Abstract
The (β/α)8 barrel is the most commonly occurring fold among protein catalysts. To lay a groundwork for engineering novel barrel proteins, we investigated the amino acid sequence restrictions at 182 structural positions of the prototypical (β/α)8 barrel enzyme triosephosphate isomerase. Using combinatorial mutagenesis and functional selection, we find that turn sequences, α-helix capping and stop motifs, and residues that pack the interface between β-strands and α-helices are highly mutable. Conversely, any mutation of residues in the central core of the β-barrel, β-strand stop motifs, and a single buried salt bridge between amino acids R189 and D227 substantially reduces catalytic activity. Four positions are effectively immutable: conservative single substitutions at these four positions prevent the mutant protein from complementing a triosephosphate isomerase knockout in Escherichia coli. At 142 of the 182 positions, mutation to at least one amino acid of a seven-letter amino acid alphabet produces a triosephosphate isomerase with wild-type activity. Consequently, it seems likely that (β/α)8 barrel structures can be encoded with a subset of the 20 amino acids. Such simplification would greatly decrease the computational burden of (β/α)8 barrel design.
The (β/α)8 barrel is the most common fold among protein catalysts, appearing in approximately 10% of all known enzyme structures (1). The barrel structure is composed of eight catenated strand-loop-helix-turn units. The β-strands are located in the interior of the protein, forming the staves of a barrel, whereas the α-helices pack around the exterior. The active sites of all known (β/α)8 barrel enzymes are located in the β→α loops (1). Recently, the catalytic activities of two naturally occurring (β/α)8 barrel enzymes were transplanted to heterologous scaffolds (2, 3). To lay the groundwork for future efforts to engineer novel (β/α)8 barrel activities, we sought to understand how an amino acid sequence organizes a catalytically active barrel conformation.
The oil-droplet and jigsaw-puzzle models offer two limiting views of how an amino acid sequence could encode a globular structure. An oil-droplet model posits that partitioning of hydrophobic and polar amino acids into oil and water phases during protein folding forces appropriate secondary and tertiary structures to form (4). In the extreme view, the pattern of hydrophobic and polar residues in an amino acid sequence (the H/P pattern) is sufficient to specify a three-dimensional conformation. Alternatively, a jigsaw puzzle model posits that amino acids in a protein structure fit together with perfect shape (and chemical) complementarity (5). Specific interactions between residues distant in sequence are presumed to pin the structure together. These interactions depend on the stereochemical details of amino acid side chains, such as shape and charge, rather than simple hydrophobic or polar character.
Existing combinatorial mutagenesis studies of enzymes favor the oil-droplet hypothesis. For example, when the hydrophobic core of the ribonuclease barnase is randomized at 13 positions, one-quarter of the variants are functional (6). Similarly, a mutant of T4 lysozyme with 10 methionine substitutions in its hydrophobic core retains 20% of the wild-type activity (7). These studies suggest that the specific identities of core residues are not essential for forming a catalytically active enzyme structure. Taken further, one might infer that novel barrel structures could be engineered by appropriately patterning hydrophobic and polar residues in an amino acid sequence. Five attempts to design (β/α)8 barrel proteins by H/P patterning have been reported (8, 9). All of the designs produced barrels with ill-defined and fluctuating tertiary structures. Similar results have been observed for essentially all globular protein designs that disregard specific side-chain interactions.
The juxtaposition of mutagenesis and design experiments suggests that amino acids outside the hydrophobic core help to determine the conformations of naturally occurring enzymes. Charged side chains on the protein surface, buried polar residues, secondary structure punctuation motifs, and shape-specific side-chain packing all have been identified as important determinants of structure in helical proteins. To investigate which, if any, of these sequence elements may help to specify the (β/α)8 fold, we mutagenized every structural residue in the canonical (β/α)8 barrel protein triosephosphate isomerase (TIM).
Materials and Methods
H/P Library Construction.
To construct a degenerate library, the codon VAA or VAG was used at phylogenetically polar positions (V = 21% A, 33% G, 46% C), and the codon NTC was used at phylogenetically hydrophobic positions (N = 15% T, 23% A, 26% C, 36% G). Codon usage was biased to match the overall amino acid composition of naturally occurring TIM sequences. Overlapping oligonucleotides covering the full yeast TIM gene sequence were synthesized and purified by denaturing acrylamide electrophoresis. Libraries of genes were assembled from the oligonucleotides by using the method of Stemmer et al. (10). Fifteen full-length unselected genes were sequenced to define the unselected pool for the degenerate library, and 5–19 functional genes were sequenced from each library (Table 3, which is published as supplemental material on the PNAS web site, www.pnas.org).
Selection for TIM Activity.
Selection assays were performed by using the pKK223f plasmid (see supplemental material). Mutant TIM genes were cut with EcoRI and HindIII and cloned into the corresponding sites of pKK223f. Plasmids were transformed into XL1Blue cells and plated on LB media containing 50 μg/ml carbenicillin. After growth overnight at 37°C, colonies were counted and scraped off the plate into 1.5 ml PBS. Plasmid DNA was isolated by standard techniques and transformed into the TIM-deficient Escherichia coli strain DF502 (11). Cells were plated on minimal M63 media supplemented with 0.2 mg/ml lactate, 0.5 μg/ml thiamine, 0.2 μg/ml uracil, 40 μg/ml histidine, and 50 μg/ml carbenicillin. DF502 carrying a wild-type plasmid produced colonies after 2 days at 37°C; mutant genes that produced colony growth within 4 days were scored as functional.
Gene Shuffling.
Seven genes containing the mutations listed in Table 1 were constructed as described above. Genes were amplified by PCR, gel-purified, and treated with DNase I (final concentrations: 1 ng/ml DNase I; 100 mM Tris, pH 8; 5 mM MgCl2) for 5 min at room temperature. The reactions were terminated by addition of 50 mM EDTA and loaded onto a 2.5% agarose gel. Fragments from 10–150 bp in length were isolated and purified. Mutant fragments were initially mixed with wild-type fragments in a ratio of 3:1 and assembled to generate libraries of shuffled genes (12). The genes were cloned into pKK223f, subjected to functional selection in DF502, and sequenced. If no genes were found to complement, shuffling was repeated by using a lower ratio of mutant to wild-type DNA fragments. All libraries were composed of >30% mutant DNA. Between 9 and 27 unique clones from both functional and unselected pools were sequenced for each shuffled library (Table 4, which is published as supplemental material). Crossover events could be observed due to variations in codon usage between mutant and wild-type genes. Genes contained an average of eight such events. χ2 analysis (13) was used to calculate the significance of observed differences in amino acid frequencies between the selected and unselected pools.
Table 1.
Structural classification of conserved residues
| Buried polar interactions | |
| Intersubunit salt bridge | D85N, K112Q |
| R189 network | Q182A, H185I, R189M, D225A, D227L |
| Cross β–β contacts | E37Q, R205M |
| Contacts to loops | N10A, T75V, D106A, T139V, Q146L, Y208F |
| Other polar interactions | Y46F, W90F, W157F, Y164F |
| Secondary structure punctuation | |
| α-Helix caps | S16A, S79A, D105A, T177A, N213A, N216A |
| α-Helix stops | G87A, G120A, P178A |
| β-Strand stops | G94A, G128A, P166A, G210A, G232A |
| β-Central residues | V7L, I40L, V61L, V91L, V123L, I127L, Y164L, I206L, F229L |
| Diamond residues | I20L, I23L, V24L, S50L, V54L, V80L, I83L, I109L, T113L, V143L, V150L, V154L, I184L, I188L, A217L, F220L, F240L, I243L, I244L, S246L |
| Barrel surface residues (nonvaline) | F5V, F11V, I40V, L93V, L125V, I127V, Y164V, I206V, Y208V, F229V |
| Barrel interior (nonvaline) | F6V, G8V, C41V, Q58V, T60V, G62V, W90V, I92V, G122V, I124V, C126V, A163V, L207V, G209V, G228V, L230V |
| Miscellaneous | |
| Hole residues | G9V, A63V, G87L, A116L, G120L, A181L |
| Solvent exposed | R3A, T4A, N35A, V51A, K107A, V142A, D183A, G233A |
| Turn packing | V36L, A200L, A201L, L204A, V226L |
| α-α Interface | Y46L, A110L, F191L |
| Barrel valines | V38L, V39L, V160L, V161L, V162L, V231I |
Underlined mutations were found to revert to wild-type sequence identity more frequently than expected by chance (P < 5%) in shuffling experiments. Italicized mutations cause at least a 10-fold decrease in in vivo activity when constructed as single substitutions. Some nonconserved positions were included to achieve complete coverage of a structural class. Residues I40, G87, W90, G120, I127, Y164, I206, Y208, and F229 fall into more than one structural class and were mutated to more than one amino acid. References and definitions for classes are given in the text.
Analysis of Single Mutants.
Sequence positions that exhibited nonrandom shuffling distributions (P ≤ 5%) were constructed as single mutants in an otherwise wild-type background by using the method of Kunkel et al. (14). All mutants were verified by DNA sequencing. Cultures of DF502 harboring mutant plasmids were grown overnight at 37°C, harvested by centrifugation, and lysed by sonication in 10% sucrose; 0.1 M Tris, pH 8; 1 mM EDTA. Cell extract concentrations were normalized by absorbance at 280 nm and assayed for activity by using a coupled enzymatic assay as described (15). Kinetic measurements were performed on a Uvikon model 9310 spectrophotometer (Kontron Instruments, Watford, U.K.) at 25°C. Results reported are the average of three independent measurements. Wild-type activity in this system corresponds to 70 μmol glyceraldehyde 3-phosphate converted to dihyroxyacetone phosphate per sec per A280 unit.
Results
Functional Selections.
Yeast TIM was used as a model (β/α)8 barrel protein for mutagenesis studies. We defined the active site to be the β→α loops (Fig. 1), which were held fixed in all experiments. Assuming that a well-folded (β/α)8 barrel is required for catalysis, selection for enzymatic function can be used as an in vivo assay of protein structure. The threshold of activity required for function in our experiments is approximately 10-4 of the wild-type activity (16). An apparent loss of in vivo activity could result from a number of effects, including a reduction in protein stability or the rate of folding, increased levels of proteolysis, or increased partitioning of protein to inclusion bodies.
Figure 1.
Phylogenetic conservation in TIM. The amino acid sequence of TIM is diagrammed according to secondary structure. The figure should be read from upper left to lower right. Secondary structure elements are designated at the left, with the following eight vertical columns corresponding to the eight β/α units of the protein. The yeast TIM sequence is shown in lowercase black letters (the first three amino acids are located at the lower right). The number of the first residue in each column is indicated at the top of the column. Conservation of a property in ≥75% of the sequences was required to assign a position to a conservation class. The green Hyd symbol indicates conservation of hydrophobic character [FILVAMGPWTYC] at that position. The cyan Pol symbol indicates conservation of polar character [HRKQNEDSTYC]. Red letters indicate conservation of a single amino acid or specific class of amino acids (symbols shown below). A blank space indicates no detectable conservation pattern. Conservation classes are defined as follows: Arom [FYW], Ncap [STDNQE], Beta [IVT], + [KRH], − [DE], Small [AG], 1mc [DN], 2mc [EQ], N-H [QNHW], φ,ψ [GP], StC [MRK], Mtl [HC], Nuc [STC].
Phylogenetic Alignment of TIM Sequences.
To guide our mutagenesis studies, we constructed an alignment of 43 unique TIM sequences from a wide range of species (Fig. 1 and supplemental material). Positions in the aligned sequences that conserved hydrophobic or polar character, but not a specific amino acid identity, were designated as phylogenetically hydrophobic or phylogenetically polar, respectively. Positions that maintained a single amino acid or class of amino acids in ≥75% of the aligned sequences were designated as phylogenetically conserved. Positions that did not conserve any amino acid or physical property were designated as phylogenetically variable.
We hypothesized that phylogenetically hydrophobic, polar, and variable positions would conform to the expectations of an oil-droplet model, whereas phylogenetically conserved positions would conform to the expectations of a jigsaw-puzzle model. To test this idea, two experiments were performed. In the first experiment, the amino acids at phylogenetically polar and hydrophobic positions were varied in a degenerate library that maintained the H/P pattern of the sequence. By comparing the amino acid composition of the library before and after selection for function, we could infer the existence of sequence preferences at specific positions. In the second experiment, phylogenetically conserved positions were divided into seven structural classes, each of which was completely mutated in a single gene. By recombining the mutant genes with a wild-type gene, selecting for functional sequences, and measuring the frequency at which each mutation reverted to the wild-type sequence identity, we could determine which mutations of conserved residues caused defects in protein function.
Mutability at Phylogenetically Hydrophobic and Polar Positions.
One prediction of an oil-droplet model is that residues in a protein should not exhibit shape or charge preferences beyond the H/P pattern. To test this prediction, we constructed a degenerate library that varied the physical properties of amino acids at phylogenetically hydrophobic and polar positions while maintaining the H/P pattern. Phylogenetically polar positions were substituted with a degenerate codon encoding lysine, glutamate, or glutamine; and phylogenetically hydrophobic positions were substituted with a degenerate codon encoding phenylalanine, isoleucine, leucine, or valine. Phylogenetically variable positions occupied by positively or negatively charged amino acids in the yeast TIM sequence were mutated, respectively, to lysine and glutamate, to preserve the total charge on the protein. All other phylogenetically variable positions were mutated to alanine. We set out to measure the fraction of sequences in the library that were functional and to determine whether all amino acid shapes and charges were tolerated equally at all sequence positions.
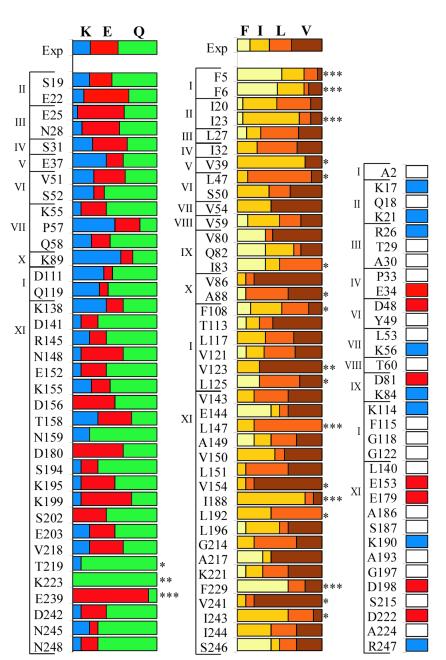
Libraries of 106 full-length degenerate genes were constructed, but contained no functional sequences after selection. It was found that the 3′ half of the degenerate library fused to a wild-type 5′ end (Fig. 2, library XI) produced functional proteins at a frequency of approximately 1 in 105. To identify functional genes with degenerate sequences in the 5′ half of the gene, further splitting of the library was required (Fig. 2, libraries I–X). By assuming that the 5′ and 3′ halves of the degenerate gene contain the same proportion of active sequences, we estimate that fewer than 1 in 1010 sequences in the full-length library are functional. This result contrasts with a previous study of barnase, which found that one in four random sequences of nonpolar amino acids were able to replace the wild-type hydrophobic core (6).
Figure 2.
Amino acid composition of functional sequences from a degenerate library. Amino acids observed in functional sequences from the degenerate library at phylogenetically polar positions (Left), phylogenetically hydrophobic positions (Center), and phylogenetically variable positions (Right). The brackets and roman numerals at the left of each graph indicate in which of the 11 libraries the position was mutated. The first row in each graph gives the average amino acid composition observed in the unselected library. Amino acids are color-coded as indicated at the top. Significant deviations from a random distribution (as determined by χ2 analysis) are indicated as follows: *, P < 5%; **, P < 1%; ***, P < 0.1%. Unconserved positions were mutated to a single amino acid. White bars indicate mutation to alanine, blue bars indicate mutation to lysine, and red bars indicate mutation to glutamate.
If all amino acids coded by a degenerate codon substitute equally well at a given position, then the amino acid composition at that position should be the same in functional sequences and in the unselected pool of sequences. Differences in amino acid composition between functional and unselected sequences presumably indicate a preference for a specific amino acid. Mutation to multiple amino acids was observed at virtually every degenerate position in the functional genes. However, six of the 40 phylogenetically hydrophobic positions showed highly significant (P < 0.1%) deviations from the distribution of amino acids in the unselected pool, whereas only one of the 36 phylogenetically polar positions showed a bias at the same level of significance.
Mutability of Conserved Residues.
Conserved positions in the phylogenetic alignment appear to violate a simple oil-droplet model and might be explained as residues that form the specific tertiary interactions predicted by a jigsaw-puzzle model. Alterations to the shape or charge of these amino acids should disrupt tertiary interactions. To test this prediction, the conserved residue positions in the aligned TIM genes were assigned to seven structural classes by examination of the yeast crystal structure (17). Because the (β/α)8 barrel is composed of a repeated structural motif, each structural class is represented by multiple residues. Analysis of the behavior of sets of residues, rather than single residues, allows more general conclusions to be drawn.
The residues in each of the seven structural classes were replaced in groups by the mutations shown in Table 1. An effort was made to construct the most conservative mutation possible at each position, so as to eliminate trivial perturbations (such as burial of a charged amino acid in the hydrophobic core or introduction of a proline into a helix). Buried polar residues were changed to hydrophobic or uncharged isosteres, conserved positions in helices were mutated to leucine because of its high helix-forming propensity, and conserved positions in β-strands were mutated to valine because of its high β-sheet propensity. Solvent exposed residues and residues involved in secondary structure punctuation were mutated to alanine.
None of the seven coordinately mutant genes was able to complement a TIM knockout in E. coli. To determine the relative detrimental effect of each substitution, the mutant genes were recombined with a wild-type gene in vitro (see Materials and Methods; we refer to this procedure below as gene shuffling). The resultant libraries were selected for functional sequences, and the frequency at which each mutation reverted to the wild-type sequence identity was scored. At 53 of 105 shuffled positions, mutant residues reverted to the wild-type sequence identity at frequencies expected by chance, suggesting that no preference for the wild-type amino acid identity exists (Table 1). Mutations at the remainder of the positions reverted to the wild-type sequence identity more frequently than expected by chance (P < 5%), suggesting that they decrease the fitness of the protein in vivo.
Single Mutant Properties.
Linkage between mutations adjacent in sequence may have influenced the apparent reversion frequencies. For example, an innocuous mutation that is located near a detrimental one will revert frequently, because crossover events between the two mutations are rare. On average, eight independent sequence segments of 30 amino acids were observed in each of the shuffled genes. Thus, mutations separated by fewer than 30 amino acids would be expected to display linkage.
To address the possibility of linkage, mutations with nonrandom reversion frequencies (P < 5%) were constructed as single substitutions in a wild-type background. The catalytic activity of each mutant was measured in a crude cellular extract (Table 2). The mutant proteins exhibited a broad, continuous range of phenotypes with most having little (<100-fold) effect on apparent specific activity. Only four single mutants (R189M, D227L, G209V, and G228V) were capable of diminishing TIM activity below the threshold required for in vivo complementation. These four positions were designated as immutable to distinguish them from the other positions. In the yeast TIM crystal structure, the guanidinium and carboxylate groups of R189 and D227 are separated by 4.8 Å (Fig. 3). To address the possibility that a single mutant at either position was stranding a charge in the hydrophobic core, the R189M/D227L double mutant was constructed. However, the double mutant was also unable to complement a TIM knockout in vivo.
Table 2.
Point mutants exhibit a broad range of phenotypes
| Not significant† | Intermediate | Deficient‡ | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R3A | W90F | Y164L | I124V | 0.7 | G87A | 3.2 | N213A/N216A | 130 | G228V | 13300 | ||||||
| T4A | V91L | Y164V | N35A | 1.0 | A201L | 3.8 | G210A | 150 | G209V | 15000 | ||||||
| V7L | L93V | P166A | A200L | 1.0 | H185I | 3.8 | G232A | 180 | D227L | 22000 | ||||||
| F11V | I109L | T177A | F240L | 1.1 | L230V | 4.5 | K107A | 210 | R189M | >22000 | ||||||
| S16A | K112Q | P178A | V24L | 1.1 | F6V | 5.8 | *** | D225A | 210 | R189M/D227L | >22000 | |||||
| I20L | − | T113L | − | A181L | I243L | 1.2 | * | Q182A | 6.3 | G9V | 230 | |||||
| E37Q | − | L125V | * | D183A | D106A | 1.2 | G8V | 6.3 | F220L | 250 | ||||||
| V39L | * | I127V | I184L | I92V | 1.3 | V36L | 6.4 | W90V | 300 | |||||||
| I40L | T139V | I188L | *** | I244L | 1.3 | − | C126V | 13 | G128A | 340 | ||||||
| I40V | V142A | F191L | L207V | 1.4 | A116L | 14 | Y208F | 1800 | ||||||||
| Y46F | V143L | − | I206L | C41V | 1.6 | G94A | 20 | G62V | 2800 | |||||||
| Y46L | Q146L | I206V | L204A | 1.7 | R205M | 28 | N10A | 3500 | ||||||||
| S50L | − | V150L | − | Y208V | I23L | 1.8 | *** | V123L | 39 | ** | A110L | 4100 | ||||
| V51A | − | V154L | * | V226L | I127L | 2.3 | F5V | 39 | *** | G122V | 4900 | A | ||||
| V54L | − | W157F | F229L | D105A | 2.3 | V38L | 39 | |||||||||
| V61L | V160L | V231I | G87L | 2.4 | A217L | 67 | − | |||||||||
| A63V | V161L | G233A | S79A | 2.6 | Q58V/T60V | 76 | A | |||||||||
| T75V | V162L | S246L | − | G120A | 2.7 | F229V | 76 | *** | ||||||||
| V80L | * | A163V | D85N | 2.7 | ||||||||||||
| I83L | * | Y164F | G120L | 2.8 | ||||||||||||
Catalytic activity of single mutants measured in crude cellular extracts expressed as fold decrease in apparent kcat/KM relative to wild type. Positions marked with asterisks were found to exhibit residue preferences in the degenerate library (Fig. 2), while positions marked with dashes tolerated multiple substitutions equally. The positions marked with A were mutated to alanine in the degenerate library.
Mutations that exhibit no preference for wild-type sequence identity in shuffling experiments.
Deficient mutants are unable to complement a TIM knockout in E. coli.
Figure 3.
Structural environment of immutable residues. Top (A) and side (B) views of TIM showing the location of immutable residues. R189 is colored blue, D227 is colored red, and G209 and G228 are colored yellow. The backbone of the region corresponding to the C-terminal subdomain of tryptophan synthase is colored green. The N and C termini are indicated by black spheres. The active site is located toward the reader in A and at the top in B.
Discussion
Combinatorial Mutagenesis as a Structural Probe.
An additive threshold model provides one simple framework for interpreting the results of the combinatorial mutagenesis presented here. In this model, proteins are represented as sets of cooperatively unfolding subdomains. Mutations have detrimental effects on the fitness of the subdomains, and the effects of multiple mutations are assumed to be additive. When the sum of the effects of mutations in any subdomain exceeds its fitness threshold, the protein will fail to function. The larger the detrimental effect of a mutation, the fewer the number of ways that it can be combined with other mutations in a functional sequence. In one limiting case, a single mutation with an individual effect that exceeds the fitness threshold of its subdomain will never appear in a functional protein. Alternatively, a mutation with zero effect on fitness can combine with other mutations to form a functional protein in as many ways as the wild-type amino acid. Therefore, the frequency at which a mutation occurs in the selected pool of sequences will be inversely proportional to the magnitude of its detrimental effect. Analysis of mutation reversion frequencies and residue compositions in combinatorial libraries allows detection of detrimental effects of mutations that, as single substitutions, would not produce observable phenotypes. If the selections reported here had been performed on an exhaustive set of point mutants, only the four immutable positions would have been discovered.
Two Distinct Hydrophobic Cores.
The hydrophobic cores of proteins are tightly packed, which has led to the suggestion that side-chain shape may play a role in determining the protein fold (5). The (β/α)8 barrel has two separate hydrophobic cores: one between the helices and the surface of the beta barrel, and a second in the interior of the β barrel.
The packing of α-helices against β-sheets has been described phenomenologically (18). Four residues designated as i, i+3, i+4, and i+7 in the α-helix form a hydrophobic diamond that packs around a single residue in the β-sheet (designated j). A small side chain is often incorporated at position i or i+7 of the helix, or at position j-2 or j+2 of the strand, to prevent a steric clash. The small residues are termed hole residues, the helix residues are termed diamond residues, and the β-sheet residue at position j is termed the β-central residue (Table 1). Analysis of the mutagenesis data reveals a high level of tolerance to shape substitutions in the α/β interface. Mutations at only two βcentral residues measurably decrease TIM activity by the shuffling assay, and only one decreases activity by more than 10-fold as a single substitution (Table 2). The data do not support a previous proposal that β-central positions require β-branched amino acids (18). Point mutations of two diamond residues decrease activity by more than 10-fold, but both substitutions lie in a putative unstable subdomain of the protein (see below). Four of the six hole residue mutations decrease protein fitness by the shuffling assay, and single substitutions of two decrease activity in cell extracts by more than 10-fold. Taken together, the data indicate that packing of the α/β interface is extremely flexible.
In contrast to the α/β interface, residues in the central core of the β barrel are extremely sensitive to substitution. Coordinate replacement of all barrel interior residues with valine would result in a volume decrease of a single methyl group. An all-valine core therefore should produce a functional protein if conservation of volume were the only constraint. However, this mutant protein is not functional, and 13 of 18 positions revert to wild-type sequence identity at high frequency in shuffling experiments (Table 1). Eight of the β-interior point mutations decrease in vivo activity between 10- and 5,000-fold, and two render the protein unable to complement a TIM knockout in E. coli (Table 2).
Secondary Structure Punctuation.
Characteristic sequence motifs at the ends of peptide α-helices have been shown to help specify the local conformation of the polypeptide chain (19). The importance of these secondary structure punctuation motifs in a protein context is still unknown. One class of motifs, α-helix caps, consists of polar residues present at the N-terminal ends of helices. These residues form hydrogen bonds to exposed amide protons (20). Of the eight helices in TIM, five have capping sequences. Only mutation of the N213/N216 capping box, however, is observed to produce greater than a 10-fold effect on activity. A second class of motifs, helix stop signals, consists of glycine and proline residues at either end of α-helices. These residues are thought to prevent propagation of helical secondary structure into adjacent sequence (19). Two glycines and a single proline in TIM fall into this category. Mutation of each of these residues reduces in vivo activity by less than 10-fold.
Glycine and proline residues also destabilize β-sheet structures and can presumably block extension of β-strands when present at strand termini. Four glycines in TIM are located at the C-terminal ends of β-strands, and mutation of each diminishes in vivo activity by more than 10-fold. These glycines have backbone dihedral angles in restricted regions of Ramachandran space. Mutation of the single proline present at the C-terminal end of β-strand 6, however, has no measurable effect. Interestingly, all structural prolines in TIM are amenable to substitution. Of the secondary structure punctuation motifs present in TIM, the glycine β-stops appear to be most important.
Polar Interactions.
Although it is generally assumed that polar residues can serve interchangeably at solvent-exposed positions in a protein structure, few studies have attempted to test this hypothesis. In contrast to the phylogenetically hydrophobic positions, only one of 36 phylogenetically polar positions in TIM shows significant sequence bias (P < 0.1%). Mutations at conserved surface positions also result in small effects, with the exception of the K107A substitution (Tables 1 and 2). Removal of the intersubunit salt bridge formed between residues D85 and K112 upon dimerization of TIM has a negligible effect on function. The data suggest that sequence requirements at the protein surface are less strict than in the core, and that surface charge patterning is not required to specify the TIM fold.
In the rare case that a polar residue appears in the hydrophobic interior of a protein, it is almost always hydrogen-bonded to other polar functional groups (21). Consequently, interactions between buried polar residues are likely candidates for the specific tertiary contacts predicted in a jigsaw-puzzle model. Mutations in a network of amino acids centered at residues R189 and D227 are found to have large detrimental effects on protein function (Table 2). Residues R189 and D227 are located on the opposite side of the barrel from the active site and are well shielded from solvent in the native structure (89% and 97% buried by solvent accessible surface area calculations). These residues appear to form a salt bridge (Fig. 3). Mutation of two polar residues buried in the β-interior (R205 and N10) also decreases considerably enzymatic activity in vivo.
A C-Terminal Subdomain in TIM?
All of the deficient mutations and seven of 15 severe mutations in Table 2 lie C-terminal to position 189, which includes the secondary structures from helix 6 to the end of the protein (Fig. 3). Fragment complementation studies of TIM (22) and N-(5′-phosphoribosyl)anthranilate isomerase (23), another (β/α)8 barrel enzyme, suggest that these proteins contain two subdomains whose boundary is located in β→α loop 6. It has been proposed that the corresponding N-terminal domain of the (β/α)8 barrel enzyme tryptophan synthase forms a folding intermediate, and that folding of the C-terminal domain is rate-limiting (24). If TIM folds analogously, mutations in the C-terminal domain might be expected to increase the lifetime of a transient, aggregation-prone species. The detrimental effects of many of the mutations identified here appear to result from aggregation during folding in vivo and in vitro (unpublished work), consistent with this hypothesis.
Implications for Engineering (β/α)8 Barrels.
Based on our data, several general observations about the TIM sequence can be made. Packing in the α/β interface, turn packing, α-helix capping, and α-helix stop signals all are found to be highly mutable. In contrast, packing of the interior of the barrel is extremely inflexible and should be granted special attention in a design effort. Glycine β-stop signals also appear to be particularly important. Identification of the R189 network indicates that buried polar residues can play a crucial role in determining (β/α)8 barrel structures. It will be interesting to see how these observations generalize to other barrel proteins.
The hydrophobic core of TIM is more sensitive to mutation than has been observed for other enzymes. Although mutations at 97 positions show no measurable effect on function, we estimate that fewer than 1 in 1010 sequences in our degenerate library are able to complement in vivo. This paradoxical observation may indicate that many local volume constraints must be satisfied to produce a functional protein. Thus, although multiple residues are suitable at each position, the combinatorial possibilities are limited. Alternatively, the fixed mutations in our library at the phylogenetically variable positions may have reduced the fitness of the protein to a point close to its complementation threshold. Given that such a small proportion of the degenerate sequences are functional, even though they contain all of the active site and phylogenetically conserved residues as well as the proper H/P pattern, it is not surprising that previous attempts to design (β/α)8 barrels failed.
Several recent studies have focused on reducing the size of the amino acid alphabet required to encode proteins (reviewed in ref. 25). A simplified alphabet could greatly decrease the computational burden of protein design. At 97 of the 182 structural positions analyzed here, mutation to one of seven amino acids (FVLAKEQ) had no measurable effect on TIM activity. At an additional 45 positions, the wild-type identity is already one of these seven residues. Thus, 142 of 182 structural positions could be readily reduced to a seven-letter alphabet. Fifteen of the remaining positions are phylogenetically variable, although our experiments do not directly measure the effects of substitutions at these positions. Given these results, it seems likely that TIM, and perhaps (β/α)8 barrels generally, could be encoded with a simplified alphabet.
Supplementary Material
Acknowledgments
We thank R. Baldwin, P. Brown, J. Frydman, D. Halpin, J. Havranek, and D. Herschlag for helpful discussions. J.S. is supported by the Paul and Mildred Berg Stanford graduate fellowship. R.B. was the recipient of a McCormick Scholars fellowship. This research was supported by a junior faculty award from the Howard Hughes Medical Institute to P.B.H., a grant from the Chicago Community Trust to P.B.H., and a Terman fellowship to P.B.H.
Abbreviations
- H/P
hydrophobic/polar
- TIM
triosephosphate isomerase
Footnotes
See commentary on page 2958.
References
- 1.Reardon D, Farber G K. FASEB J. 1995;9:497–503. doi: 10.1096/fasebj.9.7.7737457. [DOI] [PubMed] [Google Scholar]
- 2.Altamirano M M, Blackburn J M, Aguayo C, Fersht A R. Nature (London) 2000;403:617–622. doi: 10.1038/35001001. [DOI] [PubMed] [Google Scholar]
- 3.Jurgens C, Strom A, Wegener D, Hettwer S, Wilmanns M, Sterner R. Proc Natl Acad Sci USA. 2000;97:9925–9930. doi: 10.1073/pnas.160255397. . (First Published August 15, 2000; 10.1073/pnas.160255397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamtekar S, Schiffer J M, Xiong H, Babik J M, Hecht M H. Science. 1993;262:1680–1685. doi: 10.1126/science.8259512. [DOI] [PubMed] [Google Scholar]
- 5.Ponder J W, Richards F M. J Mol Biol. 1987;193:775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- 6.Axe D D, Foster N W, Fersht A R. Proc Natl Acad Sci USA. 1996;93:5590–5594. doi: 10.1073/pnas.93.11.5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gassner N C, Baase W A, Matthews B W. Proc Natl Acad Sci USA. 1996;93:12155–12158. doi: 10.1073/pnas.93.22.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka T, Kimura H, Hayashi M, Fujiyoshi Y, Fukuhara K, Nakamura H. Protein Sci. 1994;3:419–427. doi: 10.1002/pro.5560030306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Houbrechts A, Moreau B, Abagyan R, Mainfroid V, Preaux G, Lamproye A, Poncin A, Goormaghtigh E, Ruysschaert J M, Martial J A, Goraj K. Protein Eng. 1995;8:249–259. doi: 10.1093/protein/8.3.249. [DOI] [PubMed] [Google Scholar]
- 10.Stemmer W P, Crameri A, Ha K D, Brennan T M, Heyneker H L. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 11.Straus D, Gilbert W. Proc Natl Acad Sci USA. 1985;82:2014–2018. doi: 10.1073/pnas.82.7.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stemmer W P. Proc Natl Acad Sci USA. 1994;91:10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agresti A. Categorical Data Analysis. New York: Wiley; 1990. [Google Scholar]
- 14.Kunkel T A, Bebenek K, McClary J. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 15.Nickbarg E B, Knowles J R. Biochemistry. 1988;27:5939–5947. doi: 10.1021/bi00416a018. [DOI] [PubMed] [Google Scholar]
- 16.Hermes J D, Parekh S M, Blacklow S C, Koster H, Knowles J R. Gene. 1989;84:143–151. doi: 10.1016/0378-1119(89)90148-0. [DOI] [PubMed] [Google Scholar]
- 17.Davenport R C, Bash P A, Seaton B A, Karplus M, Petsko G A, Ringe D. Biochemistry. 1991;30:5821–5826. doi: 10.1021/bi00238a002. [DOI] [PubMed] [Google Scholar]
- 18.Cohen F E, Sternberg M J, Taylor W R. J Mol Biol. 1982;156:821–862. doi: 10.1016/0022-2836(82)90144-9. [DOI] [PubMed] [Google Scholar]
- 19.Gunasekaran K, Nagarajaram H A, Ramakrishnan C, Balaram P. J Mol Biol. 1998;275:917–932. doi: 10.1006/jmbi.1997.1505. [DOI] [PubMed] [Google Scholar]
- 20.Aurora R, Rose G D. Protein Sci. 1998;7:21–38. doi: 10.1002/pro.5560070103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chothia C. J Mol Biol. 1976;105:1–12. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- 22.Bertolaet B L, Knowles J R. Biochemistry. 1995;34:5736–5743. doi: 10.1021/bi00017a005. [DOI] [PubMed] [Google Scholar]
- 23.Eder J, Kirschner K. Biochemistry. 1992;31:3617–3625. doi: 10.1021/bi00129a010. [DOI] [PubMed] [Google Scholar]
- 24.Zitzewitz J A, Matthews C R. Biochemistry. 1999;38:10205–10214. doi: 10.1021/bi9909041. [DOI] [PubMed] [Google Scholar]
- 25.Plaxco K W, Riddle D S, Grantcharova V, Baker D. Curr Opin Struct Biol. 1998;8:80–85. doi: 10.1016/s0959-440x(98)80013-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.