Abstract
The human chemokine lymphotactin (Ltn) is a remarkable protein that interconverts between two unrelated native state structures in the condensed phase. It is possible to shift the equilibrium toward either conformation with selected sequence substitutions. Previous results have shown that a disulfide-stabilized variant preferentially adopts the canonical chemokine fold (Ltn10), while a single amino acid change (W55D) favors the novel Ltn40 dimeric structure. Selective noncovalent adduct protein probing (SNAPP) is a recently developed method for examining solution phase protein structure. Herein, it is demonstrated that SNAPP can easily recognize and distinguish between the Ltn10 and Ltn40 states of lymphotactin in aqueous solution. The effects of organic denaturants, acid, and disulfide bond reduction and blocking were also examined using SNAPP for the CC3, W55D, and wild type proteins. Only disulfide reduction was shown to significantly perturb the protein, and resulted in considerably decreased adduct formation consistent with loss of tertiary/secondary structure. Cold denaturation experiments demonstrated that wild-type Ltn is the most temperature sensitive of the three proteins. Examination of the higher charge states in all experiments, which are presumed to represent transition state structures between Ltn-10 and Ltn-40, reveals increased 18C6 attachment relative to the more folded structures. This observation is consistent with increased competitive intramolecular hydrogen bonding, which may guide the transition. Experiments examining the gas phase structures revealed that all three proteins can be structurally distinguished in the gas phase. In addition, the gas phase experiments enabled identification of preferred adduct binding sites.
Electronic supplementary material
The online version of this article (doi:10.1007/s13361-010-0042-3) contains supplementary material, which is available to authorized users.
Key words: h/d exchange, Protein structure, Covalent labeling
Introduction
Lymphotactin is an unusual protein in many ways. It is a human chemokine that can adopt two natively folded states, which are both functionally relevant in vivo [1]. The two native state structures are very dissimilar as shown in Scheme 1 [2]. One structure represents the typical chemokine fold, a single alpha helix adjacent to three beta-sheet strands, and is referred to as the Ltn10 structure. The other structure is known as Ltn40 and is characterized by dimerization of two proteins, with both adopting four β-sheet strands oriented in a symmetrical head to tail fashion. Although both structures share β-sheet motifs, the composition of the beta sheets is quite different between the two structural forms. Similarly, the hydrophobic cores and other tertiary and quaternary structural characteristics are essentially entirely dissimilar. Under typical biological conditions, both forms of the wild type protein exist in equilibrium and interconvert on a timescale of seconds [2]. In the Volkman laboratory, two mutants have been developed that adopt only one of the two structural motifs [3]. The CC3 mutant contains an additional disulfide bond that favors the typical Ltn10 structure. Disruption of the Ltn10 hydrophobic core in the W55D mutant pushes the structural equilibrium strongly towards the Ltn40 structure. These mutants can be utilized to examine aspects of each structure independently.
Scheme 1.
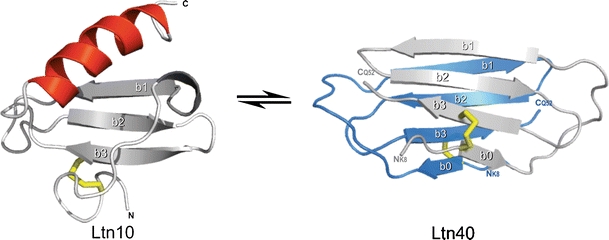
The two structures of lymphotactin
Selective noncovalent adduct protein probing (SNAPP) is a recently developed method for examining protein structure in solution [4]. This method relies on specific noncovalent interactions to probe protein structure. Experiments are conducted by introducing a reagent which binds weakly in solution, such as 18-crown-6 ether (18C6), which becomes strongly attached to the protein in the gas phase. 18C6 attaches to lysine in the gas phase due to three specific hydrogen bonds between the protonated side-chain amine and alternating oxygen atoms in the crown ether. The binding energy for this attachment is ~54 kcal/mol [5]. Less specific, although fairly strong noncovalent (42 kcal/mol) associations between 18C6 and the protonated side chain of arginine can form as well. SNAPP experiments are conducted by subjecting an aqueous solution containing protein and 18C6 to electrospray ionization (ESI). Source conditions which favor both complete desolvation and retention of noncovalent adducts must be employed. During the process of ESI, binding interactions between 18C6 and protonated side chains transition from weak to strong as the protein is rapidly desolvated. Previous experiments have determined that the local chemical environment surrounding each lysine residue strongly influences the probability for binding 18C6, with proximate salt bridge or hydrogen bonding interactions interfering the most [6]. Structural information about the protein is therefore obtained because the local chemical environment surrounding each residue is a function of protein structure [4,7]. In the end, a distribution of peaks (known as a SNAPP distribution) representing the number of 18C6s attached to the protein is observed in the mass spectrometer and is characteristic of a particular protein conformation. Changes to the protein structure will typically lead to changes in the SNAPP distribution; therefore the strength of SNAPP is in measuring structural differences for either static or dynamic systems.
SNAPP is closely related to other mass spectrometry (MS) based protein structure determination methods such as charge state distribution analysis [8–10], hydrogen/deuterium (H/D) exchange [9–11], and covalent labeling [12], but also differs in several important ways. H/D exchange monitors protein structure as a function of the exchange of amide hydrogens. The overall number, locations, and rates of exchanges can be used to interrogate structure [13]. Various covalent labeling methods can also be used to probe structure, although in this case reactions are ideally restricted to a single modification to avoid perturbation of the target structure [14]. The primary difference between SNAPP and both of these methods is that the reporting chemistry in SNAPP is highly reversible (if present at all) in solution and information is only encoded during the later stages of ESI when the protein is transitioning into the gas phase. This enables SNAPP to probe highly dynamic systems where structural changes may be occurring rapidly in solution, and on the same timescale as H/D exchange or covalent labeling. If, for example, a protein adopted an unfolded and folded state under given conditions, the SNAPP distributions for both would be determined independently during ESI and observable in different charge states (due to the different sizes) as long as the structural transition took longer than a few milliseconds (the time required to desolvate the protein). SNAPP is therefore an appropriate method to interrogate the structural features of lymphotactin, which undergoes structural changes on a significantly longer timescale [2].
Herein it is demonstrated that the Ltn10 and Ltn40 structures yield easily distinguishable SNAPP distributions. The wild type protein yields results that are most similar to the W55D variant. The addition of acid or organic denaturants does not have a large impact on the SNAPP distributions for the wild type, CC3, or W55D proteins. Reduction and blocking of the disulfide bonds affects the structures more dramatically and yields shifts in the SNAPP distributions that are consistent with significant unfolding. Data obtained from NMR experiments supports this conclusion. The effects of cold temperature on all three proteins are explored. Finally, a combination of SNAPP and radical chemistry is utilized to compare the gas phase structures of the wild type, CC3, and W55D proteins.
Experimental
Materials
Recombinant human lymphotactin proteins used in SNAPP study were expressed and purified as previously described [15]. Lymphotactin mutants CC3 and W55D were prepared by site-directed mutagenesis using the Stratagene QuickChange kit following established protocols [3]. Purified proteins were lyophilized and stored at –20 °C for subsequent study. 18-Crown-6 ether used for SNAPP studies was purchased from Alfa Aesar (Ward Hill, MA, USA). Dithiothreitol (DTT) and iodoacetamide (IAA) were purchased from Sigma Aldrich (St. Louis, MO, USA). H2O used for SNAPP experiments was purified to 18.2MΩ resistivity using Millipore Direct-Q (Billerica, MA, USA). All the solvents were obtained from Fisher Scientific (Fairlawn, NJ) and used as received. 2-(hydroxymethyliodobenzoylester)-18C6 (IBA-18C6), employed in the radical directed dissociation experiments, was prepared as follows: 0.50 mmol DCC in 5.0 mL dioxane was added to a 50 mL round bottom flask containing 0.50 mmol of 4-iodobenzoic acid and 0.50 mmol 2-hydroxymethyl-18-crown-6 ether. A catalytic amount of DMAP (~10 mg) was added. After a 12 h reaction period, a crystalline hair-like precipitate was observed. The precipitate was removed by filtration. The filtrate was then passed through celite and evaporated over nitrogen. The product was recovered as a white solid.
S–S Bond Reduction and Cysteine Protection of Ltn Proteins
To reduce disulfide bonds in the lymphotactin proteins, 60 μL solution containing 10 nmol protein and excess amount of DTT (10 mM) was incubated at 54 °C for 45 min. Free cysteine and excess DTT were treated with a stoichiometric amount of IAA (1.22 μmol for wild type Ltn and W55D, 1.24 μmol for CC3) to block all free thiol groups. The blocked proteins were purified by a protein trap (Michrom Bioresources, Inc. Auburn, CA, USA), lyophilized and stored at –20 °C for subsequent use.
SNAPP Experiments
CC3, wild type Ltn, and W55D stock solutions were diluted to 7 μM in H2O, respectively. The final concentration of 18C6 in SNAPP solutions was 84 μM. Mass spectra were obtained using an LTQ linear ion trap mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) equipped with a standard ESI-source. Protein samples mixed with 18C6 were directly infused into LTQ mass spectrometer. The electrospray parameters, such as spray voltage, sheath gas flow rates, capillary voltage, temperature, etc. were optimized and were similar to parameters described previously for SNAPP experiments [4]. Once optimized, all the parameters were maintained for all SNAPP experiments presented herein. The following are the optimized source parameters for all SNAPP experiments: spray voltage 4.8 kV, sheath gas flow rates 11, tube lens 160 V, capillary voltage 44 V and capillary temperature 275 °C.
Radical Directed Dissociation of Ltn Proteins
Five μM lymphotactin protein was mixed with 15 μM IBA-18C6 in 50/50 H2O/MeOH solution and directly infused into the LTQ linear ion trap mass spectrometer. The posterior plate of the LTQ is modified with a quartz window to transit fourth harmonic (266 nm) laser pulses from a flash lamp pumped Nd/YAG laser (Continuum, Santa Clara, CA, USA). The laser pulses are synchronized to the end of the isolation step of a typical MS2 experiment by a TTL trigger signal from the mass spectrometer to the laser via a digital delay generator (Berkeley Nucleonics, San Rafael, CA, USA). The isolation window width of MS2-MS4 experiments was set to 5 Da.
Results and Discussion
The stackplot in Figure 1a shows the mass spectra for lymphotactin alone acquired in water (front), water/methanol (middle), and water/methanol/acetic acid (back). These solvent systems are increasingly denaturing for most proteins and will typically yield substantial shifts in the charge state distribution towards higher charge states [8,16]. The increase in charge state is typically rationalized by the notion that denatured proteins are less compact and can therefore accommodate additional charges without increasing Coulombic repulsion. Interestingly, the typical shift towards higher charge states is not observed for lymphotactin. In fact, the charge state distributions barely change at all. Therefore, charge state distributions alone yield virtually no information about the structure of lymphotactin. As detailed in the Introduction, it is known that lymphotactin has two highly dissimilar native structures, which are under equilibrium. It is likely that transitioning from one structure to the other requires sampling of an unfolded state due to the large disparity between the two conformations. The data in Figure 1a is consistent with the notion that lymphotactin accesses an unfolded state even in an aqueous solution, and therefore the addition of methanol and acid does not significantly shift the charge state distribution because unfolded structures are already being sampled. From these data alone, it is clear that lymphotactin is an unusual protein.
Figure 1.
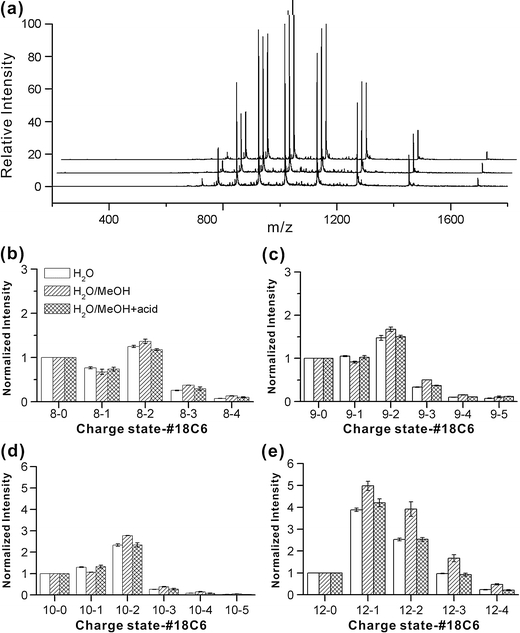
(a) ESI-MS spectra for wild type lymphotactin acquired in water, 50/50 water/methanol, and 49/49/1 water/methanol/acetic acid from front to back, respectively. SNAPP distributions for wild type lymphotactin in various charge states are shown in (b) +8, (c) +9, (d) +10, and (e) +12
In Figure 1b–d, several representative SNAPP distributions for lymphotactin in the same series of solvents are shown. These distributions have been extracted from the raw mass spectra and illustrate the number of 18C6 adducts which attach to lymphotactin, separated by charge state. The addition of 18C6 reveals another layer of structural information, allowing identical charge states to be compared with each other. The SNAPP distributions for the +8 through +10 charge states are fairly similar to each other and are also fairly constant in the various solvents. These distributions most likely represent the more folded conformations of the protein. In the +11 and +12 charge states, the SNAPP distributions begin to shift towards an increasing number of adducts (as indicated by the small relative abundance of the non-adduct peak in the +12 charge state). The average number of 18C6 adducts goes from 1.3 for the +8 charge state to 1.6 for the +12 charge state. Some differences between the various solvent systems also become notable. Interestingly, the number of crown adducts increases with the addition of methanol and then decreases again with the addition of acid. For systems where structural changes are not dominant, it is known that the addition of acid leads to a decrease in crown binding due to competitive binding by the acid [4]. This confirms that acid does not significantly influence the structure of lymphotactin. Given that the protein is highly basic (6 Lys, 8 Arg), with few acidic residues, this result is perhaps not surprising.
In Figure 2, the SNAPP distributions obtained in water for wild type Ltn, Ltn10 (CC3), and Ltn40 (W55D) are shown. There is a clear and reproducible difference between the SNAPP distributions for the CC3 and W55D mutants. Less 18C6 attachment to the W55D protein is observed in every case. SNAPP-MS is therefore easily able to distinguish the two structural variants of lymphotactin from each other when examined separately. As shown in Scheme 2, these two mutants vary in sequence by only six amino acids (W55D also contains three additional N-terminal residues, none of them are targets for 18C6, nor do they alter the Ltn40 conformation). Interestingly, one of the mutations, Ala in CC3 for Arg in W55D, creates an additional target residue for 18C6 binding. Despite this change, 18C6 attachment in W55D is less than that observed in CC3. Therefore, the differential response of SNAPP to these two proteins is clearly due to structural differences. Salt bridges are known to significantly interfere with 18C6 binding [4]. Both the Ltn10 and Ltn40 structures contain two salt bridges (Arg61-Asp64 and Arg35-Glu31 for Ltn10, Arg9-Asp50 and Glu31-Lys25 (intermolecular) for Ltn40). However, lysine is the preferred target for 18C6, and the transition for Lys25 from being freely available in Ltn10 to being incorporated into a salt bridge in Ltn40 likely accounts for some of the reduced 18C6 binding to the W55D mutant.
Figure 2.
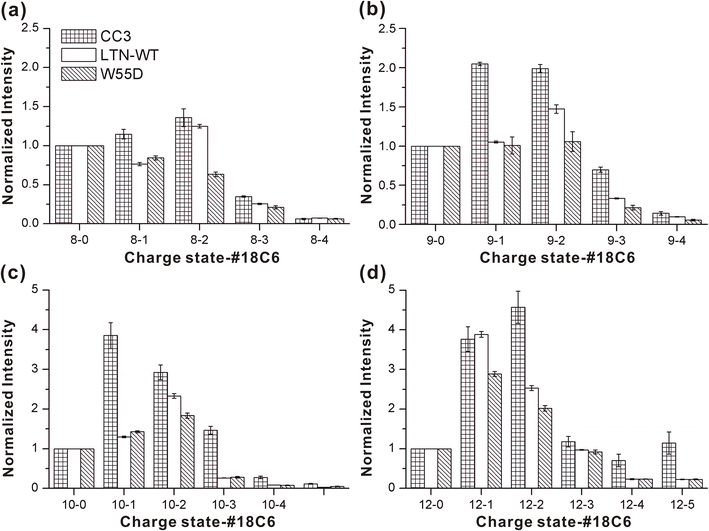
SNAPP distributions for three variants of lymphotactin sampled from water. The CC3 and W55D mutants clearly produce distinguishable distributions
Scheme 2.

Sequences for each lymphotactin variant
Although the wild type protein can adopt either the Ltn10 or Ltn40 structures, it is clear from the data in Figure 2 that the wild type Ltn SNAPP distributions are most similar to those from the W55D mutant. The similarity likely results from a combination of two factors. (1) It is known that wild type Ltn favors the Ltn40 structure at low ionic strength [17]. (2) The W55D mutant is able to freely access unfolded states which are expected to be present in wild type Ltn, but the CC3 mutant is significantly constrained by an additional disulfide bond, which likely interferes with adopting similar unfolded states. Despite similarity to W55D, wild type Ltn does present some intermediate behavior as well (for example in the 8-2 and 12-1 distributions where the wild type Ltn peaks are more similar to the CC3 values), suggesting that the equilibrium is not entirely shifted to the Ltn40 structure. It is worth noting that the dimeric association for Ltn40 known to dominate in solution is not observed in the gas phase. Presumably, this is due to loss of the hydrophobic effect as the lymphotactin dimer is desolvated, which is primarily driving the association in solution. Nevertheless, it is clear from the data in Figure 2 that the structural information encoded by attachment of 18C6 is locked in prior to the loss of the dimer during the transition to the gas phase.
Characterization of the transition between the two forms of lymphotactin is an active area of interest [18]. The ability of SNAPP to examine and potentially separate out highly dynamic structural features is an important advantage which should facilitate characterization of the transition structures. As described above, the higher charge states most likely represent structural states, which are at least partially unfolded and therefore capable of transitioning between the two structural forms. One possibility for transition is that the unfolded protein becomes entirely disordered, and subsequently refolds into either native state with probabilities determined by the nature of the solution. To investigate this theory, we obtained SNAPP distributions for proteins where the disulfide bonds had been reduced and blocked. Results from NMR experiments indicate that elimination of the disulfide bonds leads to highly disordered states (see Supporting Information). In Figure 3 the average number of 18C6 adducts for blocked and disulfide bound protein are plotted as a function of charge state. Direct comparison reveals reduced 18C6 complexation following disulfide reduction and capping for each protein.
Figure 3.
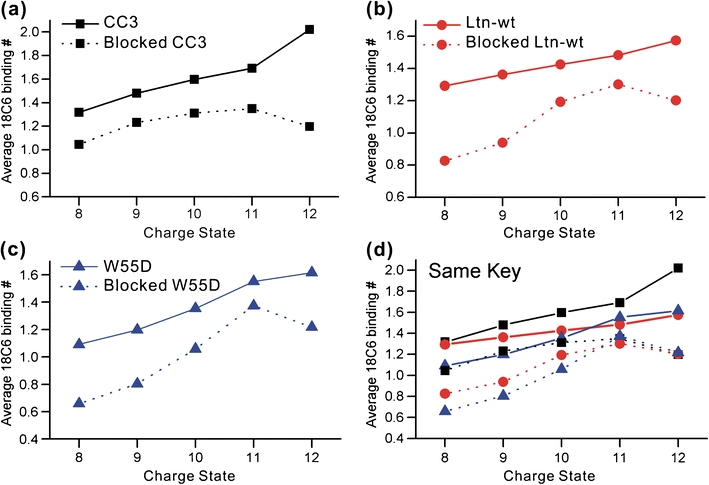
The average number of 18C6 adducts is shown as a function of charge state for (a) CC3, (b) wild type Ltn, and (c) W55D. The protected (disulfide reduced and capped) proteins are shown in dotted lines. (d) All proteins are shown together to demonstrate relative binding. The key is identical to (a)–(c)
These results can be rationalized by examining potential competitive intramolecular binding sites. Lymphotactin has few acidic residues, suggesting that the primary competition for lysine or arginine side chain binding should originate from hydrogen bonding with the backbone or side chains. When the protein is in the folded state, many backbone hydrogen binding sites will be unavailable due to involvement in secondary structure. For example, in an α helix, all of the internal amides are hydrogen bound to each other. It has been demonstrated previously that loss of secondary structure to random coil structure can lead to significantly reduced crown attachment due to competitive hydrogen bonding with the backbone [4]. In the case of lymphotactin, the reduction in crown binding is consistent with adoption of a highly disordered state where disruption of backbone hydrogen bonding leads to competitive inhibition of 18C6 binding. Comparison of the higher charge states for the blocked and unblocked structures (i.e., +11 and +12 in Figure 3) reveals significantly different behavior, suggesting that the transition structures are likely not entirely disordered. Similarly, the average binding numbers for unblocked protein are consistent with results given above and suggest that the wild type Ltn and W55D proteins access similar transition state structures, while CC3 does not (Figure 3d). For the blocked proteins, all three have virtually identical average 18C6 binding in the +11 and +12 charge states (Figure 3d), suggesting absence of any structural differences for the unfolded, disordered states.
Another interesting variable, which can produce structural change and is known to influence lymphotactin, is temperature. To examine temperature effects, we simply gathered data on identical solutions at room temperature and with the addition of an ice pack to the syringe containing the protein solution. The temperature of the syringe is ~0 °C, although the solution will warm slightly passing through the line to the electrospray tip. Results were acquired after 20 min equilibration time and are shown in Figure 4. Both the CC3 and W55D distributions do not undergo any compelling shifts in 18C6 adduction after application of the ice pack. Interestingly, wild type Ltn does exhibit a decrease in 18C6 binding at lower temperature. It is therefore clear that temperature induced structural shifts can be observed by SNAPP. Conversely, merely dropping the temperature does not necessarily influence SNAPP distributions since the CC3 and W55D distributions remain unchanged. The results suggest that out of the three variants examined, wild type Ltn is most susceptible to temperature denaturation. The drop in number of 18C6 adducts at lower temperatures is similar to the trend observed in Figure 3 and suggests cold denaturation is responsible for the shift in the SNAPP distribution, rather than some other type of structural shift (for example skewing the equilibrium in favor of the CC3 structure, which would lead to an increase in 18C6 attachment). It should be remembered that wild type Ltn is the only form of the protein, which is capable of simultaneously interchanging between the Ltn-10 and Ltn-40 structures. It is likely that this ability necessitates that any particular folded state be in a rather shallow well of stability, making susceptibility to denaturation more likely.
Figure 4.
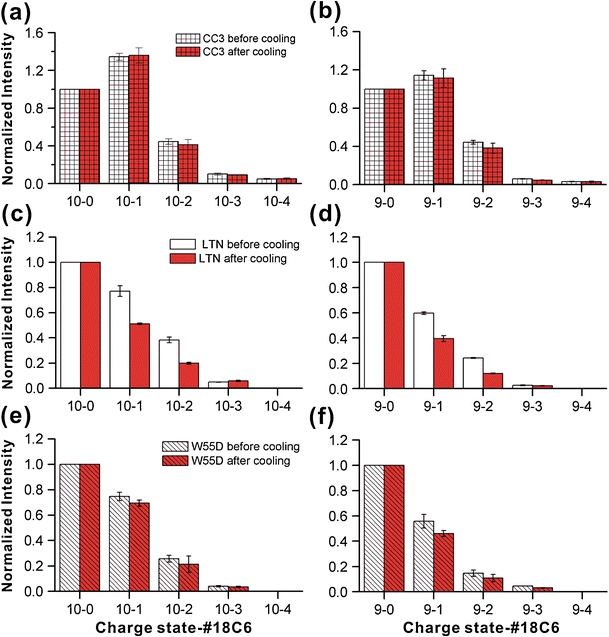
SNAPP distributions acquired in water at room temperature and after the addition of an ice pack. Only the wild type Ltn distribution shifts to any significant extent
The SNAPP experiments reported up to this point were designed to investigate the solution phase structure of lymphotactin, even though ultimate detection took place in the gas phase. It is also possible that information about solution phase structures may be obtained by actually examining molecules in the gas phase [19,20]. This would be possible in at least two scenarios; (1) if the solution phase structure was retained in the gas phase, direct analysis would apply, or (2) if differences in the solution phase structures led to differences in the gas phase structures, it would be possible to monitor changes in structure. The second scenario would of course only reveal differences in structure rather than information about the structures themselves. It is beyond the scope of the present experiments to attempt to determine if the solution and gas phase structures for lymphotactin are the same; however, we did examine the gas phase structures of the wild type Ltn, CC3, and W55D proteins to determine if they were distinguishable from each other. This was accomplished by using IBA-18C6, which is an 18C6 based molecule with a highly efficient radical precursor attached. Following photoactivation with 266 nm light, a radical is generated by dissociation of the carbon–iodine bond. This radical is subsequently transferred to the protein. As we have demonstrated previously, radical migration and consequent dissociation in gaseous protein ions is largely determined by the overall protein structure [21]. Therefore, differences in protein structure would be expected to yield differences in dissociation. Interestingly with respect to SNAPP, this experiment also potentially offers information about preferred binding sites for 18C6.
The results from radical directed dissociation of all three proteins in the +7 charge state are shown in Figure 5. It is clear that all three dissociation spectra are distinct, although the wild type Ltn and W55D spectra share similarities. For CC3, there is an abundant cleavage between N67 and T68, as shown in Figure 5a. In addition, there is modest dissociation observed along the remainder of the c-terminal portion of the protein. Not surprisingly, there is no dissociation observed between the portions of the protein which are linked by disulfide bonds. This does not mean that there is no dissociation occurring in the region, as such dissociation could not be observed unless two backbone bonds were broken. There is also a very modest amount of dissociation on the N-terminal side of the disulfide linked portion, suggesting that indeed some segments of the protein within the disulfide bonds are likely accessible to the radical. The wild type Ltn in Figure 5b yields a significantly different distribution. Dissociation at several residues in close proximity to Lys76 is abundant, suggesting that this may be a favorable site for attachment of IBA-18C6. In comparison with CC3, several peaks towards the C-terminal portion of the protein are missing. W55D yields results similar to those for wild type Ltn, but even less dissociation is observed. Importantly, even though there is an M to V substitution in the region where dissociation is observed for the W55D mutant (see Scheme 2), the mutation does not appear to affect dissociation, which is actually observed at that location in all three proteins.
Figure 5.
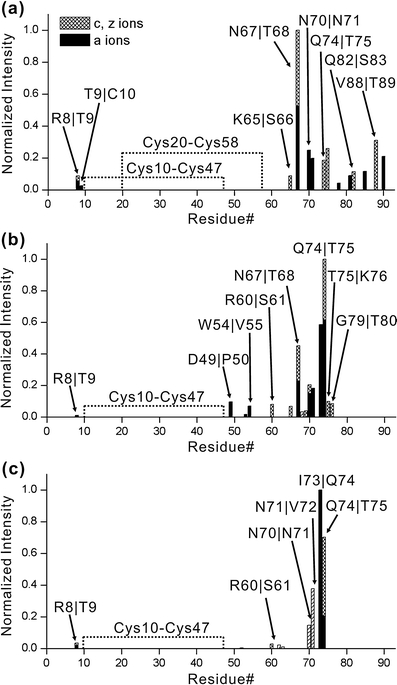
Dissociation points are shown as a function of sequence for experiments probing the gas phase structures of the +7 charge state for (a) CC3, (b) wild type Ltn, and (c) W55D (the numbering of residues for W55D has been shifted to be consistent with the other proteins, for actual numbering see Scheme 2). Differences in dissociation indicate differences in structure as described in the text
Investigation of higher charge states, where tertiary structure is lost in the gas phase due to Coulombic repulsion, reveals that most of the dissociation is centered near Lys76. In the absence of tertiary structure, dissociation is expected to be most abundant in the vicinity of the starting location of the radical. This suggests that Lys76 is a preferred binding site for 18C6. Due to the cyclic nature of all three proteins, which all contain one or more disulfide bonds, it cannot be determined whether there are other equally preferred binding sites. In any case, the differences between the dissociation spectra clearly indicate that the gas phase structures for the proteins are dissimilar. Since the sequences of these proteins are all very similar, the differences most likely result from either retention or memory of the solution phase structures.
For both the Ltn10 and Ltn40 structures, portions of the C-terminal tail of the protein are largely disordered as determined by NMR (see italicized residues in Scheme 2). These sections are therefore absent in the structures shown in Scheme 1, but they are still present and can contribute to the results which are obtained in SNAPP experiments. These regions contain several charged residues, which are potential 18C6 targets and the results from RDD suggest that at least one lysine in this region is a favorable binding site. Fortunately, SNAPP is capable of extracting structural information even for proteins which are completely disordered [22]. In the case of lymphotactin, some of the differences between the observed SNAPP results and those that would be predicted by the known structures may be attributable to structural shifts in these disordered regions.
Conclusions
The conformationally distinct Ltn10 and Ltn40 native states of lymphotactin can clearly be distinguished and structurally characterized by SNAPP-MS. The results also suggest that the distribution of structures occupied by lymphotactin is not significantly influenced by the addition of methanol or acid. In contrast, reduction and blocking of the disulfide bonds produce a denatured protein. Cold temperature experiments suggest that out of three sequence variants examined, the wild type protein is the most susceptible to denaturation. With regards to the structural transition between the two native states of lymphotactin, the data reveal that slightly increased 18C6 binding is observed for the transition structures relative to folded conformations. This observation is consistent with a small increase in intramolecular hydrogen bonding interactions that enhances the availability of lysine or arginine for 18C6 binding. These intra-chain interactions may coordinate the transition between Ltn10 and Ltn40. In the gas phase, the sequence variants of lymphotactin also adopt distinct structures, as revealed by radical directed dissociation. It is unclear whether the differences result from retention of solution phase structure in the gas phase or some other source. Overall, the results indicate that SNAPP is a useful method for probing structure in proteins that undergo slow global conformational transitions.
Electronic Supplementary Materials
Below is the link to the electronic supplementary material.
Supporting Information (PDF 237 kb)
Acknowledgments
The authors wish to thank the NIH for funding this research (for BFV, grant R01 AI063325, and for RRJ, grant R01 GM084106).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Tuinstra RL, Peterson FC, Kutlesa S, Elgin ES, Kron MA, Volkman BF. Interconversion between two unrelated protein folds in the lymphotactin native state. PNAS. 2008;105(13):5057–5062. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Volkman BF, Liu TY, Peterson FC. Lymphotactin Structural Dynamics. Methods Enzymol. 2009;461:51–69. doi: 10.1016/S0076-6879(09)05403-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuinstra RL, Peterson FC, Elgin ES, Pelzek AJ, Volkman BF. An engineered second disulfide bond restricts Lymphotactin/XCL1 to a Chemokine-like conformation with XCR1 agonist activity. Biochemistry. 2007;46:2564–2573. doi: 10.1021/bi602365d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ly T, Julian RR. Using ESI-MS to probe protein structure by site-specific Noncovalent Sttachment of 18-Crown-6. J. Am. Soc. Mass Spectrom. 2006;17:1209–1215. doi: 10.1016/j.jasms.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Rodgers, M. T.; Chen, Y. Noncovalent Molecular Recognition of Protonated Peptidomimetic Bases by 18-Crown-6: Structure versus Energetics. Proceedings of the 58th ASMS Conference on Mass Spectrometry and Allied Topics; Salt Lake City, Utah.
- 6.Liu Z, Cheng S, Gallie DR, Julian RR. Exploring the mechanism of SNAPP mass spectrometry utilizing site directed mutagenesis to examine Ubiquitin. Anal. Chem. 2008;80:3846–3852. doi: 10.1021/ac800176u. [DOI] [PubMed] [Google Scholar]
- 7.Ly T, Pujanauski BG, Sarpong R, Julian RR. Surveying Ubiquitin structure by Noncovalent attachment of distance constrained Bis(Crown) Ethers. Anal. Chem. 2008;80:5059–5064. doi: 10.1021/ac800177s. [DOI] [PubMed] [Google Scholar]
- 8.Grandori R. Origin of the conformation dependence of protein charge-state distributions in Electrospray ionization mass spectrometry. J. Mass Spectrom. 2003;38:11–15. doi: 10.1002/jms.390. [DOI] [PubMed] [Google Scholar]
- 9.Kaltashov IA, Eyles SJ. Studies of biomolecular conformations and conformational dynamics by mass spectrometry. Mass Spectrom. Rev. 2002;21:37–71. doi: 10.1002/mas.10017. [DOI] [PubMed] [Google Scholar]
- 10.Yan X, Watson J, Ho PS, Deinzer ML. Mass spectrometric approaches using electrospray ionization charge states and hydrogen–deuterium exchange for retermining protein structures and their conformational changes. Mol. Cell. Proteom. 2003;3:10–23. doi: 10.1074/mcp.R300010-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Jones LM, Zhang H, Vidavsky I, Gross ML. Online, high-pressure digestion system for protein characterization by hydrogen–deuterium exchange and mass spectrometry. Anal. Chem. 2010;82:1171–1174. doi: 10.1021/ac902477u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konermann L, Stocks BB, Czarny T. Laminar flow effects during laser-induced oxidative labeling for protein structural studies by mass spectrometry. Anal. Chem. 2010;82:6667–6674. doi: 10.1021/ac101326f. [DOI] [PubMed] [Google Scholar]
- 13.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom. Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 14.Mendoza VL, Vachet RW. Probing protein structure by amino acid-specific covalent labeling and mass spectrometry covalent labeling. Mass Spectrom. Rev. 2009;28:785–815. doi: 10.1002/mas.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterson FC, Elgin ES, Nelson TJ, Zhang F, Hoeger TJ, Linhardt RJ, Volkman BF. J. Biol. Chem. 2004;279:12598–12604. doi: 10.1074/jbc.M311633200. [DOI] [PubMed] [Google Scholar]
- 16.Dobo A, Kaltashov IA. Detection of multiple protein conformational ensembles in solution via deconvolution of charge-state distributions in ESI MS. Anal. Chem. 2001;73:4763–4773. doi: 10.1021/ac010713f. [DOI] [PubMed] [Google Scholar]
- 17.Kuloglu ES, McCaslin DR, Markley JL, Volkman BF. Structural rearrangement of human Lymphotactin, a C-Chemokine. under physiological solution conditions. J. Biol. Chem. 2002;277(20):17863–17870. doi: 10.1074/jbc.M200402200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camilloni C, Sutto L. Lymphotactin: how a protein can adopt two folds. J. Chem. Phys. 2009;131:245105–254106. doi: 10.1063/1.3276284. [DOI] [PubMed] [Google Scholar]
- 19.Clemmer DE, Jarrold MF. Ion mobility measurements and their applications to clusters and biomolecules. J. Mass Spectrom. 1997;32:577–592. doi: 10.1002/(SICI)1096-9888(199706)32:6<577::AID-JMS530>3.0.CO;2-4. [DOI] [Google Scholar]
- 20.Danell AS, Parks JH. FRET Measurements of trapped oligonucleotide duplexes. Int. J. Mass Spectrom. 2003;229:35–45. doi: 10.1016/S1387-3806(03)00253-7. [DOI] [Google Scholar]
- 21.Ly T, Julian RR. Elucidating the tertiary structure of protein ions in Vacuo with site specific photo-initiated radical reactions. J. Am. Chem. Soc. 2010;132:8602–8609. doi: 10.1021/ja910665d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ly T, Julian RR. Protein–metal interactions of calmodulin and α-Synuclein monitored by selective noncovalent adduct protein probing mass spectrometry. J. Am. Soc. Mass Spectrom. 2008;19:1663–1672. doi: 10.1016/j.jasms.2008.07.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information (PDF 237 kb)


