Abstract
Ubiquitin proteasomal pathway (UPP) is the principle mechanism for protein catabolism and affects cellular processes critical for survival and proliferation. Levels of tumor suppressor protein p53 are very low in cells due to its rapid turnover by UPP-mediated degradation. While p53 is mutated in human cancers, most human melanomas maintain wild-type conformation. In this study, to investigate the effects of UPP inhibitor in vitro and in vivo, we used a genetically-engineered mouse model (GEMM) that has the same genetic alterations as those of human melanomas. Melanoma cells were established from mouse tumors and named 8B20 cells. Treatment of 8B20 cells with the UPP inhibitors, MG132 and clasto-lactacystin-β-lactone, led to an increase in levels of p53 while treatment with non-proteasomal inhibitors did not alter p53 levels. UPP inhibitors induced formation of heavy molecular weight ubiquitinated proteins, a hallmark of UPP inhibition, and p53-specific poly-ubiquitinated products in 8B20 cells. To further decipher the mechanism of p53 stabilization, we investigated half-life of p53 in cells treated with cycloheximide to block de novo protein synthesis. Treatment of 8B20 cells with MG132 led to an increase in the half-life of p53. Further analysis revealed that p53 stabilization was not mediated by phosphorylation of Ser-15 and Ser-20 residues. In vivo studies showed that MG132 induced p53 overexpression and reduced tumor growth, suggesting an important role of p53 stabilization in controlling melanoma. Taken together, our studies provide a proof of principle for using a GEMM to address the mechanisms of action and efficacy of melanoma treatment.
Keywords: Skin Cancer, p53, Tumor suppressor, Proteasomal degradation, Ubiquitination. Proteasomal inhibitor MG132
Introduction
Several studies have shown that repeated exposures of UV irradiation to human skin results in both melanoma and nonmelanoma skin cancers [1–4]. The tumor suppressor protein p53 is extensively investigated because of its role as a major tumor suppressor in humans and other mammals [5] (for a review see ref. 6). Loss or mutation of p53 is strongly associated with an increased susceptibility to cancer or neoplasia [7]. When normal cells are subjected to stress signals, such as DNA damage or oxidative stress, p53 is activated, resulting in transcription of downstream genes that coordinate either growth arrest or apoptosis of cells [8] to prevent proliferation and clonal expansion of damaged cells. Therefore, controlling p53-dependent pathway is a critical step in tumor initiation and progression. The loss of p53 activity is a major mechanism through which tumors become resistant to cell death, escape the local microenvironment and become recalcitrant to various types of therapeutic interventions.
Majority of cancers harbor point mutations in p53 which are likely in the central region of the protein responsible for DNA binding [9]. However, unlike other solid tumors, melanomas typically lack p53 mutations and retain the expression of wild-type p53 [10–13]. This is a surprising phenomenon given the highly malignant nature of the melanoma and its resistance to therapeutic interventions [14]. Melanoma development has been strongly associated with the deletions or mutations in the cyclin-dependent kinase inhibitor 2A (CDKN2A) [15,16], leading to inactivation of p16INK4a/cyclin dependent kinases (CDK) 4 and 6/retinoblastoma protein (p16INK4a/CDK4, 6/pRb) and p14ARF/human double minute 2/p53 (p14ARF/HDM2/p53) tumor suppressor pathways. The identification of small molecules and proteins that increase p53 stability and thereby protect cells against cancer progression is an active area of research [17].
In the current study, we explored whether the tumors with wild-type p53 such as melanoma, may benefit from the treatment strategies that activate p53. Here we utilize, for the first time, a genetically-engineered mouse model (GEMM) of melanoma with melanocyte specific mutant ras, deletion of CDKN2A and wild-type p53 that have the same genetic alterations as those of human melanomas to study the effects of proteosomal inhibitor MG132 on the melanoma both in vitro and in vivo.
Experimental
Chemicals and Reagents
Cysteine protease inhibitor E-64, serine protease inhibitor PMSF, cycloheximide and bovine serum albumin were purchased from Sigma-Aldrich (St. Louis, MO). Protein A/G-agarose and clasto-lactacystin-β-lactone were obtained from Calbiochem/EMD Chemicals, Inc. (San Diego, CA). MG132 was purchased from BIOMOL International, L.P. (Plymouth Meeting, PA). Protease inhibitor cocktail tablets (cat # 11 836 153 001) were obtained from Roche Diagnostics (Mannhein, Germany). All other chemicals and reagents used in this study were of highest purity grade available commercially.
Antibodies
Anti-ubiquitin (rabbit polyclonal) and anti-actin (mouse monoclonal) antibodies were purchased from Sigma-Aldrich. Anti-p53 (rabbit polyclonal) antibody was obtained from Novocastra Laboratories Ltd. (Newcastle upon Tyne, UK). Phospho-p53 (ser15) and phospho-p53 (ser20) antibodies were from Cell Signaling Technology (Danvers, MA). Horseradish peroxidase-conjugated secondary antibodies were purchased from Jackson Immuno Research Laboratories, Inc. (West Grove, PA).
Animals
Animals used in the current study were Tyr-H-rasG12V transgenic mice (FVB/N background) and Ink4a/Arf knockout (FVB/N background) obtained from National Cancer Institute at Frederick (Frederick, MD). These animals were cross-bred twice to generate transgenic mice with melanocyte-specific H-rasG12V expression on an Ink4a/Arf-deficient background. Animals were housed at UCD vivarium under specific pathogen-free conditions according to the National Institutes of Health Animal Care Guidelines under an Institutional protocol reviewed and approved by the Institutional Animal Care and Use Committee (IACUC). These transgenic mice spontaneously develop cutaneous melanoma with high penetrance (74%) at around 3 to 4 months of age. The melanomas are amelanotic but express melanocyte pigment genes such as MART-1 and TRP-2.
Cell Lines
8B20 mouse melanoma cells were established from a melanoma tumor spontaneously arising in the transgenic mice with melanocyte-specific H-rasG12V expression on an Ink4a/Arf-deficient background. B16F10, a murine melanoma cell line, was obtained from the American Type Culture Collection (ATCC; Manassas, VA).
SDS-PAGE, Immunoprecipitation and Immunoblotting Analysis
Exponentially growing cells were seeded at ~2 ×106 in 100-mm plates, and 12 hours later the cells were harvested. For immunoprecipitation studies, medium was removed and cells were washed with phosphate-buffered saline (PBS, pH 7.4), lysed on ice in RIPA buffer (50mM Tris-Hcl, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 20 mM NaF, 100 mM Na3VO4, 0.5 % NP-40, 1 % Triton X-100 and 1 mM PMSF (pH 7.4) with a freshly added protease inhibitor cocktail (Roche Diagnostics), and then centrifuged at 13,200 g for 10 minutes. Ten μg of the appropriate antibody was added to the supernatants and the mixture was gently rotated at 4°C for 12 hours. Protein A/G-agarose (40 μL) was then added, and the incubations were continued for additional 90 minutes. The protein A/G-agarose was collected by centrifugation and the beads were washed three times with 50 mM Tris-HCl, pH 8.0, containing 150 mM NaCl, 1 % (v/v) Nonidet P-40. Protein A/G- agarose beads were then suspended in 2× Laemmli [18] SDS sample buffer and heated to 90°C for 5 minutes. Immunoprecipitated proteins were analyzed by SDS-PAGE (12 % gels) followed by transfer to a polyvinylidene difluoride membrane (0.4 μM) in 25 mM Tris, 192 mM glycine, and 20 % methanol at 110 V for 1 hour. Membranes were blocked overnight in 10 mM Tris-HCl, pH 8.0, 125 mM NaCl and 0.2 % (v/v) Tween 20 containing 5 % (w/v) nonfat dry milk (TBST-M5).
Immunoblot analysis of the total polyubiquitinated proteins and p53 specific polyubiquitinated forms were analysed using anti-ubiquitin antibodies. For the determination of cellular levels of proteins, cells were scraped after the respective treatments, washed with PBS, lysed in RIPA buffer and then sonicated on ice. The supernatant was obtained by centrifugation at 13,200 g for 5 minutes, and the protein concentration was determined by the method of Lowry [19]. Protein bands were visualized using enhanced chemiluminescence as described by the manufacturer (Pierce Biotechnology, Inc., Rockford, IL). Densitometric analysis of the membranes was performed using GelDoc 200 (Bio-Rad Laboratories, Hercules, CA).
Inhibitor Studies
Cells were grown to 85 % confluency in 60-mm dishes in complete minimal essential medium. Fresh medium containing various inhibitors MG132, clasto-lactacystin-β-lactone, E-64 and PMSF were added to each plate and the medium was replaced with fresh dose of the inhibitor 3 hours later. After 6 hours of incubation with various inhibitors, cells were washed in PBS and then lysed in ice-cold RIPA buffer. Protein concentrations were determined and target proteins were analyzed by immunoblotting.
p53 Protein Turnover Studies
p53 protein stability was determined in the presence or absence of proteasomal inhibitor MG132 using 8B20 cells. Approximately 2 ×107 cells were plated into 100-mm plates, and 12 hours later fresh medium containing cycloheximide (50 μg/mL) was added. At indicated time intervals, cells were scraped and pelleted by centrifugation at 13,200 g for 10 minutes. Cells were lysed in RIPA buffer and supernatant collected as described above. To assess the p53 turnover in the presence of proteasomal inhibitor MG132, cells were treated with MG132 (10 μM) and cycloheximide at a concentration of 50 μg/mL simultaneously to block the de novo protein synthesis. Proteasomal inhibitor and cycloheximide were present in the medium during the entire duration of the experiment.
Cell Cycle Analysis
Cells were synchronized by growth confluence, harvested by trypsinization and re-plated in 100 mm plates. After 24 hours cells were treated with varying concentrations of the proteosomal inhibitor MG132 (0–10 μM) for 24 hours. Cells were then washed with PBS and an aliquot of cells was stained with Krishan’s stain [20] at 4°C for 24 hours. Cell cycle distribution was analyzed by a FACSCalibur™ and determined with Cell Quest software in the FACS analysis core of the University of Colorado Denver, Cancer Center (Aurora, CO).
Melanoma Growth in vivo
Transgenic mice with melanocyte-specific H-rasG12V expression on an Ink4a/Arf-deficient background develop melanoma spontaneously. When visible tumors (around 3 mm in diameter) were identified, eight mice were randomly assigned into 2 cohorts: control group (untreated) or treatment group (daily intraperitoneal treatment with 5 μg/kg MG132 for 10 days). Tumor size was measured every 2 days by caliper measurements using the following formula: tumor volume = [length × width2] / 2. Mice were euthanized after the treatment.
Immunohistochemical Analysis
Formalin-fixed and paraffin-embedded samples were sectioned, deparaffinized and rehydrated. Antigen retrieval was achieved by boiling the sections in citric acid buffer (pH 6.0), and endogenous peroxidase activity was quenched with 3 % H2O2. Sections were incubated with the anti-p53 antibody, followed by biotinylated anti-rabbit antibody conjugated with horseradish peroxidase labeled streptavidin. Color development was achieved by incubation with 3, 3′-diaminobenzidine, and the sections were counterstained with Harris hematoxylin.
Data Analysis
All of the experiments were replicated at least twice. The results were analyzed by Student’s unpaired t test to compare two groups. The data are presented as the means ± S.E., and differences were considered significant if p < 0.05.
Results
p53 Protein in 8B20 Mouse Melanoma Cells Is a Target of Ubiquitin Proteasomal Pathway (UPP)
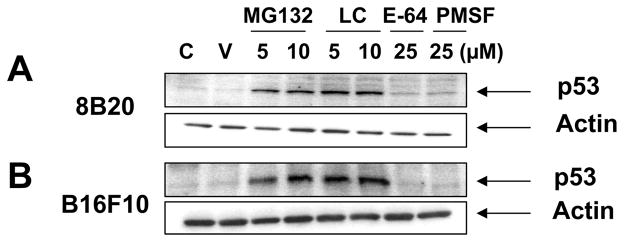
The effect of UPP inhibitors (MG132 and clasto-lactacystin-β-lactone) and non-proteasomal inhibitors (PMSF and E-64) on the p53 stability was examined using 8B20 mouse melanoma cells obtained from the melanoma tumor derived from a GEMM of melanoma. In addition, B16F10 mouse melanoma cells were used as a control since this cell line is the most commonly used mouse melanoma cell line and has a wild-type p53 [21]. p53 levels were increased by the addition of MG132 or clasto-lactacystin-β-lactone in both melanoma cell lines (Fig. 1A & 1B). Treatment of these cell lines either with E-64 or PMSF had no effect on the degradation of p53 by the UPP. These results confirm the predominant role of the UPP in the targeted destruction of p53 in the melanoma cell lines.
Figure 1. Proteosomal inhibitors block the degradation of p53 in (A) 8B20 and (B) B16F10 melanoma cell lines.
Exponentially growing 8B20 and B16F10 cell lines were either untreated (C: control), or treated with ethanol (V: vehicle control), proteosomal inhibitors (MG132 and LC: clasto-lactacystin-β-lactone) or non-proteosomal inhibitors (E-64 and PMSF) at the indicated concentrations for 6 hours. Cell lysate (100 μg) was loaded on a 12 % polyacrylamide gel and subjected to SDS-PAGE and immunoblot analysis. Membranes were probed with anti-p53 antibody (mouse specific). β-Actin levels are shown as loading controls. The data are one of three representative experiments.
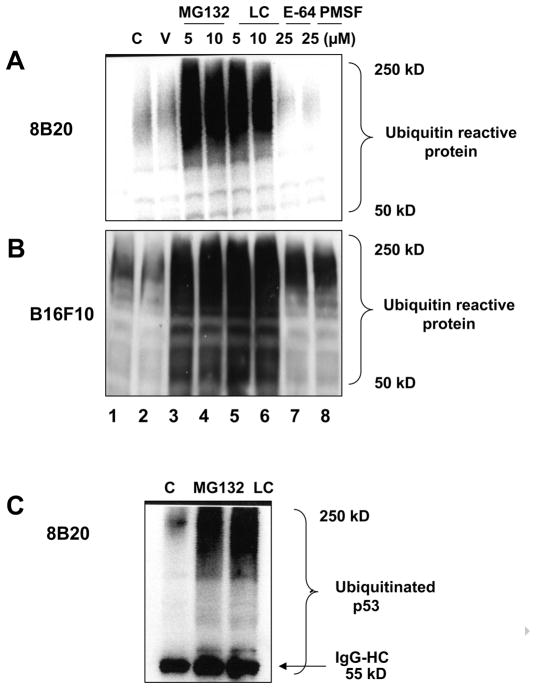
One of the hallmarks of UPP mediated degradation of its target proteins is the build up of high molecular weight ubiquitin positive protein adducts. Samples treated with the proteasomal inhibitors were tested for the appearance of high molecular weight polyubiquitinated proteins using an anti-ubiquitin antibody in 8B20 and B16F10 mouse melanoma cell lines (Fig. 2A & 2B). The accumulation of the polyubiquitinated protein species was greatly enhanced when the UPP was inhibited either by the addition of MG132 or clasto-lactacystin-β-lactone (Fig. 2A & 2B, lanes 3–6). No forms of polyubiquitinated proteins were detected following the treatment with E-64 or PMSF (Fig. 2A & 2B, lanes 7 and 8).
Figure 2.
(A, B) Treatment of murine melanoma cells with proteosomal inhibitors MG132 and clasto-lactacystin-β-lactone results in the generation of high molecular weight ubiquitin reactive products Exponentially growing 8B20 and B16F10 mouse melanoma cells were either untreated (C: control) or treated with ethanol (V: vehicle control), proteosomal inhibitors (MG132 and LC: clasto-lactacystin-β-lactone) or non-proteosomal inhibitors (E-64 and PMSF) at the indicated concentrations for 6 hours. Cell lysate (100 μg) was loaded on a 12 % polyacrylamide gel and subjected to SDS-PAGE and immunoblot analysis. Membranes were probed with the anti-ubiquitin antibody. (C) Detection of p53 specific ubiquitinated products in 8B20 cells treated with proteosomal inhibitors. 8B20 cells were untreated (C: control) or treated with either MG132 (10 μM) or lasto-lactacystin-β-lactone (LC) (10 μM) for 6 hours. Cell lysate containing 500 μg of protein was used for immunoprecipitation using anti-p53 antibody (10 μg) followed by SDS-PAGE and immunoblotting analysis with an anti-ubiquitin antibody. Lower bands represent IgG-heavy chain (IgG-HC) at 55kD. β-Actin levels are shown as loading controls. The data are one of three representative experiments.
Consistent with the UPP-mediated degradation of p53 in 8B20 melanoma cells, immunoprecipitation using anti-p53 antibody followed by the immunoblots using the anti-ubiquitin antibody revealed the presence of polyubiquitinated p53 protein in cell lysates from 8B20 cells treated with proteasomal inhibitors (Fig. 2C).
p53 Protein Half-life Increases in Response to UPP Inhibitors
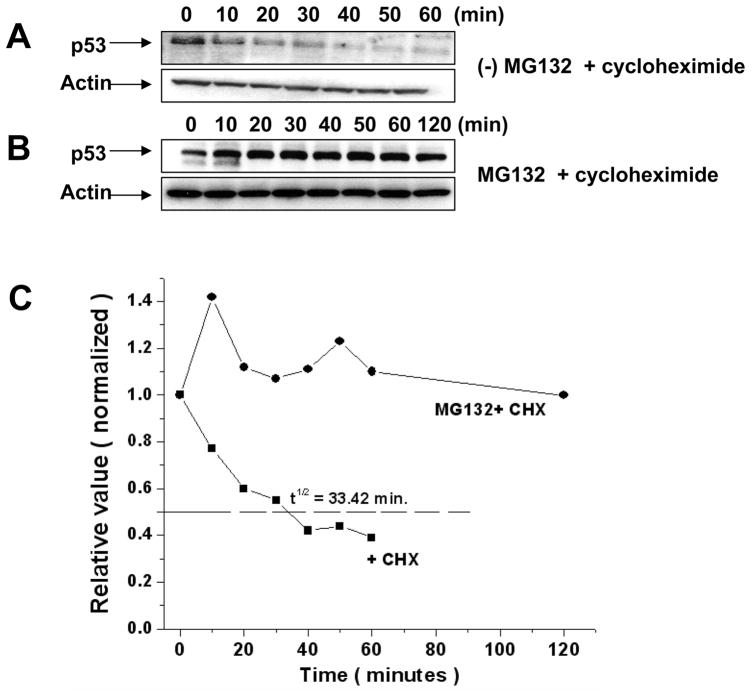
Since the p53 protein is efficiently degraded by the UPP, inhibitors of the UPP such as MG132 are likely to increase the half-life of p53 protein. Therefore the half-life of p53 protein in the 8B20 melanoma cells was investigated. Cells were pretreated with cycloheximide to inhibit de novo protein synthesis, and at the indicated time intervals cells were collected and lysed as described in methods. Resulting immunoblots were probed for the p53. The half-life of the p53 protein in 8B20 cells was found to be approximately thirty three minutes (Fig. 3A & 3C). On the contrary, treatment of the 8B20 cells with both cycloheximide and the UPP inhibitor MG132 led to an increased stability of p53 protein in these cells (Fig. 3B & 3C).
Figure 3. Effect of proteosomal inhibitor MG132 on the stability of wild-type p53 in 8B20 cells.
Exponentially growing 8B20 cells were treated with cycloheximide (50 μg/mL) alone to block the protein synthesis (A) or with MG132 (10 μM) and cycloheximide (50 μg/mL) (B). At the indicated time intervals cell samples were collected and processed as described in the methods. p53 analysis on the immnunoblots was carried out using anti-p53 antibody. β-Actin levels are shown as loading controls. The data are one of three representative experiments. (C). Densitometric analysis of p53 normalized with actin level is shown. Cells were treated with cycloheximide (CHX) in the presence and absence of MG132. Calculated half-life of p53 is shown.
Proteosomal Inhibitors MG132 and Clasto-lactacystin-β-lactone Mediated Stabilization of p53 Is Not Mediated by the Phosphorylation of Serine 15 and 20 Residues of p53
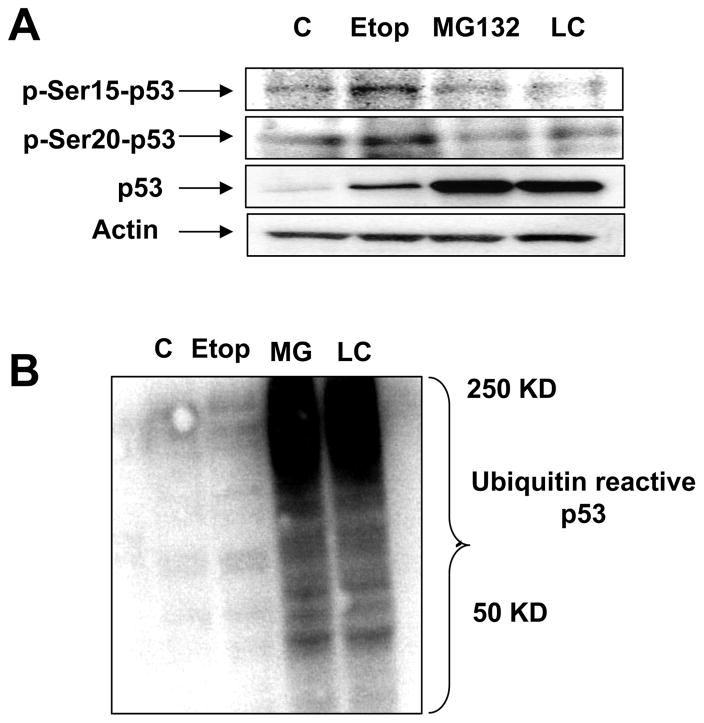
Serine 15 and serine 20 residues are critical for the binding of MDM2, which controls p53 destruction via UPP pathway. DNA damaging agent such as etoposide results in the upregulation of p53 by phosphorylation of p53. In order to investigate whether p53 stabilization by proteasomal inhibitors is mediated by protein phospohorylation, 8B20 cells were treated with either MG132, clasto-lactacystin-β-lactone or etoposide for the indicated durations. Cell lysates were immunoblotted for the phosphorylated forms of Ser-15 and Ser-20 residues of the p53 protein. Immunoblots revealed that the etoposide treatment of the 8B20 melanoma cells led to the increased reactivity of Ser-15 and Ser-20 residues of p53 whereas the treatment with MG132 and clasto-lactacystin-β-lactone did not result in the phosphorylation of Ser-15 and Ser-20 residues of p53 (Fig. 4A). All three treatments resulted in the upregulation of the total p53 protein compared to the untreated samples; however, the amount of total p53 upon the treatment with etoposide was much less compared to the treatment with the proteasomal inhibitors (Fig. 4A). These data indicate that, in 8B20 melanoma cells, p53 is predominantly degraded by the UPP, and that the inhibition of UPP leads to the accumulation of p53 protein.
Figure 4. Differential mechanisms of p53 upregulation by proteosomal inhibitors and DNA damaging drug in 8B20 cells.
8B20 cells were untreated (C: control) or treated with DNA damaging drug Etoposide (Etop) (50 μM) for 24 hours or the proteosomal inhibitors MG132 (10 μM) or clasto-lactacystin-β-lactone (LC) (10 μM) for 6 hours. After the treatment, cells were harvested and the lysates were subjected to SDS-PAGE. (A) Immunoblots were probed for the phosphorylated forms of serine 15 or serine 20 residues of p53 protein. Total p53 levels and β- Actin levels are also shown. (B) Immunoblots were probed with an anti-ubiquitin antibody. The data are one of three representative experiments.
Etoposide Treatment of 8B20 Melanoma Cells Does Not Result in the Inhibition of UPP
Since we found that etoposide treatment resulted in the upregulation of p53 albeit to a lesser extent compared to the proteosomal inhibitors (MG132 and clasto-lactacystin-β-lactone) (Fig. 4A), we further investigated if etoposide treatment led to the inhibition of UPP. When the 8B20 cell lysates were analyzed for the ubiquitin reactive heavy molecular weight proteins, it was found that etoposide treatment of 8B20 cells did not result in the inhibition of UPP, as observed in Fig. 4B.
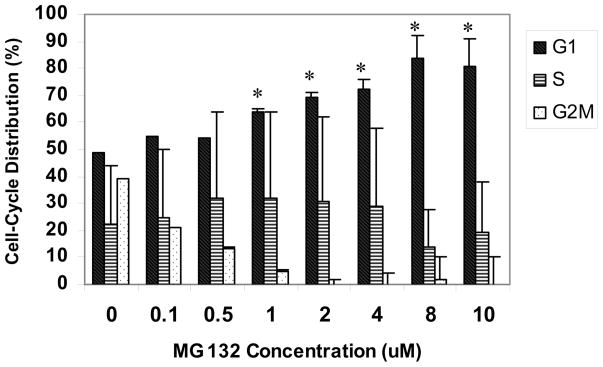
Treatment of 8B20 Cells with MG132 Results in G1 Cell Cycle Arrest
8B20 cells were treated with varying concentrations of MG132 (0–10 μM) for 24 hours. Cells were then washed with PBS and an aliquot of cells was stained with Krishan’s stain at 4°C for 24 hours. Cell cycle distribution was analyzed by the flow cytometry. As shown in Fig. 5, DNA flow cytometric analysis revealed that treatment with MG132 significantly induced G1 phase arrest of the cell cycle in a dose-dependent manner.
Figure 5. Effect of MG132 on the cell cycle in 8B20 cells.
8B20 murine melanoma cells were exposed to varying concentrations of MG132 (0–10 μM) for 24 hours. Cell cycle analysis was performed by flow cytometry as described in Materials and Methods. Percentages of G1, S and G2/M phase are shown. Data show mean ± S.E. (n = 3). The error bars are from one experiment done in triplicate, and the experiments were replicated twice. * p<0.05
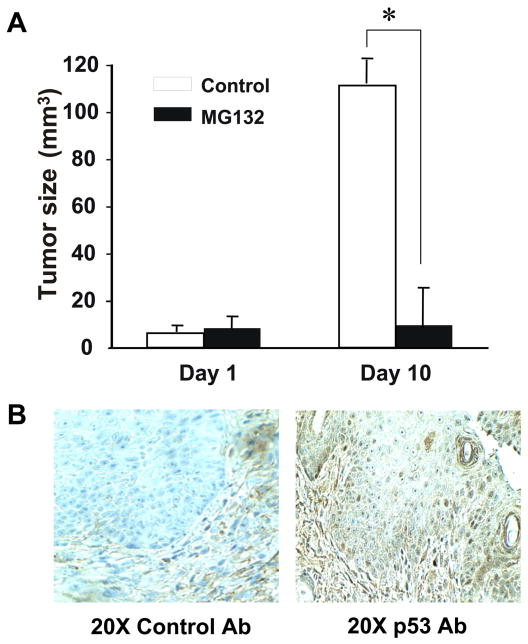
MG132 Treatment Decreases Tumor Growth in vivo
In an attempt to further investigate the effect of proteosomal inhibitor MG132 in controlling the tumor growth, we performed in vivo experiments using a GEMM that carries a melanocyte specific H-rasG12V expression on Ink4a/Arf-deficient background. These mice spontaneously develop cutaneous melanoma. Visible tumors were identified on ear (3 mm in diameter), the mice were either untreated or treated with MG132 at a concentration of 5 μg/Kg of the body weight intraperitoneally daily for 10 days [22]. Whereas continuous tumor growth was observed in the untreated mice, the tumor size remained small in the MG132 treated mice (Fig. 6A). Immunohistochemistry of the MG132-treated tumors showed increased expression of p53 (Fig. 6B).
Figure 6. Effect of the proteosomal inhibitor MG132 on tumor growth in mice.
Mice with melanocyte specific H-ras G12V expression on Ink4a/Arf-deficient background that develop spontaneous melanoma were treated with proteosomal inhibitor MG132 at a concentration of 5 μg/kg of the body weight. Tumor size was followed for ten days. (A) Tumor sizes of MG132-treated or untreated mouse tumors on Day 1 and Day 10 are shown as a comparison. Data show mean ± S.E. (n = 4). The error bars are from one experiment using 4 mice, and the experiments were replicated twice. * p<0.05 compared with the control tumors. (B) Tumors were stained with a control Ig-G or anti-mouse p53 antibody. Staining for p53 is shown in brown. >Effects of proteasomal inhibitors on p53 were investigated both in vivo and in vitro.> Mouse melanoma cells treated with proteasomal inhibitors increased p53 levels. > Increased p53 levels were not mediated by phosphorylation of serines.> MG132 induced p53 resulted in decreased tumor volume.
Discussion
The UPP is multi-catalytic protease machinery responsible for most non-lysosomal intracellular protein degradation [23–25]. Interestingly, multiple pro-oncogenic factors are controlled by the proteasome; those include transcriptional factors, cyclins, CDK inhibitors, cytokines and a series of pro- and anti-apoptotic factors [26]. Therefore, inhibition of proteasome is emerging as a potent cellular tool to selectively kill tumor cells by exploiting their altered genetic background [27,28]. Proteasome inhibitors have been widely used to address mechanistic analyses of protein stability and regulation in a variety of essential cellular processes. Classical aldehyde-based agents such as MG132 have been instrumental in identifying key mediators of apoptosis, cell cycle progression and response to oxidative stress in normal and tumor cells [29].
In the current study, we have demonstrated that the wild-type p53 in 8B20 mouse melanoma cells is a target for degradation by the UPP. The biochemical approach to understand the mechanism of p53 degradation and stabilization of p53 by the UPP inhibitors has demonstrated that p53 in the mouse melanoma cells is targeted to degradation by the UPP but not by other proteases such as cysteine protease or serine protease present in the cells. The build up of p53 protein in cell lines upon inhibition of UPP is in agreement with earlier observations from human non-small cell lung cancer [30], human diploid fibroblasts [31] and human metastatic melanoma cell lines [32]. One of the hallmarks of UPP inhibition is the formation of heavy molecular weight ubiquitin reactive protein upon UPP inhibition. Indeed, we found that UPP inhibition by the proteasomal inhibitors leads to the formation of heavy molecular weight ubiquitin positive protein. Furthermore, immunoprecipitation studies have shown that inhibition of UPP pathway by the proteasomal inhibitors can yield p53 specific ubiquitinated forms. These observations are consistent with earlier publications which demonstrated that other substrates for the UPP pathway e.g. NADPH: Quinone oxidoreductase 1 (NQO1) could also be stabilized by the UPP inhibitors MG132 and clasto-lactacystin-β-lactone, resulting in the ubiquitinated forms of NQO1 [33,34].
Since changes in p53 protein levels have been attributed to increase in protein stability [35–37], we investigated the effect of MG132 on p53 half-life. 8B20 mouse melanoma cells treated with cycloheximide alone were shown to have a half-life of about thirty three minutes, in close agreement with an earlier publication where the half-life of p53 was shown to be thirty minutes [38]. In contrast, when cells were treated with MG132 followed by cycloheximide to block the de novo protein synthesis, p53 half-life became longer than 2 hours. A four fold and eight-fold increase in the half-life of p53 protein upon treatment with MG132 has been reported in human diploid fibroblasts [31] and a mouse keratinocyte cell line [38], respectively.
Changes in the phosphorylation state of p53 are important in leading to its stabilization and activation [35,36]. For example, DNA damage leads to post-translational modifications such as phosphorylation of p53, resulting in its stabilization and activation [39]. Phosphorylation occurs at serines 6, 9, 15, 20, 33, 37 and 392 and threonine 18 after the DNA damage [40–45]. When 8B20 mouse melanoma cells were treated with UPP inhibitors, no significant changes in the phosphorylation of p53 Ser-15 and Ser-20 residues were observed, while treatment of the same cells with a DNA damaging agent etoposide resulted in the phosphorylation of Ser-15 and Ser-20 residues of the p53 protein. The phosphorylation of Ser-15 and Ser-20 residues on the p53 led to a marginal stabilization of the total p53 protein, albeit to a much lesser extent than the UPP inhibitors. Further analysis of the p53 protein treated with UPP inhibitors and etoposide showed that etoposide treatment does not result in the inhibition of the UPP pathway, indicating that the mechanism of p53 stabilization by DNA damaging agents is different from that of the UPP inhibitors.
The treatment of 8B20 mouse melanoma cells with MG132 led to the growth arrest in the G1 phase of the cell cycle. Furthermore, when the 8B20 mouse melanoma cells were treated with MG132 for 24 hours, we observed cell death in a dose dependent manner, using a Vi-cell cell viability counter for the trypan blue staining (data not shown). It is possible that the growth arrest and cell death might have been in part as a result of p53-induction by the proteasomal inhibitor MG132. The cell cycle in eukaryotes is regulated by CDKs. The cyclin, members of the cell cycle regulators, binds to and activates CDKs. Sequential formation, activation and subsequent inactivation of CDKs and cyclins are critical for the control of cell cycle [46]. p53 activates the expression of CDK inhibitors such as p21WAF1/CIP1 [47] and/or downregulates Cdc25A, a phosphatase that activates CDK2, a kinase essential for G1 to S transition [48], resulting in cell cycle arrest in the G1 stage. P53 also mediates apoptosis through a pathway involving bax activation, cytochrome c release and caspase-9 activation [49]. MG132 has been shown to exert similar effects in gastric carcinoma cells and A549 lung cancer cells [50].
Furthermore, the above in vitro findings were confirmed by in vivo studies using a GEMM with melanocyte specific mutant H-ras, deletion of CDKN2A and wild-type p53. While melanoma tumors continued to grow without treatment, those with the proteasomal inhibitor (MG132) treatment showed little growth, indicating a therapeutic potential of these inhibitors for tumor inhibition. Upregulation of p53 in the tumor tissues treated with the proteasomal inhibitor suggests the important role of p53 stabilization in controlling the tumor growth.
Taken together, we used a GEMM of melanoma to decipher the mechanisms of action and assess the efficacy of proteasomal inhibitors in melanoma. The proteasome is an ubiquituous enzyme complex that plays a critical role in the degradation of many proteins involved in cell cycle regulation, apoptosis and angiogenesis. Since these pathways are fundamental for cell survival and proliferation, particularly in cancer cells, the inhibition of proteasome is an attractive potential anticancer-therapy which needs to be fully exploited in the treatment of various cancers including melanoma.
Acknowledgments
We greatly appreciate Dr. Helge Riemann (Department of Dermatology, UCD, Aurora, CO) for his help with the analysis of tumor data, and Dr. Yiqun Shellman (Department of Dermatology, UCD, Aurora, CO) for helpful discussions. AA was supported by a training grant from NIH (5 T32 AR007411-25). A part of this study was also supported by a grant from Skin Cancer Foundation (AA & MF).
Abbreviations
- UPP
ubiquitin proteasomal pathway
- MG132
Z-Leu-Leu-Leu-CHO
- LC
clasto-lactacystin-β-lactone
- E-64
trans-epoxysuccinyl-L-leucylamido-(4-guanidino)butane
- PMSF
phenylmethylsulfonyl fluoride
- PBS
phosphate-buffered saline
- RIPA
radioimmune precipitation
- SDS-PAGE
sodium dodecyl sulphate polyacrylamide gel electrophoresis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yusuf N, Irby C, Katiyar SK, Elmets CA. Photodermatol Photoimmunol Photomed. 2007;23:48–56. doi: 10.1111/j.1600-0781.2007.00262.x. [DOI] [PubMed] [Google Scholar]
- 2.Reagan-Shaw S, Afaq F, Aziz MH, Ahmad N. Oncogene. 2004;23:5151–5160. doi: 10.1038/sj.onc.1207666. [DOI] [PubMed] [Google Scholar]
- 3.Katiyar SK, Afaq F, Azizuddin K, Mukhtar H. Toxicol Appl Pharmacol. 2001;176:110–117. doi: 10.1006/taap.2001.9276. [DOI] [PubMed] [Google Scholar]
- 4.Anwar A, Mallikarjuna Gu, Brady S, Qamar L, Behbakht K, Shellman YG, Agarwal R, Norris DA, Horwitz LD, Fujita M. Photochemistry and Photobiology. 2008;84:477–483. doi: 10.1111/j.1751-1097.2007.00288.x. [DOI] [PubMed] [Google Scholar]
- 5.Royods JK, Lacopetta B. Cell Death Differ. 2006;13:1017–1026. doi: 10.1038/sj.cdd.4401913. [DOI] [PubMed] [Google Scholar]
- 6.Vousden KH, Lane DP. Nat Rev. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 7.Steele RJ, Thompson AM, Hall PA, Lane DP. Br J Surg. 1998;85:1460–1467. doi: 10.1046/j.1365-2168.1998.00910.x. [DOI] [PubMed] [Google Scholar]
- 8.Stewart ZA, Ietenpol JA. Chem Res Toxicol. 2001;14:243–263. doi: 10.1021/tx000199t. [DOI] [PubMed] [Google Scholar]
- 9.Vousden KH, Lu XX. Nature Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 10.Satyamoorthy K, Chehab NH, Waterman MJ, Lein MC, El-Deiry WS, Herlyn M, Halazonetis TD. Cell Growth Differ. 2000;11:467–474. [PubMed] [Google Scholar]
- 11.Weiss J, Schwechheimer K, Cavenee WK, Herlyn M, Arden KC. Int J Cancer. 1993;54:693–699. doi: 10.1002/ijc.2910540427. [DOI] [PubMed] [Google Scholar]
- 12.Haluska FG, Hodi FS. J Clin Oncol. 1998;16:670–682. doi: 10.1200/JCO.1998.16.2.670. [DOI] [PubMed] [Google Scholar]
- 13.Kichina JV, Rauth S, Das Gupta TK, Gudkiv AV. Oncogene. 2003;22:4911–4917. doi: 10.1038/sj.onc.1206741. [DOI] [PubMed] [Google Scholar]
- 14.Smalley KS, Contractor R, Haas NK, Kulp AN, Atilla-Gokcumen GE, Williams DS, Bregman H, Flaherty KT, Soengas MS, Meggers E, Herlyn M. Cancer Res. 2007;67:209–217. doi: 10.1158/0008-5472.CAN-06-1538. [DOI] [PubMed] [Google Scholar]
- 15.Chin L. Nat Rev Cancer. 2003;3:559–570. doi: 10.1038/nrc1145. [DOI] [PubMed] [Google Scholar]
- 16.Sharpless NE, Kannan K, Xu J, Bosenberg MW, Chin L. Oncogene. 2003;22:5055–5059. doi: 10.1038/sj.onc.1206809. [DOI] [PubMed] [Google Scholar]
- 17.Hupp TR, Lane DP, Ball KL. Biochem J. 2000;352:1–17. [PMC free article] [PubMed] [Google Scholar]
- 18.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 20.Krishan A. J Cell Biol. 1975;66:188–193. doi: 10.1083/jcb.66.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Thor A, Shtivelman E, Cao Y, Tu G, Heath TD, Debs RJ. J Biol Chem. 1999;274:13338–13344. doi: 10.1074/jbc.274.19.13338. [DOI] [PubMed] [Google Scholar]
- 22.Bonuccelli G, Sotgia F, Schubert W, Park DS, Frank PG, Woodman SE, Insabato L, Cammer M, Minetti C, Lisanti MP. Am J Pathol. 2003;163:1663–1675. doi: 10.1016/S0002-9440(10)63523-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glickman MH, Ciecanhover A. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 24.Bashir T, Pagano M. Adv Cancer Res. 2003;88:101–144. doi: 10.1016/s0065-230x(03)88305-7. [DOI] [PubMed] [Google Scholar]
- 25.Liu CW, Corby GN, Demartino MJ, Thomas PJ. Science. 2003:408–411. doi: 10.1126/science.1079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jesenberger V, Jentsch S. Nat Rev Mol Cell Biol. 2002;3:112–121. doi: 10.1038/nrm731. [DOI] [PubMed] [Google Scholar]
- 27.Adams J. Nat Rev Cancer. 2004;4:349–360. doi: 10.1038/nrc1361. [DOI] [PubMed] [Google Scholar]
- 28.Rajkumar SV, Richardson PG, Hideshima T, Anderson KC. J Clin Oncol. 2005;23:630–639. doi: 10.1200/JCO.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 29.Pickart CM, Cohen RE. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- 30.Fernández Y, Miller TP, Denoyelle C, Estaban JA, Tang WH, Bengston AL, Soengas MS. J Biol Chem. 2006;281:1107–1117. doi: 10.1074/jbc.M511607200. [DOI] [PubMed] [Google Scholar]
- 31.Maki CG, Huibregtse JM, Howley PM. Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- 32.Amiri KI, Horton LW, LaFleur BJ, Sosman JA, Richmond A. Cancer Res. 2004;64:4912–4918. doi: 10.1158/0008-5472.CAN-04-0673. [DOI] [PubMed] [Google Scholar]
- 33.Anwar A, Siegel D, Kepa JK, Ross D. J Biol Chem. 2002;277:14060–14067. doi: 10.1074/jbc.M111576200. [DOI] [PubMed] [Google Scholar]
- 34.Anwar A, Dehan D, Siegel D, Kepa JK, Tang LJ, Pietenpol JA, Ross D. J Biol Chem. 2003;278:10368–10373. doi: 10.1074/jbc.M211981200. [DOI] [PubMed] [Google Scholar]
- 35.Liu M, Dhanwada KR, Birt DF, Hecht S, Pelling JC. Carcinogenesis. 1994;15:1089–1092. doi: 10.1093/carcin/15.6.1089. [DOI] [PubMed] [Google Scholar]
- 36.Chernov MV, Ramana CV, Adler VV, Stark GR. Proc Natl Acad Sci USA. 1998;95:2284–2289. doi: 10.1073/pnas.95.5.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McVean M, Xiao H, Isobe K, Pelling JC. Carcinogenesis. 2000;21:633–639. doi: 10.1093/carcin/21.4.633. [DOI] [PubMed] [Google Scholar]
- 38.Bean LJ, Stark GR. Oncogene. 2001;20:1076–10784. doi: 10.1038/sj.onc.1204204. [DOI] [PubMed] [Google Scholar]
- 39.Higashimoto Y, Saito S, Tong XH, Hong A, Sakaguchi K, Apella E, Anderson CW. J Biol Chem. 2000;275:23199–23203. doi: 10.1074/jbc.M002674200. [DOI] [PubMed] [Google Scholar]
- 40.Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Apella E. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shieh SY, Ikeda M, Taya Y, Prives C. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 42.Kapoor M, Lazano G. Proc Natl Acad Sci USA. 1998;95:2834–2837. doi: 10.1073/pnas.95.6.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu H, Taya Y, Ikeda M, Levine AJ. Proc Natl Acad Sci USA. 1998;95:6399–6402. doi: 10.1073/pnas.95.11.6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waterman MJ, Stavridi ES, Waterman JL, Halazonetis TD. Nat Genetics. 1998;19:175–178. doi: 10.1038/542. [DOI] [PubMed] [Google Scholar]
- 45.Kastan MB, Bartek J. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 46.Agarwal ML, Taylor WR, Stark GR. Proc Natl Acad Sci USA. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rother K, Kirschner R, Sanger K, Boglig L, Mossner J, Engeland K. Oncogene. 2007;26:1949–1953. doi: 10.1038/sj.onc.1209989. [DOI] [PubMed] [Google Scholar]
- 48.Akhtar RS, Geng Y, Klocke BJ, Roth KA. Cell Death Differ. 2006;13:1727–1739. doi: 10.1038/sj.cdd.4401879. [DOI] [PubMed] [Google Scholar]
- 49.Zhang W, Tong Q, Li S, Wang X, Wang Q. Cancer Invest. 2008;26:1032–1036. doi: 10.1080/07357900802104997. [DOI] [PubMed] [Google Scholar]
- 50.Zhao P, Zhong W, Ying X, Yuan Zo, Fu J, Zhou Z. Toxicology. 2008;250:39–46. doi: 10.1016/j.tox.2008.05.016. [DOI] [PubMed] [Google Scholar]