Abstract
Buforin II (BF2) is a histone-derived antimicrobial peptide that causes cell death by translocating across membranes and interacting with nucleic acids. It contains one proline residue critical for its function. Previous research found that mutations replacing proline lead to decreased membrane translocation and antimicrobial activity as well as increased membrane permeabilization. This study further investigates the role of proline in BF2’s antimicrobial mechanism by considering the effect of changing proline position on membrane translocation, membrane permeabilization, and antimicrobial activity. For this purpose, four mutants were made with proline substitution (P11A) or relocation (P11A/G7P, P11A/V12P, P11A/V15P). These mutations altered the amount of α-helical content. Although antimicrobial activity correlated with the α-helical content for the peptides containing proline, membrane translocation did not. This observation suggests that factors in BF2’s bactericidal mechanism other than translocation must be altered by these mutations. To better explain these trends we also measured the nucleic acid binding and membrane permeabilization of the mutant peptides. A comparison of mutant and wild type BF2 activity revealed that BF2 relies principally on membrane translocation and nucleic acid binding for antimicrobial activity, although membrane permeabilization may play a secondary role for some BF2 variants. A better understanding of the role of proline in the BF2 antimicrobial mechanism will contribute to the further design and development of BF2 analogues. Moreover, since proline residues are prevalent among other antimicrobial peptides, this systematic characterization of BF2 provides general insights that can promote our understanding of other systems.
Keywords: buforin II, histone-derived antimicrobial peptide, proline, membrane translocation, nucleic acid binding
1. INTRODUCTION
Antimicrobial peptides have received increased attention in recent years due to their potential therapeutic applications [25]. Buforin II (BF2) is a 21 amino acid, cationic antimicrobial peptide derived from the naturally occurring peptide buforin I [20]. BF2 shares complete sequence identity with histone H2A and is the best studied histone-derived antimicrobial peptide (HDAP) [4]. While many antimicrobial peptides target the bacterial cell membrane [9, 35], BF2 is part of a growing group of peptides that appear to have an intracellular target [6]. In particular, BF2 is thought to cause cell death by translocating across the cell membrane and binding to nucleic acids [19, 34].
When associated with a lipid membrane, the structure of BF2 consists of a N-terminal random coil region and a C-terminal α-helix (Val 12 to Lys 21). The helix is distorted by a proline hinge (Pro 11), leading to the formation of an extended helix (Gly 7 – Pro 11) [11, 36]. Proline residues are prevalent in several AMPs [15, 27], such as peptides in the family of proline-rich antimicrobial peptides (PRAPs) [18]. Moreover, mutating proline residues markedly alters the properties of several antimicrobial peptides [1, 3, 12, 13, 22, 26, 28, 30, 31, 37, 38]. This is also true for BF2, as previous studies demonstrate that its sole proline plays a critical role in its function, affecting the peptide’s membrane translocation, membrane permeabilization, and antimicrobial activity [10, 11, 21]. For example, proline substitution in the BF2 analogue P11A significantly reduced translocation and increased membrane permeabilization [11, 21]. Similarly, studies have shown that P11A and P11L mutations decrease the antimicrobial activity of BF2 [11, 21]. While these studies have highlighted the importance of the presence of proline for BF2 function, it is unknown how sensitive BF2 function is to the position of proline within the peptide.
In this study, we investigate the effect of proline’s position on the antimicrobial mechanism of BF2. For this purpose, we created three proline relocation mutants, P11A/G7P, P11A/V12P and P11A/V15P (Table 1). Two of our peptides moved the proline one helical turn towards the N-terminus (P11A/G7P) or C-terminus (P11A/V15P), which should provide a respectively longer or shorter α-helical region than wild type BF2 while maintaining the presence of a proline residue. Another peptide (P11A/V12P) moved the proline by a single residue, placing it on a different helical face while having a minimal effect on α-helicity. This variant was particularly interesting to consider since the location of proline on a particular helical face is important for the antimicrobial activity of some peptides [28].
TABLE 1.
Sequences of peptides used in this study and results from radial diffusion assays. Residues modified from the wild type BF2 sequence are highlighted as bold underline. The average diameter of clearance of E. coli in radial diffusion assays. The average diameter for each peptide was taken over 48 different wells on plates from at least two separate experiments with uncertainty expressed as standard error.
| PEPTIDE | PEPTIDE SEQUENCE | DIAMETER OF CLEARANCE (MM) |
|---|---|---|
| BF2 | TRSSRAGLQFPVGRVHRLLRK | 11.5 ± 0.2 |
| P11A | TRSSRAGLQFAVGRVHRLLRK | 10.05 ± 0.09 |
| P11A/G7P | TRSSRAPLQFAVGRVHRLLRK | 12.0 ± 0.2 |
| P11A/V12P | TRSSRAGLQFAPGRVHRLLRK | 10.0 ± 0.1 |
| P11A/V15P | TRSSRAGLQFAVGRPHRLLRK | 9.1 ± 0.2 |
USING THESE MUTANTS, WE WERE ABLE TO OBTAIN FURTHER INSIGHT INTO THE ROLE OF THE PROLINE KINK IN BF2 MEMBRANE TRANSLOCATION, MEMBRANE PERMEABILIZATION AND ANTIMICROBIAL ACTIVITY. IN ORDER TO BETTER EXPLAIN TRENDS IN ANTIMICROBIAL ACTIVITY, WE ALSO CONSIDERED DNA BINDING AS AN ADDITIONAL PROPERTY IN BF2’S ANTIMICROBIAL MECHANISM. TOGETHER, THESE RESULTS IMPLY THAT BF2’S ANTIMICROBIAL MECHANISM RELIES PRINCIPALLY ON MEMBRANE TRANSLOCATION AND NUCLEIC ACID BINDING.
2. MATERIALS AND METHODS
2.1 MATERIALS
Regular and biotinylated wild type BF2, P11A, P11A/G7P, P11A/V12P, and P11A/V15P peptides were synthesized by GenScript (Piscataway, NJ) or EZ-Biolabs (Westfield, IN) and obtained at >95% purity. Other chemicals were obtained from Sigma, unless otherwise noted.
2.2 LARGE UNILAMELLAR VESICLE (LUV) PREPARATION AND CONCENTRATION DETERMINATION
Lipid cake composed of 75% POPC and 25% POPG was desiccated overnight and rehydrated in buffer. For translocation measurements, TBS buffer (0.05 M Tris chloride, 0.1 M NaCl, pH 7.4) was used. For circular dichroism measurements, HEPES buffer (10 mM HEPES, 45 mM NaCl, 1 mM EDTA, pH 7.4) was used. The vortexed suspension was subjected to five freeze-thaw cycles using liquid nitrogen and extruded through 100 nm polycarbonate filters 21 times. The LUV concentration was determined in duplicate or triplicate by phosphorous analysis (WWW.SIGMAALDRICH.COM).
2.3 CIRCULAR DICHROISM SPECTROSCOPY
CD spectra of each peptide were collected on a Jasco J-810 using a 1-mm path-length quartz cell. The spectra of 25 µM solutions of each peptide in phosphate buffer (10 mM sodium phosphate, 45 mM NaCl, 1 mM EDTA, pH 7.4) were measured in the presence of 3:1 POPC:POPG LUVs (1 mM). For each sample, ten scans from 250 to 195 nm were averaged at 25 °C.
2.4 TRANSLOCATION MEASUREMENT BY BIOTIN-AVIDIN INTERACTION ASSAY
Determination of peptide translocation into vesicles was performed analogous to the method to measure histone translocation developed by Loyter and co-workers [7, 23, 24]. Biotinylated peptide solution (5 µM) was incubated with 3:1 POPC: POPG lipid vesicles (10 mM) in a total volume of 200 µL for 1h at 25 °C to allow peptide translocation. To remove untranslocated peptide, vesicles were pelleted by centrifugation at 200,000 g for 20 min using a Beckman airfuge and resuspended in 133 µL avidin (2 mg/mL in TBS) to block untranslocated surface peptide. Excess avidin was blocked by centrifugation and pellet resuspension in 133 µL biocytin (5 mg/mL in TBS). Following 15 min incubation, the sample was pelleted again and resuspended in 200 µL of 1% Triton (w/v). 10 µl of this suspension was added to MaxiSorp plate wells (Nalge Nunc, Rochester, NY) containing 190 µl of Triton gelatin buffer (.2% Triton (w/v), .05% gelatin (w/v) in TBS). The plates were incubated overnight at 4 °C to allow protein binding to the well bottom. After three washes using TBS buffer, each well was filled with 200 µL Triton gelatin buffer and incubated for 5 min at 37 °C to block the unoccupied space on the bottom surface of the well. Then, following another three washes with TBS buffer, each well was filled with 200 µL of Tween gelatin solution (0.2% Tween (w/v, Fisher), 0.1% gelatin (w/v) in TBS) containing alkaline phosphatase-streptavidin (0.002 mg/mL) and incubated for 1 h at 37 °C.. After three more washes with TBS buffer, each well was filled with 200 µL phosphatase substrate solution (phosphatase substrate 1.1 mg/ml, 0.1 M diethanolamine, 5 mM MgCl2, in TBS, pH 9.5). The concentration of colored product of phosphatase enzymatic activity was measured using a Molecular Devices SpectraMax 340PC384 Microplate Spectrophotometer at the absorption wavelength of 405 nm.
In addition to the experimental condition, two control conditions were used to quantify all membrane-associated peptide and measure the background absorbance of the intact vesicle containing translocated peptides. To quantitate all membrane-associated peptide, including peptide that did not translocate to the inside of the vesicle, an identical peptide/vesicle sample was exposed to TBS without avidin before vesicle disruption. To quantitate the background absorbance of an undisrupted vesicle solution, another identical peptide/vesicle sample was not exposed to Triton. All three of these conditions were performed for each peptide trial. The percent of membrane associated peptide that translocated was determined according to Equation 1, where A is equal to the absorbance of the experimental sample, ANA is the absorbance of the sample without avidin, and ANT is the absorbance of sample without Triton. To control for any potential differences between vesicle preparations, each trial measured percent translocation for wild type and a mutant peptide using an identical vesicle solution. Thus, results are presented as percent translocation relative to wild type BF2. A minimum of three independent trials was performed for each sample.
| (1) |
2.5 BACTERIAL CULTURE PREPARATION
Frozen Top10F E. coli cells transformed with the pET45b plasmid to confer ampicillin resistance were grown overnight (14–16 h) in Luria Broth (LB) containing ampicillin (25 l/mL). Following 1:1000 dilution in 50 mL of LB, the culture was incubated at 37 °C until reaching mid-log phase. The resulting culture was centrifuged for 10 min at 1000 g at 4°C, washed once with buffer, pelleted again, and resuspended in phosphate buffer (10 mM sodium phosphater/45 mM NaCl/1mM EDTA, pH 7.4) to a final concentration of 1.25×108 CFU/mL.
2.6 RADIAL DIFFUSION ASSAY
The antibacterial activity of BF2 variants was measured using the radial diffusion assay of Lehrer and co-workers [29]. 4x106 CFU E. coli cells were mixed by vortexing with 10 mL of molten underlay agar (1:100 TSB, 1% agarose w/v, 10 mM sodium phosphate, pH 7.4). The bacteria - containing agar was poured into a Petri dish and allowed to gel. Wells were formed in the solid media using a pipette attached to a bleach trap, and 2.5 µL of sterile peptide solutions (4x10−4 M) were added to wells. After incubating for 3 h at 37 °C, the plates were covered with a layer of overlay agar (6% w/v TSB, 1% w/v agarose) and incubated overnight at 37 °C. After overnight incubation, the diameter of the zone without bacterial growth around each well was measured, and measurements from 48 different wells on plates from at least two separate experiments were averaged for each peptide.
2.7 MEMBRANE PERMEABILITY MEASUREMENT BY PI UPTAKE ASSAY
The fluorescence intensity of a 2 mL E. coli (1.25×108 CFU/mL) culture containing 1 mg/mL propidium iodide (PI) was measured using a Varian Cary Eclipse fluorescence spectrophotometer (excitation wavelength: 535 nm; emission wavelength: 617 nm) and normalized to 0% fluorescence. Peptide was added to the mixture to achieve 2 µM effective peptide concentration. The permeability strength of each proline mutant was quantitated using Equation 2, where F5min is the fluorescence intensity induced 5 minutes after peptide addition. Each peptide was tested in at least three independent experiments using bacteria from different overnight cultures.
| (2) |
2.8 DNA BINDING MEASUREMENT BY FLUORESCENT INTERCALATOR ASSAY
The DNA binding strength of BF2 variants was measured using a fluorescent intercalator displacement assay [8, 33]. The fluorescence intensity of STE buffer (10 mM Tris, 50 mM NaCl, 1 mM EDTA, pH 8.0) was measured using a Varian Cary Eclipse fluorescence spectrophotometer (excitation wavelength: 509 nm; emission wavelength: 527 nm) and normalized to 0% relative fluorescence. The dsDNA (IDT, Coralville, IA) used was derived from DNA adjacent to the histone H2A region identical to the BF2 sequence in a histone•DNA crystal structure (AAATACACTTTTGGT) [14]. This dsDNA sequence was used in previous studies of BF2 DNA binding [34]. A 1 mL mixture of dsDNA (1.10 µM base pair concentration) and thiazole orange (0.55 µM) was made in STE buffer. Maximum fluorescence intensity was measured 5 min after DNA addition. The solution was then titrated with a 78 µM peptide solution. The decrease in fluorescence was measured 5 min after each addition of peptide until the fluorescence intensity decreased to ~50% of the maximum fluorescence (C50). The peptide concentration at which fluorescent intensity was decreased to half was extrapolated and the peptide’s relative DNA binding constant (K) was calculated by taking its reciprocal. The DNA binding constant was measured in at least three independent experiments for each peptide.
3. RESULTS
3.1 CD SPECTROSCOPY
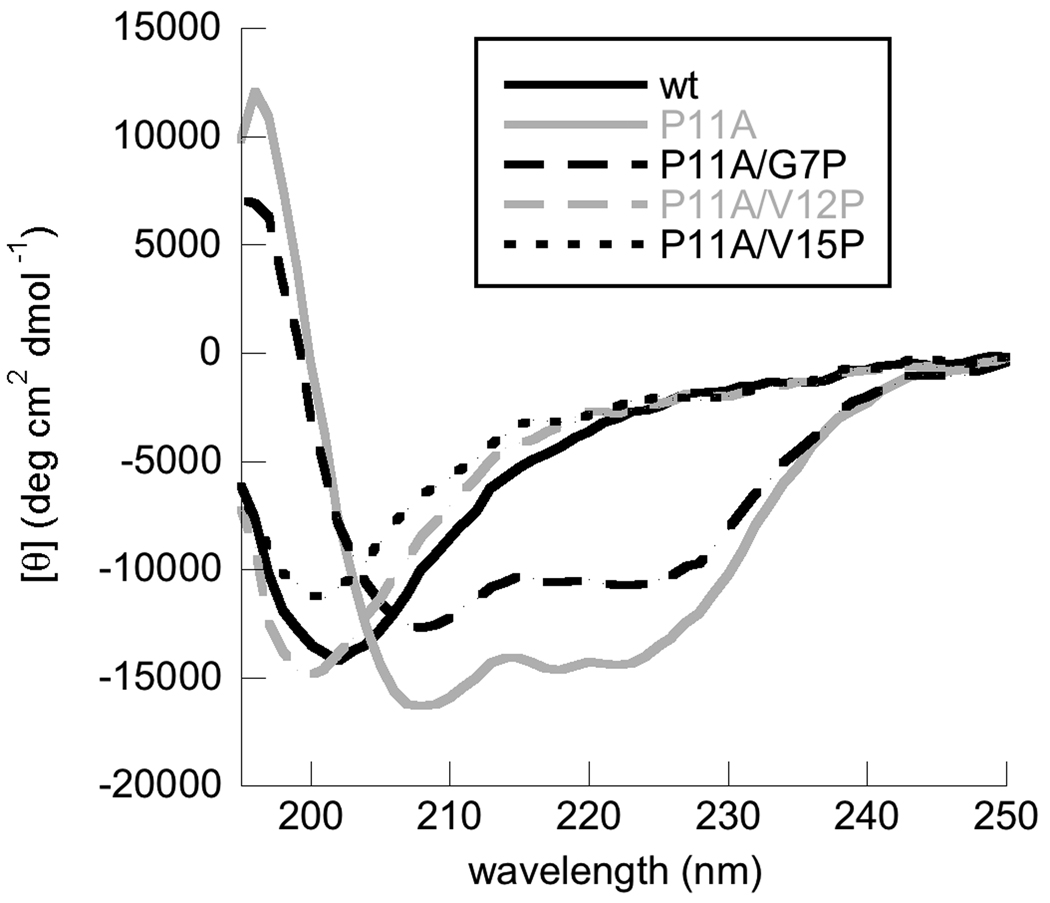
We initially measured the secondary structure of our peptides associated with 1 mM 3:1 POPC:POPG lipid vesicles using CD spectroscopy (Fig. 1). CD spectra for wildtype and P11A were consistent with those obtained previously for these peptides in the presence of pure POPG vesicles [11]. Previously, researchers observed that the spectrum for wild type BF2 in the presence of anionic vesicles is identical to its spectrum in the presence of 1:1 TFE:water [11]. An NMR structure in this TFE:water mixture showed that the peptide assumes a C-terminal helical region with a proline kink under these conditions [36]. As seen in past work [11], our spectra for P11A had significantly more helical structure than wild type. The spectra for our other proline relocation mutants followed our expected trend. Moving proline towards the N-terminus (P11A/G7P) increased the amount of α-helicity relative to wild type. Conversely, moving proline towards the C-terminus (P11A/V15P) led to an apparent decrease in the amount of α-helicity relative to wild type. Moving proline by a single residue (P11A/V12P) did not have a significant effect on secondary structure.
Fig. 1.
Circular dichroism (CD) spectra of wild type (black solid), P11A (gray solid), P11A/G7P (black dashed), P11A/V12P (gray dashed), and P11A/V15P (black dotted) BF2 peptides in the presence of 1 mM POPC:POPG (3:1) vesicles. The concentration of all peptides was 25 µM.
3.2 ANTIMICROBIAL ACTIVITY
Antimicrobial activities of the BF2 proline mutants were measured and compared to the wild type peptide using a radial diffusion assay. Consistent with previous studies [11], the proline substitution mutant (P11A) exhibited a lowered antimicrobial activity despite containing a longer helical region. However, among the variants with a proline present (wild type, P11A/G7P, P11A/V12P, and P11A/V15P) there was a correlation between bactericidal ability and helical content (Table 1). This observation was similar to that in previous work using truncation mutants of BF2 [21].
3.3 MEMBRANE TRANSLOCATION
Membrane translocation into lipid vesicles was measured using an ELISA-based assay adapted from Rosenbluh et al. [23]. Although this method requires the use of biotinylated peptide, previous studies have shown that BF2 with a biotin at the N-terminus maintains its ability to readily translocate into bacterial cells [21]. As observed in past studies [11], removing proline entirely with a P11A mutant significantly reduced translocation relative to wild type (Table 2). Replacing proline a single residue away from its wild type position in P11A/V12P effectively restored the normal translocation behavior of BF2 (Table 2). Relocating proline by one turn in either the N- (P11A/G7P) or C-terminal (P11A/V15P) direction partially restored wild type translocation.
TABLE 2.
Relative percent translocation of membrane associated peptides into POPC:POPG (3:1) lipid vesicles, relative percent fluorescence increase in propidium iodide (PI) uptake experiments with E. coli, and relative DNA binding constants (K) measured using a fluorescent intercalator displacement assay. Since all values reported for proline mutants are relative to wild type, the relative values for wild type BF2 were 1. For all experiments, averages were obtained over at least three independent experiments for each peptide, with uncertainty expressed as standard error.
| RELATIVE PERCENT TRANSLOCATION |
RELATIVE MEMBRANE PERMEABILIZATION |
RELATIVE DNA BINDING |
|
|---|---|---|---|
| P11A | 0.36 ± 0.05 | 2.6 ± 0.5 | 1.15 ± 0.11 |
| P11A/G7P | 0.66 ± 0.12 | 2.8 ± 0.7 | 1.83 ± 0.14 |
| P11A/V12P | 1.04 ± 0.13 | 1.3 ± 0.4 | 0.91 ± 0.03 |
| P11A/V15P | 0.76 ± 0.08 | 1.5 ± 0.4 | 1.03 ± 0.04 |
While we observed a correlation between antimicrobial activity and helicity for the relocation mutants (P11A/G7P, P11A/V12P, and P11A/V15P), the mutants’ translocation abilities do not correlate with their helical content. Removing proline drastically reduces translocation, but relocating proline by one turn towards either the N- or C- terminus (P11A/G7P and P11A/V15P) results in intermediate translocation that is less effective than BF2 but greater than P11A (Table 2). Relocating proline to a different helical face one residue away from its original position with P11A/V12P restores the wild type translocation behavior (Table 2), implying that moving proline to a different helical face does not affect translocation. Thus, it appears that having the deformation from proline in a different position in the helix does promote translocation relative to the peptide without a proline residue. However, the wild type peptide appears to have the deformation in an ideal position for effective translocation, since moving proline by one helical turn in either direction significantly decreases its ability to cross the vesicle membranes.
Previous results had showed a correlation between cell entry and α-helical propensity in truncated analogs of BF2 [21]. However, our results show that in full-length BF2 the position of the proline is more important than the overall amount of α-helical structure for translocation since P11A/G7P and P11A/V15P have quite similar translocation ability despite the fact that P11A/G7P has significantly more helical structure. The differences in cell entry previously observed upon truncation may have resulted from both a relative repositioning of the helical deformation in the peptide and the removal of C-terminal residues that were apparently important for translocation.
3.4 MEMBRANE PERMEABILIZATION
The ability of the BF2 proline mutants to cause membrane permeabilization was determined using a propidium iodide (PI) uptake assay in which an E. coli culture was incubated with the fluorescent intercalator PI and subsequently exposed to peptides. The assay utilizes the fluorescence increase caused by PI-DNA interaction that occurs following peptide-induced membrane leakage to quantitate the membrane permeabilization strengths of the BF2 mutants. PI has been used to measure membrane permeabilization in previous studies of BF2 [21]. While one potential concern with this method is that BF2 could potentially compete with PI for DNA binding, this competition should not occur to an appreciable extent in our experiments because of the high DNA binding constant of PI and the presence of PI at a 750-fold higher concentration than peptide.
As observed in past studies [11], proline removal in the P11A mutant significantly increased leakage relative to wild type (Table 2). Relocating proline one residue away from its original position with P11A/V12P restored the wild type leakage behavior (Table 2). Extending the helical region by one turn in P11A/G7P effectively increases leakage activity to a level similar to that of P11A. However, reducing the helical content by one turn in P11A/V15P has relatively little effect on permeabilization. Thus, it appears that wild type BF2 exhibits a basal level of permeabilization that can be increased by mutations that increase α-helical structure but cannot be decreased by mutations that decrease the helical region. Notably, this basal level of permeabilization is relatively low compared to most membrane-active antimicrobial peptides. For example, previous studies established that BF2 exhibits more than 100-fold weaker membrane leakage than magainin [11], an antimicrobial peptides that relies on membrane permeabilization to induce cell death [16].
3.5 DNA BINDING
The results described in the previous sections show that the mutants’ translocation properties do not fully correlate with their antimicrobial activity. In particular, we had expected that membrane translocation would correlate with antimicrobial activity since translocation is thought to be an integral part of BF2’s antimicrobial mechanism. However, this is not true for all mutants. For example, P11A/G7P exhibits lower translocation despite having achieved comparable antimicrobial performance as wild type BF2 (Tables 1 and 2). The discrepancy between peptide function and antimicrobial activity suggests the involvement of processes other than translocation in BF2’s antimicrobial mechanism. Since past research had shown a relationship between BF2 DNA affinity and antibacterial activity [34], we studied nucleic acid binding as a potential component to the antimicrobial activity of our proline relocation mutants.
The DNA binding strengths of the proline mutants were studied using a fluorescent intercalator displacement (FID) assay [8, 33]. Removing proline with P11A did not dramatically alter DNA binding (Table 2), suggesting the presence of proline is not critical for peptide-DNA interaction. Similarly, proline relocation by one residue (P11A/V12P) or one turn (P11A/V15P) toward the C-terminus did not significantly alter DNA binding (Table 2). However, relocating proline one turn toward the N-terminus in P11A/G7P did increase DNA binding (Table 2). These observations suggest that the nucleic acid binding of BF2 variants is related to their specific conformation and not to overall trends in α-helical structure. Notably, the DNA binding of the buforin mutants did not correlate with their translocation ability, as some mutations only altered one property (e.g. P11A and P11A/V15P) or had an opposite effect on the two properties (e.g. P11A/G7P). As discussed below, this allowed us to observe that the antibacterial activity of BF2 mutants was generally based on a balance of these two properties.
4. DISCUSSION
The ability of BF2 to translocate across lipid membranes without causing significant membrane disruption has led it to become the best studied histone-derived antimicrobial peptide (HDAP) [4]. However, the role of its sole proline residue in translocation is not well understood on the molecular level. The results of this study support the importance of proline for BF2 translocation, as translocation ability was partially rescued by reintroducing a proline residue into a P11A mutant of BF2. Our results also show that the BF2 proline residue is in an apparently ideal region in the peptide sequence to promote translocation, since moving it one turn towards either the N- or C-terminus of the peptide decreases translocation, while moving it a single residue leaves translocation essentially unaltered. Interestingly, MD simulations performed in our group have implied that the deformation caused by proline can promote formation of BF2•lipid hydrogen bonds that could help facilitate translocation [5].
Since the proline mutants considered in this study affect different properties of BF2 (Fig. 2 and Table 3), we can consider whether their antibacterial activity can be explained by the currently accepted BF2 antimicrobial mechanism. In this mechanism, the peptide translocates into the cell and interacts with intracellular nucleic acids (Fig. 2) [19]. If this mechanism is correct, mutations that alter either translocation or nucleic acid binding should alter the antibacterial efficacy of BF2. However, based on the proposed mechanism, one would expect changes in membrane permeabilization to have less of an effect on the antibacterial activity of BF2.
FIG. 2.
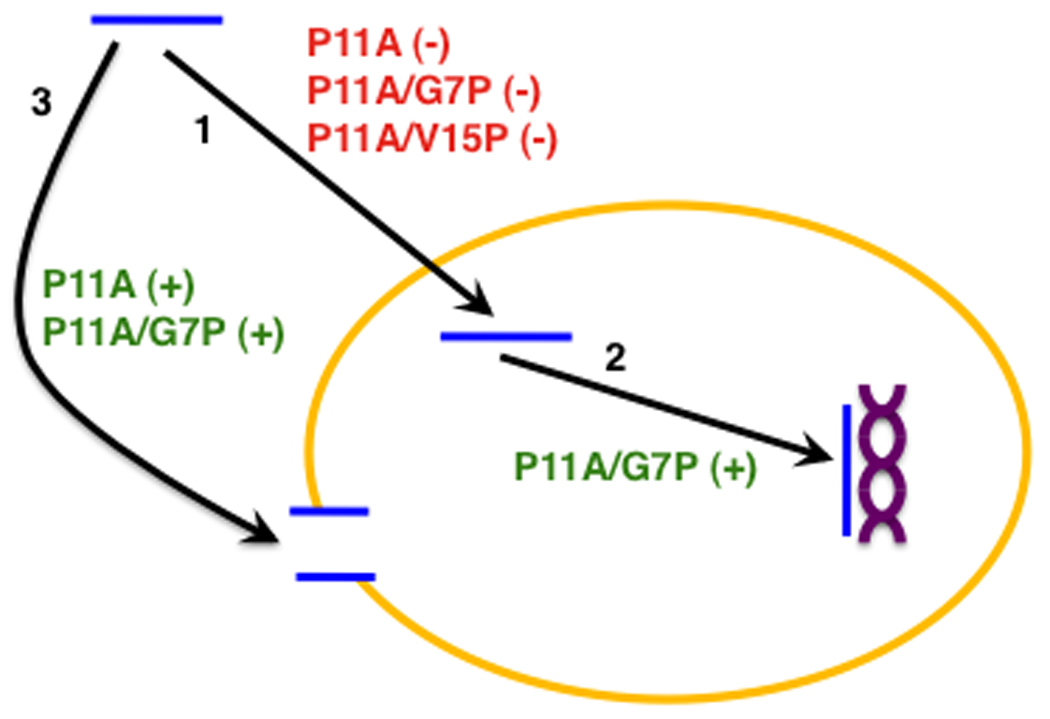
Schematic of the currently accepted antimicrobial mechanism of BF2 in which the peptide (blue) translocates (arrow 1) across the cell membrane (yellow) and interacts (arrow 2) with nucleic acids (purple). As well, some BF2 mutants have an increased ability to induce membrane permeabilization (arrow 3) relative to wild type. The ability of proline mutations considered in this study to increase (green) or decrease (red) the efficiency of each of these processes is noted on the figure.
TABLE 3.
Properties of BF2 proline mutants compared to those of the wild type.
| P11A | P11A/G7P | P11A/V12P | P11A/V15P | |
|---|---|---|---|---|
| TRANSLOCATION | LOWER | LOWER | SIMILAR | LOWER |
| DNA BINDING | SIMILAR | HIGHER | SIMILAR | SIMILAR |
| MEMBRANE PERMEABILIZATION | HIGHER | HIGHER | SIMILAR | SIMILAR |
| ANTIMICROBIAL ACTIVITY | LOWER | SIMILAR | LOWER | LOWER |
The pattern of characteristics observed for our proline mutants is consistent with the currently accepted BF2 mechanism (Table 3 and Fig. 2). A qualitative comparison of the mutant peptide properties with those of BF2 shows that antimicrobial performance principally depends on membrane translocation and DNA binding, since a change in either of these properties leads to a change in antimicrobial activity (Table 3). Conversely, membrane permeabilization seems to play a relatively minor role in the activity of BF2. For example, due to lower membrane translocation, P11A exhibits lower antimicrobial activity despite causing almost twice the membrane permeabilization of wild type (Table 3), indicating that greater membrane permeabilization did not compensate for translocation inefficiency. On the other hand, higher DNA binding does compensate for the lower translocation of P11A/G7P, allowing the mutant to achieve a similar level of antimicrobial performance as BF2 (Table 3). Similarly, P11A/V15P’s lower translocation results in its lower antimicrobial activity, since its DNA binding and membrane permeabilization strengths are similar to that of BF2. Our data does imply that membrane permeabilization may play a secondary role in the activity of some BF2 variants as the greater antimicrobial activity of P11A compared to P11A/V15P could be due to its increased membrane permeabilization (Tables 1 and 2). Nonetheless, our overall results suggest that translocation and DNA binding have dominating influences on the peptides’ overall antimicrobial activity and therefore are likely to constitute the main components of BF2’s antimicrobial mechanism.
The P11A/V12P mutant provides an intriguing exception to these trends. Despite having similar levels of translocation, permeabilization, and DNA binding as the wild type, P11A/V12P nevertheless shows a decreased antimicrobial activity. Interestingly, this peptide places proline on a different helical face. While the helical face on which prolines are located has been observed to affect the activity of other antimicrobial peptides [28], it did not affect any of the properties of BF2 measured in this study. This indicates that there could be other factors affecting BF2’s antimicrobial mechanism besides those considered in this study, which would be interesting to pursue in future work.
While these studies provide important systematic insights into the role of proline in BF2, they can also help researchers consider the role of proline more broadly in antimicrobial peptides. For example, our group recently reported the design of three novel HDAPs that each contain a single proline residue [32], and it will be interesting to consider whether proline plays a similar role in the function of these peptides as it does in BF2. Our results also provide a useful starting point for a more thorough consideration of how the position of proline affects the function of other naturally occurring antimicrobial peptides. This is particularly important since proline residues are prevalent not only in other HDAPs isolated from natural sources, such as hipposin [2], but also in other groups of antimicrobial peptides, such as the proline-rich antimicrobial peptides [15, 18].
Our results also imply that proline relocation may be a useful approach in efforts to engineer the function of known peptides that contain proline. In BF2, moving the proline residue apparently allows the "tuning" of membrane properties, and a similar effect may be seen in other peptides. This relocation of proline residues may allow for more subtle changes in peptide properties than the introduction or removal of prolines, which has been used to alter membrane properties of antimicrobial peptides in previous studies [3, 17, 28, 37].
ACKNOWLEDGEMENTS
The authors would like to thank Rachel B. Nelson for input on lipid vesicle assays, the Department of Chemistry at Brandeis University for use of their circular dichroism spectrometer, and Anne Gerschenson for assistance with circular dichroism. Funding was provided by National Institute of Allergy and Infectious Diseases (NIH-NIAID) Award R15AI079685 and a Research Corporation Cottrell College Science Award. Additional student support was provided by the Arnold and Mabel Beckman Foundation, the National Science Foundation (CHE-0353813), the Howard Hughes Medical Institute, and the Staley fund.
ABBREVIATIONS
- BF2
Buforin II
- CD
Circular Dichroism
- CFU
Colony Forming Unit
- EDTA
Ethylenediaminetetraacetic Acid
- ELISA
Enzyme-Linked Immunosorbent Assay
- HDAP
Histone-Derived Antimicrobial Peptide
- HEPES
4-(2-Hydroxyethyl)-1-Piperazineethanesulfonic Acid
- LB
Luria Broth
- LUV
Large Unilamellar Vesicle
- MD
Molecular Dynamics
- PI
Propidium Iodide
- POPC
Palmitoyloleoylphosphatidylcholine
- POPG
Palmitoyloleoylphosphatidylglycerol
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Ambroggio EE, Separovic F, Bowie JH, Fidelio GD, Bagatolli LA. Direct visualization of membrane leakage induced by the antibiotic peptides: maculatin, citropin, and aurein. Biophys J. 2005;89:1874–1881. doi: 10.1529/biophysj.105.066589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birkemo GA, Lüders T, Andersen Ø, Nes IF, Nissen-Meyer J. Hipposin, a histone-derived antimicrobial peptide in Atlantic halibut (Hippoglossus hippoglossus L.) Biochim Biophys Acta. 2003;1646:207–215. doi: 10.1016/s1570-9639(03)00018-9. [DOI] [PubMed] [Google Scholar]
- 3.Chia BC, Carver JA, Mulhern TD, Bowie JH. Maculatin 1.1, an anti-microbial peptide from the Australian tree frog, Litoria genimaculata solution structure and biological activity. Eur J Biochem. 2000;267:1894–1908. doi: 10.1046/j.1432-1327.2000.01089.x. [DOI] [PubMed] [Google Scholar]
- 4.Cho JH, Sung BH, Kim SC. Buforins: Histone H2A-derived antimicrobial peptides from toad stomach. Biochim Biophys Acta. 2009;1788:1564–1569. doi: 10.1016/j.bbamem.2008.10.025. [DOI] [PubMed] [Google Scholar]
- 5.Fleming E, Maharaj NP, Chen JL, Nelson RB, Elmore DE. Effect of lipid composition on buforin II structure and membrane entry. Proteins. 2008;73:480–491. doi: 10.1002/prot.22074. [DOI] [PubMed] [Google Scholar]
- 6.Hale JD, Hancock RE. Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert Rev Anti Infect Ther. 2007;5:951–959. doi: 10.1586/14787210.5.6.951. [DOI] [PubMed] [Google Scholar]
- 7.Hariton-Gazal E, Rosenbluh J, Graessmann A, Gilon C, Loyter A. Direct translocation of histone molecules across cell membranes. J Cell Sci. 2003;116:4577–4586. doi: 10.1242/jcs.00757. [DOI] [PubMed] [Google Scholar]
- 8.Jenkins TC. Optical absorbance and fluorescence techniques for measuring DNA-drug interactions. Methods Mol Biol. 1997;90:195–218. doi: 10.1385/0-89603-447-X:195. [DOI] [PubMed] [Google Scholar]
- 9.Jenssen H, Hamill P, Hancock RE. Peptide antimicrobial agents. Clin Microbiol Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi S, Chikushi A, Tougu S, Imura Y, Nishida M, Yano Y, et al. Membrane translocation mechanism of the antimicrobial peptide buforin 2. Biochemistry. 2004;43:15610–15616. doi: 10.1021/bi048206q. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi S, Takeshima K, Park CB, Kim SC, Matsuzaki K. Interactions of the novel antimicrobial peptide buforin 2 with lipid bilayers: proline as a translocation promoting factor. Biochemistry. 2000;39:8648–8654. doi: 10.1021/bi0004549. [DOI] [PubMed] [Google Scholar]
- 12.Lee K, Shin SY, Kim K, Lim SS, Hahm KS, Kim Y. Antibiotic activity and structural analysis of the scorpion-derived antimicrobial peptide IsCT and its analogs. Biochem Biophys Res Commun. 2004;323:712–719. doi: 10.1016/j.bbrc.2004.08.144. [DOI] [PubMed] [Google Scholar]
- 13.Luders T, Birkemo GA, Nissen-Meyer J, Andersen O, Nes IF. Proline conformation-dependent antimicrobial activity of a proline-rich histone h1 N-terminal Peptide fragment isolated from the skin mucus of Atlantic salmon. Antimicrob Agents Chemother. 2005;49:2399–2406. doi: 10.1128/AAC.49.6.2399-2406.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 15.Markossian KA, Zamyatnin AA, Kurganov BI. Antibacterial proline-rich oligopeptides and their target proteins. Biochemistry (Mosc) 2004;69:1082–1091. doi: 10.1023/b:biry.0000046881.29486.51. [DOI] [PubMed] [Google Scholar]
- 16.Matsuzaki K. Magainins as paradigm for the mode of action of pore forming polypeptides. Biochim Biophys Acta. 1998;1376:391–400. doi: 10.1016/s0304-4157(98)00014-8. [DOI] [PubMed] [Google Scholar]
- 17.Oh D, Shin SY, Lee S, Kang JH, Kim SD, Ryu PD, et al. Role of the hinge region and the tryptophan residue in the synthetic antimicrobial peptides, cecropin a(1–8)-magainin 2(1–12) and its analogues, on their antibiotic activities and structures. Biochemistry. 2000;39:11855–11864. doi: 10.1021/bi000453g. [DOI] [PubMed] [Google Scholar]
- 18.Otvos L., Jr The short proline-rich antibacterial peptide family. Cell Mol Life Sci. 2002;59:1138–1150. doi: 10.1007/s00018-002-8493-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park CB, Kim HS, Kim SC. Mechanism of action of the antimicrobial peptide buforin II: buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem Biophys Res Commun. 1998;244:253–257. doi: 10.1006/bbrc.1998.8159. [DOI] [PubMed] [Google Scholar]
- 20.Park CB, Kim MS, Kim SC. A novel antimicrobial peptide from Bufo bufo gargarizans. Biochem Biophys Res Commun. 1996;218:408–413. doi: 10.1006/bbrc.1996.0071. [DOI] [PubMed] [Google Scholar]
- 21.Park CB, Yi KS, Matsuzaki K, Kim MS, Kim SC. Structure-activity analysis of buforin II, a histone H2A-derived antimicrobial peptide: the proline hinge is responsible for the cell-penetrating ability of buforin II. Proc Natl Acad Sci. 2000;97:8245–8250. doi: 10.1073/pnas.150518097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pukala TL, Brinkworth CS, Carver JA, Bowie JH. Investigating the importance of the flexible hinge in caerin 1.1: solution structures and activity of two synthetically modified caerin peptides. Biochemistry. 2004;43:937–944. doi: 10.1021/bi035760b. [DOI] [PubMed] [Google Scholar]
- 23.Rosenbluh J, Hariton-Gazal E, Dagan A, Rottem S, Graessmann A, Loyter A. Translocation of histone proteins across lipid bilayers and Mycoplasma membranes. J Mol Biol. 2005;345:387–400. doi: 10.1016/j.jmb.2004.10.046. [DOI] [PubMed] [Google Scholar]
- 24.Rosenbluh J, Singh SK, Gafni Y, Graessmann A, Loyter A. Non-endocytic penetration of core histones into petunia protoplasts and cultured cells: a novel mechanism for the introduction of macromolecules into plant cells. Biochim Biophys Acta. 2004;1664:230–240. doi: 10.1016/j.bbamem.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Rotem S, Mor A. Antimicrobial peptide mimics for improved therapeutic properties. Biochim Biophys Acta. 2009;1788:1582–1592. doi: 10.1016/j.bbamem.2008.10.020. [DOI] [PubMed] [Google Scholar]
- 26.Shin SY, Park EJ, Yang ST, Jung HJ, Eom SH, Song WK, et al. Structure-activity analysis of SMAP-29, a sheep leukocytes-derived antimicrobial peptide. Biochem Biophys Res Commun. 2001;285:1046–1051. doi: 10.1006/bbrc.2001.5280. [DOI] [PubMed] [Google Scholar]
- 27.Sitaram N. Antimicrobial peptides with unusual amino acid compositions and unusual structures. Curr Med Chem. 2006;13:679–696. doi: 10.2174/092986706776055689. [DOI] [PubMed] [Google Scholar]
- 28.Song YM, Yang ST, Lim SS, Kim Y, Hahm KS, Kim JI, et al. Effects of L- Or D-Pro incorporation into hydrophobic or hydrophilic helix face of amphipathic alpha-helical model peptide on structure and cell selectivity. Biochem Biophys Res Commun. 2004;314:615–621. doi: 10.1016/j.bbrc.2003.12.142. [DOI] [PubMed] [Google Scholar]
- 29.Steinberg DA, Lehrer RI. Designer assays for antimicrobial peptides. Disputing the 'one-size-fits-all' theory. Methods Mol Biol. 1997;78:169–186. doi: 10.1385/0-89603-408-9:169. [DOI] [PubMed] [Google Scholar]
- 30.Suh JY, Lee YT, Park CB, Lee KH, Kim SC, Choi BS. Structural and functional implications of a proline residue in the antimicrobial peptide gaegurin. Eur J Biochem. 1999;266:665–674. doi: 10.1046/j.1432-1327.1999.00917.x. [DOI] [PubMed] [Google Scholar]
- 31.Thennarasu S, Nagaraj R. Specific antimicrobial and hemolytic activities of 18-residue peptides derived from the amino terminal region of the toxin pardaxin. Protein Eng. 1996;9:1219–1224. doi: 10.1093/protein/9.12.1219. [DOI] [PubMed] [Google Scholar]
- 32.Tsao HS, Spinella SA, Lee AT, Elmore DE. Design of novel histone-derived antimicrobial peptides. Peptides. 2009;30:2168–2173. doi: 10.1016/j.peptides.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 33.Tse WC, Boger DL. A fluorescent intercalator displacement assay for establishing DNA binding selectivity and affinity. Acc Chem Res. 2004;37:61–69. doi: 10.1021/ar030113y. [DOI] [PubMed] [Google Scholar]
- 34.Uyterhoeven ET, Butler CH, Ko D, Elmore DE. Investigating The nucleic acid interactions and antimicrobial mechanism of buforin II. FEBS Lett. 2008;582:1715–1718. doi: 10.1016/j.febslet.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 35.Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 36.Yi GS, Park CB, Kim SC, Cheong C. Solution structure of an antimicrobial peptide buforin Ii. FEBS Lett. 1996;398:87–90. doi: 10.1016/s0014-5793(96)01193-3. [DOI] [PubMed] [Google Scholar]
- 37.Zhang L, Benz R, Hancock RE. Influence of proline residues on the antibacterial and synergistic activities of alpha-helical peptides. Biochemistry. 1999;38:8102–8111. doi: 10.1021/bi9904104. [DOI] [PubMed] [Google Scholar]
- 38.Zhu WL, Lan H, Park Y, Yang ST, Kim JI, Park IS, et al. Effects of Pro --> peptoid residue substitution on cell selectivity and mechanism of antibacterial action of tritrpticin-amide antimicrobial peptide. Biochemistry. 2006;45:13007–13017. doi: 10.1021/bi060487+. [DOI] [PubMed] [Google Scholar]