This study compared mtDNA damage accumulation with aging and age-related macular degeneration (AMD) in the human RPE. The findings support the hypothesis that mitochondrial defects are an underlying pathologic event in AMD.
Abstract
Purpose.
Increasing evidence suggests a central role for mitochondrial (mt) dysfunction in age-related macular degeneration (AMD). Previous proteomic data from the retinal pigment epithelium (RPE) revealed significant changes to mt proteins, suggesting potential functional defects and damage to mitochondrial DNA (mtDNA) with AMD progression. The present study tests the hypothesis that mtDNA damage increases with aging and AMD.
Methods.
Genomic DNA was isolated from the macular region of human donor RPE graded for stages of AMD (Minnesota Grading System [MGS] 1–4). Region-specific mtDNA damage with normal aging was evaluated in 45 control subjects (ages 34–88 years, MGS 1) and AMD-associated damage in diseased subjects (n = 46), compared with that in age-matched control subjects (n = 26). Lesions per 10 kb per genome in the mtDNA and nuclear DNA were measured with long-extension polymerase chain reaction (LX PCR). The level of deleted mtDNA in each donor was measured with quantitative real-time PCR (qPCR).
Results.
With aging, an increase in mtDNA damage was observed only in the common deletion region of the mt genome. In contrast, with AMD, mtDNA lesions increased significantly in all regions of the mt genome beyond levels found in age-matched control subjects. mtDNA accumulated more lesions than did two nuclear genes, with total damage of the mt genome estimated to be eight times higher.
Conclusions.
Collectively, the data indicate that mtDNA is preferentially damaged with AMD progression. These results suggest a potential link between mt dysfunction due to increased mtDNA lesions and AMD.
Age-related macular degeneration (AMD) is a progressive eye condition that is the leading cause of legal blindness in individuals older than 65 in the developed world.1 By affecting the central (macular) region of the human retina, AMD degrades the central visual function that subserves reading, driving, writing, and face recognition. The loss of these abilities significantly affects daily function and quality of life.2 Currently approved treatment options for AMD are available to a limited number of patients with advanced wet-type AMD, in which the treatments limit vision loss by inhibition of vascular leakage3,4 but do not address disease pathogenesis. In addition, the inconvenience, cost, and quality of life that result from monthly injections increase the burden on patients as well as the health care system. Despite recent improvements, many patients still progress to the advanced stages of AMD. The next major steps in sight preservation include strategies centered on early detection, treatment of the early stages, and prevention. Importantly, studying the early disease process is the first step toward the development of new treatments.
Many investigators believe that the pathogenesis of AMD begins in the retinal pigment epithelium (RPE), a monolayer of cells between the neural retina and the retinal basement membrane (Bruch's membrane). The RPE maintains retinal health and homeostasis by photoreceptor phagocytosis, nutrient transport, and secretion of growth factors.5 Drusen, the earliest clinical sign of AMD, appear beneath the RPE as yellow, lipoproteinaceous deposits. Drusen features (e.g., size, character, quantity, and shape) provide the basis for the grading system used in this study (Minnesota Grading System, MGS6). This system replicates a widely used clinical research classification system7 by linking common definitions from clinics to eye bank tissue (ex vivo). Thus, the use of MGS provides a methodology to directly link biochemical changes in human donor eyes at distinct stages of AMD to clinical phenotypes in patients.6
Converging evidence from several recent studies implicate mitochondrial (mt) damage in the AMD disease process.8 Severe disruptions to mt cristae structure and a decreased number of mitochondria were reported in a morphologic analysis of AMD donor RPE.9 Our first proteomic analysis of the RPE identified altered mt proteins with AMD progression,10 thus prompting an in-depth investigation of the mt proteome. The second proteomic analysis suggested potential damage to mtDNA with AMD.11 Since aging is a strong risk factor for AMD, the present study evaluated the extent of mtDNA damage in human RPE with both aging and AMD progression. To our knowledge, this is the first study to distinguish damage associated with normal from pathologic aging (i.e., AMD). Our data revealed low mtDNA damage with normal aging compared with AMD and elevated damage preceding significant macular degeneration and vision loss. Collectively, these results suggest a role for mtDNA damage in AMD.
Methods
Tissue Procurement and Grading
Human donor eyes were obtained from the Minnesota Lions Eye Bank (Minneapolis, MN) in accordance with the criteria outlined in the Declaration of Helsinki and as approved by the Institutional Review Board at the University of Minnesota. Globes were stored at 4°C in a moist chamber after enucleation until they were photographed and processed for evaluation of ocular disease. Tissue dissection was performed as outlined elsewhere.12,13 Macular RPE cells were isolated with a 6-mm trephine punch positioned over the macula. Criteria established by the Minnesota Grading System (MGS) were used to classify the globes into 1 of 4 stages.6 MGS 1 represents the control group. MGS 2, 3, and 4 are early, intermediate, and advanced stages of AMD, respectively. Advanced AMD (MGS 4) is characterized as the end stage, with central geographic atrophy (GA; dry AMD) or choroidal neovascularization (CNV; wet AMD). Of the 11 donors in the MGS 4 category in the present study (Fig. 1, Table 1), 3 exhibited characteristics of both wet and dry AMD, 4 had wet AMD, 3 had dry AMD, and 1 had no type specified. All tissues were dissected fresh and were frozen at −80°C until analysis. Exclusion criteria for the present study included a history of diabetes or glaucoma; clinical symptoms of diabetic retinopathy, advanced glaucoma, and myopic degeneration; or atypical debris in the eyes.
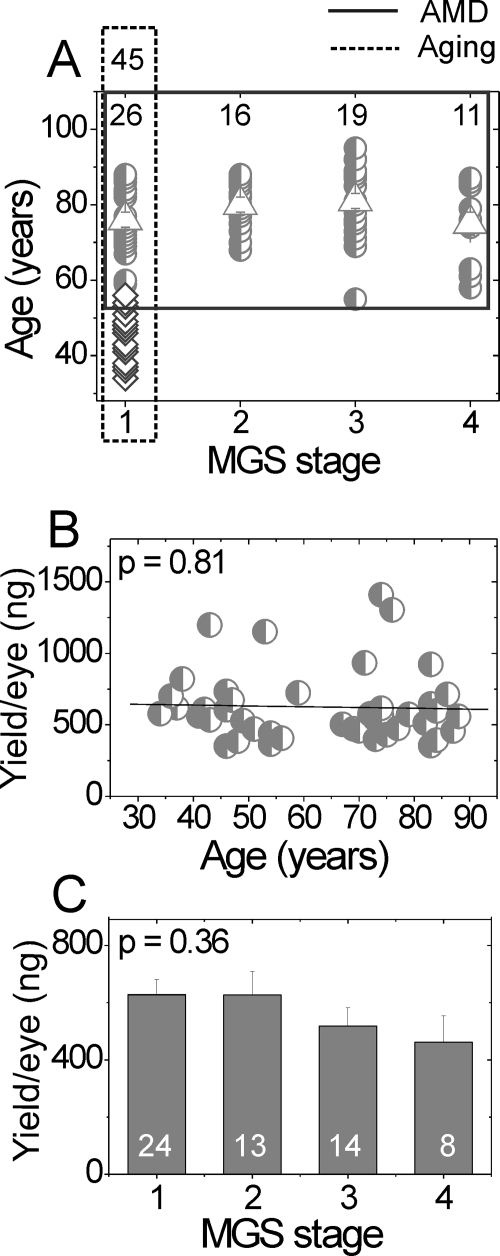
Figure 1.
Donor ages and DNA yield in aging and AMD comparisons. (A) Age of donors. Dashed line: outlines the control donors used in the aging comparison (n = 45). Diamonds: donors aged 34 to 56 years (n = 19); circles: donors aged 59 to 88 (n = 26). These donors (circles) are also the age-matched control subjects for comparison with the AMD group. Solid line: outlines the age-matched donors (half-filled circles) used in the AMD-comparison (n = 72). The number of donors for each MGS stage is shown at top. Triangles: mean age of each age-matched MGS group. No significant difference was detected for the mean age of each MGS stage (one-way ANOVA, P = 0.18). (B, C) DNA yield for control (MGS 1) donors in the aging comparison (B) and the AMD comparison (C). Data are the mean ± SEM. DNA was isolated from a macular punch. No significant difference was detected for DNA yield with either aging (regression analysis, P = 0.81) or MGS stage (one-way ANOVA, P = 0.36).
Table 1.
Donor Demographics and Clinical Information*
| MGS Grade | Sample (n) | Sex |
Age (y) |
Cause of Death (n)† | ||
|---|---|---|---|---|---|---|
| M | F | Mean ± SD | Range | |||
| 1 (young) | 19 | 12 | 7 | 45 ± 6 | 34–56 | Cancer (12), CVA (1), organ failure (1), renal disease (1), respiratory (2), sepsis (2) |
| 1‡ | 26 | 13 | 13 | 76 ± 8 | 59–88 | ALS (1), cardiogenic shock (1), cancer (9), organ failure (2), CVA (1), hemorrhage (2), myocardial infarct (2), respiratory (4), sepsis (4) |
| 2 | 16 | 9 | 7 | 80 ± 6 | 68–88 | Cancer (4), CVA (2), hemorrhage (1), head injury (1), myocardial infarct (2), PF (1), renal disease (1), respiratory (2), sepsis (2) |
| 3 | 19 | 11 | 8 | 81 ± 10 | 55–95 | ACE (3), cardiomyopathy (1), cancer (6), myocardial infarct (2), renal failure (3), respiratory (4) |
| 4 | 11 | 3 | 8 | 75 ± 11 | 58–87 | Cancer (4), organ failure (1), CVA (1), renal disease (3), respiratory (1), hemorrhage (1) |
CVA, cardiovascular accident; ALS, amyotrophic lateral sclerosis; PF, pulmonary fibrosis; ACE, acute cardiac event. Organ failure includes multiple organ failure, and cardiac failure.
Information supplied by the Minnesota Lions Eye Bank.
The number of donors for each cause of death indicated in parentheses.
These donors were used as the age-matched control in the AMD comparison and were also included in the age comparison.
DNA Isolation and Quantification
Total genomic DNA was isolated from the macular RPE or cultured cells (QIAamp DNA mini kit; Qiagen, Valencia, CA) and quantified (Picogreen dsDNA Assay Kit; Invitrogen, Carlsbad, CA), according to the manufacturers' protocols. The dye fluorescence was measured with a multiwell plate reader (CytoFluor; PerSeptive Biosystems, Inc., Framingham, MA) with a 485-nm excitation filter and 535-nm emission filter. Lambda DNA provided with the kit was used to construct a standard curve of known DNA concentrations. Because of low DNA yield from some donors, all samples could not be included in each PCR analysis. Thus, there may be differences in sample numbers per group between analyses.
Cell Culture
Human ARPE-19 cells were grown to confluence in 75-cm2 flasks in DMEM:F12 growth medium containing 0.5 mM sodium pyruvate, 50 U/mL penicillin, 50 mg/mL streptomycin, and 10% fetal bovine serum. The human osteosarcoma 143B and cytoplasmic hybrid (cybrid) ΔH2–1 cell lines were generous gifts from Carlos T. Moraes (University of Miami) and Edgar A. Arriaga (University of Minnesota), respectively. The 143B cells were cultured in 0.22-μm filtered MEM medium (pH 7.4) containing 10% (vol/vol) fetal bovine serum. ΔH2–1 cells were cultured in high-glucose DMEM containing 10% (vol/vol) fetal bovine serum, 50 μg/mL uridine, and 10 μg/mL gentamicin. All cell types were maintained at 37°C and 10% CO2.
Long Extension PCR
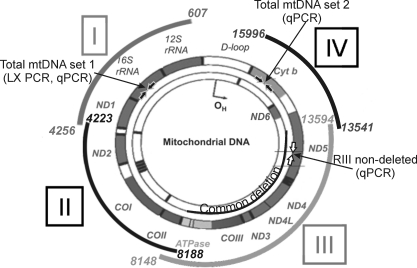
The LX PCR assay was performed as previously described14 (GeneAmp XL PCR kit; Applied Biosystems, Inc. [ABI], Foster City, CA). Conditions for each primer set (i.e., template amount, magnesium concentration, and cycle number) were empirically optimized to ensure that the assay was in the linear range. Half-template samples and no-DNA control samples were run on each plate, to verify linearity and as a negative control, respectively. The PCR amplification profile included an initial denaturation for 1 minute at 94°C, followed by 20 cycles for regions I, III, IV and 22 cycles for region II of denaturation for 15 seconds at 94°C, annealing/extension at 66°C for 12 minutes, and a final extension at 72°C for 10 minutes. A small 191-bp mtDNA fragment (total mtDNA set 1, Fig. 2, Table 2) in the 16S rRNA gene was amplified to correct for differences in total mtDNA copies in each sample. An additional measurement of the total mtDNA content was achieved by amplifying the Cyt b gene (222 bp) of the mtDNA (total mtDNA set 2; Fig. 2, Table 2) that was used to validate the total measurement for each sample. The amplification profile for both small fragments included an initial denaturation for 1 minute 94°C; followed by 20 cycles of 15 seconds at 94°C, 45 seconds at 66°C, and 45 seconds at 72°C; and a final extension at 72°C for 10 minutes. The coefficient of variation for LX PCR determined for four samples assayed on four different days was 11% ± 0.2%.
Figure 2.
Primer locations within the mt genome. Primers were designed to amplify discrete regions (I-IV) of the mt genome. The area spanned by each region is shown as a curved segment outside the mt genome, and base pair locations are provided. The primer set amplifying a 191-bp region that gives a measure of the total amount of mtDNA (total mtDNA set 1) is shown within the 16S rRNA gene (filled block arrows). A second set of primers amplifying a 222-bp region in the Cyt b gene provided a second measure of total mtDNA content for this study (total mtDNA primer set 2, solid block arrows). The RIII-ND primer set, which binds within the CD, is shown in the ND5 gene (open block arrows). The 4977-bp CD is shown within the mt genome. 12S and 16S rRNA are genes for mt ribosomal RNA. Genes for electron transport proteins are ND1–6, COI-III, ATPase 6, ATPase 8, and Cyt b. D-loop is the noncoding region. The schematic of the mt genome was modified with permission from Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Gen. 2005;6:389–402.
Table 2.
Primer Locations and Sequences for PCR Amplification
| Primer Target (Direction)* | Location (bp) | Sequence (5′–3′) | Length† (bp) | Amplicon‡ (bp) |
|---|---|---|---|---|
| Region I (F) | 607 | CAC TGA AAA TGT TTA GAC GGG CTC ACA | 27 | 3,649 |
| Region I (R) | 4,256 | GAG GGG GAA TGC TGG AGA TTG TAA TG | 26 | |
| Region II (F) | 4,223 | CCA TAC CCA TTA CAA TCT CCA GCA TTC C | 28 | 3,965 |
| Region II (R) | 8,188 | CTC CAC AGA TTT CAG AGC ATT GAC CG | 26 | |
| Region III (F) | 8,148 | GAC CGG GGG TAT ACT ACG GT | 20 | 5,446 |
| Region III (R) | 13,594 | TGT CAG GGA GGT AGC GAT GA | 20 | |
| Region IV (F) | 13,541 | CAT ACA CAA ACG CCT GAG CCC TAT CT | 26 | 2,454 |
| Region IV (R) | 15,996 | GCT TTG GGT GCT AAT GGT GGA GTT AAA | 27 | |
| Total mtDNA set 1 (F)§ | 2,671 | CAG TGA AAT TGA CCT GCC CGT GAA | 24 | 191 |
| Total mtDNA set 1 (R)§ | 2,862 | TCT TAG CAT GTA CTG CTC GGA GGT | 24 | |
| Total mtDNA set 2 (F)§ | 14,619 | CCC CAC AAA CCC CAY YAC YAA ACC CA | 26 | 222 |
| Total mtDNA set 2 (R)§ | 14,841 | TTT CAT CAT GCG GAG ATG TTG GAT GG | 26 | |
| Beta globin (F)‖ | 48,510 | CGA GTA AGA GAC CAT TGT GGC AG | 23 | 13,500 |
| Beta globin (R)‖ | 62,029 | GCA CTG GCT TAG GAG TTG GAC T | 22 | |
| HPRT (F)‖ | 11,525 | TGC CTG CTG TAT AGC ACT ATG CCT | 24 | 13,472 |
| HPRT (R)‖ | 24,973 | GCT CTA CCC TAT CCT CTA CCG TCC | 24 | |
| Total mtDNA set 1 template (F)¶ | 2,410 | TAC CCT CAC TGT CAA CCC AAC ACA | 24 | 627 |
| Total mtDNA set 1 template (R)¶ | 3,061 | GTG CAG CCG CTA TTA AAG GTT CGT | 24 | |
| RIII-ND template (F)¶ | 12,203 | TAC CCT CAC TGT CAA CCC AAC ACA | 24 | 583 |
| RIII-ND template (R)¶ | 12,810 | GTG CAG CCG CTA TTA AAG GTT CGT | 24 | |
| Region III-ND (F)# | 12,355 | TAA CCA CCC TAA CCC TGA CTT CCC TA | 26 | 191 |
| Region III-ND (R)# | 12,547 | TGT GGC TCA GTG TCA GTT CGA GAT | 24 | |
| Beta globin short (F)** | 62,367 | CTT GGG TTT CTG ATA GGC AC | 20 | 147 |
| Beta globin short (R)** | 62,514 | CTT AGG GTT GCC CAT AAC AG | 20 |
Primer target, primer target region on the mitochondrial genome (refer to Fig. 2).
Length, length of primer.
Amplicon, length of amplified product.
Primer set 1 in the 16S rRNA and set 2 in the Cyt b genes of the mtDNA.
Primer sets used to quantify damage in the nuclear genome.
Primer sets amplifying template fragments for total mtDNA and RIII-ND primer sets.
Region III nondeleted (ND) primer set (Fig. 2).
Obtained from rtprimerdb.org, ID: 3897.
Damage to the nuclear genome was quantified by amplifying 13.5-kb fragments of the human β-globin and hypoxanthine phosphoribosyltransferase (HPRT) genes. The PCR profiles for these fragments were the same as those for the large mtDNA fragments, except that the cycle number was 26 and the annealing/extension temperature was 64°C. A small nDNA fragment (147 bp) in the β-globin gene was amplified to normalize for differences in total amounts of nDNA. The same PCR program as the small mtDNA fragment was used except the annealing temperature was 55°C. All PCR reactions were performed in triplicate. All reactions were periodically run on agarose gels, to confirm that the amplicon size was correct.
Calculation of DNA Lesion Frequency
Lesion frequency was calculated according to the Poisson equation, assuming a random distribution of lesions.15,16 The equation f(x) = e−λ λx/ x! for the 0-class molecules (x = 0 for molecules exhibiting no damage) calculates lesion frequency (λ) per length of DNA amplified. This equation can be redefined for x = 0, λ = −ln AD/AO, where AD is the amplification of damaged templates and AO is the amplification of undamaged templates. Lesion frequency is presented as lesions per 10 kb per genome (both strands) of the mtDNA. Genomic DNA from ARPE-19 cells was used as an undamaged template for the age comparison due to the unavailability of a very young donor, which would be the optimal control (no damage) for our analysis. Since the ARPE-19 cells gave the highest amplification for each region, suggesting lower levels of damage compared with the donor RPE, we felt they were a valid estimate of an undamaged template. For the AMD comparison, average relative amplification of age-matched control subjects (MGS 1) was used as the denominator for the calculation.
Quantitative Real-Time PCR
Reactions for qPCR were performed (Q5 Multicolor Real-Time PCR Detection System; Bio-Rad, Hercules, CA). Three sets of primers were used in the qPCR analysis (Table 2; total mtDNA set 1, region III-nondeleted [ND], and β-globin short). The RIII-ND primer set is located within the 4977-bp common deletion (CD). It amplifies only if that region is present, thus giving a measure of intact (nondeleted) mt genomic copies. Total mtDNA copies were quantified by amplifying 16S rRNA mt gene (total mtDNA) and normalized to the copies of invariable β-globin nuclear gene (147 bp). For each primer set, the reaction was conducted in triplicate in a 25-μL final volume containing 1.0 mM Mg2+ (IMMOLASE; Bioline), 200 μM dNTP (ABI), 1× buffer (IMMOLASE; Bioline), 10 nM fluorescein (USB Corp., Cleveland, OH), 0.15× SYBR green (Invitrogen), 800 nM forward and reverse primer (Integrated DNA Technologies, Coralville, IA), 0.4 U/μL DNA polymerase (IMMOLASE; Bioline), and 0.25 mg/mL bovine serum albumin (Sigma-Aldrich, St. Louis, MO). Reactions were performed with the following parameters: 10 minutes at 95°C, followed by 30 cycles of 94°C for 30 seconds, 55°C for 45 seconds, and 72°C for 1 minute. A final extension was performed at 72°C for 5 minutes. An RPE genomic sample was included in each run as the calibrator. A standard curve was included for each primer set to confirm high-amplification efficiency. The efficiency for all primer sets tested was in the 92% to 100% range. Normalized change in expression was determined using the modified Livak (2−ΔΔCt) method. Verification of a single product was accomplished by including a melting curve with each PCR program and periodically running reactions on agarose gels.
To calculate the percentage of deleted mt genomes in our samples, we used the primer sets total mtDNA set 1 and RIII-ND to measure total mt genomes and nondeleted mt genomes, respectively. To accurately quantify copies of total and nondeleted genomes, we constructed standard curves using qPCR as follows. Template fragments for total mtDNA set 1 and RIII-ND were generated by the template primer sets shown in Table 2. These fragments were amplified from ARPE-19 genomic DNA, run on agarose gels, excised, and purified with a gel extraction kit according to the manufacturer's protocol (QIAquick; Qiagen). To verify that amplicon sequence matches the expected regions of the mt genome, we sequenced the gel-purified fragments using dye termination chemistry (Prism BigDye Terminator ver. 3.1 Cycle Sequencing Kit; ABI). Subsequently, template fragments were quantified with a dye fluorescence assay (Picogreen; Invitrogen) as described previously and the number of copies were calculated based on the approximate molecular weight of a base pair (650 Da) and the length of our fragments. Standard curves were constructed for both the total mtDNA set 1 and RIII-ND by serially diluting 103 to 107 template copies. The amplification efficiency for both dilution curves was ∼90%. As positive and negative control subjects in this assay, we used genomic DNA from cybrid ΔH2–1 cells (∼75% of mt genomes contain a 7,522-bp deletion spanning 7,982–15,504) and 143B human osteosarcoma cells, respectively.17
Statistical Analysis
Before statistical analyses, data were examined for statistical outliers (i.e., values outside three interquartile ranges from the 25th and 75th percentiles of the data distribution; NCSS software; NCSS 2001, Kaysville, UT). Data acquired from LX PCR and qPCR were analyzed with two statistical models. Lesion accumulation with aging was analyzed by linear regression (Origin Lab 7.5; OriginLab Corp., Northampton, MA). Data from donors graded for specific stages of AMD were analyzed using one-way ANOVA and Tukey-Kramer post hoc test for means comparison. When normality assumptions were violated, the Kruskal-Wallis nonparametric test and the Kruskal-Wallis z-test were used. The results are expressed as the mean ± SEM (AMD data). P ≤ 0.05 was considered statistically significant.
Results
Donor Characteristics
A summary of the donor demographics and clinical information is presented in Table 1. The average time from death to tissue cryopreservation for the donors used in this study was 17.3 ± 4.3 hours (mean ± SD, n = 91) and was not significantly different between the MGS donor groups (P = 0.45). As shown in the distribution of donor ages (Fig. 1A), 45 donors (34–88 years) categorized as MGS 1 were used to assess DNA damage with normal aging. Age-matched donors from each of the four MGS categories were used for investigating DNA damage with AMD. There was no significant difference in average age between the four MGS donor groups (78 ± 9 years) used in the AMD comparison (P = 0.18, n = 72).
Decrease in Total mtDNA with Aging but Not with AMD
Total genomic DNA isolated from the RPE macular region yielded 589 ± 29 ng per eye (n = 78, mean ± SEM). There was no significant difference in total DNA yield with either aging (P = 0.81, n = 43, Fig. 1B) or between MGS groups (P = 0.36, n = 59, Fig. 1C). It is important to note that the variability in DNA yield observed between samples could mask age- or MGS stage–specific differences. This possibility is especially true of the MGS comparison. For example, the MGS 4 category contains a mixture of donors exhibiting characteristics of either CNV or GA. Yields (mean nanograms ± SEM) for donors with each characteristic were as follows: CNV, 195 ± 105 (n = 2); GA, 637 ± 177 (n = 3, includes both central and noncentral GA); both CNV and GA 529 ± 12 (n = 2); or unknown 337 ng (n = 1). Unfortunately, the limited availability of MGS 4 donors prevents separate analysis of these different end-stage clinical phenotypes.
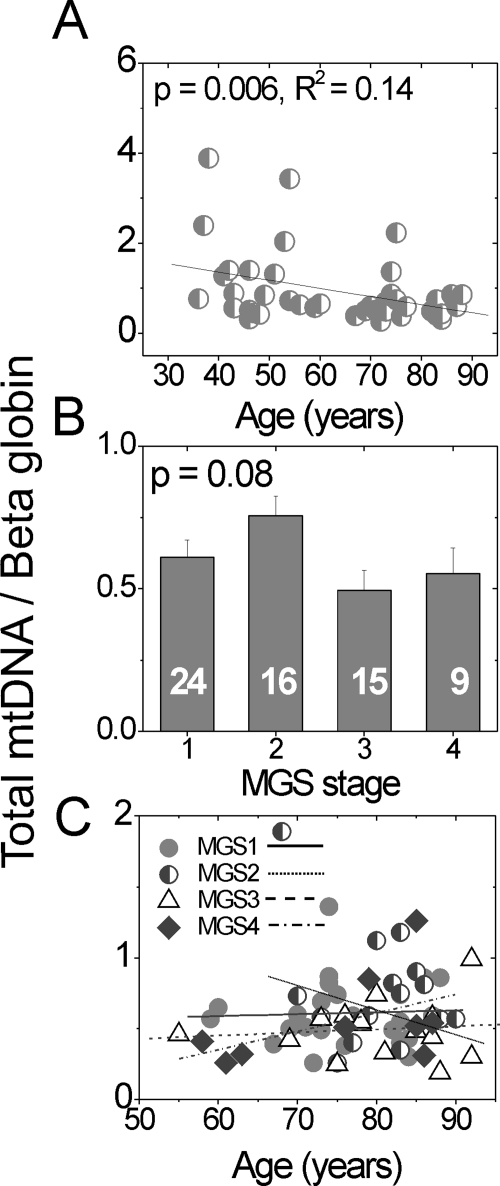
Since genomic copies of the β-globin nuclear gene are constant in diploid cells, amplification of the β-globin nuclear gene (147 bp) and the 16S rRNA mt gene (191 bp; Fig. 2) by qPCR was used to quantify total mtDNA copies per RPE cell (16S rRNA/ β-globin) in each donor. With aging, total mtDNA content significantly decreased in the RPE macula (Fig. 3A). In contrast, AMD donors showed no significant stage-specific differences (Fig. 3B) nor any change as a function of age within each MGS stage (Fig. 3C).
Figure 3.
The total mtDNA content decreases with aging, not with AMD progression. Total mtDNA content compared to β-globin was calculated from amplification of small regions of the mt genome (set 1, 16S rRNA; Fig. 2) and nuclear β-globin gene using qPCR. Total mtDNA content (A) of each MGS 1 (control) donor, ages 34 to 88 years (n = 44); (B) of each MGS stage (mean ± SEM); and (C) as a function of age for each MGS category. Linear regression analysis demonstrated no significant relationship between age and mtDNA content (C, P > 0.12) for any MGS stage. The number of samples in each MGS category is shown in the respective columns. The assay was preformed in triplicate.
Increase in Region-Specific Damage in the mtDNA with AMD
To identify areas of the mt genome that are preferentially affected with aging and AMD, primers were designed to amplify discrete regions of the mtDNA by using LX PCR (Fig. 2; Table 2). The LX PCR assay is based on the principle that DNA lesions (i.e., abasic sites, strand breaks, and thymine dimers) can slow down or block the progression of a thermostable DNA polymerase and prevent complete product synthesis.15,18,19 Thus, the amount of amplification in this assay is proportional to the amount of undamaged templates.
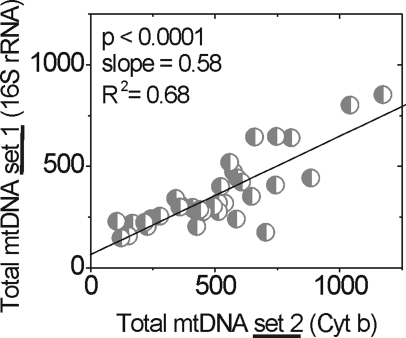
Amplification was normalized to total mtDNA content by using the 16S mt rRNA amplification product. Since the probability of lesion occurrence in a very small region is low, this measurement can be used to estimate the total mtDNA molecules in each sample. The validity of this measurement was confirmed by the significant correlation with amplification of a second small fragment (222 bp; Fig. 2, Table 2) located within the Cyt b gene (Fig. 4).
Figure 4.
Comparison of two independent measures of total mtDNA content in donor samples. Two small fragments, 191 bp of 16S rRNA (total mtDNA set 1) and 222 bp of Cyt b gene (total mtDNA set 2), of the mt genome were amplified by using LX PCR. Comparison of these measures by linear regression shows highly significant correlation between the two measurements (n = 38).
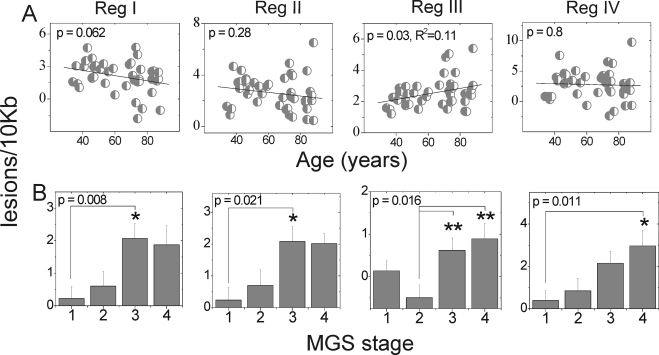
With aging, increased mtDNA damage was observed only in region III (Fig. 5A), which encompasses the well-characterized CD (Fig. 2). To determine whether there is an age effect within region III in our AMD donors, we performed linear regression analysis for each MGS category. We found no significant relationship between lesion frequency in region III and age for any MGS stage (P > 0.12, data not shown). We also tested other regions and found no significant age effect in any MGS stage (P > 0.08, data not shown).
Figure 5.
mtDNA damage increased with AMD progression. Lesion frequency (lesions/10 kb/double strands) calculated for regions I, II, III, and IV of the mt genome using LX PCR was compared with age and AMD progression. (A) Comparison of lesion accumulation with aging (n = 43) normalized to the relative amplification of ARPE-19 DNA. (B) Comparison of mean lesion frequency (mean ± SEM) at stages of AMD (sample size: MGS 1, 25–26; MGS 2, 16–17; MGS 3, 17–18; and MGS 4, 10–11) normalized to average relative amplification of MGS 1 age-matched control subjects. Significance was set at P ≤ 0.05. All reactions were performed in triplicate for each sample. *Significantly different from MGS 1. **Significantly different from MGS 2.
In contrast to the limited damage observed with aging, donors with AMD exhibited increased lesion frequency in all regions of the mt genome (Fig. 5B). Notably, at MGS 3, a disease stage that precedes vision loss, donors showed a significant increase in lesion frequency in regions I and II. In addition, lesion frequency in region IV showed a significant linear correlation with disease progression (P = 0.001; Fig. 5B), suggesting that damage accumulates throughout the disease process.
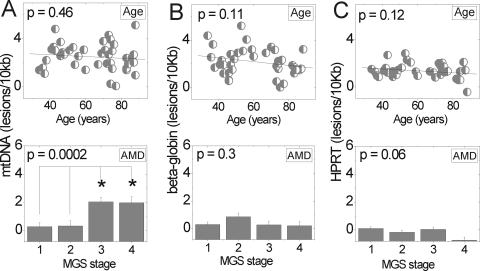
mt-Targeted Damage with AMD
Lesion frequencies in the mt and nuclear genomes were compared to determine whether mtDNA was more susceptible to damage (Fig. 6). Total lesion frequency of the mt genome was calculated by taking an average of lesion frequency for all four regions tested. Lesions in the nuclear genome were estimated from amplification of a 13.5-kb fragment of the β-globin and HPRT genes. Amplification of nuclear genes was normalized to the amplification of a 147-bp fragment within the β-globin gene to account for differences in the amount of nuclear DNA between samples.
Figure 6.
mtDNA but not nDNA showed increased damage with AMD. (A) Lesion frequency for each region of the mt genome (Fig. 4) was averaged to determine total mtDNA lesions per genome for each donor. Lesion frequency for nDNA was calculated from amplification of (B) β-globin and (C) HPRT genes. A small fragment (147 bp) of the β-globin gene was amplified to normalize amplification to total amount of nuclear DNA in each sample. Top: comparison of lesion frequency with aging (n = 43, mtDNA, n = 40, β-globin, n = 43, HPRT). Bottom: total lesion frequency for each stage of AMD (mean ± SEM). Significance was set at P ≤ 0.05.
With aging, lesion frequency did not change in either mtDNA (Fig. 6A) or nuclear DNA (Figs. 6B, 6C). Donors with AMD showed a significant lesion increase in mtDNA by later stages of AMD but not in the nuclear DNA. Note that at MGS 3, the mean lesion frequency was approximately eight times higher in mtDNA than in nuclear DNA. Previous reports of cultured RPE cells subjected to H2O2 exposure resulted in damage to mt DNA, but not to nuclear DNA.20 Similarly, our results demonstrate that damage was targeted to the mtDNA with AMD and supports a mechanism of oxidative damage in disease progression.
Increase in Deleted mt Genomes with Aging and AMD
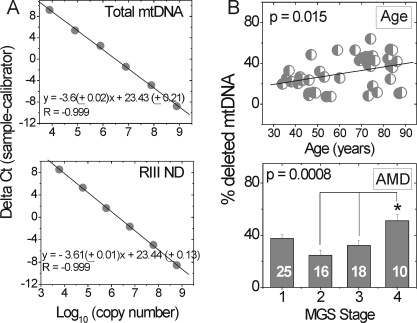
The 4977-bp CD in the major arc of the mt genome has been reported to increase in an age-dependent manner in a variety of postmitotic tissues.21 To provide a quantitative measure of the percent mtDNA containing deletions in the major arc, we developed a qPCR assay that used standard curves of known copy numbers (Fig. 7A). The coefficient of variation for this measurement was 10% ± 0.8%. To validate the accuracy of this assay, we determined the percentage of deleted genomes in Delta H2–1 cybrid cells reported to contain 80% ± 13% deleted mtDNA by using a multiplex qPCR assay.17 In agreement with previous data, our assay measured 78% ± 5% deletions in this cell line. Determining the amount of deleted mtDNA in the RPE is important, because the tolerance for this mt defect is tissue-specific. For example, a study of brains of individuals with Parkinson showed that the healthy substantia nigra contains 30% to 50% deleted mtDNA from ages 20 to 95 years, demonstrating the high tolerance for deletions in these neurons.22 In addition, relative levels of deleted mt genomes have not been determined for RPE with AMD and will provide important details about the amount of deletion that can be tolerated by RPE with aging and disease.
Figure 7.
Deletions increased with aging and AMD. (A) The percentage of deleted mtDNA in each individual was quantified using standard curves generated for each primer set by serially diluting a known amount of copies. Template DNA for total mtDNA (primer set 1) and RIII ND primer sets were produced from ARPE-19 DNA. Equations shown were averaged from measurements repeated on 3 days. (B) Level of deleted mtDNA increased with aging (P = 0.015, R2 = 0.17, n = 44) and with AMD (P = 0.0008). The number of samples used for each MGS stage is reported in the respective columns. All reactions were performed in triplicate for each sample. *Significantly different from MGS 2 and 3.
In the absence of AMD (MGS 1), the deletion load in donors aged 34 to 88 years increased from ∼20% to 40% (Fig. 7B). In age-matched donors, there was a small but statistically significant increase of ∼14% between MGS 2 and end-stage AMD (MGS 4, Fig. 7B). To determine whether deletions accumulated as a function of age in the AMD donors, we performed linear regression analysis. We found no significant correlation between deletion accumulation and age in any MGS stage (P > 0.11, data not shown). Taken together, these results suggest that RPE cells can withstand high levels of deletion with physiologic aging and that there is a small increase with end-stage AMD but no significant accumulation of deletions when compared with the normal control.
Discussion
Results from several different experimental approaches have produced compelling evidence implicating mt dysfunction as a potential pathologic feature of AMD. For example, recent studies have shown a significant association between mtDNA haplogroups and AMD prevalence.23–25 Of importance, the variants associated with higher risk for AMD occur within the mt region encoding for proteins of the electron transport chain, suggesting the potential for altered bioenergetics with AMD. Proteomic analysis of the retina,26 RPE,10,11 and the choroid/Bruch's membrane27 each identified altered content of mt proteins in AMD donor eyes. Ultrastructural analysis of RPE mitochondria showed a decrease in the number of mitochondria and in structural integrity in AMD donors.9 Data from the present study using a PCR analytical approach showed increased global mtDNA damage in AMD donor RPE. Collectively, these results support the hypothesis that mt defects could be an underlying pathologic event in AMD.8
Reports in the literature have shown increased damage to mtDNA with normal aging in the retina and RPE, suggesting that these age-related changes in the mitochondria increase the risk of AMD.28–30 However, no published reports have focused on both aging and AMD, to directly establish the mtDNA damage associated with each process. This study included a novel investigation of regional damage in the mtDNA with physiologic and pathologic (i.e., AMD) aging using clinically graded human tissue. Our findings demonstrated, for normal aging, decreased mtDNA copy number and damage limited to only one region that encompasses the CD. With AMD, however, all regions tested showed increased mtDNA damage by later stages (i.e., MGS 3 or 4) of the disease. Collectively, our data indicate a potential mechanism for RPE dysfunction with AMD that is directly linked to measurable mtDNA damage.
Analysis of mtDNA damage using LX PCR has many advantages.31 First, this PCR strategy requires only a small amount of DNA, which is highly relevant for studies in which the availability of human tissue is limited. Second, this method is quantitative because analysis is performed within the exponential phase, where amplification is directly proportional to the starting amount of undamaged DNA. Third, there is no need to purify mitochondria or isolate mtDNA, because sequence-specific primers are used. This method is highly desirable because damage can occur during isolation procedures, leading to an overestimation of the damage.32 Finally, this method allows the amplification of large template fragments and is therefore sufficiently sensitive to detect damage as low as 1 lesion/105 bp from 15 ng of total DNA.14 Although the LX PCR technique is not sensitive to all types of DNA damage (e.g., 8-hydroxy-guanosine) and cannot distinguish between types of damage, the results are nonetheless important in establishing the presence of mtDNA damage in human RPE. Furthermore, the low DNA yield from a pair of human macular RPE limits detection of some DNA damage such as 8-hydroxy-guanosine using ELISA and HPLC approaches, thus establishing the value of LX PCR.
An important finding in this study was that total mtDNA content decreased with age but showed no significant AMD-dependent change. As demonstrated by earlier work, changes in total mtDNA copy number with age are tissue specific. For example, mtDNA content decreased with age in skeletal muscle,33 liver,34 and neurons,35 whereas heart, spleen, kidney, lung,36 and brain37 showed an increase. Because there is a correlative decrease in ATP production and mt-encoded proteins in skeletal muscle with a decline in mtDNA content,33 the decrease we observed with physiologic aging may have consequences for ATP production and mt function in aged RPE.
Using two independent PCR measures, we observed an age-dependent increase in DNA damage in region III, which encompasses the CD (Figs. 5, 7). The increased damage and deletions observed in region III agree with findings in other reports of an age-dependent increase in CD in rodent RPE and human neural retina and RPE.28–30 As reported for numerous other postmitotic tissues such as neurons, muscle, and cochlear tissue, CD is an apparent hallmark of aging.21,38 One proposed mechanism for the formation of mtDNA deletions involves DNA repair subsequent to double-strand DNA breaks (DSBs).39 DSBs are frequently generated as a consequence of oxidative damage.40 Repair initiates with 3′ to 5′ exonuclease activity at the DSBs, which exposes regions of direct repeats that can misanneal, resulting in the degradation of the unbound strands. The ends are then re-ligated to produce a deleted mtDNA molecule. Thus, the age-dependent accumulation of deletions is probably due to ongoing repair that occurs over an individual's lifetime. Deleted mt species increased slightly with AMD but was not significantly different from control levels.
The critical threshold of deletions necessary to cause a functional defect in postmitotic cells has been examined in only a few cell types. For example, the abundance of mtDNA deletions must reach >85% in muscles of Kearns-Sayre syndrome patients for functional defects to manifest.41 In substantia nigra, the critical threshold is between 48% and 67%. This estimate is based on the report that neurons containing a 48% deletion load had normal cytochrome c oxidase (COX) activity, whereas neurons containing 67% deletions had decreased COX activity.22 Our results suggest that RPE cells are capable of withstanding at least ∼40% deletion loads without clinically obvious RPE dysfunction with physiological aging (Fig. 6).
Our data highlight marked differences in damage accumulation with aging and disease. Age-related damage appeared to be limited to region III, whereas AMD was associated with lesions affecting multiple regions of the mt genome. These results are consistent with our previous proteomic analysis of mt proteins that suggested that AMD-related mt dysfunction results from increased mtDNA damage.11 Notably, damage is significantly increased as early as MGS stage 3, before macular atrophy or neovascularization (Figs. 5, 6).
There are several potential mechanisms that could explain the increased mtDNA damage with AMD. One is that more damage is produced from increased reactive oxygen species (ROS), a hypothesis that has been proposed previously.42 The other mechanisms that have not been experimentally tested thus far in AMD donor tissue include diminished mtDNA repair and decreased autophagy with AMD. Autophagy, or more specifically mitophagy, is the mechanism for lysosomal elimination of damaged mitochondria from the cell. Previous work has shown that an impaired clearing mechanism leads to an accumulation of mtDNA damage.43
Since postmortem tissue is not well-suited for functional analyses, the consequences of RPE mtDNA damage are postulated from studies of cultured cells. In studies of cultured endothelial cells, increased mtDNA lesions correlated with altered mt membrane potential and apoptosis.44 A second study in endothelial cells showed that increased mtDNA damage results in decreased mtDNA-encoded transcripts, cellular ATP levels, and redox function.45 In cultured RPE cells, oxidant-induced mtDNA damage also results in increased reactive oxygen/nitrogen species and decreased mt membrane potential.46
Damage of the nDNA was approximately eight times lower than that of the mt genome (Fig. 6), indicating that mtDNA preferentially accumulates damage during the disease process. This result is in agreement with previous observations in cultured RPE cells where nDNA was rapidly repaired, whereas mtDNA sustained damage when subjected to an oxidant.42 Increased susceptibility of the mt genome to damage could be due to several factors, including that mtDNA is more exposed and lacks protective protein complexes such as histones that cover and protect nDNA. The location of mtDNA is in close proximity to the electron transport chain, which is a source of ROS. In addition to electrons that are uncoupled from the respiratory chain, the presence of localized metal ions in the mt inner membrane may function as catalysts in the generation of ROS. Finally, mt repair processes are fewer and seem to be less efficient than in the nuclear genome.19,47 Together, these factors, along with our data, show the increased vulnerability of mtDNA with AMD progression.
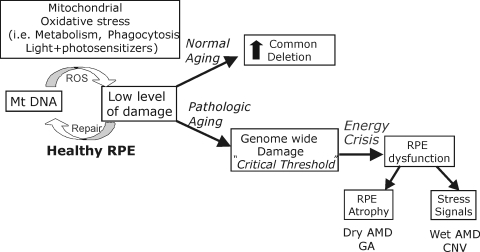
In summary, we used two quantitative PCR strategies to investigate mtDNA damage in a comparative study of normal aging versus pathologic aging in the RPE of human tissue with AMD. Our observations identified mtDNA as a site of damage that is preferentially affected during the course of AMD and also established that these changes occur by the intermediate (MGS 3) stage of the disease. Our data are consistent with the idea that increased mtDNA damage could be one factor leading to RPE dysfunction (Fig. 8). ROS generated under normal conditions, such as light (especially short-wavelength blue light), phagocytosis, and metabolism, place the RPE under considerable metabolic demand and oxidative stress. With normal aging, mtDNA damage is maintained at low levels with an increase only in the CD region. With AMD, however, we see mt genome-wide damage that could exceed a critical threshold that reduces proper bioenergetic function. Impaired RPE can lead to an imbalance in signaling factors (i.e., increased VEGF,48) and apoptosis49 resulting in end-stage AMD (Fig. 8). Thus, protecting mtDNA integrity via therapeutics targeted to the mitochondria early in AMD could stop or ameliorate the progression to vision loss.
Figure 8.
Putative model of the pathologic course of AMD. Under normal conditions, RPE mtDNA damage is maintained at low levels. With normal aging, mtDNA damage occurs in the CD region, affecting perhaps up to 40% of mtDNA genomes by age 90. With AMD, damage accumulates across the mt genome, affecting mt metabolism and RPE function. The damage can manifest in RPE atrophy and an imbalance in signaling factors, resulting in TGA and CNV that are associated with AMD.
Acknowledgments
The authors thank the Minnesota Lions and Minnesota Lions Eye Bank (MLEB) personnel for their assistance in procuring eyes and Kathy Goode at the MLEB for processing eye tissue.
Footnotes
Supported in part by National Institute on Aging Grant AG025392, an unrestricted grant from Research to Prevent Blindness to the Departments of Ophthalmology at the University of Minnesota and Emory University, and a generous donation from the Daniel and Helen Lindsay Family.
Disclosure: P.P. Karunadharma, None; C.L. Nordgaard, None; T.W. Olsen, None; D.A. Ferrington, None
References
- 1. Friedman DS, O'Colmain BJ, Munoz B, et al. Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol. 2004;122:564–572 [DOI] [PubMed] [Google Scholar]
- 2. Brown MM, Brown GC, Sharma S, et al. The burden of age-related macular degeneration: a value-based analysis. Curr Opin Ophthalmol. 2006;17:257–266 [DOI] [PubMed] [Google Scholar]
- 3. Brown DM, Kaiser PK, Michels M, et al. Ranibizumab versus verteporfin for neovascular age-related macular degeneration. N Engl J Med. 2006;355:1432–1444 [DOI] [PubMed] [Google Scholar]
- 4. Rosenfeld PJ, Brown DM, Heier JS, et al. Ranibizumab for neovascular age-related macular degeneration. N Engl J Med. 2006;355:1419–1431 [DOI] [PubMed] [Google Scholar]
- 5. Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845–881 [DOI] [PubMed] [Google Scholar]
- 6. Olsen TW, Feng X. The Minnesota Grading System of eye bank eyes for age-related macular degeneration. Invest Ophthalmol Vis Sci. 2004;45:4484–4490 [DOI] [PubMed] [Google Scholar]
- 7. Age-Related Eye Disease Study Research Group The Age-Related Eye Disease Study system for classifying age-related macular degeneration from stereoscopic color fundus photographs: the Age-Related Eye Disease Study Report Number 6. Am J Ophthalmol 2001;132:668–681 [DOI] [PubMed] [Google Scholar]
- 8. Jarrett SG, Lin H, Godley BF, Boulton ME. Mitochondrial DNA damage and its potential role in retinal degeneration. Prog Retin Eye Res. 2008;27:596–607 [DOI] [PubMed] [Google Scholar]
- 9. Feher J, Kovacs I, Artico M, et al. Mitochondrial alterations of retinal pigment epithelium in age-related macular degeneration. Neurobiol Aging. 2006;27:983–993 [DOI] [PubMed] [Google Scholar]
- 10. Nordgaard CL, Berg KM, Kapphahn RJ, et al. Proteomics of the retinal pigment epithelium reveals altered protein expression at progressive stages of age-related macular degeneration. Invest Ophthalmol Vis Sci. 2006;47:815–822 [DOI] [PubMed] [Google Scholar]
- 11. Nordgaard CL, Karunadharma PP, Feng X, Olsen TW, Ferrington DA. Mitochondrial proteomics of the retinal pigment epithelium at progressive stages of age-related macular degeneration. Invest Ophthalmol Vis Sci. 2008;49:2848–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Decanini A, Nordgaard CL, Feng X, Ferrington DA, Olsen TW. Changes in select redox proteins of the retinal pigment epithelium in age-related macular degeneration. Am J Ophthalmol. 2007;143:607–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ethen CM, Feng X, Olsen TW, Ferrington DA. Declines in arrestin and rhodopsin in the macula with progression of age-related macular degeneration. Invest Ophthalmol Vis Sci. 2005;46:769–775 [DOI] [PubMed] [Google Scholar]
- 14. Santos JH, Meyer JN, Mandavilli BS, Van Houten B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol Biol. 2006;314:183–199 [DOI] [PubMed] [Google Scholar]
- 15. Santos JH, Mandavilli BS, Van Houten B. Measuring oxidative mtDNA damage and repair using quantitative PCR. Methods Mol Biol. 2002;197:159–176 [DOI] [PubMed] [Google Scholar]
- 16. Ayala-Torres S, Chen Y, Svoboda T, Rosenblatt J, Van Houten B. Analysis of gene-specific DNA damage and repair using quantitative polymerase chain reaction. Methods. 2000;22:135–147 [DOI] [PubMed] [Google Scholar]
- 17. Poe BG, Navratil M, Arriaga EA. Absolute quantitation of a heteroplasmic mitochondrial DNA deletion using a multiplex three-primer real-time PCR assay. Anal Biochem. 2007;362:193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kalinowski DP, Illenye S, Van Houten B. Analysis of DNA damage and repair in murine leukemia L1210 cells using a quantitative polymerase chain reaction assay. Nucleic Acids Res. 1992;20:3485–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A. 1997;94:514–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ballinger SW, Van Houten B, Jin GF, Conklin CA, Godley BF. Hydrogen peroxide causes significant mitochondrial DNA damage in human RPE cells. Exp Eye Res. 1999;68:765–772 [DOI] [PubMed] [Google Scholar]
- 21. Meissner C, Bruse P, Mohamed SA, et al. The 4977 bp deletion of mitochondrial DNA in human skeletal muscle, heart and different areas of the brain: a useful biomarker or more? Exp Gerontol. 2008;43:645–652 [DOI] [PubMed] [Google Scholar]
- 22. Bender A, Krishnan KJ, Morris CM, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–517 [DOI] [PubMed] [Google Scholar]
- 23. Canter JA, Haas DW, Kallianpur AR, et al. The mitochondrial pharmacogenomics of haplogroup T: MTND2*LHON4917G and antiretroviral therapy-associated peripheral neuropathy. Pharmacogenomics J. 2008;8:71–77 [DOI] [PubMed] [Google Scholar]
- 24. Jones MM, Manwaring N, Wang JJ, Rochtchina E, Mitchell P, Sue CM. Mitochondrial DNA haplogroups and age-related maculopathy. Arch Ophthalmol. 2007;125:1235–1240 [DOI] [PubMed] [Google Scholar]
- 25. SanGiovanni JP, Arking DE, Iyengar SK, et al. Mitochondrial DNA variants of respiratory complex I that uniquely characterize haplogroup T2 are associated with increased risk of age-related macular degeneration. PLoS One. 2009;4:e5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ethen CM, Reilly C, Feng X, Olsen TW, Ferrington DA. The proteome of central and peripheral retina with progression of age-related macular degeneration. Invest Ophthalmol Vis Sci. 2006;47:2280–2290 [DOI] [PubMed] [Google Scholar]
- 27. Yuan X, Gu X, Crabb JS, et al. Quantitative proteomic comparison of the macular Bruch's membrane/choroid complex from age-related macular degeneration and normal eyes. Mol Cell Proteomics. 2010;9:1031–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barreau E, Brossas JY, Courtois Y, Treton JA. Accumulation of mitochondrial DNA deletions in human retina during aging. Invest Ophthalmol Vis Sci. 1996;37:384–391 [PubMed] [Google Scholar]
- 29. Barron MJ, Johnson MA, Andrews RM, et al. Mitochondrial abnormalities in ageing macular photoreceptors. Invest Ophthalmol Vis Sci. 2001;42:3016–3022 [PubMed] [Google Scholar]
- 30. Wang AL, Lukas TJ, Yuan M, Neufeld AH. Increased mitochondrial DNA damage and down-regulation of DNA repair enzymes in aged rodent retinal pigment epithelium and choroid. Mol Vis. 2008;14:644–651 [PMC free article] [PubMed] [Google Scholar]
- 31. Kovalenko OA, Santos JH. Analysis of oxidative damage by gene-specific quantitative PCR. Curr Protoc Hum Genet. 2009;Chapter 19:Unit 19.1 [DOI] [PubMed] [Google Scholar]
- 32. Richter C. Oxidative damage to mitochondrial DNA and its relationship to ageing. Int J Biochem Cell Biol. 1995;27:647–653 [DOI] [PubMed] [Google Scholar]
- 33. Short KR, Bigelow ML, Kahl J, et al. Decline in skeletal muscle mitochondrial function with aging in humans. Proc Natl Acad Sci U S A. 2005;102:5618–5623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barazzoni R, Short KR, Nair KS. Effects of aging on mitochondrial DNA copy number and cytochrome c oxidase gene expression in rat skeletal muscle, liver, and heart. J Biol Chem. 2000;275:3343–3347 [DOI] [PubMed] [Google Scholar]
- 35. Blokhin A, Vyshkina T, Komoly S, Kalman B. Variations in mitochondrial DNA copy numbers in MS brains. J Mol Neurosci. 2008;35:283–287 [DOI] [PubMed] [Google Scholar]
- 36. Masuyama M, Iida R, Takatsuka H, Yasuda T, Matsuki T. Quantitative change in mitochondrial DNA content in various mouse tissues during aging. Biochim Biophys Acta. 2005;1723:302–308 [DOI] [PubMed] [Google Scholar]
- 37. Barrientos A, Casademont J, Cardellach F, Estivill X, Urbano-Marquez A, Nunes V. Reduced steady-state levels of mitochondrial RNA and increased mitochondrial DNA amount in human brain with aging. Brain Res Mol Brain Res. 1997;52:284–289 [DOI] [PubMed] [Google Scholar]
- 38. Markaryan A, Nelson EG, Hinojosa R. Quantification of the mitochondrial DNA common deletion in presbycusis. Laryngoscope. 2009;119:1184–1189 [DOI] [PubMed] [Google Scholar]
- 39. Krishnan KJ, Reeve AK, Samuels DC, et al. What causes mitochondrial DNA deletions in human cells? Nat Genet. 2008;40:275–279 [DOI] [PubMed] [Google Scholar]
- 40. Breen AP, Murphy JA. Reactions of oxyl radicals with DNA. Free Radic Biol Med. 1995;18:1033–1077 [DOI] [PubMed] [Google Scholar]
- 41. Sciacco M, Bonilla E, Schon EA, DiMauro S, Moraes CT. Distribution of wild-type and common deletion forms of mtDNA in normal and respiration-deficient muscle fibers from patients with mitochondrial myopathy. Hum Mol Genet. 1994;3:13–19 [DOI] [PubMed] [Google Scholar]
- 42. Liang FQ, Godley BF. Oxidative stress-induced mitochondrial DNA damage in human retinal pigment epithelial cells: a possible mechanism for RPE aging and age-related macular degeneration. Exp Eye Res. 2003;76:397–403 [DOI] [PubMed] [Google Scholar]
- 43. Terman A, Dalen H, Eaton JW, Neuzil J, Brunk UT. Aging of cardiac myocytes in culture: oxidative stress, lipofuscin accumulation, and mitochondrial turnover. Ann N Y Acad Sci. 2004;1019:70–77 [DOI] [PubMed] [Google Scholar]
- 44. Xie L, Zhu X, Hu Y, et al. Mitochondrial DNA oxidative damage triggering mitochondrial dysfunction and apoptosis in high glucose-induced HRECs. Invest Ophthalmol Vis Sci. 2008;49:4203–4209 [DOI] [PubMed] [Google Scholar]
- 45. Ballinger SW, Patterson C, Yan CN, et al. Hydrogen peroxide- and peroxynitrite-induced mitochondrial DNA damage and dysfunction in vascular endothelial and smooth muscle cells. Circ Res. 2000;86:960–966 [DOI] [PubMed] [Google Scholar]
- 46. Gramajo AL, Zacharias LC, Neekhra A, et al. Mitochondrial DNA damage induced by 7-ketocholesterol in human retinal pigment epithelial cells in vitro. Invest Ophthalmol Vis Sci 51:1164–1170 [DOI] [PubMed] [Google Scholar]
- 47. Sawyer DE, Roman SD, Aitken RJ. Relative susceptibilities of mitochondrial and nuclear DNA to damage induced by hydrogen peroxide in two mouse germ cell lines. Redox Rep. 2001;6:182–184 [DOI] [PubMed] [Google Scholar]
- 48. Bertram KM, Baglole CJ, Phipps RP, Libby RT. Molecular regulation of cigarette smoke induced-oxidative stress in human retinal pigment epithelial cells: implications for age-related macular degeneration. Am J Physiol Cell Physiol. 2009;297:C1200–C1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jin GF, Hurst JS, Godley BF. Hydrogen peroxide stimulates apoptosis in cultured human retinal pigment epithelial cells. Curr Eye Res. 2001;22:165–173 [DOI] [PubMed] [Google Scholar]