Abstract
Dnmt1 and Dnmt3a, two major DNA methyltransferases, are expressed in postmitotic neurons, but their function in the central nervous system (CNS) is unclear. We generated conditional mutant mice that lack either Dnmt1, or Dnmt3a, or both exclusively in forebrain excitatory neurons and found only double knockout (DKO) mice exhibited abnormal hippocampal CA1 long-term plasticity and deficits of learning and memory. While no neuronal loss was found, the size of hippocampal neurons in DKO was smaller; furthermore, DKO neurons showed a deregulation of gene expression including class I MHC and Stat1 that are known to play a role in synaptic plasticity. In addition, we observed a significant decrease in DNA methylation in DKO neurons. We conclude that Dnmt1 and Dnmt3a are required for synaptic plasticity, learning and memory through their overlapping roles in maintaining DNA methylation and modulating neuronal gene expression in adult CNS neurons.
The long-lasting changes in synaptic plasticity underlying learning and memory require changes in neuronal gene expression1. Epigenetic mechanisms such as histone modification2 and DNA methylation are thought to contribute to this adaptive neuronal gene expression3. It has been shown that different histone modifications are associated with various neuronal gene expression states2, 4; moreover, increasing histone acetylation by histone deacetylase inhibitors promotes recovery of learning and memory in a mouse model of neurodegeneration5, 6. However, the function of DNA methylation in the nervous system is still unclear. The substantial expression of two DNA methyltransferases, maintenance enzyme Dnmt1 and de novo DNA methyltransferase Dnmt3a, in non-dividing adult neurons7-9 is intriguing because they are normally functioning in fast dividing cells to establish and/or maintain DNA methylation patterns. For instance, during cell proliferation, Dnmt1 localizes to the replication foci and primarily methylates the unmethylated daughter strand DNA after DNA synthesis so that the parental methylation pattern would be maintained in daughter cells. In contrast, Dnmt3a is more actively involved in methylation unmethylated DNA to establish new methylation patterns as seen in early mouse embryogenesis10.
Since mice with the mutation of any of the three functional Dnmts (Dnmt1, Dnmt3a, or Dnmt3b) are not viable10, 11, conditional knockout mice were generated to study the function of DNA methylation in the central nervous system (CNS)12-15. Deletion of Dnmt1 in neural progenitor cells causes hypomethylation in postmitotic neurons in the CNS (estimated between 30-50% by HPLC analysis, our unpublished data). These mutant neural progenitor cells undergo precocious astroglial differentiation16, cell death, and have defects in neuronal maturation and synaptic transmission. While these studies provide ample evidence that NA methylation is essential in neuronal development and function, they did not reveal the role of Dnmt expression in postmitotic neurons. Our earlier study of Dnmt1 gene deletion in postmitotic CNS neurons failed to reveal any obvious neuronal phenotypes, such as a loss of DNA methylation in cultured cerebellar neurons or cortical and hippocampal neurons in vivo. Nevertheless, we did observe a moderate effect on cortical neuronal survival in a rodent stroke model when Dnmt1 was reduced by 50% in postmitotic cortical and hippocampal neurons17, 18. Recently, studies with Dnmt inhibitors in either in vitro brain slices or hippocampus in vivo 19-21 suggest that DNA methylation may target specific genes involved in synaptic plasticity and learning and memory. However, the potential toxic effects of Dnmt inhibitors on protein translation in non-dividing neurons22 could complicate the interpretation of those results. The lack of selectivity of Dnmt inhibitors also makes it difficult to correlate specific neural effects with individual Dnmts. Unlike histone modifications, DNA methylation is presumably a more static epigenetic mark and whether DNA methylation undergoes turnover in postmitotic neurons is still under debate23. However, evidence of selective DNA demethylation in selective neuronal gene promoters3, 24-26 in the CNS argues for the presence of demethylation activity in neurons.
Circumstantial evidences suggest that de novo methyltransferases Dnmt3a and Dnmt3b are important for neural development and function. ICF (or Immunodeficiency, Centromere instability and Facial anomalies) syndrome, a rare human genetic disorder, is caused by a recessive mutation of the DNMT3B gene10, 27. A significant portion of ICF patients suffer mental retardation28. Gene expression studies showed that Dnmt3b is highly expressed in the murine neural tube between E7.5-9.510, suggesting a potential role for Dnmt3b in early stages of neurogenesis. We have shown that Dnmt3a is present in both developing and mature CNS8, suggesting a potential involvement of Dnmt3a in methylation regulation in both embryonic and adult CNS neurons. More recently, Nguyen et al. (2007) showed that conditional mutant mice (Nes-Cre1; Dnmt3a) lacking Dnmt3a in the entire CNS were born normal, but die in young adulthood15. These Nes-Cre1; Dnmt3a mutant mice exhibited a loss of motor neurons in the hypoglossal nucleus and morphological defects in neuromuscular junctions of the diaphragm muscle, suggesting a role for Dnmt3a in the maintenance of motor neuron survival and neuromuscular endplate structure15.
In this study, we compared the phenotypes of conditional mutant mice deficient for either Dnmt1 or Dnmt3a (or both) exclusively in the forebrain postnatal postmitotic neurons. We aim to address whether Dnmt1 and Dnmt3a play a redundant role in the maintenance of DNA methylation in postmitotic neurons. Furthermore, we determine the effects of Dnmt deficiencies on neuronal gene expression as well as synaptic function and learning and memory behaviors.
RESULTS
Smaller cell size and deficits in neural plasticity in the hippocampus of DKO mice
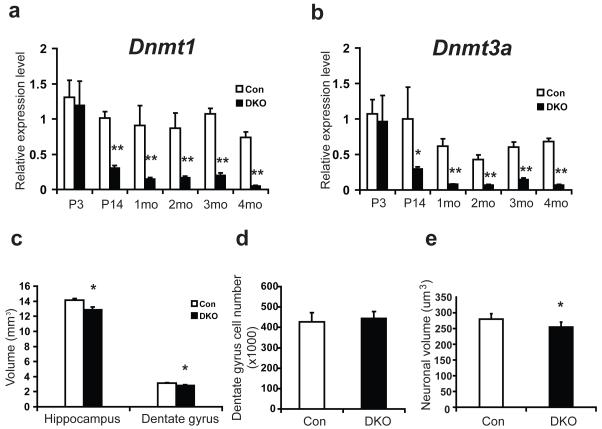
Calcium/calmodulin-dependent protein kinase IIα (CamKIIα) is expressed in postmitotic neurons postnatally29. We therefore used the CamKIIα promoter-driven Cre recombinase (CamK-Cre93) to induce Dnmt gene deletion in postmitotic neurons in the CNS29, 30. This CamK-Cre93 transgene can induce specific gene deletion in excitatory neurons in the mouse forebrain shortly after birth since its expression peaks within 2-3 weeks postnatally12, 29. Our previous study with a conditional deletion of Dnmt1 in postmitotic neurons did not show any significant alteration in DNA methylation in repetitive elements12. To determine if Dnmt1 and Dnmt3a play redundant roles in the CNS, we made Dnmt1 and Dnmt3a double conditional knockouts (DKO) in addition to single Dnmt1 or Dnmt3a conditional knockouts (SKO). Consistent with the CamKIIα expression pattern, Dnmt1 and Dnmt3a gene deletion was not detected at postnatal day 3 (Fig. 1a, b). However, the levels of Dnmt transcripts were significantly decreased at postnatal day 14 and remained low beyond P30 (Fig. 1a, b). Immunostaining of Cre recombinase in DKO confirmed that Cre expression is restricted to mature neurons, which is consistent with previous findings showing neuron-specific conditional gene deletion by the CamK-Cre93 transgene12(Supplementary Fig. 1a). Southern blot analysis also confirmed that the efficiency of the Dnmt gene deletion (Supplementary Fig. 1b) is at a constant level (approximately 50% from 1-4 months old as reported)12.
Figure 1.
Mice with conditional deletion of Dnmt1 and Dnmt3a show small hippocampus without cell loss. a, b, Real time PCR analysis showed decreased expression of Dnmt1(a) and Dnmt3a(b) in DKO forebrain. Relative expression levels of Dnmt1(a) and Dnmt3a(b) in DKO and wildtype littermate control (Con) were compared at postnatal day 3 (P3), P14, 1 month (mo), 2 mo, 3 mo, and 4 mo. Three pairs of samples were used for each time point. c, Stereology analysis identified reduced volume of hippocampus as well as dentate gyrus in DKO as compared with control mice. d, DKO had similar number of dentate gyrus cells as control mice. e, DKO dentate gyrus neuronal cell body is significantly smaller as compared with control mice. 5 control and 6 DKO male mice at 3 mo were used in c, d and e. Data are presented as means±s.e.m. *indicates P<0.05, **indicates P<0.01.
DKO mice have normal lifespans and do not exhibit obvious behavioral defects in their home cages. Gross histological examination suggested that all brain structures are intact (Supplementary Fig. 2a). Using stereological protocols, we measured hippocampal volumes in control and mutant mice. Though we did not see any abnormalities in the brains of SKO mice (data not shown), we found that DKO mice have significantly smaller hippocampi (9.31% average volume reduction, P<0.05) when compared with control wildtype (Fig. 1c, control: 14.16±0.19 mm3 and DKO: 12.84±0.41 mm3 in mean±s.e.m. P<0.05). The dentate gyrus in DKO showed a similar volume reduction (9.77% on average, Fig. 1c, control: 3.12±0.07 mm3 and DKO: 2.82±0.11 mm3 in mean±s.e.m. P<0.05). These observations suggest that the volume reduction of the DKO dentate gyrus is directly proportional to the volume reduction of the DKO hippocampus. We used an unbiased optical fractionator and found that the total number of dentate gyrus granule cells is similar between DKO and control mice (Fig. 1d, control: 427.87±44.10 and DKO: 443.72±34.40 in mean±s.e.m, P>0.05). However, quantitative measurement of individual neuronal volume in dentate gyrus indicated that DKO neurons exhibited a statistically significant decrease in neuronal size when compared to control mice (Fig. 1e, control: 280.08±6.98 μm3 and DKO: 254.95±7.17 μm3 in mean±s.e.m, P<0.05). Dentate gyrus is one of the two CNS regions exhibiting adult neurogenesis. We performed an adult neurogenesis assay by BrdU labeling of adult neural stem cells and newly born neurons in dentate gyrus and found no difference between DKO and control mice (suppl. Fig 2b). Taken together, our results suggest that the hippocampal volume difference between control and DKO is most likely due to smaller neuronal size in DKO.
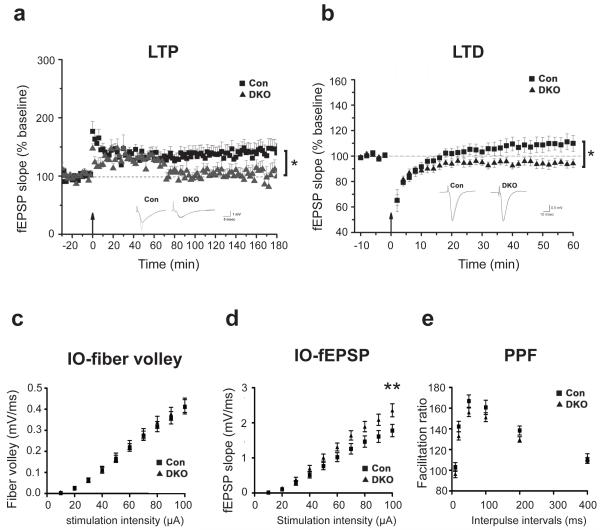
Next, we examined the impact of Dnmt1 and Dnmt3a deficiency within forebrain excitatory neurons on synaptic function. We first examined hippocampal plasticity in Schaffer collateral-CA1 synapses. CA1 long-term potentiation (LTP) and long-term depression (LTD) were studied in littermate controls, DKO as well as SKO animals. We found that Schaffer Collateral LTP induced by a 100 Hz tetanus was significantly attenuated (P<0.05) in DKO (Fig. 2a). In addition, DKO showed enhanced induction of LTD using a stimulation protocol (1 Hz for 15 minutes) that normally fails to induce LTD in adult wildtype mice (P<0.05, Fig. 2b). Instead this protocol induced a small potentiation in controls (P<0.05, Fig. 2b). Intriguingly, SKO mice showed neither LTP nor LTD abnormalities (Supplementary Fig. 3a-c for Dnmt1 SKO and Supplementary Fig. 3a and d for Dnmt3a SKO). Furthermore, the basal synaptic transmission from the DKO mice and controls (Con) are virtually identical as shown by plotting varying stimulus intensity (10–100μA) against the presynaptic fiber volley amplitudes and postsynaptic fEPSP slopes (Figure 2c-d). Paired-pulse facilitation (PPF) studies across different inter-stimulus intervals (ISIs) revealed no differences between DKO and Con group (Figure 2e). Although it is still unclear whether the abnormalities in synaptic plasticity are mediated by pre- or post-synaptic mechanisms, our data indicate that Dnmt1 and Dnmt3a are required for normal synaptic plasticity.
Figure 2.
Impaired neural plasticity in DKO mice. a, LTP was reduced in adult DKO mice (* P<0.05). Field EPSP (fEPSP) slopes in control (square symbols) versus DKO mice (triangle symbols) were recorded before and after tetanic stimulation (100Hz, 1sec). Scale bar, 5 msec/1mV. b, LTD was enhanced in adult DKO mice (* P<0.05). fEPSP slopes were recorded before and after stimulation (1Hz, 15min). Scale bar, 10 msec/0.5mV. Representative recordings are shown in insets (a, b). 28 slices from 9 Con and 13 slices from 7 DKO were used in a. 10 slices from 5 Con and 8 slices from 4 DKO were recorded in b. The basal synaptic transmission from the DKO mice and Con are identical as shown by plotting varying stimulus intensity (10–100μA) against the presynaptic fiber volley amplitudes (c) and postsynaptic fEPSP slope (d). n =33 slice from 8 mice for DKO group and n =28 slice from 8 mice for Con group. **indicates P<0.01. e, Paired-pulse facilitation (PPF) studies across different inter-stimulus intervals (ISIs) revealed no difference between DKO and Con group. n =16 slice from 4 mice for DKO group and n =14 slice from 4 mice for Con group. Slice numbers were used for statistical analysis.
Deficits in hippocampal learning and memory in DKO mice
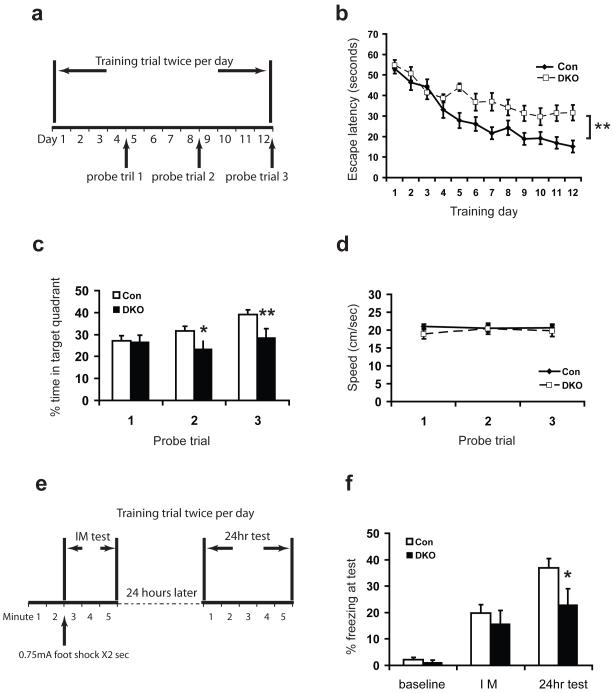
Since deficits in CA1 LTP are known to disrupt spatial learning, we next tested the mice in the hidden-platform version of the Morris water maze, a hippocampus-dependent learning and memory task. In this task, animals were trained to search for a submerged platform in a pool of water. After 4, 8 and 12 days of training (2 trials/day), spatial learning was assessed with probe trials in which the platform was removed from the pool, and the mice were allowed to search for 60 s (Fig. 3a). While Dnmt1 or Dnmt3a SKO mice performed normally in this spatial learning task (Supplementary Fig. 4), DKO mice took longer to find the platform during training (genotype F(1, 308)=6.25, P=0.019; day F(11, 308)=18.26, P<0.0001; genotype X day F(11, 308)=1.37, P=0.185. Fig. 3b). When the platform was removed during the post-training probe trials, DKO mice spent less time in the target quadrant compared to littermate control (P<0.05, Fig. 3c), indicating impaired spatial learning and memory ability. However, swimming speed was normal in DKO mice compare to wildtype (Fig. 3d).
Figure 3.
Impaired learning and memory in DKO mice. a-d, Morris water maze test. a, Schematic drawing of the Morris water maze test design. b, Escape latency time to find the hidden platform plotted versus training day. Control mice improved significantly faster than DKO mice. c, Percentage time spent in target quadrant during three probe trials demonstrated that the DKO spent less time in the target quadrant. n = 13 mice for DKO group and n =17 mice for Con group. d, The swimming speeds of the DKO were indistinguishable from controls. 17 control and 13 DKO mice at 3 mo were used in b-d. e-f, Contextual fear conditioning test. e, Schematic drawing of the contextual fear conditioning test design. Mice were trained and tested immdediately (IM/average for 3 min) and 24h later in a conditioning chamber. f, Contexual memory consolidation was impaired in DKO mice as demonstrated by decreased freezing frequency at 24 h. The acquisition was normal in DKO immediately after shock. Baseline, freezing before shock presentation; IM, freezing immediately after shock presentation. n = 21 mice for DKO group and n =13 mice for Con group in figure f. Data are presented as means±s.e.m. * indicates P<0.05. **indicates P<0.01.
Having determined that DKO exhibit deficits of hippocampus-dependent memory, we further performed contextual fear-conditioning tests to examine whether DKO exhibit defects in acquisition or consolidation of memory. We found that control and DKO behaved similarly in terms of memory acquisition (Fig. 3e and 3f, IM test), but DKO showed impairment in memory consolidation (Fig. 3f, 24hr test). Sigificantly, we did not observe any behavioral defects in Dnmt1 or Dnmt3a SKO in the fear-conditioning test (Supplementary Fig. 5), consistent with the idea that Dnmt1 and Dnmt3a have redundant roles in the regulation of learning and memory in adult animals.
Up-regulation of class I Major Histocompatibility Complex (MHC I), Stat1, and other genes in DKO forebrain
The late stages of long term plasticity as well as learning and memory consolidation require changes30 in gene expression. Previous studies suggested that Reelin and protein phosphatase 1(PP1) are modulated by DNA methylation; furthermore, infusion of Dnmt inhibitors 5′azadeoxylcytidine and zebularine paired with behavioral memory training trigger the up-regulation of these two learning and memory-associated genes19-21. We therefore performed qRT-PCR analysis to determine whether expression of Reelin and PP1 are altered in the absence of both Dnmt1 and Dnmt3a. However, we found that the expression levels of these genes is normal in DKO hippocampi, suggesting that alterations in the expression of these two genes are not the basis for the abnormalities in synaptic plasticity and learning and memory in DKO animals (Supplementary Fig. 6). To directly identify novel target genes that are deregulated in DKO forebrain, we performed mouse genome-wide gene expression microarray analysis. Bioinformatics analysis showed that 84 (0.26%) genes were upregulated more than 1.5 fold, whereas 7 genes (0.02%) showed higher than 1.5 fold down regulation.
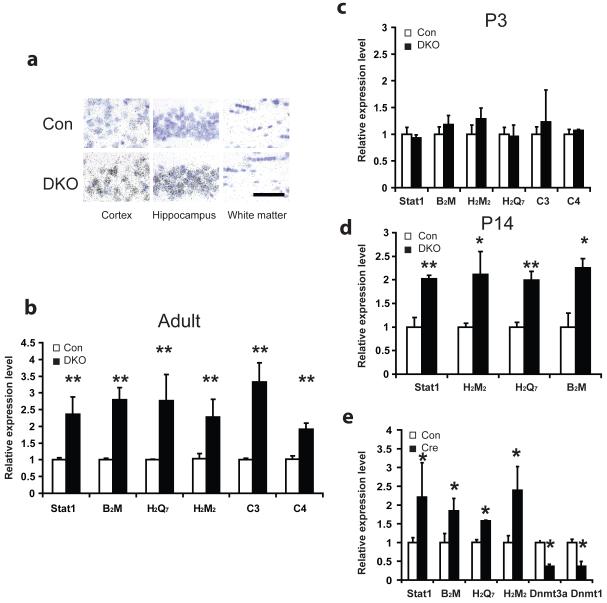
Interestingly, the DKO gene-expression analyses demonstrated that the highly up-regulated genes are immune genes including MHC I and complement system, as well as Stat1 that is activated in interferon-gamma induced immune response (Table 1). These immune genes are known to be important for synaptic function and learning and memory in the CNS31-34. The up-regulation of MHC I and other immune genes’ expression was confirmed by qRT-PCR in adult hippocampi (Fig. 4b) as well as cortical tissues (data not shown). In situ hybridization experiments also showed that enhanced MHC I transcripts are within neuronal cells (Fig. 4a). Similarly, by using phosphor-Stat1 immunohistochemistry34, we found that the active form of Stat1 was also concentrated in neuronal cell types within DKO mouse brain (Fig. 5a). The induction of these immune genes appears to be a direct consequence of Dnmt1 and Dnmt3a deficiency in neuronal cells because their expression change was noticed soon after CamKIIα cre-activated deletion of Dnmt1 and Dnmt3a two weeks after birth (Fig. 4d), but not at P3 when Dnmt deletion has not occurred yet (Figs 1a, b, 4c). Single Dnmt1 or Dnmt3a conditional mutant mice did not show altered gene expression compared to wildtype (Supplementary Fig. 7a.b), which supports the notion that Dnmt1 and Dnmt3a play redundant roles in repressing these genes in neuronal cells.
Table 1.
List of upregulated genes within DKO mouse brain
| Gene Symbol | ID | Gene Name | Fold Change |
|---|---|---|---|
| MHCI-mediated immunity | |||
| B2m | NM_009735 | beta-2 microglobulin | 3 |
| H2-D | AK083376 | clone:C920025E04 product:weakly similar to H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-37 ALPHA CHAIN PRECURSOR | 2.32 |
| H2-D1 | NM_010380 | histocompatibility 2, D region locus 1 | 3.14 |
| H-2K | NM_019909 | MHC (A.CA/J(H-2K-f) class I antigen (LOC56628) | 3.34 |
| H2-M2 | AY302216 | strain P/J MHC class Ib antigen | 1.94 |
| H2-M3 | NM_013819 | histocompatibility 2, M region locus 3 | 1.93 |
| H2-Q1 | NM_010390 | histocompatibility 2, Q region locus 1 | 2.54 |
| H2-Q5 | NM_010393 | histocompatibility 2, Q region locus 5 | 2.4 |
| H2-Q7 | NM_010394 | histocompatibility 2, Q region locus 7 | 2.85 |
| H2-Q8 | NM_023124 | histocompatibility 2, Q region locus 8 | 3.9 |
| H2-DMb1 | NM_010387 | histocompatibility 2, class II, locus Mb1 | 2.79 |
| H2-Q5 | TC1514992 | clone:A130027C03 product:histocompatibility 2, Q region locus 4, full insert sequence, partial (79%) | 3.27 |
| H2-T23 | NM_010398 | histocompatibility 2, T region locus 23 | 2.4 |
|
| |||
| mRNA transcription regulation | |||
| Sox21 | NM_145464 | SRY-box containing gene 21 | 2.79 |
| Stat1 | NM_009283 | signal transducer and activator of transcription 1 | 1.91 |
|
| |||
| Interferon-related immunity | |||
| Cxcl10 | NM_021274 | chemokine (C-X-C motif) ligand 10 | 9.71 |
| Gbp2 | NM_010260 | guanylate nucleotide binding protein 2 | 3.44 |
| Gbp4 | NM_018734 | guanylate nucleotide binding protein 4 | 3.81 |
| Ifi204 | NM_008329 | interferon activated gene 204 | 3.04 |
| Ifi27 | NM_029803 | interferon, alpha-inducible protein 27 | 3.66 |
| Ifi44 | NM_133871 | interferon-induced protein 44 | 3.23 |
| Ifi47 | NM_008330 | interferon gamma inducible protein 47 | 3.15 |
| Ifitm7 | NM_028968 | interferon induced transmembrane protein 7 | 2.04 |
| Igtp | NM_018738 | interferon gamma induced GTPase | 5.55 |
| Iigp1 | NM_021792 | interferon inducible GTPase 1 | 7.23 |
| Iigp2 | NM_019440 | interferon inducible GTPase 2 | 5 |
| Oas1f | NM_145153 | 2′-5′ oligoadenylate synthetase 1F | 2.56 |
| Oasl2 | NM_011854 | 2′-5′ oligoadenylate synthetase-like 2 | 5.55 |
|
| |||
| Complement-mediated immunity | |||
| C3 | NM_009778 | complement component 3 | 3.85 |
| C4 | NM_009780 | complement component 4 (within H-2S) | 3.64 |
|
| |||
| Cell communication | |||
| Ccl12 | NM_011331 | chemokine (C-C motif) ligand 12 | 2.51 |
| Ccl5 | NM_013653 | chemokine (C-C motif) ligand 5 | 4.05 |
| Cxcl5 | BC033508 | chemokine (C-C motif) ligand 5, mRNA (cDNA clone MGC:35989 IMAGE:4925413) | 3.35 |
| Lgals3bp | NM_011150 | lectin, galactoside-binding, soluble, 3 binding protein | 2.71 |
| Mell1 | NM_013783 | mel transforming oncogene-like 1 | 1.73 |
| Mgl1 | NM_010796 | macrophage galactose N-acetyl-galactosamine specific lectin 1 | 2.03 |
Figure 4.
Induction of immune genes in DKO mouse brain. a, Neuronal induction of MHC I gene expression in DKO. In situ hybridization of H2D was post-stained with cresyl violet to reveal the nuclei which appeared purple. Silver grains appeared black. In the cortex and the hippocampus of DKO, MHC I signal is highly concentrated in the neurons whose nuclei are large and lightly stained in cresyl violet. Whereas in the white matter, where most cells are glia and whose nuclei are small and dark stained, there were no significant difference compare to wildtype. Scale bar at bottom right, 100um, applied to all panels. b, Real time PCR analysis confirmed upregulation of immune genes Stat1, B2M, H2M2, H2Q7, C3 and C4 within DKO hippocampi of 2-3 months adults. The induction was also found as early as postnatal day 14 (P14) (d) but no significant difference in expression was noticed at P3 hippocampi (c). 3-4 pairs of samples were used for each experiment. e, Real time PCR analysis showed significant induction of Stat1, B2M, H2M2, H2Q7 as well as decreased expression of Dnmt3a and Dnmt1 in Dnmt12lox/2lox Dnmt3a2lox/2lox mouse hippocampal neuronal cultures 5 days after adeno-cre virus infection as compared with sham control cultures with adeno-GFP viruses infection. Data are presented as means±s.e.m. *indicates P<0.05. **indicates P<0.01.
Figure 5.
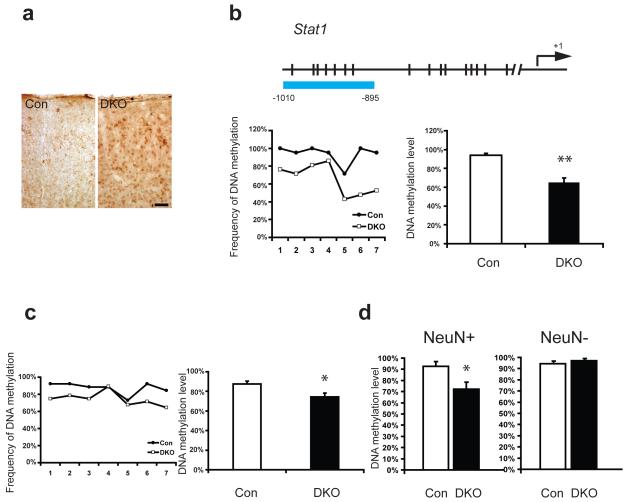
Protein increase of Stat1 within DKO mouse brain is associated with promoter demethylation in neuronal cells. a, Immunohistochemistry of phosphorylated Stat1 demonstrated its increased expression within DKO cortex, mainly in cells of neuronal shape. Scale bar at bottom right, 50um, applied to both panels. b, Bisulfite sequencing of Stat1 promoter showed demethylation within DKO as compared with control samples at 3month old. Data as shown were from genomic DNA of wildtype and DKO mouse forebrains. Schematic gene promoter structure is shown on top with arrow pointing out transcription starting site (+1). Each CpG site is marked with a vertical slash. The blue bar highlights the promoter region that underwent methylation analysis with the relative location from the transcription starting site. A summary of methylation frequency at individual CpG sites was shown in a line chart and a summary of mean DNA methylation levels of individual alleles was shown in column chart26. c, Bisulfite sequencing of Stat1 promoter showed enhanced DNA demethylation within DKO at P14. d, Stat1 promoter demethylation within DKO mouse brain existed only in neurons. Bisulfite sequencing was performed on DNA samples from FACS sorted NeuN positive and negative nucleus sub-populations of both DKO and control mouse forebrains. Total 30 clones from 3 samples were analyzed. Data are presented as means±s.e.m. Unpaired student t-test. *indicates P<0.05, **indicates P<0.01.
To further determine whether conditional deletion of Dnmt1 and Dnmt3a leads to cell-autonomous changes in the up-regulation of MHC I and other immune molecules in the CNS, we examined gene expression in pureDnmt12lox/2lox; Dnmt3a2lox/2lox hippocampal neuronal cultures by conditionally deleting both Dnmt1 and Dnmt3a with infection of adeno-cre viruses (Fig. 4e). We found that deficiency of both Dnmt1 and Dnmt3a in pure hippocampal neuron cultures also induced MHC I and other immune genes. Thus, our data suggest that up-regulation of MHC I molecules in DKO forebrain neurons is likely a cell intrinsic event.
Significant demethylation of neuronal genes in DKO forebrain neurons
The neuronal up-regulation of Stat1 and MHC I-related immune genes in DKO mouse forebrain could be due to direct effect of promoter demethylation as a consequence of Dnmt1 and Dnmt3a deficiency in neurons or due to an indirect route through the up-regulation of other positive transcription activator(s) or down-regulation of transcription repressor(s) for immune genes. We therefore examined whether DNA demethylation occurs in postmitotic neurons in the DKO forebrain tissues with only cortex and hippocampus. We first examined the candidate gene Stat1, a key factor in neural plasticity34, as well as in the interferon (IFN) signaling pathway that can induce MHC I gene expression in neurons35, 36. Our previous study also indicated that Stat1 expression is repressed by DNA methylation8. We performed bisulfite sequencing analysis of the Stat1 gene promoter. While the methylation level at −400bp to −750bp of the Stat1 promoter was similar between DKO and control mice (Supplementary Fig. 8b), we found a demethylation domain at −895 to −1010 bp of the Stat1 promoter in DKO (Fig. 5b). However, Dnmt SKO did not show any decrease in methylation in the Stat1 promoter (Supplementary Fig. 8a), suggesting Dnmt1 and Dnmt3a can compensate for each other in maintaining DNA methylation in SKO. We further found that DNA demethylation in this Stat1 promoter domain occurs as early as at P14 when deletion of both Dnmt1 and Dnmt3a was just completed (Fig 5c). This result indicates that Dnmt1 and Dnmt3a play a role in maintaining DNA methylation in CNS neurons from an early postnatal stage.
To determine whether demethylation in the Stat1 gene promoter is exclusive to neuronal populations, we further isolated NeuN+ nuclei through NeuN live staining and FACS-sorting for methylation analysis (Supplementary Fig. 9). Consistent with the neuronal specific deletion of the Dnmt genes, our data showed that significant demethylation only appeared in neuronal populations with the NeuN marker, but not in NeuN negative controls (Fig. 5d).
Having seen selective demethylation in the Stat1 gene promoter in DKO but not SKOs, we decided to quantify potential reductions of total DNA methylation in DKO cortex and hippocampus through mass spectrometer analysis. We found that DKO cortex and hippocampus exhibited approximately 20% reduction of total methylcytosine levels (n=4 P<0.01) (Fig. 6a), suggesting that demethylation in DKO neurons could be widespread. Additionally, this difference was not found in any Dnmt SKO brain (Supplmentary Fig. 10), which again supports the notion about the complementary roles of Dnmt1 and Dnmt3a on DNA methylation pattern maintenance in the CNS. To further systematically identify what kind of neuronal genes are significantly demethylated in DKO forebrain neurons, we next carried out a DNA methylation profiling experiment using Methylated DNA Immunoprecipitation (MeDIP). Using 5-methylcytosine antibody for DNA immunoprecipitation and cross-hybridization to the Agilent promoter arrays (covering 15,561 proximal gene regions mainly at −800bp to +200bp37), we identified 161 candidate loci that would exhibit demethylation (based on established criteria37: Log2 >0.2 and t-test P <0.01of hybridization signals of control and DKO. Supplementary Table 1). Using PANTHER classification categories (http://www.pantherdb.org), gene ontology analysis indicated that these 161 loci were enriched within promoters of genes of cytokine signaling, signal transduction or oxidoreductase gene categories (Fig. 6b). It is worth noting that the 161 potentially demethylated promoters did not show up in the list of 84 up-regulated genes of DKO mice. We suspect that the minimal overlap could be due to the technical limitation of the current MeDIP microarray that mainly focused on the −800bp to +200bp proximal gene promoters, potentially missing out many more demethylated regions outside of this 1 kb region. A perfect case is the absence of the corresponding array probe for the −895 to −1010bp domain on the Stat1 promoter, whose demethylation is correlated with an increase in Stat1 mRNA. Alternatively, it is also possible that moderate demethylation may not be sufficient to trigger gene up-regulation as we previously discovered with Dnmt1−/− embryonic stem cells37. Nevertheless, the identification of a list of potentially demethylated gene promoters allowed us to validate whether the selective demethylation takes place in a subset of neuronal gene promoters. We therefore selected eight candidate genes for the confirmation of methylation changes through sodium bisulfite methylation sequencing analysis. The choice of these eight genes is based on the potential roles for these genes in neuronal signaling or ion-channel functions. We found that six gene promoters (75% of accuracy) indeed underwent demethylation changes in DKO mouse brain (Fig. 6c-f, and unpublished data). Specifically, Kcne1, Dhh, Pten (analyses in two regions) genes each showed 10%-30% less methylation from three pairs of mutant DKO and littermate control mice. Together with mass spectrometry data, our MeDIP and bisulfite sequencing analysis supports the notion that Dnmt1 and Dnmt3a are essential for keeping proper methylation patterns at certain genomic loci in postmitotic CNS neurons.
Figure 6.
Gene ontology and bisulfite sequencing verification of MeDIP-chip analysis. a, Quantitative analysis of 5-methylcytosine using LC-ESI-MS MS. A representative chromatogram following injection of DNA hydrolysis of DKO and WT forebrain samples showing the MRM response for 5mdC (m/z 242.1-126.0) is shown. 5-Methylcytosine content is expressed as the percentage of 5-methylcytosine in the total cytosine pool. Data are the mean ± s.e.m. from replicates of 4 separate experiments (n = 16). b, Gene ontology classifications of MeDIP-chip result based on categories of biological process and molecular function. The ontology term is on the y-axis and the P value of enrichment significance is on the x-axis. c-f, Four gene loci Dhh (c), Kcne1 (d), Pten region1(e) and Pten region2(f)) were verified for DNA demethylation change. The results are displayed in the same manner as in figure 5. Data as shown were from 2-3 month old wildtype control and DKO mouse forebrains. Data are presented as means±s.e.m. Unpaired student t-test. *indicates P<0.05, **indicates P<0.01.
DISCUSSION
By conditionally deleting Dnmt1 and Dnmt3a exclusively in postnatal postmitotic neurons, our experiments provided the first direct evidence that DNA methyltransferases are required for the maintenance of neuronal DNA methylation, for proper control of gene expression, and consequently for synaptic plasticity, as well as learning and memory. Because Dnmt1 or Dnmt3a SKOs do not show the defects in DKOs, our current study demonstrated that Dnmt1 and Dnmt3a have redundant roles in postmitotic neurons. Previously, the overlapping functions of different members of Dnmts were only observed in fast dividing cancer or embryonic stem cells. For example, DNMT3B and DNMT1 are known for their cooperative role in maintaining DNA methylation in colon cancer cells38, 39. Within mouse embryonic stem cells, Dnmt1 cooperates with Dnmt3a and Dnmt3b in the maintenance of DNA methylation 40, 41. In dividing cells, Dnmt1 is concentrated in the replicating foci during S-phase to maintain DNA methylation following DNA synthesis. Dnmt3a and Dnmt3b are frequently associated with heterochromatin regions and co-localize with heterochromatin protein 1 alpha (HP1) in a cell cycle-independent manner42. However, the localization of Dnmt1 and Dnmt3a in postmitotic CNS neurons appears to be in a diffuse pattern (8 and unpublished data) as neuronal cells are arrested at the G0 phase of the cell cycle. How Dnmt1 and Dnmt3a play an overlapping role is still unclear. It is possible that Dnmt1 and Dnmt3a are co-localized or in the same methylation complex to maintain methylation patterns in postmitotic neurons. Alternatively, either Dnmt1 or Dnmt3a alone can recognize newly demethylated CpGs and carry out re-methylation function. Future studies with genome-wide location analysis of Dnmt1, Dnmt3a or Dnmt complex analysis in adult CNS neurons may resolve this question.
Our current study demonstrated that Dnmt1 and Dnmt3a deficiency in forebrain neurons causes defects in synaptic plasticity as well as learning and memory. In humans, mutations in DNA methylation machinery are associated with mental retardation disorders. The notable examples include DNMT3B mutations for ICF syndrome and MECP2 mutations for Rett syndrome. Thus, identifying the crucial learning and memory genes affected by Dnmt1 and Dnmt3a deficiency in mutant mice would be beneficial to understand human disorders as well. Among those deregulated genes in DKO mice, we suspect that up-regulation of immune genes, such as class I MHC32 molecule and Stat134, in neurons may be relevant to the defects in learning and memory identified here. Neurophysiological abnormalities associated with MHC I-deficient mice have been reported, including enhanced LTP and loss of LTD32, 43. The DKO phenotype in this study includes up-regulation of immune genes, loss of LTP and facilitated LTD, which is opposite to what has been observed in MHC I deficient mice. In addition, the proposed roles of immune molecules on neuronal dendrite pruning and elimination43, 44 are also consistent with our current observation of smaller cell size in the DKO mouse brain, which could result from a defect in dendritic pruning. It is still unclear how Dnmt1 and Dnmt3a deficiency in the forebrain leads to up-regulation of MHC I and other immune genes. We note that demethylation-induced MHC I gene expression also takes place in cultured fibroblasts, suggesting that DNA hypomemethylation-mediated MHC I up-regulation is not unique to postmitotic neurons45. It is found that MHC I expression can be activated in neurons via the interferon gamma (IFNγ) pathway35, 36. Our current finding that the Stat1 gene is demethylated and up-regulated in Dnmt-deficient neurons is consistent with the possibility that the cytokine signaling pathway may be involved in MHC I up-regulation in neurons.
Our study showed that DNA demethylation takes place in postmitotic neurons in the absence of both Dnmt1 and Dnmt3a in vivo. How DNA demethylation takes place in postmitotic neurons is also uncertain. In plants, a DNA glycocylase-based demethylation mechanism has been identified46. In mammals, several DNA demethylation pathways have been proposed (e.g. via MBD247, Gadd45a48 and Gadd45b26) and the most likely pathway of DNA demethylation in non-dividing neurons appears to be through base-excision repair49. Indeed, it has been proposed that Dnmt1 in neuronal cells plays a role in maintenance of DNA methylation after DNA repair50. In neuronal cells, high levels of oxidative stress can cause oxidative deamination of 5′methylcytosine (5′mC), leading to G:T mismatch. The G:T mismatich can activate the DNA base-excision\repair pathway and subsequently result in unmethylated C:G base-pairing. More recently, it was shown that neurons, but not non-neuronal cells, contain 5′hydroxyl-methylcytosine (5′HO-mC)51. 5′HO-mC in neurons is the product of the oxidation of 5′methylcytosine (5′mC) through the Tet1 enzymes52. Interestingly, we found that levels of total 5′mC and 5′HO-mC in control and DKO brain tissues are proportional to the total 5′mC levels (Supplementary Fig.10), supporting the notion that 5′HO-mC is derived from hydroxylation of 5′mC substrate. Presumably, 5′HO-mC can be replaced with cytosine via a repair mechanism, leading to demethylation. Considering our data showing that Dnmt1 and Dnmt3a play a redundant role in CNS neurons, we hypothesize that either Dnmt1 or Dnmt3a can prevent potential demethylation in neurons caused by DNA oxidation/repair pathways.
METHODS
Transgenic mice
Generation of Dnmt12lox/2lox mice has been described previously12. The Dnmt3a2lox/2lox mice were generated by En Li’s laboratory at Novartis Institute of Biomedical Research53. The transgenic mice were backcrossed with 129Jae mice for several generations to generate a common genetic background. We crossed Dnmt12lox/2lox mice with Dnmt3a2lox/2lox mice to produce Dnmt1 and Dnmt3a floxed homozygous offspring. Subsequent matings of three kinds of Dnmt floxed homozygotes with CamKIIαCre mice lead to CamKIIαCre; Dnmt12lox/2lox Dnmt3a2lox/2lox double knockout (DKO), CamKIIαCre; Dnmt12lox/2lox, and CamKIIαCre; Dnmt3a2lox/2lox single knockout (SKO) and CamKIIαCre negative littermate control. All genotypes were confirmed by PCR. All SKOs and DKO mice are fertile and develop normally. All conditional mutants have a normal lifespan compared to wildtype littermates. The animal protocols used in the studies here were approved by the UCLA Institutional Animal Research Committee.
Real time PCR analysis
Total RNA was extracted using Trizol reagent (Invitrogen). 1ug RNA samples were treated with Dnase I (Invitrogen) and reverse transcription was performed using iScript RT kit (Bio-Rad). Then, the SybrGreen supermix kit (Bio-Rad) was used for real-time PCR. Threshold cycle (Ct) was determined on the linear phase. Results were normalized by Ct of 18s. Relative gene expression fold difference was calculated by 2− Δnormalized Ct .
PCR primers used in this experiment are:
Dnmt1 F: CCT AGT TCC GTG GCT ACG AGG AGA A
Dnmt1 R: TCT CTC TCC TCT GCA GCC GAC TCA
Dnmt3a F: GCC GAA TTG TGT CTT GGT GGA TGA CA
Dnmt3a R: CCT GGT GGA ATG CAC TGC AGA AGG A
18S F: GCCCTGTAATTGGAATGAGTCCACTT
18S R: CTCCCCAAGATCCAACTACGAGCTTT.
β2M F: CCGTCTACTGGGATCGAGAC
β2M R: GCTATTTCTTTCTGCGTGCAT
H2M2 F: TTTAAAGGATCCCACTCCTTG
H2M2 R: CTGTCCCATCTGTTCCATCC
H2Q7 F: CCCTGACTTGGCAGTTGAAT
H2Q7 R: GTATGGAGGAGGCTCCCATC
Stat1 F: GGAGGTGAACCTGACTTCCA
Stat1 R: ATCGGTTCTGGTGCTTCCTT
C3 F: GCAGGTCAATAGCCTTCCTG
C3 R: CGTTGTAGAGCTGCTGGTCA
C4 F: GCTGACCTGGAGAAGCTGAC
C4 R: TACAGGACAGCACTGGATGG
PP1b F: TGTTGTCATGGAGGACTGTCA
PP1b R: ACACCACGGTCATTTTCTCC
PP1g F: ACC ACT CAA GAT ATG TGG TGA CA
PP1g R: CTA TTG ATG CTG GCG CAC T
Reelin F: GAA AGC TTC CAA GGT GAC GA
Reelin R: GGC AGC TTG CCT TAT CTG AC
Immunohistochemistry
Mouse brains were fixed by 4% PFA via cardiac perfusion under deep anesthesia. Overnight 4% PFA post-fixation was followed. The samples were cryoprotected by 30% sucrose immersion. After embedding in OCT (Tissue-Tek), 10um thick sections were cut and placed on Superfrost slides (Fisher). Tissue sections were washed with PBS and were permeablized and blocked with 0.5% triton X and 5% normal goat serum in PBS. Tissue sections were then incubated in primary antibodies at 4 °C overnight unless stated otherwise. The primary antibodies used in this experiment are rabbit anti-Cre (Covance, 1:3000) and mouse anti-NeuN (Chemicon, 1:300). The next day, the tissue sections were rinsed three times in PBS and then were incubated with fluorochrome-conjugated goat secondary antibodies (Jackson Immunoresearch) for 1 hour at room temperature. Hoechst staining was carried out in the middle of three PBS washes. The coverslips were mounted with MW 488 mounting media. Activated phosphor-Stat1(Abcam, 1:500) immunohistochemistry was carried out as previously described34.
Southern blot analysis
DNA samples were extracted from brain tissues. 10 ug DNA samples were digested by SpeI restriction enzyme overnight and were subjected to electrophoresis and transferred to a positive charged Hybond membrane. DNA probe12 was labeled with 32P dCTP using Prime-It II Random Primer Labeling Kit (Stratagene). Hybridization of the blot was performed by Quickhyb protocol (Stratagene). PhosphorImager was then used for hybridization detection.
BrdU labeling
BrdU (Sigma) was i.p. injected every two hours for 5 times at 10mg/kg body weight. 12 hours later after the first injection, the mice were cardiac perfused with 4% paraformaldehyde under deep anesthesia. Then they were postfixed overnight and cryoprotected in 30% sucrose. Coronal and sagittal sections were cut and stained for Brdu antibody by using VECTASTAIN Elite ABC kits (Vector) with DAB as substrate. The BrdU positive cells at sub granular zone of dentate gyrus were counted in accordance with previously described54 In brief, four sections from each mouse at comparable rostrocaudal levels were counted. Four mice per genotype were used.
Stereology
Mouse brains were fixed by cardiac perfusion of 4% paraformaldehyde under deep anesthesia. Then they were postfixed overnight in 4% paraformaldehyde and cryoprotected in 30% sucrose. 40 μm thick sections of hippocampi were cut coronally and kept in cryoprotection solution. A series of sections (10-12 sections per brain) were randomly picked with 240 μm apart (one-in-six) and mounted onto Fisher brand superfrost slides. The slides went through Cresyl Violet staining under regular protocol. The structures were then traced and contoured under a 5X objective. The volume was determined by using Cavalieri protocol (StereoInvestigator; MicroBrightField). The absolute granule cell number was also estimated using optical fractionator with reference of previous publications 55, 56. Counting frames of 15 μm × 15 μm × 40 μm were used in a 100 μm × 100 μm matrix, they were randomly superimposed onto the dentate gyrus. The nucleator program was used for dentate gyrus neuronal cell body size measuring according to previous publication57. The counting frame used was 15 μm ×15 μm, sampling grid was 100 μm ×100 μm. The coefficient of sampling error (Gundersen’s CE, m=1) was used for determining the precision of the estimates or sampling variation within each brain. The CE was controlled to be less than 0.10 for all experiments.
Electrophysiology
Hippocampus Slice Preparation— Hippocampal slices were prepared as described previously58, 59. Animals were anesthetized and sacrificed using a rodent guillotine. The brain was immersed in ice-cold cutting saline (CS (in mM): 110 sucrose, 60 NaCl, 3 KCl, 1.25 NaH2PO4, 28 NaHCO3, 0.5 CaCl2, 7 MgCl2, 5 glucose, 0.6 ascorbate) prior to isolation of the caudal portion containing the hippocampus and entorhinal cortex. Transverse slices (400 μm) were prepared with a Vibratome (The Vibratome Company, St. Louis, MO). During isolation, slices were stored in ice-cold CS. After isolation, cortical tissue was removed and hippocampal slices were equilibrated in a mixture of 50% CS and 50% artificial cerebrospinal fluid (ACSF (in mM): 125 NaCl, 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 2 CaCl2, 1 MgCl2, 25 glucose) at room temperature. Slices were further equilibrated in 100% ACSF for 45 min at room temperature, followed by a final incubation in 100% ACSF at 32 °C for 1 h. All solutions were saturated with 95%/5% O2/CO2.
Slice Electrophysiology— Extracellular field recordings were obtained as described previously58, 59. Electrophysiology was performed in an interface chamber (Fine Science Tools, Foster City, CA). Oxygenated ACSF (95%/5% O2/CO2) was perfused into the recording chamber at a rate of 1 ml/min. Electrophysiological traces were digitized and stored using Digidata (models 1200 and 1320A) and Clampex software (Axon Instruments, Union City, CA). Extracellular stimuli were administered on the border of areas CA3 and CA1 along the Schaffer-collaterals using Teflon-coated, bipolar platinum electrodes. fEPSPs were recorded in stratum radiatum with an ACSF-filled glass recording electrode (1–3MΩ). The relationship between fiber volley and fEPSP slopes over various stimulus intensities was used to assess baseline synaptic transmission. All subsequent experimental stimuli were set to an intensity that evoked a fEPSP that had a slope of 50% of the maximum fEPSP slope. Slices were stimulated for at least 10 minutes to generate baseline values before long term potentiation (LTP) or depression (LTD) induction. LTP was induced with 1,100 Hz tetani (1 s). LTD was elicited with 1 Hz protocol (900 pulses, 15 min). The results were analyzed via two-way ANOVA with repeated measures. Post-hoc comparisons after two-way ANOVA were made using the method of Bonferroni. Significance for all tests was set at p < 0.05.
For synaptic transmission experiments, all test stimuli and tetanus pulses were 100 μs in duration and 1/2–2/3 maximal stimulation strength. LTP was induced using 100 Hz/1 s stimulation protocols. For measurement of paired-pulse facilitation (PPF), we used different interstimulus intervals (ISIs) of 10, 20, 50, 100, 200, and 400 ms. All recordings were done using the same setup and in the same time span. Statistical comparisons were made with n equalling the number of slices.
Morris water maze behavioral test
During the training trials of the hidden-platform version of the Morris water maze test, the platform location was fixed and submerged under opaque water. Mice were trained with two trials per day (1 min per trial) for 12 days. During each trial, the mouse was released into the water maze facing the wall of the pool at a pseudo-random starting position. The trial ended when either the mouse climbed onto the platform or a maximum of 60 seconds elapsed. The mouse was then kept on the platform for 5 seconds at the end of each trial. A 60-second probe test was conducted at the end of training on days 4, 8 and 12, in which the platform was removed from the pool. The search pattern of the mice was recorded by a camera and analyzed with the VHS software.
Contextual fear conditioning
Training consisted of placing the mice in a conditioning chamber and presenting a shock (2 s, 0.75 mA) 2 min later. Mice remained in the chamber for an additional 3-minutes. The mice were placed back in the same chamber and tested for contextual fear conditioning 24 h later. Our index of memory, freezing (the cessation of all movement except for respiration), was assessed via an automated scoring system (Med Associates) with a 30 frames per s sampling; the mice needed to freeze continuously for at least 1 s before freezing was counted.
Microarray analysis
Gender matched 2-3 months old mice forebrain (cortex and hippocampus) tissues were dissected and extracted for total RNA by using Trizol reagent (Invitrogen). RNA quality and quantity were determined by RNA6000 Nano Labchip kit (Agilent 5065-4776). CRNAs were converted and fluorescently labeled by using Agilent low RNA input fluorescent linear amplification kit. Briefly, cDNA was first synthesized from total RNA, and then Cyanine-3 CTP for control sample or Cyanine-5 CTP for mutant sample was used for in vitro transcription for fluorescent cRNA synthesis. The product was purified using the RNeasy kit from Qiagen following instructions. Then cRNAs were quantified by NanoDrop ND-1000 spectrophotometer. Yield and specific activity were calculated following instructions. Only cRNA with yield >750ng and specific activity >8.0pmol Cy3 or Cy5 per ug were used for hybridization. The resulting labeled cRNA probes were combined and hybridized to the same Agilent 44K mouse gene expression array. The assembled slide chamber was placed in rotisserie in a hybridization oven set to 65°C with rotation speed at 4rpm. After 17 hours of hybridization, the slide chamber was dissembled and microarray slide was washed as instructed. Then the slide was scanned using Agilent scanner. The resultant data were retrieved by Agilent feature extraction software 8.5 and analyzed by FOCUS software.
In situ hybridization
Riboprobes were labeled with [α-35S]UTP by using riboprobe combination system kit (Promega) and then purified with RNeasy Kit (Qiagen) following instructions. The probe plasmid was a gift from Dr. Stan Nelson at UCLA. Cryostat sections (15 μm) fixed on slides were washed/incubated in the following series of solutions at room temperature: 1) 0.1 M glycine in 0.1M PB twice for 3 min each, 2) 0.1M PB for 15 min, 3) acetic anhydride/TEA buffer for 10 min, 4) 2× sodium chloride-sodium citrate (SSC) twice for 15 min each, 5) 50, 70, 95, and 100% ethanol for 1 min each, 6) chloroform for 5 min, 7) 100 and 95% ethanol for 1 min each. Then probes were diluted in hybridization solution and added onto the slides. Hybridized slides were attached onto coverslips and sealed with DPX mountant (Gallard Schlesinger, Plainview, NY), and incubated at 60 °C for 16-20 hours. Following hybridization, slides were washed in the following series of solutions: 1) 4x SSC twice for 20 min each at 60 °C, 2) RNase A in RNase buffer for 30 min at 45 °C, 3) 2× SSC 4 times for 20 min at RT, 4) 0.5× SSC twice for 20 min at 60 °C, 5) 0.1× SSC for 20 min at 60 °C, 6) 0.1× SSC for 20 min at RT. Slides were rinsed in H2O and then air dried. Hybridized slides were pressed against a film for 4-5 days prior to dipping in Kodak NTB2 emulsion. Following exposure to emulsion at 4 °C the slides were developed, stained with Cresyl Violet, attached onto coverslips with Permount, and photographed in bright and dark field optics (Nikon Eclipse).
Hippocampal neuron cultures and adeno-virus infection
Hippocampal neurons were isolated from mouse embryos at E15.5 as previously described60. Briefly, after treatment with papain and DNase for 20 min at 37°C and mechanical dissociation, hippocampal cells were plated on coverslips coated with poly-DL-ornithine and laminin. The hippocampal neurons were cultured in Neurobasal medium supplemented with glutamine, penicillin–streptomycin and B27 (Invitrogen). The hippocampal cultures were infected with adeno-cre virus at 3 DIV with adeno-GFP virus infection as control12.
Methylation analysis
Genomic DNA was digested by Bgl II enzyme and treated with sodium bisulfite afterwards8. After DNA cleaning with Wizard DNA cleanup kit (Promega), either nested or touchdown PCRs were carried out. The primers were designed by Methprimer program. The PCR products were purified by PCR SV gel and PCR cleanup kit (Promega). TOPO cloning was performed by following the manufacture instruction (Invitrogen). Normally 10 minipreps were set up per sample and sent in for sequencing to detect methylation sequencing patterns.
The PCR primers used in this experiment are:
Dhh outside F: TTTATTTGGTAGTTGATTTTTAGTT
Dhh inside F: TTTTTTTGTTATAGGTATTGAATTTT
Dhh inside R: ACCAAAAACCAAACACAAATCTTAC
Dhh outside R: AAAAAAAACATCCTATTCTTTA
Kcne1 outside F: AGGTGTGAGTTAATAGGGGTAGTTT
Kcne1 inside F: GGTTTGGTTAGGTTGGTAGATGA
Kcne1 inside R: TTCTCTAAACCCAATACCTCTCTTTAA
Kcne1 outside R: TTTAACTTTAAACATCAAAATTTTAATTAT
Pten regionA outside F: GAGGTGTTTTTTGGAGAAAGTATGA
Pten regionA inside F: TGGGTAAAGGTGTGTGTTATTAAATT
Pten regionA inside R: CCCTCAACAAAAAAACTCAATCTAA
Pten regionA outside R: CTCTAACCCCTCAACAAAAAAACT
Pten regionB outside F: TTGGAGTTAAGATTGAAGTAGAAGATTTAG
Pten regionB inside F: TGAAGTAGAAGATTTAGTAGTTAAAGTGAA
Pten regionB inside R: AATCCCAACAACCACATAATAACTC
Pten regionB outside R: CTAATAAAATAACTCAACAATTAAAAACAC
Stat1 outside F: GAGAAATTTGAATTGTTTAAGATTAAGT
Stat1 inside F: TTTAGATTGGTATGATATGATTAGT
Stat1 inside R: AAACCAACTATACTCAAAACACAAA
Stat1 outside R: AATTACCCTATCCTTAAATATCTACTA
PCR conditions used in this experiment were the same as described before8.
Neuronal DNA isolation
We followed the protocol as described 61. Briefly, fresh mouse forebrain samples were homogenized by douncing 50 times in 5 ml nuclei extraction buffer (0.32 M Sucrose,5 mM CaCl2,3 mM Mg(Ac)2,0.1 mM EDTA, 10 mM Tris-HCl (pH8)) with 0.1% Triton X-100. After douncing, brain homogenates were transferred into 14 ml ultracentrifuge tubes (Beckman) over a 9 ml of sucrose cushion. Ultracentrifugation was done at 24400 rpm for 2.5 hrs at 4°C (Beckman, L8-70M, SW28 rotor). After centrifugation, 1200 μl of chilled 1 × PBS was added over the nuclei pellet and incubated for 20 minutes on ice. The nuclei were then dissociated and tagged by a mouse anti-NeuN antibody that was already incubated with anti-mus IgG (Alexa 488) antibody. Next, the fluorescent nuclei ran through a FACS machine (BD Biosciences) with proper gates settings. A small portion of the NeuN+ nuclei were re-run on the FACS machine to validate the purity. Immunonegative (NeuN−) nuclei were collected in parallel. The nuclei were then harvested by centrifuge and lysed for DNA under regular protocol.
Quatitation of total methylcytosine through mass spectrometry
The DNA hydrolysis protocol was as described62. Typically, 1 μg of genomic DNA was first denatured (100 °C, 3 min) and then chilled on ice. A 1/10 volume of 0.1 M ammonium acetate (pH 5.3) and 2 units of nuclease P1 (Roche Molecular Biochemicals Indianapolis, IN) were added and the sample was incubated (45 °C, 2 h). Then a 1/10 volume of 1 M ammonium bicarbonate and 0.002 units of venom phosphodiesterase I (Sigma Chemical Co.) were added followed by another incubation (37 °C, 2 h). Subsequently, 0.5 unit of alkaline phosphatase (Fermentas, Hanover, MD) was added, and the mixture was incubated at 37 °C for another 1h. LC-ESI-MS MS was performed as described63 with some modifications.2 DNA hydrolysis samples (20 μl) were injected directly without further treatment onto a reverse phase HPLC column (Aquasil C18 2.1 × 200 mm, 5 μ particle size, Thermo Scientific, San Jose, CA) with flow rate of 220 μL/min and equilibrated in Buffer A (0.1% formic acid in water v/v). The analytes were eluted by an increasing concentration of Buffer B (0.1% formic acid in methanol v/v). The effluent from the column was directed to an electrospray ion source connected to a triple quadrupole mass spectrometer (AppliedBiosystems 4000Q Trap) operating in the positive ion multiple reaction monitoring mode using previously optimized conditions, and the intensity of specific MH+→fragment ion transitions were recorded (5mdC m/z 242.1→126.1 and dC m/z 228.1→112.1). The 5mdC content in each sample was calculated from the MRM peak area by extrapolation from the response obtained following injection of known quantities of the authentic compound, and expressed as a percentage of the total cytosine pool assuming equivalent molar responses.
Methylated DNA immunoprecipitation and microarray experiment (MeDIP-chip)
2 μg of genomic DNA from 3 month old DKO and wildtype littermate forebrain tissue was sonicated to an average size of 350-400 bp. 20 μg mouse anti 5 methyl-cytosine antibody (Eurogentec) was added into the genomic DNA sample and the mixture was rotated at 4 degree overnight. 100 μl Dynabeads M-280 sheep anti-mouse IgG beads were washed 5 times with FB buffer (10mM Tris-HCl, pH 7.5; 50mM NaCl; 1mM EDTA). The beads were resuspended in FB buffer, added to DNA-Antibody mixture and rotated at 4 degree for 8 hours. The beads were washed four times with FB buffer, one time each with FB containing 150 mM or 300 mM NaCl. Serial elution with 200 μl FB buffer of 1.5% SDS, 0.5% SDS and 0.1% SDS were made and each elution was vortexed for 1 minute. All three elutions were pooled together and the DNA was purified by using phenol/chloroform. The eluted DNA was resuspended in 80 μl water. BioPrime array CGH genomic labeling kit (Invitrogen) was used to label equal amount of pull down DNA with Cy5 (wt) or Cy3 (KO). Agilent promoter array hybridization was set up as previously described37.
Statistical analysis
Data are presented as mean±s.e.m. Unless otherwise noted, the Student’s t-test (two-tailed) and the two-way ANOVA test were used for statistical analysis. Statistical significance was determined at P <0.05.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. X-H. Lu and Dr. X. W. Yang for help with stereology analysis; T. Chailangkarn and Dr. S. Fouse for technical support; Dr. P. Golshani for critical reading of the paper. We also thank all members of the Fan laboratory for help and advice. Funding was provided by grants fromNational Institute of Health to G.F. (RO1 NS051411); to A.J.S.(P50 MH0779720 and R01AG13622); and to J.D.S (NIA, NIMH and NINDS). G.F. is a Carol Moss Spivak Scholar in Neuroscience.
Footnotes
Accession number. Microarray data are available at GEO (Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/prejects/geo/index.cgi) with accession number GSE19367.
Note: Supplementary information is available on the Nature Neuroscience website.
References
- 1.Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science (New York, N.Y. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- 2.Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nature reviews. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- 3.Martinowich K, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science (New York, N.Y. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 4.Colvis CM, et al. Epigenetic mechanisms and gene networks in the nervous system. J Neurosci. 2005;25:10379–10389. doi: 10.1523/JNEUROSCI.4119-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan JS, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- 7.Goto K, et al. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation. 1994;56:39–44. doi: 10.1046/j.1432-0436.1994.56120039.x. [DOI] [PubMed] [Google Scholar]
- 8.Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- 9.Inano K, et al. Maintenance-type DNA methyltransferase is highly expressed in post-mitotic neurons and localized in the cytoplasmic compartment. J Biochem. 2000;128:315–321. doi: 10.1093/oxfordjournals.jbchem.a022755. [DOI] [PubMed] [Google Scholar]
- 10.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 11.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 12.Fan G, et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci. 2001;21:788–797. doi: 10.1523/JNEUROSCI.21-03-00788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Golshani P, Hutnick L, Schweizer F, Fan G. Conditional Dnmt1 deletion in dorsal forebrain disrupts development of somatosensory barrel cortex and thalamocortical long-term potentiation. Thalamus Relat Syst. 2005;3:227–233. doi: 10.1017/S1472928807000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutnick LK, et al. DNA hypomethylation restricted to the murine forebrain induces cortical degeneration and impairs postnatal neuronal maturation. Human molecular genetics. 2009;18:2875–2888. doi: 10.1093/hmg/ddp222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen S, Meletis K, Fu D, Jhaveri S, Jaenisch R. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Dev Dyn. 2007;236:1663–1676. doi: 10.1002/dvdy.21176. [DOI] [PubMed] [Google Scholar]
- 16.Fan G, et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development (Cambridge, England) 2005;132:3345–3356. doi: 10.1242/dev.01912. [DOI] [PubMed] [Google Scholar]
- 17.Endres M, Fan G, Meisel A, Dirnagl U, Jaenisch R. Effects of cerebral ischemia in mice lacking DNA methyltransferase 1 in post-mitotic neurons. Neuroreport. 2001;12:3763–3766. doi: 10.1097/00001756-200112040-00032. [DOI] [PubMed] [Google Scholar]
- 18.Endres M, et al. DNA methyltransferase contributes to delayed ischemic brain injury. J Neurosci. 2000;20:3175–3181. doi: 10.1523/JNEUROSCI.20-09-03175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson ED, Kavalali ET, Monteggia LM. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J Neurosci. 2008;28:395–406. doi: 10.1523/JNEUROSCI.3796-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levenson JM, et al. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. The Journal of biological chemistry. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- 21.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 22.Juttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci U S A. 1994;91:11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 24.Weaver IC, et al. Epigenetic programming by maternal behavior. Nature neuroscience. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 25.Weaver IC, et al. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma DK, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science (New York, N.Y. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu GL, et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 28.Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genet. 2000;1:11–19. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- 29.Ouimet CC, McGuinness TL, Greengard P. Immunocytochemical localization of calcium/calmodulin-dependent protein kinase II in rat brain. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:5604–5608. doi: 10.1073/pnas.81.17.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wayman GA, Lee YS, Tokumitsu H, Silva A, Soderling TR. Calmodulin-kinases: modulators of neuronal development and plasticity. Neuron. 2008;59:914–931. doi: 10.1016/j.neuron.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boulanger LM, Huh GS, Shatz CJ. Neuronal plasticity and cellular immunity: shared molecular mechanisms. Curr Opin Neurobiol. 2001;11:568–578. doi: 10.1016/s0959-4388(00)00251-8. [DOI] [PubMed] [Google Scholar]
- 32.Huh GS, et al. Functional requirement for class I MHC in CNS development and plasticity. Science (New York, N.Y. 2000;290:2155–2159. doi: 10.1126/science.290.5499.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stevens B, et al. The classical complement cascade mediates CNS synapse elimination. Cell. 2007;131:1164–1178. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 34.Tropea D, et al. Gene expression changes and molecular pathways mediating activity-dependent plasticity in visual cortex. Nat Neurosci. 2006;9:660–668. doi: 10.1038/nn1689. [DOI] [PubMed] [Google Scholar]
- 35.Neumann H, Cavalie A, Jenne DE, Wekerle H. Induction of MHC class I genes in neurons. Science (New York, N.Y. 1995;269:549–552. doi: 10.1126/science.7624779. [DOI] [PubMed] [Google Scholar]
- 36.Neumann H, Schmidt H, Cavalie A, Jenne D, Wekerle H. Major histocompatibility complex (MHC) class I gene expression in single neurons of the central nervous system: differential regulation by interferon (IFN)-gamma and tumor necrosis factor (TNF)-alpha. The Journal of experimental medicine. 1997;185:305–316. doi: 10.1084/jem.185.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fouse SD, et al. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell stem cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhee I, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 39.James SR, Link PA, Karpf AR. Epigenetic regulation of X-linked cancer/germline antigen genes by DNMT1 and DNMT3b. Oncogene. 2006;25:6975–6985. doi: 10.1038/sj.onc.1209678. [DOI] [PubMed] [Google Scholar]
- 40.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang G, et al. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol Cell Biol. 2002;22:480–491. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bachman KE, Rountree MR, Baylin SB. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. The Journal of biological chemistry. 2001;276:32282–32287. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- 43.Boulanger LM, Shatz CJ. Immune signalling in neural development, synaptic plasticity and disease. Nature reviews. 2004;5:521–531. doi: 10.1038/nrn1428. [DOI] [PubMed] [Google Scholar]
- 44.Perry VH, O’Connor V. C1q: the perfect complement for a synaptic feast? Nature reviews. 2008;9:807–811. doi: 10.1038/nrn2394. [DOI] [PubMed] [Google Scholar]
- 45.Jackson-Grusby L, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nature genetics. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 46.Zhu JK. Epigenome sequencing comes of age. Cell. 2008;133:395–397. doi: 10.1016/j.cell.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 48.Barreto G, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 49.Ma DK, Guo JU, Ming GL, Song H. DNA excision repair proteins and Gadd45 as molecular players for active DNA demethylation. Cell cycle (Georgetown, Tex. 2009;8:1526–1531. doi: 10.4161/cc.8.10.8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brooks PJ, Marietta C, Goldman D. DNA mismatch repair and DNA methylation in adult brain neurons. J Neurosci. 1996;16:939–945. doi: 10.1523/JNEUROSCI.16-03-00939.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science (New York, N.Y. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science (New York, N.Y. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dodge JE, et al. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. The Journal of biological chemistry. 2005;280:17986–17991. doi: 10.1074/jbc.M413246200. [DOI] [PubMed] [Google Scholar]
- 54.Han YG, et al. Hedgehog signaling and primary cilia are required for the formation of adult neural stem cells. Nat Neurosci. 2008;11:277–284. doi: 10.1038/nn2059. [DOI] [PubMed] [Google Scholar]
- 55.Lu XH, et al. Bacterial artificial chromosome transgenic mice expressing a truncated mutant parkin exhibit age-dependent hypokinetic motor deficits, dopaminergic neuron degeneration, and accumulation of proteinase K-resistant alpha-synuclein. J Neurosci. 2009;29:1962–1976. doi: 10.1523/JNEUROSCI.5351-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hong SM, et al. Reduced hippocampal neurogenesis and skill reaching performance in adult Emx1 mutant mice. Exp Neurol. 2007;206:24–32. doi: 10.1016/j.expneurol.2007.03.028. [DOI] [PubMed] [Google Scholar]
- 57.Cui L, et al. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 58.Zhou Y, et al. Interactions between the NR2B receptor and CaMKII modulate synaptic plasticity and spatial learning. J Neurosci. 2007;27:13843–13853. doi: 10.1523/JNEUROSCI.4486-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levenson JM, et al. Regulation of histone acetylation during memory formation in the hippocampus. The Journal of biological chemistry. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- 60.Ethell IM, et al. EphB/syndecan-2 signaling in dendritic spine morphogenesis. Neuron. 2001;31:1001–1013. doi: 10.1016/s0896-6273(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 61.Jiang Y, Matevossian A, Huang HS, Straubhaar J, Akbarian S. Isolation of neuronal chromatin from brain tissue. BMC Neurosci. 2008;9:42. doi: 10.1186/1471-2202-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crain PF. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods in enzymology. 1990;193:782–790. doi: 10.1016/0076-6879(90)93450-y. [DOI] [PubMed] [Google Scholar]
- 63.Song L, J S, Kazim L, Karpf AR. Specific method for the determination of genomic DNA methylation by liquid chromatography-electrospray ionization tandem mass spectrometry. Anal Chem. 2005;77:504–510. doi: 10.1021/ac0489420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.