtained by a Zeiss Axioimager LSM-501 confocal microscope and analysed with LSM image analysis software (Carl Zeiss AG, Germany).
Results
Characterization of the AtCSP4 gene
AtCSP4 (AtGRP2b; At2g21060) and AtCSP2 (AtGRP2/CSDP2; At4g38680) are highly homologous to one another and encode smaller sized proteins relative to the other two Arabidopsis CSPs (AtCSP3; At2g17870 and AtCSP1/CSDP1;At4g36020). The AtCSP4 gene encodes a 603 bp transcript which lacks introns. AtCSP4 is a small sized protein with a highly conserved N-terminal CSD and three repeated glycine-rich domains which are interspersed by two C-terminal CCHC zinc finger motifs (Supplementary Fig. S1A at JXB online). AtCSP4 shows 86% amino acid homology to AtGRP2 and their CSD regions are highly conserved (Supplementary Fig. S1B). Due to this high similarity in sequence, it is possible that AtCSP4 may function redundantly with AtCSP2.
To determine if AtCSP2 expression is affected by AtCSP4, AtCSP2 transcript levels were studied in a T-DNA insertion mutant and AtCSP4 overexpression plants (Supplementary Fig. S1C). As expected, the T-DNA insertion mutant and overexpression plants showed a loss and accumulation of AtCSP4 transcript, respectively (Supplementary Fig. S1C). In order to assess if AtCSP4 levels affect AtCSP2 transcript accumulation, experiments were also carried out to test for alterations of AtCSP2 transcript abundance, and none were detected. These data indicate that AtCSP4 and AtCSP2 do not participate in any feedback regulation of one another.
AtGRP4 expression analyses
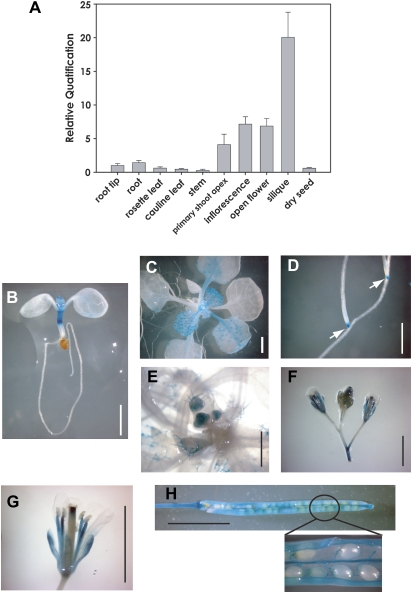
The Genevestigator (http://www.genevestigator.ethz.ch/) public microarray database confirmed that AtCSP4 mRNA is present in all tissues of Arabidopsis but is enriched in reproductive and meristematic regions. AtCSP4 and AtCSP2 transcripts are expressed at higher levels relative to the two additional AtCSP genes (Nakaminami et al., 2009). Comparative analysis of AtCSP4 expression using microarray and real-time PCR data confirmed that the AtCSP4 transcript is highly abundant in reproductive tissues, especially in carpels and siliques. To characterize the tissue specificity of the AtCSP4 transcript, a Taq-Man probe qRT-PCR assay was employed. As shown in Fig. 1A, the AtCSP4 mRNA is expressed in all tissues similar to the pattern of AtCSP2/AtGRP2 (Fusaro et al., 2007; Nakaminami et al., 2009). The AtCSP4 gene transcript is much less abundant in mature leaf tissues where cell divisions are reduced and tissue identity has already been determined. In contrast, the AtCSP4 transcript preferentially accumulates in reproductive and meristematic tissues such as inflorescences and shoot apices. In particular, AtCSP4 is highly abundant in mature siliques harbouring full-grown seeds.
Fig. 1.
Tissue-specific gene expression of AtCSP4. (A) Quantitative real-time PCR analysis of AtCSP4 mRNA expression in different tissues. All data points for AtCSP4 expression levels were calibrated with the respective normalized value of cDNA from root tips. The presented data represent the average and standard deviation of three replicates. Actin2 was used as an internal control for normalization. (B–H) pAtCSP4:GUS expression in transgenic Arabidopsis. A –2 kb promoter region of AtCSP4 was inserted into the pCAMBIA1303 binary vector (pAtCSP4:GUS). (B) pAtCSP4:GUS expression in a 7 DAG seedling, (C) 21 DAG, (D) lateral root from 21 DAG, (E) inflorescence meristem from 28 DAG, (F) floral bud and early flower from a 35 DAG plant, (G) flower of a 35 DAG plant, (H) fully matured silique of a 42 DAG plant. After taking the picture in (H), the silique was opened to visualize seeds in the magnified region. GUS staining was performed from seedlings grown on MS plates (B–D) or soil (E–H) under long-day conditions. The scale bar indicates 0.5 cm. The white arrow in (D) indicates staining in root tips.
To monitor the tissue-specific gene expression of AtCSP4 in planta, transgenic plants were developed by transforming wild-type plants with the T-DNA binary vector containing the AtCSP4 promoter fused to the GUS reporter gene (Supplementary Fig. S2A at JXB online). T2 generation transgenic plants were used for GUS expression assays. GUS staining results revealed that AtCSP4 is expressed in all tissues in good accordance with data obtained by qRT-PCR analyses. Specifically, the AtCSP4 promoter is highly active in reproductive and meristematic tissues. In root tissues, root tip areas of primary and lateral roots were strongly stained, while root vascular tissues were weakly stained. At 21 days after germination (DAG), lateral root tips contained high levels of GUS (Fig. 1D). In young seedlings at 7 DAG, the hypocotyl contained strong GUS gene expression relative to other tissues (Fig. 1B). Tissues exhibiting high numbers of cell divisions such as young leaves, leaf primordia, and shoot apices showed high GUS gene expression (Fig. 1C). In leaf blades, GUS expression was particularly accumulated at the trichome base on the younger leaf surface area. In contrast, mature and older leaves no longer maintained high levels of GUS expression. Vascular tissue also exhibited GUS expression in young leaves, but its expression disappeared gradually following leaf expansion. The apical meristem exhibits high activity of the AtCSP4 promoter, resulting in dark blue staining in the centre of shoot apices from both primary and lateral shoots (Fig. 1C) and the inflorescence meristem (Fig. 1E, F). Reproductive tissues exhibit stronger AtCSP4 promoter activity relative to vegetative tissues (Fig. 1F–H). Siliques with mature seeds showed the highest level of GUS gene expression relative to any other tissue (Fig. 1H). Specifically, the dehiscence area and two valves in siliques stained very strongly (Fig. 1H). Taken together, the qRT-PCR and GUS staining results indicate that AtCSP4 is expressed in multiple tissues but accumulates preferentially in meristemetic and reproductive tissues.
Morphological analysis of an AtCSP4 overexpression and a T-DNA insertion mutant
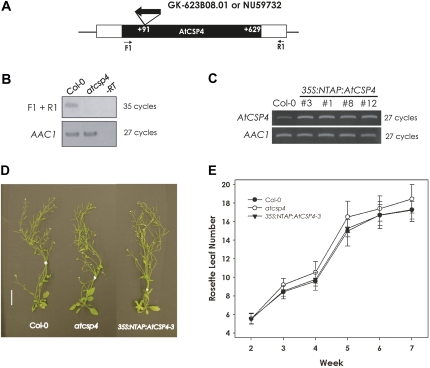
To characterize AtCSP4 gene function in planta, an AtCSP4 overexpression and a T-DNA insertional mutant line was functionally characterized. The T-DNA insertion line was obtained from the GABI-kat collection (GK-623B08.01). A homozygous line was obtained and the precise location of the T-DNA insertion was determined with sequence analysis with a T-DNA-flanking and gene-specific primer pair. From these genotyping results, it was confirmed that the T-DNA insertion occurs within the cold shock domain-coding region at +91 bases from the first initiation codon (Fig. 2A). When total RNA was isolated and converted into cDNA, AtCSP4 transcript was not detected in GK-623B08.01 with a primer pair that was designed to amplify full-length AtCSP4 (Fig. 2B: F1+R1), indicating disruption of the AtCSP4 locus.
Fig. 2.
Morphological phenotype of a T-DNA insertion allele and overexpression of AtCSP4 in vegetative tissues. (A) Genetic map of the T-DNA insertion position within the AtCSP4 locus. A white box represents the AtCSP4 exon and black boxes represent its 5′- and 3'-untranslated regions. With respect to the open reading frame, this GABI-Kat line has an insertion at +91 bp. (B) Semi-quantitative RT-PCR analysis of AtCSP4 expression in leaves from plants at 28 DAG of the wild type (Col-0) and AtCSP4 in the GABI-Kat T-DNA insertion line. PCR was performed for the described cycle numbers with primers flanking the insertion and additional non-flanking primers. AAC1 was used as an internal control. All semi-quantitative RT-PCR figures are representative images from three replicate reactions. (C) Semi-quantitative RT-PCR analysis of AtCSP4 expression in leaves from 28 DAG of the wild type (Col-0) and three different independent 35S:NTAP:AtCSP4 transgenic lines exhibiting the defective seed phenotype. (D) Comparison of whole plants at 42 DAG. All plants were grown in soil under long-day conditions (bar=2 cm). (E) Total leaf number in wild-type, atcsp4, and 35S:NTAP:AtCSP4-3 plants. Leaf number was counted weekly post-germination (n=30). All plots show the means, and error bars represent the standard deviation.
An overexpression line of AtCSP4 fused to an N-terminal TAP-tag that was driven by a 35S promoter (35S:NTAP:AtCSP4) was generated as a means to observe the functional effect of ectopic overexpression of AtCSP4 (Supplementary Fig. S2B at JXB online). Seventeen independent gentamycin-resistant lines were isolated from the T1 generation. Four representative lines were characterized in greater detail, with all exhibiting embryo-lethal and abortive phenotypes (Fig. 3). AtCSP4 transcript abundance was visualized with semi-quantitative RT-PCR, confirming elevated levels of AtCSP4 transcript in overexpression lines relative to the wild type (Fig. 2C).
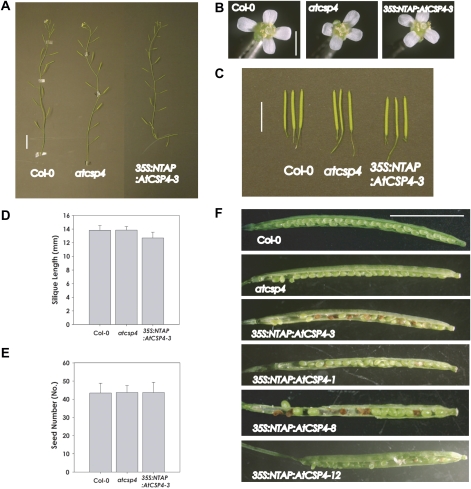
Fig. 3.
Morphological phenotype of the T-DNA insertion allele and overexpression of AtCSP4 in Arabidopsis vegetative tissues. All plants used in this test were grown in soil under long-day conditions. (A) Silique formation on the primary stem at 49 DAG (bar=2 cm). (B) Comparison of flower size. Opened flowers were collected from primary bolts at 35 DAG (bar=0.2 cm). (C) Comparison of silique size. Fully matured siliques were detached at 49 DAG (bar=1 cm). (D) Silique size comparison. Silique sizes were determined with the longest siliques from 50 different plants from wild-type, atcsp4, and 35S:NTAP:AtCSP4-3 plants (n=50). (E) Seed number comparison. Seeds were counted from the same siliques which were used in D. All plots in D and E show means ±SD. (F) Seed formation and maturation of the wild type and atcsp4, and the T3 generation of 35S:NTAP:AtCSP4-3, 1, 8, and 12 lines. All siliques were opened from fully matured siliques at 49 DAG (bar=0.5 cm).
To gain insight into the function of AtCSP4, the phenotypes of T-DNA insertion mutant and overexpression lines of AtCSP4 were compared with the wild type under long-day light conditions. As shown in Fig. 3, the 35S:NTAP:AtCSP4 overexpression lines exhibit typical phenotypes in mature seedlings. The seedling height of both homozygous atcsp4 and heterozygous 35S:NTAP:AtCSP4 lines did not differ from those observed in the wild type under long-day conditions (Fig. 2D). In addition, total leaf number (Fig. 2E) and leaf size (not shown) did not differ significantly from wild type. With respect to flowering time, all lines initiated flowering at 23 DAG, similar to wild-type plants (data not shown). Four independent transgenic plants harbouring the 35S:NTAP empty vector were also characterized and did not exhibit any phenological or growth abnormalities (data not shown). Collectively, these data demonstrate that overexpression or disruption of AtCSP4 does not affect vegetative development or flowering time.
Although atypical phenotypes were not observed during vegetative growth, brown and shrunken seeds were observed within mature siliques of 35S:NTAP:AtCSP4 overexpression lines. It was not possible to obtain 35S:NTAP:AtCSP4 homozygous plants, even after screening up to the T4 generation using gentamycin resistance as a selectable marker (data not shown). These data strongly support the hypothesis that the embryo-lethal phenotype arrests normal seed development in 35S:NTAP:AtCSP4 homozygous lines.
Reproductive tissue morphology of the wild type, the homozygous atcsp4 T-DNA insertion mutant, and the representative heterozygous 35S:NTAP:AtCSP4-3 overexpression line was compared for all tests. The distance between individual siliques on the primary stem of atcsp4 and 35S:NTAP:AtCSP4 plants was similar to that of the wild type (Fig. 3A), and the size and structures of atcsp4 and 35S:NTAP:AtCSP4 flowers were not altered (Fig. 3B). However, the length of mature siliques in the 35S:NTAP:AtCSP4-3 line was shorter than that of the wild-type and atcsp4 (Fig. 3C, D). Overexpression of the NTAP:AtCSP4 fusion protein resulted in an abortive or embryo-lethal phenotype (Fig. 3F).
The extent of embryo lethality varied among the four characterized lines. Specifically, the 35S:NTAP:AtCSP4-12 line possessed white immature seeds as well as embryo-lethal seeds. 35S:NTAP:AtCSP4-3 and 35S:NTAP:AtCSP4-8 lines showed a very similar distribution of embryo-lethal seeds and the 35S:NTAP:AtCSP4-1 line contained the lowest number of embryo-lethal seeds. The proportion of the 35S:NTAP:AtCSP4-3 plants generating short siliques harbouring embryo-defective seed to the plants generating wild-type silique and seeds is 55%:45% in T3 plants that were grown in soil without antibiotic selection (n=36). The numbers of defective seeds and normal wild-type seeds in short siliques were also counted to determine the segregation ratio in self-fertilized T3 generation plants of 35S:NTAP:AtCSP4-3. The total number of defective seed and wild-type seed in the plants showing an embryo-lethal phenotype segregated to 51%:49% (Table 1). However, the atcsp4 T-DNA insertion mutant exhibited typical silique morphology and contained mature seeds. Taken together, these data confirm that the AtCSP4 gene alone is not an essential gene for completion of the reproductive stage. This hypothesis is supported further by the fact that AtCSP4 is 1000-fold down-regulated in the Ler ecotype relative to Col-0 (Nakaminami et al., 2009). However, ectopic overexpression of AtCSP4 induces short siliques that harbour ∼50% embryo-defective seeds. Overexpression of AtCSP4 disrupts normal seed and silique development via an unknown functional mechanism.
Table 1.
Segregation of homozygous atcsp4 and heterozygous 35S:NTAP:AtCSP4-3 plants
| Plant genotype | Defective seeds | Wild-type seed | Total no. of seeds analysed | No. of siliques analysed |
| Col-0 | 0.58% | 99.42% | 868 | 20 |
| atcsp4 | 0.69% | 99.31% | 874 | 20 |
| 35S:NTAP: AtCSP4-3 | 51.66% | 49.34% | 2228 | 50 |
Image analysis of embryo development
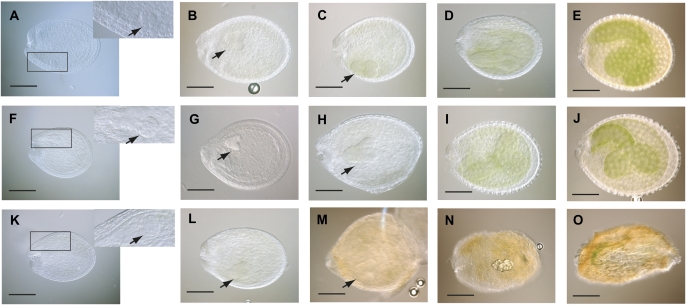
Further studies were conducted to identify the precise stage in embryo development which is impaired by overexpression of AtCSP4. Seeds were cleared and extracted from early maturing stages of siliques derived from 35S:NTAP:AtCSP4-3, atcsp4 T-DNA insertion mutant, and wild-type plants. Each silique stage was referred to according to the stages of embryo development as described in the AtGenExpress website (http://www.genomforschung.uni-bielefeld.de/GF-research/AtGenExpress-SeedSiliques.html). As shown in Fig. 4F–J, the atcsp4 T-DNA insertion mutant exhibits normal embryo and endosperm development in seeds. No morphological defects were observed in the external seed shape and size in atcsp4 mutants. Since 35S:NTAP:AtCSP4-3 exhibits two different seed phenotypes (embryo-lethal and wild-type) within the same silique, all seeds were carefully observed until the embryo-defective phenotype appeared. No abnormalities were observed in seeds from 35S:NTAP:AtCSP4-3 plants until after the early heart stage (as seen in Fig. 4K and L). After the heart stage, 35S:NTAP:AtCSP4-3 seeds did not develop to the torpedo stage and the endosperm began shrinking and the seed coat rapidly turned brown (Fig. 4M–O). In constrast, seeds from the atcsp4 T-DNA insertion mutant and wild type maintained a green colour and normal shape (Fig. 4C–E, H–J). Wild-type-appearing seeds in 35S:NTAP:AtCSP4-3 plants continued developing normally and did not differ morphologically from Wild type (data not shown). Thus, it is concluded that the embryo-lethal mutation in homozygous 35S:NTAP:AtCSP4-overexpressing seeds results from defects occurring during the late heart stage of embryo development.
Fig. 4.
Comparative analysis of stages of silique development in the T-DNA insertion and overexpression lines of AtCSP4. Seeds were detached and cleared from 49 DAG siliques. (A–E) Wild-type seeds, (F–J) atcsp4, and (K–O) 35S:NTAP:AtCSP4-3. A, F, and K show globular stage embryos; B, G, and L show early heart stage embryos; C, H, and M show late heart stage embryos; D, I, and N are torpedo stage embryos; and E, J, and O show bent cotyledon stage embryos (bar=100 μm). Note the browning of embryo from 35S:NTAP:AtCSP4-3 plants beginning at the late heart stage. Right upper images on A, F, and K are enlarged pictures of the area marked with a rectangle on the respective panel. Black arrowheads point to developing embryos from globular to late heart stages.
Embryogenesis-related genes and MADS-box protein gene expression during early silique development stages
To elucidate the relationship between defective seed maturation and related gene expression in the 35S:NTAP:AtCSP4-3 line, the gene expression levels of both initial floral organogenesis- and seed development-related genes were studied in 35S:NTAP:AtCSP4 plants. As shown in Fig. 4M, embryo development in 35S:NTAP:AtCSP4-3 is impaired from the late heart stage. Publicly available microarray data of silique development revealed that AtCSP4 peaks in gene expression in the carpel developmental stage, which occurs during early silique development (http://www.genevestigator.ethz.ch/). Therefore, our initial focus was on gene expression of candidate genes controlling early stages of silique development.
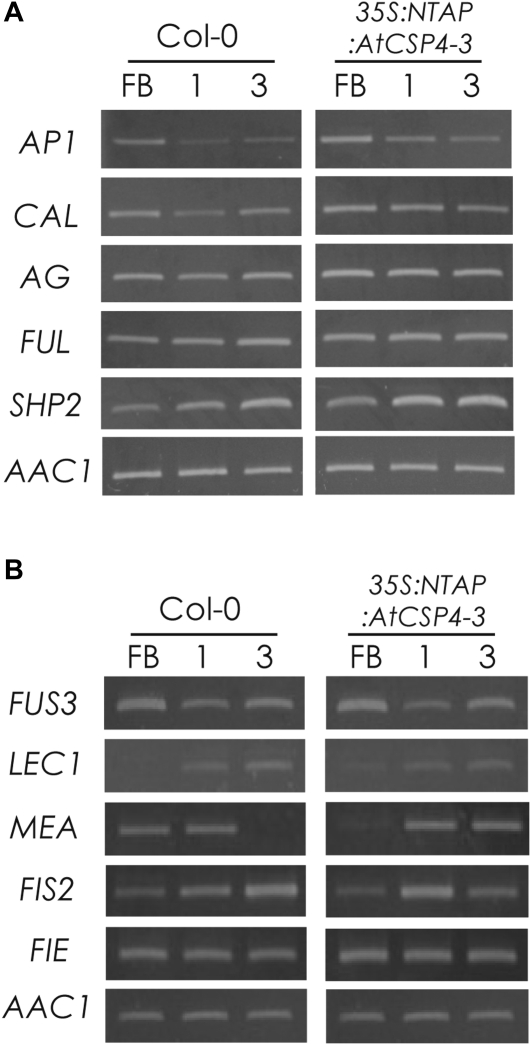
MADS-box proteins have been functionally implicated in early silique development, ovule development, and seed embryogenesis (Lehti-Shiu et al., 2005). Since one of the characteristic mutant phenotypes of 35S:NTAP:AtCSP4 plants was a shortened silique length, the expression of MADS-box protein genes was monitored in developing flowers and two stages of silique development. Additionally, the transcript expression of pistil and fruit development genes such as FUL and SHP2 was also determined. In floral buds and stage 1 siliques, transcripts of AP1, CAL, FUL, and AG exhibited slight accumulation in 35S:NTAP:AtCSP4-3 plants relative to the wild type. The expression pattern for SHP2 exhibited the most noticeable shift, with accumulation occurring earlier in overexpression plants. With the exception of SHP2, gene expression patterns were similar between wild-type and overexpression plants (Fig. 5A). In summary, overexpression of AtCSP4 predominantly affects the mRNA accumulation of SHP2 during floral and silique development with minimal effect on AP1, CAL, AG, and FUL. Shortened siliques in overexpression plants may result from the collective alterations in gene expression patterns for several MADS-box protein genes at the onset of the early stages of silique development.
Fig. 5.
Semi-quantitative RT-PCR analysis of genes involved in silique development and seed embryogenesis. (A) Semi-quantitative RT-PCR analysis of MADS-box genes. AP1, CAL, AG, and SHP2 were affected by overexpression of AtCSP4 in floral buds (FB), and stage 1 and stage 3 embryos. (B) Semi-quantitative RT-PCR of seed embryogenesis-related genes. LEC1 and FUS3 were used as marker genes related to embryo development. MEA, FIS2, and FIE were used as maker genes related to endosperm development. Overexpression of AtCSP4 affects endosperm development genes such as MEA and FIS2 but not FIE. Arabic numbers denote the silique stages. AAC1 was used as an internal control. All semi-quantitative RT-PCR figures are representative images from three replicate reactions.
The relationship between the impairment of silique development (shortened size) and the development of abnormal seeds was also studied by comparison of MEA, FIS2, and FIE transcripts. Transcripts for FUS3 and LEC1 genes were not altered in overexpression plants relative to wild type (Fig. 5B). The ABI3 transcripts could not be detected in the amplification assays even after 40 cycles of amplification (data not shown). Among the genes involved in endosperm development, MEA mRNA was highly accumulated during the early stage of embryogenesis in 35S:NTAP:AtCSP4-3 plants (Fig. 5B). Consistent with previously reported data, the MEA transcript was detected in inflorescences and open flowers before fertilization and it disappeared as embryo development proceeded in wild-type plants (Baroux et al., 2006). In contrast, the MEA transcript in 35S:NTAP:AtCSP4-3 was expressed at a much greater level relative to that of the wild type in the later stage of embryo development (Fig. 5B). Two other endosperm developmental genes, FIS2 and FIE, were expressed at similar levels in floral buds in wild-type and 35S:NTAP:AtCSP4-3 plants, but FIS2 expression was reduced during stage 3 in 35S:NTAP:AtCSP4-3 plants. In summary, expression of MEA and FIS2 is affected during the floral bud and early stages of silique development in 35S:NTAP:AtCSP4-3 plants. MEA transcripts were most notably affected during stage 3 of silique develoment. Expression of LEC1, FUS3, and FIE was not affected by overexpression of NTAP:AtCSP4, indicating that these genes are not likely to contribute to the defective seed phenotype observed in 35S:NTAP:AtCSP4-3 plants.
Subcellular localization of AtCSP4 in onion cells
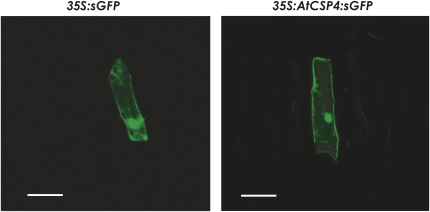
The subcellular localization of AtCSP4 was examined by making a C-terminal fusion to sGFP(s65T) (Supplementary Fig. S2C at JXB online). The 35S:sGFP and 35s:AtCSP3:sGFP plasmids were transformed into onion epidermal cells by particle bombardment. 35S:sGFP was expressed in both the nucleus and cytosol (Fig. 6), and 35S:AtCSP4:sGFP-transformed onion cells also exhibited a nuclear and cytoplasmic localization.
Fig. 6.
AtCSP4 subcellular localization in onion epidermal cells. Transient analysis of the subcellular localization of AtCSP4 in onion epidermal cells (bar=125 μm). Similar to the 35S:GFP control, note that AtCSP4 localized to the cytosol and the nuclei.
Discussion
Putative redundant function of AtCSP4/AtGRP2b and AtCSP2/AtGRP2
CSPs are commonly found in plants (Karlson and Imai, 2003); however, despite recent functional studies in wheat, Arabidopsis, and rice, their precise functional roles remain poorly understood (Karlson et al., 2002; Karlson and Imai, 2003; Nakaminami et al., 2006; Fusaro et al., 2007; Sasaki et al., 2007; Chaikam and Karlson, 2008; Kim et al., 2009; Nakaminami et al., 2009). Arabidopsis possesses four CSPs which can be sorted into two groups based on similarity of amino acid sequence and motifs. AtCSP2/AtGRP2/CSDP2 and AtCSP4/AtGRP2b proteins contain ∼200 amino acids, and AtCSP3 and AtCSP1/CSDP1 contain ∼300 amino acids. The amino acid identity and similarity between AtCSP2 and AtCSP4 is 86% and 95%, respectively (Supplementary Fig. S1B at JXB online). Detailed tissue-specific expression analyses confirmed that both of these genes are highly expressed in meristemic and reproductive tissues (Nakaminami et al., 2009). Although the AtCSP4 transcript is less abundant than AtCSP2, AtCSP4 exhibits a similar trend of expression during development (Nakaminami et al., 2009). Comparative analysis of gene expression between the Ler and Col-0 ecotype revealed that AtCSP4 expression is 1000-fold reduced in Ler relative to the Col-0 ecotype, suggesting that a loss of AtCSP4 is not critical for plant survival (Nakaminami et al., 2009). This hypothesis is further supported by T-DNA mutant analysis where knockout of t he AtCSP4 gene does not appear to have adverse effects on plant growth and development (Fig. 2). Interestingly, when AtCSP4 is overexpressed, severe development effects occur. AtCSP2 transcript is unaffected in atcsp4 knockout and overexpression mutant lines. The functional and interactive relationship between AtCSP4 and AtCSP2 is not known at this time.
qRT-PCR analyses (Fig. 1A) (Nakaminami et al., 2009) and GUS reporter data confirm that AtCSP4 is predominantly expressed in meristemic and reproductive tissues, especially in siliques (Fig. 1B–H). These GUS data are also in good agreement with AtCSP2 gene expression data previously obtained by in situ hybridization and GUS analysis (Fusaro et al., 2007; Sasaki et al., 2007; Nakaminami et al., 2009). When a homozygous atcsp4 T-DNA insertional mutant was characterized, no atypical phenotypes were observed. These data are in contrast to those previously observed for RNA interference (RNAi) mutant lines of AtCSP2 (Fusaro et al., 2007). Taken together with these aforementioned findings from RNAi and the confirmed 1000-fold reduction of AtCSP4 in the Ler ecotype, the lack of any aberrant phenotype in vegetative and reproductive tissues in the atcsp4 T-DNA insertion mutant (Figs 2–4), it is hypothesized that AtCSP2 has greater functional importance in planta (Fusaro et al., 2007; Sasaki et al., 2007; Nakaminami et al., 2009). Furthermore, the similarities in the expression patterns of AtCSP4 and AtCSP2 suggest AtCSP4 and AtCSP2 are functionally redundant.
Overexpression of AtCSP4 reduces silique lengths and induces embryo lethality
In an effort to understand in planta functional roles for AtCSP4, an atcsp4 homozygous T-DNA insertion mutant and four independent 35S:NTAP:AtCSP4 overexpression lines were characterized. The atcsp4 T-DNA insertion mutant does not exhibit any atypical morphological or developmental phenotypes (Figs 2, 3). Flowering time and whole seedling size did not differ from those observed in wild-type plants. Unlike the atcsp4 T-DNA insertion mutant, an RNAi mutant of AtCSP2 showed early flowering and abnormal flower generation (Fusaro et al., 2007). On the other hand, overexpression of AtCSP4 resulted in atypical phenotypes in reproductive tissues such as shortened silique size and defective seed maturation. However, the developmental timing of vegetative and reproductive tissues was not affected by ectopic overexpression of AtCSP4 (Fig. 2). In heterozygous 35S:NTAP:AtCSP4-3 plants, mature siliques contained ∼50% defective seeds (Table 1), resembling a FIS gene mutant with impaired endosperm development (Grossniklaus et al., 1998). For this parent-of-origin effect, the embryo-lethal seed phenotype in a heterozygous mea mutant, a representative FIS gene, also shows a 50% proportion in self-fertilization. The other fis mutants such as mis1 also exhibited a similar ratio of defective seed (Kohler et al., 2003a, b). Embryo formation in 35S:NTAP:AtCSP4-3 arrests at the late heart stage (Fig. 4) which is the same time point where defects in embryo formation are observed for a fis mutant allele (Ohad et al., 1996; Chaudhury et al., 1997; Grossniklaus et al., 1998; Kiyosue et al., 1999). The defective seed formation in AtCSP4 overexpression lines probably results from a parent-of-origin effect on the endosperm development process.
Expression of MADS-box and seed embryogenesis-related genes is altered by overexpression of AtCSP4
Microscopic observation of developing embryos in 35S:NTAP:AtCSP4-3 seeds revealed that early shrinkage of endosperm tissue initiates at the late heart stage (Fig. 4M). It is important to note that animal CSPs have also been implicated in embryo development. For example, mouse YB-1 and MSY4 proteins are highly expressed during embryogenesis, and a double knockout in mice cells prevent cell senescence, suggesting both genes share function in proliferative tissue in higher vertebrates (Lu et al., 2006). In Arabidopsis, ChIP analysis confirmed that AGL15, a well-studied MADS-box protein expressed in developing embryos, binds to the promoter regions of AtCSP4 and AtCSP2 (Nakaminami et al., 2009). These correlative data suggest that AtCSPs may also play an important role in silique development similar to their animal counterparts (Nakaminami et al., 2009). Therefore, it is of interest to further understand the functional relationship of plant CSPs during silique development.
MADS-box proteins commonly function in relation to floral transition, fruit development, and ovule development (Lehti-Shiu et al., 2005; Seymour et al., 2008). Silique development is established from carpel identity determination, which is regulated by AG. AP1 and FUL function redundantly in floral meristem identity determination but their function is divergent in relation to lateral development involving fruit development (Fernandez et al., 2000). The ful mutant generates a very small silique size due to defects in silique valve cell differentiation (Ferrandiz et al., 2000). Also, FUL interferes with SHP1 and SHP2 gene expression, but AG induces their gene expression in early stages of silique development. Overexpression of AtCSP4 resulted in slight up-regulation of AP1, CAL, AG, and FUL in floral bud and stage 1 silique tissues. SHP2 expression exhibited the greatest shift due to AtCSP4 overexpression.
As shown in Fig. 4, early embryo formation and endosperm development in 35S:NTAP:AtCSP4-3 is not retarded before the late heart embryo formation stage. However, its development ceased from the late heart stage of embryo formation (Fig. 4M). In 35S:NTAP:AtCSP4-3, transcripts of early embryo formation genes such as FUS3 and ABI3 could not be detected, while the expression of MEA and FIS2 that are involved in endosperm development was affected during early seed development stages. Collectively, the defective seed in 35S:NTAP:AtCSP4-3 may be caused by an impairment of late embryogenesis and endosperm development. MEA, FIE, and MSI1 form a huge complex (∼650 kDa) which may combine with FIS2 (Kohler et al., 2003b; Chanvivattana et al., 2004). A loss-of-function mutant of the FIS gene perturbs endosperm development in a similar manner that is consistent with overexpression of AtCSP4. Gene expression of MEA and FIS2, which are FIS class PcG proteins in Arabidopsis, is increased after fertilization in the AtCSP4 overexpression line (Fig. 5).
MEA functions in relation to DNA methylation in the chromatin complex for sustaining paternal gene imprinting. MEA silencing in vegetative tissues is mediated by H3K27 methylation (Jullien et al., 2006a, b). Expression of the MEA gene, which encodes a PcG protein, is also decreased at the floral bud stage, with a dramatic decrease at silique stage 3. Judging from the previously published phenotype of various MADS-box protein mutants, it is presumed that overexpression of AP1, AG, and SHP2 does not result in embryo lethality (Mizukami and Ma, 1992; Mandel and Yanofsky, 1995; Liljegren et al., 2000).
The direct functional link between the ectopic overexpression of AtCSP4 and MEA is not known at this time. In animals, CSPs function in diverse processes at both the transcriptional and post-transcriptional level, such as transcriptional activation/repression, alternative splicing, RNA stability, and RNA masking, among others. In plants, the most well characterized CSP is NAB1, which functions in RNA masking of LHCBM mRNA at the post-transcriptional level (Mussgnug et al., 2005). Other plant CSPs have been suggested to function as RNA-binding proteins based on the results of nucleic acid binding assays in wheat and Arabidopsis (Fusaro et al., 2007; Sasaki et al., 2007; Nakaminami et al., 2009). In the case of winter wheat WCSP1 and Arabidopsis AtCSP1 (Nakaminami et al., 2006; Kim et al., 2007), RNA chaperone activity has been proposed on the basis of in vivo and in vitro assays; however, no data have yet indicated direct functions as transcriptional activators or repressors. Subcellular localization data revealed that AtCSP4 is a nucleocytoplasmic protein (Fig. 6), providing correlative evidence which suggests that AtCSP4 does not function directly to regulate transcription of target genes but rather exerts a functional effect via post-transcriptional regulation. Taken together with morphological and semi-quantitative RT-PCR analyses, shortened silique and embryo lethality may be the result of an independent alteration of gene transcripts for MADS-box protein and endosperm development-related genes in AtCSP4 overexpression mutants.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Secondary protein structure of AtCSP4 and redundancy with AtCSP2.
Figure S2. Schematic representation for AtCSP4 gene functional analysis.
Table S1. Primer list for AtCSP4 transcript detection and amplicon length of the PCR product with the R1 primer.
Table S2. Primer list for semi-quantitative RT-PCR analysis.
Table S3. List of context sequence and amplicon lengths for Taq-Man probes.
Acknowledgments
The authors would like to thank Drs Kentaro Nakaminami and Joseph Morton (West Virginia University) for assistance with microscopy work. The authors also thank GABI-Kat for kindly supplying the T-DNA insertion mutant seedline. This project was supported by a National Science Foundation grant (IBN-0416945) to DK. West Virginia Agriculture and Forestry Experiment Station Scientific Article No. 3082.
References
- Aida M, Vernoux T, Furutani M, Traas J, Tasaka M. Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development. 2002;129:3965–3974. doi: 10.1242/dev.129.17.3965. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Liljegren SJ, Pelaz S, Gold SE, Burgeff C, Ditta GS, Vergara- Silva F, Yanofsky MF. MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. The Plant Journal. 2000;24:457–466. doi: 10.1046/j.1365-313x.2000.00891.x. [DOI] [PubMed] [Google Scholar]
- Bader AG, Vogt PK. Inhibition of protein synthesis by Y box-binding protein 1 blocks oncogenic cell transformation. Molecular and Cellular Biology. 2005;25:2095–2106. doi: 10.1128/MCB.25.6.2095-2106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroux C, Gagliardini V, Page DR, Grossniklaus U. Dynamic regulatory interactions of Polycomb group genes: MEDEA autoregulation is required for imprinted gene expression in Arabidopsis. Genes and Development. 2006;20:1081–1086. doi: 10.1101/gad.378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun RE. Temporal control of protein synthesis during spermatogenesis. International Journal of Andrology. 2000;23(Suppl 2):92–94. doi: 10.1046/j.1365-2605.2000.00027.x. [DOI] [PubMed] [Google Scholar]
- Chaikam V, Karlson D. Functional characterization of two cold shock domain proteins from Oryza sativa. Plant, Cell and Environment. 2008;31:995–1006. doi: 10.1111/j.1365-3040.2008.01811.x. [DOI] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development. 2004;131:5263–5276. doi: 10.1242/dev.01400. [DOI] [PubMed] [Google Scholar]
- Chaudhury AM, Berger F. Maternal control of seed development. Seminars in Cell and Developmental Biology. 2001;12:381–386. doi: 10.1006/scdb.2001.0267. [DOI] [PubMed] [Google Scholar]
- Chaudhury AM, Koltunow A, Payne T, Luo M, Tucker MR, Dennis ES, Peacock WJ. Control of early seed development. Annual Review of Cell and Developmental Biology. 2001;17:677–699. doi: 10.1146/annurev.cellbio.17.1.677. [DOI] [PubMed] [Google Scholar]
- Chaudhury AM, Ming L, Miller C, Craig S, Dennis ES, Peacock WJ. Fertilization-independent seed development in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 1997;94:4223–4228. doi: 10.1073/pnas.94.8.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- de Folter S, Busscher J, Colombo L, Losa A, Angenent GC. Transcript profiling of transcription factor genes during silique development in Arabidopsis. Plant Molecular Biology. 2004;56:351–366. doi: 10.1007/s11103-004-3473-z. [DOI] [PubMed] [Google Scholar]
- Evdokimova V, Ruzanov P, Anglesio MS, Sorokin AV, Ovchinnikov LP, Buckley J, Triche TJ, Sonenberg N, Sorensen PH. Akt-mediated YB-1 phosphorylation activates translation of silent mRNA species. Molecular and Cellular Biology. 2006;26:277–292. doi: 10.1128/MCB.26.1.277-292.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova V, Ruzanov P, Imataka H, Raught B, Svitkin Y, Ovchinnikov LP, Sonenberg N. The major mRNA-associated protein YB-1 is a potent 5′ cap dependent mRNA stabilizer. EMBO Journal. 2001;20:5491–5502. doi: 10.1093/emboj/20.19.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes and Development. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- Fernandez DE, Heck GR, Perry SE, Patterson SE, Bleecker AB, Fang SC. The embryo MADS domain factor AGL15 acts postembryonically. Inhibition of perianth senescence and abscission via constitutive expression. The Plant Cell. 2000;12:183–198. doi: 10.1105/tpc.12.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandiz C, Liljegren SJ, Yanofsky MF. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science. 2000;289:436–438. doi: 10.1126/science.289.5478.436. [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jurgens G. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- Fusaro AF, Bocca SN, Ramos RL, et al. AtGRP2, a cold-induced nucleo-cytoplasmic RNA-binding protein, has a role in flower and seed development. Planta. 2007;225:1339–1351. doi: 10.1007/s00425-006-0444-4. [DOI] [PubMed] [Google Scholar]
- Galweiler L, Guan C, Muller A, Wisman E, Mendgen K, Yephremov A, Palme K. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;282:2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jurgens G. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell. 2003;112:219–230. doi: 10.1016/s0092-8674(03)00003-5. [DOI] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jurgens G, Palme K. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature. 2001;413:425–428. doi: 10.1038/35096571. [DOI] [PubMed] [Google Scholar]
- Goldberg RB, de Paiva G, Yadegari R. Plant embryogenesis: zygote to seed. Science. 1994;266:605–614. doi: 10.1126/science.266.5185.605. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB. Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science. 1998;280:446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- Gu Q, Ferrandiz C, Yanofsky MF, Martienssen R. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development. 1998;125:1509–1517. doi: 10.1242/dev.125.8.1509. [DOI] [PubMed] [Google Scholar]
- Guitton AE, Page DR, Chambrier P, Lionnet C, Faure JE, Grossniklaus U, Berger F. Identification of new members of fertilisation independent seed polycomb group pathway involved in the control of seed development in Arabidopsis thaliana. Development. 2004;131:2971–2981. doi: 10.1242/dev.01168. [DOI] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development. 2004;131:657–668. doi: 10.1242/dev.00963. [DOI] [PubMed] [Google Scholar]
- Heck GR, Perry SE, Nichols KW, Fernandez DE. AGL15, a MADS domain protein expressed in developing embryos. The Plant Cell. 1995;7:1271–1282. doi: 10.1105/tpc.7.8.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien PE, Katz A, Oliva M, Ohad N, Berger F. Polycomb group complexes self-regulate imprinting of the Polycomb group gene MEDEA in Arabidopsis. Current Biology. 2006a;16:486–492. doi: 10.1016/j.cub.2006.01.020. [DOI] [PubMed] [Google Scholar]
- Jullien PE, Kinoshita T, Ohad N, Berger F. Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. The Plant Cell. 2006b;18:1360–1372. doi: 10.1105/tpc.106.041178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens G. Apical–basal pattern formation in Arabidopsis embryogenesis. EMBO Journal. 2001;20:3609–3616. doi: 10.1093/emboj/20.14.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens G. Axis formation in plant embryogenesis: cues and clues. Cell. 1995;81:467–470. doi: 10.1016/0092-8674(95)90065-9. [DOI] [PubMed] [Google Scholar]
- Karlson D, Imai R. Conservation of the cold shock domain protein family in plants. Plant Physiology. 2003;131:12–15. doi: 10.1104/pp.014472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlson D, Nakaminami K, Toyomasu T, Imai R. A cold-regulated nucleic acid binding protein of winter wheat shares a domain with bacterial cold shock proteins. Journal of Biological Chemistry. 2002;277:35248–35256. doi: 10.1074/jbc.M205774200. [DOI] [PubMed] [Google Scholar]
- Kim JS, Park SJ, Kwak KJ, Kim YO, Kim JY, Song J, Jang B, Jung CH, Kang H. Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in. Escherichia coli. Nucleic Acids Research. 2007;35:506–516. doi: 10.1093/nar/gkl1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MH, Sasaki K, Imai R. Cold shock domain protein 3 regulates freezing tolerance in Arabidopsis thaliana. Journal of Biological Chemistry. 2009;284:23454–23460. doi: 10.1074/jbc.M109.025791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T, Ohad N, Yadegari R, et al. Control of fertilization independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 1999;96:4186–4191. doi: 10.1073/pnas.96.7.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C, Makarevich G. Epigenetic mechanisms governing seed development in plants. EMBO Reports. 2006;7:1223–1227. doi: 10.1038/sj.embor.7400854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C, Hennig L, Bouveret R, Gheyselinck J, Grossniklaus U, Gruissem W. Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO Journal. 2003a;22:4804–4814. doi: 10.1093/emboj/cdg444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C, Hennig L, Spillane C, Pien S, Gruissem W, Grossniklaus U. The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes and Development. 2003b;17:1540–1553. doi: 10.1101/gad.257403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M. The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays. 2003;25:691–698. doi: 10.1002/bies.10300. [DOI] [PubMed] [Google Scholar]
- Laux T, Wurschum T, Breuninger H. Genetic regulation of embryonic pattern formation. The Plant Cell. 2004;16(Suppl):S190–S202. doi: 10.1105/tpc.016014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti-Shiu MD, Adamczyk BJ, Fernandez DE. Expression of MADS-box genes during the embryonic phase in Arabidopsis. Plant Molecular Biology. 2005;58:89–107. doi: 10.1007/s11103-005-4546-3. [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature. 2000;404:766–770. doi: 10.1038/35008089. [DOI] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;93:1195–1205. doi: 10.1016/s0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- Lu ZH, Books JT, Ley TJ. Cold shock domain family members YB-1 and MSY4 share essential functions during murine embryogenesis. Molecular and Cellular Biology. 2006;26:8410–8417. doi: 10.1128/MCB.01196-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZH, Books JT, Ley TJ. YB-1 is important for late-stage embryonic development, optimal cellular stress responses, and the prevention of premature senescence. Molecular and Cellular Biology. 2005;25:4625–4637. doi: 10.1128/MCB.25.11.4625-4637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luerssen H, Kirik V, Herrmann P, Misera S. FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. The Plant Journal. 1998;15:755–764. doi: 10.1046/j.1365-313x.1998.00259.x. [DOI] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A. Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proceedings of the National Academy of Sciences, USA. 2000;97:10637–10642. doi: 10.1073/pnas.170292997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Koltunow A, Dennis ES, Peacock WJ, Chaudhury AM. Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 1999;96:296–301. doi: 10.1073/pnas.96.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MA, Yanofsky MF. A gene triggering flower formation in Arabidopsis. Nature. 1995;377:522–524. doi: 10.1038/377522a0. [DOI] [PubMed] [Google Scholar]
- Mayer U, Jurgens G. Pattern formation in plant embryogenesis: a reassessment. Seminars in Cell and Develomental Biology. 1998;9:187–193. doi: 10.1006/scdb.1997.0210. [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Ma H. Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell. 1992;71:119–131. doi: 10.1016/0092-8674(92)90271-d. [DOI] [PubMed] [Google Scholar]
- Mussgnug JH, Wobbe L, Elles I, et al. NAB1 is an RNA binding protein involved in the light-regulated differential expression of the light-harvesting antenna of Chlamydomonas reinhardtii. The Plant Cell. 2005;17:3409–3421. doi: 10.1105/tpc.105.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaminami K, Hill K, Perry SE, Sentoku N, Long JA, Karlson DT. Arabidopsis cold shock domain proteins: relationships to floral and silique development. Journal of Experimental Botany. 2009;60:1047–1062. doi: 10.1093/jxb/ern351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaminami K, Karlson DT, Imai R. Functional conservation of cold shock domains in bacteria and higher plants. Proceedings of the National Academy of Sciences, USA. 2006;103:10122–10127. doi: 10.1073/pnas.0603168103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad N, Yadegari R, Margossian L, Hannon M, Michaeli D, Harada JJ, Goldberg RB, Fischer RL. Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. The Plant Cell. 1999;11:407–416. doi: 10.1105/tpc.11.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad N, Margossian L, Hsu YC, Williams C, Repetti P, Fischer RL. A mutation that allows endosperm development without fertilization. Proceedings of the National Academy of Sciences, USA. 1996;93:5319–5324. doi: 10.1073/pnas.93.11.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Valon C, Kohara A, Misera S, Giraudat J. The ABSCISIC ACIDINSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. The Plant Cell. 1997;9:1265–1277. doi: 10.1105/tpc.9.8.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenicova L, de Folter S, Kieffer M, et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. The Plant Cell. 2003;15:1538–1551. doi: 10.1105/tpc.011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Harada JJ. Arabidopsis embryogenesis. Methods in Molecular Biology. 2008;427:3–16. doi: 10.1007/978-1-59745-273-1_1. [DOI] [PubMed] [Google Scholar]
- Park SJ, Kwak KJ, Oh TR, Kim YO, Kang H. Cold shock domain proteins affect seed germination and growth of Arabidopsis thaliana under abiotic stress conditions. Plant and Cell Physiology. 2009;50:869–878. doi: 10.1093/pcp/pcp037. [DOI] [PubMed] [Google Scholar]
- Raffetseder U, Frye B, Rauen T, Jurchott K, Royer HD, Jansen PL, Mertens PR. Splicing factor SRp30c interaction with Y-box protein-1 confers nuclear YB-1 shuttling and alternative splice site selection. Journal of Biological Chemistry. 2003;278:18241–18248. doi: 10.1074/jbc.M212518200. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Kim MH, Imai R. Arabidopsis COLD SHOCK DOMAIN PROTEIN2 is a RNA chaperone that is regulated by cold and developmental signals. Biochemical and Biophysical Research Communications. 2007;364:633–638. doi: 10.1016/j.bbrc.2007.10.059. [DOI] [PubMed] [Google Scholar]
- Schrick K, Mayer U, Horrichs A, Kuhnt C, Bellini C, Dangl J, Schmidt J, Jurgens G. FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes and Development. 2000;14:1471–1484. [PMC free article] [PubMed] [Google Scholar]
- Seymour G, Poole M, Manning K, King GJ. Genetics and epigenetics of fruit development and ripening. Current Opinion in Plant Biology. 2008;11:58–63. doi: 10.1016/j.pbi.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proceedings of the National Academy of Sciences, USA. 2001;98:11806–11811. doi: 10.1073/pnas.201413498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Ruiz RA, Lohner A, Jurgens G. The GURKE gene is required for normal organization of the apical region in the Arabidopsis embryo. The Plant Journal. 1996;10:1005–1016. doi: 10.1046/j.1365-313x.1996.10061005.x. [DOI] [PubMed] [Google Scholar]
- Wilkinson MF, Shyu AB. Multifunctional regulatory proteins that control gene expression in both the nucleus and the cytoplasm. Bioessays. 2001;23:775–787. doi: 10.1002/bies.1113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.