Abstract
S-nitrosylation, the selective and reversible addition of nitric oxide (NO) moiety to cysteine (Cys) sulfur in proteins, regulates numerous cellular processes. In recent years, proteomic approaches have been developed that are capable of identifying nitrosylated Cys residues. However, the features underlying specificity of Cys modification with NO remain poorly defined. Previous studies suggested that S-nitrosylated Cys may be flanked by an acid-base motif or hydrophobic areas, and show high reactivity, low pKa and high sulfur atom exposure. In the current study, we prepared an extensive, manually curated dataset of proteins with S-nitrosothiols, accounting for a variety of biochemical functions, organisms of origin and physiological responses to NO. Analysis of this generic NO-Cys dataset revealed that proximal acid-base motif, Cys pKa, sulfur atom exposure, Cys conservation or hydrophobicity in the vicinity of the modified Cys do not define the specificity of S-nitrosylation. Instead, this analysis revealed a revised acid-base motif, which is located more distantly to the Cys and has its charged groups exposed. We hypothesize that, rather than being strictly employed for direct activation of Cys, the modified acid-base motif is engaged in protein-protein interactions whereby contributing to trans-nitrosylation as an important and widespread mechanism for reversible modification of Cys with NO moiety. For proteins lacking the revised motif, we discuss alternative mechanisms including a potential role of nitrosoglutathione as a transacting agent.
Keywords: Cysteine S-nitrosylation, redox thiols, post-translational modifications, acid-base motif, cysteine reactivity, trans-nitrosylation, nitric oxide, oxidative modification
Introduction
S-nitrosylation, the covalent addition of nitric oxide (NO) moiety to the sulfur atom of cysteine (Cys) residues, is a reversible protein posttranslational modification; its function in signal transduction was initially established in smooth vascular muscle cells where upon binding to the heme iron of guanylate cyclase [1], and consequent cGMP-dependent protein kinase-mediated activation of potassium channels [2], NO was found to be involved in a vasodilatative process. In the last decade, S-nitrosylation has drawn much attention from the biomedical community and led to a growing understanding of its direct effect in a variety of signaling pathways, under both physiological and pathological conditions [3–6].
Several methods, each with its own advantages and weaknesses, have been developed to characterize nitrosylated proteins, such as Saville–Griess method, gas phase chemiluminescence, mass spectrometry, and the biotin-switch method [7–10]. However, some of these methods (e.g., Saville–Griess and gas phase chemiluminescence) are unable to identify the modified Cys in S-nitrosylated proteins. Nevertheless, over the past decade, the number of identified S-nitrosylation sites has grown from a few, usually identified via laborious analyses, to hundreds, identified by sophisticated high-throughput proteomic approaches [11–13]. In particular, the biotin-switch method has recently been extensively used for identification of targets of S-nitrosylation in various model systems [14–17].
Taking advantage of a growing number of NO-Cys sites, bioinformatics analyses were then carried out based on several reported features of these sites initially observed by manual analyses. Although no linear sequence motifs were identified, these studies revealed a recurrence (even if not uniform) of an acid-base motif in the sequences flanking S-nitrosylated residues [18]. A recent study [19] analyzed common sequence and tertiary structure features in a set of 18 S-nitrosylated Cys, revealing that if the acid-base motif could be a general feature of S-nitrosylated Cys [20,21], it is not at the sequence level. By analyzing available structures of proteins in the dataset (i.e., 4 proteins out of 18 for which crystal structures were available), the authors found the motif within 7 Å of the modified Cys. However, it was also pointed that, given the different chemistries probably involved in S-nitrosylation in vivo, the motif is not a conserved feature of all Cys residues that are modified with NO.
Another feature often invoked for the structural environment of NO-Cys sites is hydrophobicity around the modified Cys. It was shown that hydrophobic protein surfaces can concentrate lipophilic NO and molecular oxygen, allowing formation of efficient nitrosating species (N2O3) precisely at the site of hydrophobic thiol, e.g., Cys 132 in argininosuccinate synthetase [22]. Thus, selective targeting of Cys 132 by S-nitrosylation could be achieved due to specific structural features of the environment around Cys. An overall hydrophobic content and a nearby positive charge (e.g., His 116 in argininosuccinate synthetase) could facilitate thiol deprotonation, promoting the formation of thiolate, a target of NO. A further aspect often suggested to play a role in S-nitrosylation is a low pKa of Cys, but no detailed analysis has been reported. Indeed, formation of the NO adduct to Cys also occurs at higher pH (around 9) in thioredoxin 1 (Trx1). All these features, as well as physiological aspects of S-nitrosylation, have been discussed in detail in several reviews [23, 24].
Studies conducted thus far with regard to the general features of NO-Cys sites invariantly suggest a role of tertiary structure around the modifiable Cys residues. But no extensive studies have been reported that analyze various classes of S-nitrosylated proteins. Likewise, tools capable of predicting new NO-Cys sites are not available. To examine whether NO-Cys sites have common properties, we built a dataset of 55 non-redundant proteins, containing 70 NO-Cys; the analysis of this dataset for general features of NO-Cys sites is described in this paper.
Results
Reference datasets
We first built a dataset of proteins containing S-nitrosylated Cys residues based on literature reports (Supplementary information, Table S1). We only considered proteins with established crystal or NMR structures and proteins which could be modeled by standard homology modeling approaches using Swiss Model server, and used non-redundant proteins with less than 50% sequence identity to any other protein in the dataset. We also required the proteins in our dataset to have established position for the modifiable sites (i.e., NO-Cys site experimentally demonstrated). Finally, proteins found to contain NO-Cys based on the experiments that employed the concentration of NO-sylating agents in the mM range were excluded, as high levels of these agents could result in artifacts (i.e., NO-Cys sites that are not biologically relevant or do not occur under physiological conditions). This restriction, even if it may exclude some naturally occurring NO-Cys sites, was used to limit false positives. Nevertheless, a potential weakness of the published studies is that they employed exogenous NO or S-nitrosylating agents to discover proteins and their sites of modifications and that many of them have not been confirmed in vivo. In view of the absence of endogenous S-nitrosocysteine proteomes, we focused on the data generated by in vitro approaches.
Our searches resulted in a set of 55 proteins from various organisms containing 70 NO-Cys sites (Table S1), including Trx1, cyclic nucleotide gated channel alpha 2 (CNG2), mitogen-activated protein kinase kinase kinase 5 (ASK1), mitogen-activated protein kinase (JNK1), dimethylarginine dimethylaminohydrolase (DDAH), Bcl2, FADD-like apoptosis regulator isoform 1 (FLIP), caspase 3, caspase 1, calpain 2, GST theta (GSTt), GST pi (GSTp), serine/threonine protein phosphatase (Ser/Thr protein phosphatase), 14-3-3 protein zeta/delta, malate dehydrogenase, pyruvate kinase, triosephosphate isomerase, vesicle-fusing ATPase C-terminal domain (NSF C-terminal domain), vesicle-fusing ATPase N-terminal domain (NSF N-terminal domain), ADP/ATP translocase, sodium/potassium-transporting ATPase, tubulin alpha, GSTmu (GSTm), semaphorin 4D, glucokinase, annexin 6, S-adenosylmethionine synthetase (MAT), matrix metalloproteinase 9 (MMP9), iron-responsive element binding protein (IRP2), inhibitor of nuclear factor kappa-B kinase (IKKB), peroxiredoxin 2 (PRX2), oxidative stress transcriptional regulator (OXYR), Ras p21, creatine kinase, GAPDH, vinculin, T-complex protein 1, Ras-related protein Rab, peptidylprolyl isomerase, myosin heavy chain 9, stress 70 protein (GRP75), COPA protein, annexin 2, annexin 11, elongation factor alpha (EF1A), protein tyrosine phosphatase (PTP), syntaxin 1A, myeloid differentiation primary response protein (MyD88), hemoglobin subunit beta, tubulin beta, histone deacetylase 2, and Parkinson’s disease protein 7 (DJ1), Dynamin-2, SG15 ubiquitin-like modifier (ISG15), and G-protein coupled receptor kinase 2 (GRK2). The majority of these proteins contained one reported nitrosylation site, but several had multiple modification sites (e.g., tubulin alpha and beta, GAPDH) as reported in detail in Table S1.
As our goal was to analyze NO-Cys sites for general features, we employed a broad set of proteins without separating them for different physiological roles and regulatory, signaling and stress functions. In addition, we did not consider subsets of NO-Cys sites based on the chemical form and concentration of nitrosylating agents, and biochemical pathways involved. As we discuss in more detail later in the text, the use of these parameters would require significantly more confirmed NO-Cys sites. Also, due to the scope of our work (i.e., distinguishing common features of NO-Cys sites and utilizing this information for predictive purposes), choosing a broad protein dataset was important, but subsets of proteins in our dataset could also be defined based on the parameters discussed above, and utilized in the future studies.
NO-Cys pKa, exposure and conservation
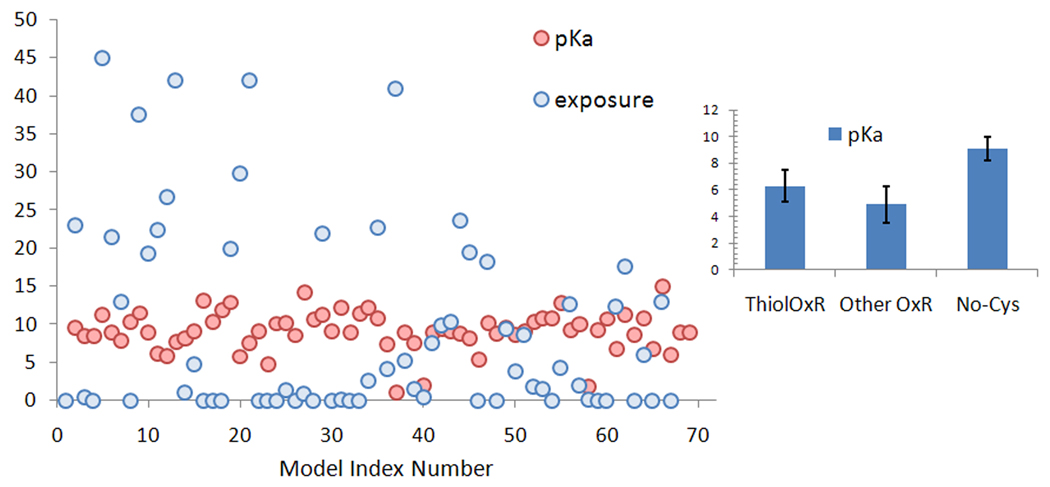
We first tested our dataset for parameters most often discussed in regard to posttranslational Cys modifications: pKa, exposure, and conservation. We computed, with the program PropKa, pKa values of Cys residues that are targets for S-nitrosylation and found them to be slightly higher (average 9.1) than those of redox and non-redox catalytic Cys residues (average pKa 5.5, calculated with the same program) (Fig 1), but consistent with the average pKa of Cys residues in proteins.
Figure 1. Calculated pKa and sulfur atom exposure of known NO-Cys sites.
The model index number (X-axis) follows the order of proteins listed in the text and detailed in Table S1, given as Supplementary Information. Sulfur atom exposure (related numerical values in the Y-axis, in Å2) is shown by red circles and pKa values (in pK units) by blue circles. In the right panel, the calculated average pKa for NO-Cys (with standard deviation) is compared with those of catalytic Cys in thiol oxidoreductases (ThiolOxR) and other enzymes containing catalytic Cys (Other OxR).
We next assessed Cys exposure (i.e., S atom exposure). While some NO-Cys sites were exposed, others were not (Figure 1). Approximately 48% of NO-Cys sites had exposure higher than 1 Å2 when a 1.4 Å probe (to mimic the water molecule) was employed, and 65% when a 1.2 Å probe was used (to account for the slightly smaller NO molecule). Thus, although some enrichment in sulfur exposure was detected for NO-Cys, about 35% of sulfur atoms of NO-Cys sites were predicted to be buried (i.e., with exposure values ≤1.0 Å2) and not accessible even to small molecular probes.
We further analyzed conservation of NO-modified Cys in proteins in our dataset. PSI-Blast search of the NCBI non-redundant database revealed an average conservation of 62% for NO-Cys sites, but also this parameter greatly varied among proteins in the dataset (Figure S2 and Table S1). Indeed, NO-Cys can be based on both highly (e.g., PRX2, GAPDH) and poorly (e.g., Cys 50 in GSTt, Cys 73 in TRX1) conserved Cys, reflecting low significance of the average value (standard deviation 34%). Thus, Cys conservation does not define NO-Cys sites either.
Sequence analysis: amino acid composition and hydrophobicity
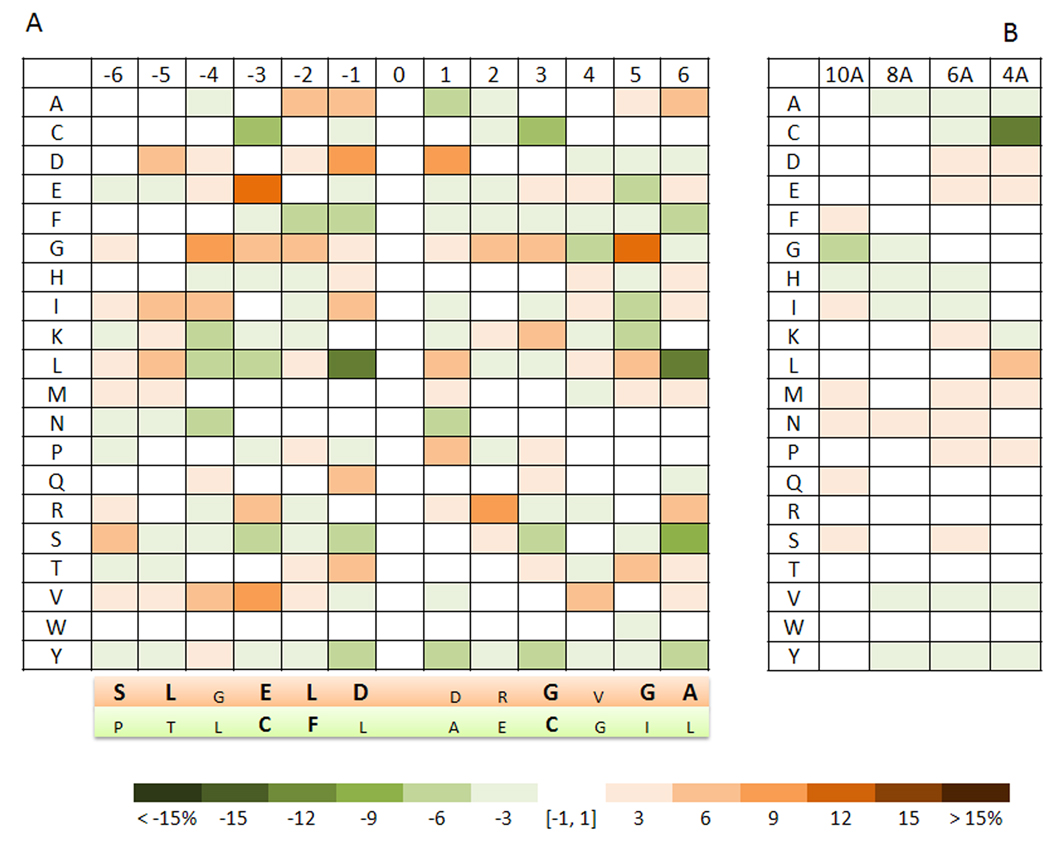
To further analyze common features of NO-Cys sites, we focused on sequence analysis of Cys residues in the dataset. Amino acid composition of NO-Cys-flanking sequences is shown in Figure 2A. The data obtained for 70 NO-Cys sites were compared to those of a reference set of Cys residues made up of 1,000 randomly chosen eukaryotic proteins from PDB. An overrepresentation of negatively charged residues was detected for NO-Cys sites, including aspartate in positions −1 and +1, and glutamate in position −3. The presence of a nearby (at the sequence level) acid residue is regarded as one of the features of a putative motif for NO-Cys, and in at least one case (KCNQ1 channel, NO-Cys 445), the presence of the acidic amino acid was found to be necessary for NO-sylation of Cys [25]. However, in our analysis, even if the flanking acidic residues were over-represented (Figure 2), they did not represent a feature characteristic of all or even a majority of NO-Cys sites. This observation did not exclude a possibility that subsets of NO-Cys sites, such as those exclusively involved in signaling or derived from a particular group of organisms, could be defined by the presence of a flanking to NO-Cys acidic residue. This feature, however, does not apply to the entire group of NO-Cys sites. In this regard, an important case study is ryanodine receptor with its NO-Cys 3635. This protein contains 12 Cys flanked by an acidic residue, however, its only nitrosylated Cys is not flanked (i.e., the flanking residues are Ala 3634 and Phe 3636).
Figure 2. Amino acid composition of NO-Cys sites.
A. Sequences flanking NO-Cys sites. Cys residues that form NO-Cys sites are located in the zero position with 6 residues upstream and 6 downstream shown in the form of relative occurrence of 20 common amino acids in proteins (designated by one-letter code on the left). A scale bar (below the two panels) indicates the percent differential occurrence of amino acids in the NO-Cys set, compared to a reference set of Random Cys. The variation (shown by intensity of blue and brown colors in the scale bar panel) refers to the percentage value by which a residue occurs more often (brown) or less often (green) than the corresponding residue in the reference Random Cys set. Below the main panel, shaded in orange are the amino acids found to be most overrepresented around NO-Cys, shown for each position (amino acids in bold corresponds to both the most frequent residue for that position and the most over-represented). In lower case, the amino acids are shown, which although being overrepresented in respect to Random Cys, are not the most frequent residues for the indicated position. In turn, shaded in green (second panel, below the orange shaded one) are the amino acids which are the most under-represented around NO-Cys (in bold are amino acids being both the most under-represented and the less frequent for that position) B. Tertiary structure and compositional analysis of NO-Cys are shown for regions within 4, 6, 8, and 10 Å from the sulfur atom of NO-Cys. The color scheme is as in panel A.
We discuss in detail the actual tertiary structure of NO-Cys-flanking acidic residues later in the text, while describing the occurrence and distribution of a potential revised acid-base motif for all proteins in our dataset. Additional sequence features of NO-Cys sites included a rare occurrence of an additional Cys nearby (the 13 residue window was considered; particularly striking is the lack of second Cys in positions +2 and −2, which are also the positions particularly enriched for Cys residues that flank catalytic redox and metal-binding Cys) and under-representation of leucine, especially in positions −1 and +6. However, attempts to build a NO-Cys profile and a signature sequence derived from it (Figure 2A) revealed the lack of statistical significance, indicating that these factors could not be employed for prediction of NO-Cys sites. These observations are in line with previous reports [14,15] and suggest that sequence analysis of NO-Cys sites is not sufficiently robust for reliable identification of S-nitrosylated Cys residues.
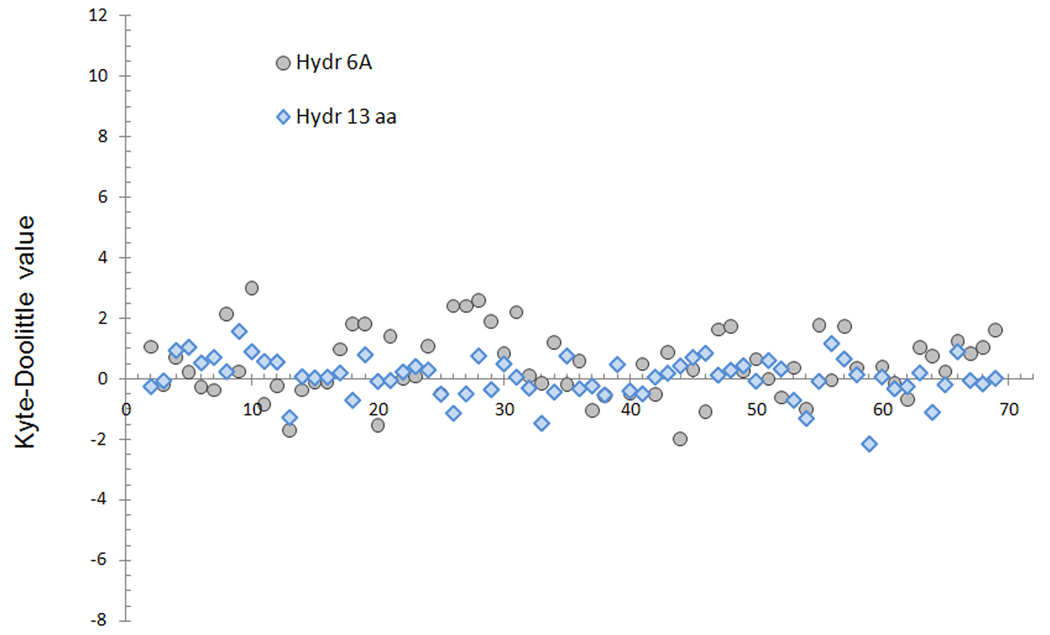
We analyzed the sequences flanking NO-Cys for hydrophobicity as defined by the Kyte-Doolittle scale (Figure 3). The average hydrophobicity of NO-Cys sites was slightly increased (an average value of 0.03 +/− 0.7), but considerable variation existed among NO-Cys sites. Moreover, hydrophobic environment is a feature commonly found around Cys residues. In fact, the set of randomly chosen Cys residues (Random Cys) showed the average value of 0.09 +/− 0.8. Thus, the sequences flanking NO-Cys do not significantly vary from those containing random Cys, and do not permit differentiation of NO-Cys and other Cys sites.
Figure 3. Hydrophobicity analysis of NO-Cys sites.
The NO-Cys dataset was analyzed according to the Kyte-Doolittle scale (i.e., on the Y axis, positive values correspond to hydrophobic and negative to hydrophilic environment). In the X axis, models are ordered according to their order in Table S1, given as supplementary information. Values for the primary structure profiles are shown in blue rhombi, and gray circles show the values for the structural regions around the NO-Cys sites (defined as residues within 6 Å from the sulfur atom of NO-Cys).
Structure-based analysis: amino acid composition and hydrophobicity
We further performed structural profile analyses of NO-Cys-containing proteins using a previously described method [26]. Structural profile analyses build a signature sequence of amino acids located around a specified residue: in our work, segments of amino acids located within 8 Å of each NO-Cys were extracted from the structure and combined into single contiguous sequences, generating structural profiles. However, this approach also did not yield common features among proteins containing NO-Cys (Figure S3). It should be noted that the structural profile analysis performs well for catalytic Cys in proteins [23,27], and in fact has been designed for this purpose. The evolutionary constraints act to a greater extent on the active sites than on pos t-translational modification sites (the former often being significantly more conserved).
As a next step, we analyzed the amino acid composition and hydrophobic content of the regions surrounding NO-Cys. For this purpose, we considered regions that fall inside spheres with defined radii (4 to 10 Å) and centered at the sulfur atom of each NO-Cys. As a reference, we employed an unbiased set of representative structures from the Random Cys dataset. Hydrophobicity in the NO-Cys set was only slightly higher (average Kyte-Doolittle value of 0.4 +/− 0.5 for the 6 Å region), with significant variation among NO-Cys sites (Figure 2). These values were not significantly different from those of the Random Cys set (0.5 +/− 0.5). Further analyses at other distances (i.e., 4 Å, 8 Å, 10 Å) showed similar behavior.
Turning our attention to the amino acid composition, we analyzed the average frequency of each residue at various distances (4 to 10 Å) from the modified Cys (Figure 2B). Once again, the low occurrence of other Cys residues near NO-Cys was the most obvious feature (with an average occurrence for the closest distance (4 Å) of 1.1% +/− 1.9% for NO-Cys, compared with 11% +/− 7.5% for the reference set). In addition, slight but insignificant overrepresentation of negatively charged residues and leucine was observed, in accordance with the sequence analysis (Figure 2B). Interestingly, low occurrence of His was evident from the structure-based approach, whereas this feature was not detected at the sequence level (Figure 2A). This trend, even if intriguing, is, however, not statistically significant. Altogether, the results indicated considerable heterogeneity within the NO-Cys set.
However, at a closer look, only 4% of proteins in our dataset (3 out of 70) did not have any charged residues within 6 Å from NO-Cys. In contrast, nearly one fourth (26%) of random Cys was characterized by the absence of net charge in close proximity (6 Å) to the Cys. Among the closest charged residues located near NO-Cys, 82% had a positive charge (and 18% negative), whereas the values for the Random Cys reference set were 64% and 36%, respectively.
With regard to the concomitant occurrence of at least one negatively and one positively charged residue (i.e., the acid-base motif) within the 6 Å region from NO-Cys, only 26% of NO-Cys and 19% of random Cys satisfied this requirement. These observations are important because they indicate that the acid-base motif, often referred in the literature as being a characteristic feature of NO-Cys sites, does not actually define these modification sites, at least within 6 Å of NO-Cys.
We also considered a possibility that the previously reported features, even if being unreliable descriptors of NO-Cys sites, may be used for predictive purposes if used in a combination. We carried out principal component analysis (PCA) that included Cys exposure, pKa, hydrophobicity, conservation and sequence features (detailed in Table S1); based on the results of PCA, however, we did not find any combination of parameters capable of describing the NO-Cys sites as whole. Taken together, our data indicate significant NO-Cys heterogeneity in all parameters considered, which is consistent with the idea that other mechanisms (or additional components) are behind specific S-nitrosylation. Thus, besides the direct nitrosylation chemistry (i.e., direct reaction of Cys with NO), various trans-nitrosylation processes might potentially account for many of the observed NO-Cys sites. For example, caspase 3 does not undergo nitrosylation in the absence of TRX1 [28]. The interaction between these proteins is driven by two essential and oppositely charged amino acids (Glu 70, Lys 72), which are exposed to solvent in the molecular surface proximal to the NO-Cys 73 site (the actual trans-nitrosylating residue) of TRX1; however, this acid-base motif does not point toward the sulfur atom and while the basic residue is within 6 Å (with some atoms, but not the charged one), the acidic Glu 70 residue is outside the range. It should be mentioned that TRX1 may have a more complex but still specific interaction with caspase 3 [29, 30] due to NO transfer in both directions between these proteins. Given the potential importance of mandatory protein-protein interactions and the occurrence of the acid-base motif, we further examined these features in more detail.
A distant acid-base motif: occurrence and distribution
When longer distances (up to 8 Å) were considered, approximately 90% of NO-Cys sites had both positively and negatively charged residues (e.g., there were at least one acidic and one basic residue within 8 Å). However, this feature was again insufficient in defining the NO-Cys set: 85% of control proteins also had such ionizable residues within 8 Å. To address the relationship between NO-Cys and charged residues, we evaluated their positioning in respect to the modifiable Cys sulfur atoms by examining whether these residues point their charged groups (GrF) toward the NO-Cys sulfur. In fact, if the negatively and positively charged residues are involved in the reaction between Cys and NO, one would expect their actual active atoms (i.e., atoms of GrF) to be located in proximity to the sulfur atom.
We calculated the distances between each charged atom (Lys, Arg, His, Glu and Asp within 8 Å from NO-Cys) and the sulfur atom (S) of Cys; for simplicity, we further refer to this measurement as the GrF-S distance. These distances were compared with those of the approximate center of mass (CM) of each residue (S-CM distance). When the GrF-S distance was smaller than the S-CM distance, the charged functional group was considered as pointing toward the NO-Cys sulfur atom. On the other hand, when GrF-S was greater than S-CM (i.e., the center of mass was closer to S than GrF), the charged functional group was considered to point outward.
First, we took a closer look at the positioning of negatively charged residues relative to the Cys, as these residues were more frequently found in the sequence analysis discussed before. All NO-Cys sites flanked by an acidic residue (i.e., with Asp or Glu in positions +1 or −1 in the sequence) were separately analyzed, and, remarkably, in the vast majority of these cases the acidic residue had the charged atoms pointing away from the sulfur atoms. This observation is illustrated in Figure S4. These data are in agreement with the previously discussed results: acidic residues, even when they are near the NO-Cys in the sequence, tend to stay distant from the reactive sulfur atom in the structure.
Recently, human DPR-1 was found to possess a single NO-modification site, Cys 644, which upon modification led to a series of toxic cellular events associated with Alzheimer’s disease [31]. In addition, this NO-Cys site had flanking acidic residues (Asp 643 and Glu 645). We modeled the structure of human DPR-1 and analyzed the orientation of these residues; once again both acidic residues flanking the NO-Cys site were clearly pointing outward with respect to Cys 644 (Figure S4, panel R).
Considering the NO-Cys dataset as a whole, only several proteins (DDAH Cys 249, creatine kinase Cys 283; Ras p21 Cys 118, MAT Cys 121; histone deacetylase 2 Cys 274) had positively and negatively charged groups pointing toward the Cys (~7% of the dataset). In addition, both GrF were closer than 6 Å to the NO-Cys sulfur atom only in three cases: MAT (positive GrF charge was found at 5.3 Å and negative at 5 Å), Ras p21 (positive GrF at 5 Å, negative GrF at 5.8 Å) and histone deacetylase (positive GrF at 3.9 Å and negative GrF at 3.1 Å).
When all other non-modifiable Cys (i.e., all Cys in the dataset that are not known to be S-nitrosylated) were used as a reference set, both types of charged functional groups pointed toward the Cys in 24% of control Cys (Figure S5). These data suggested that the presence of charged residues (i.e., the acid-base motif) that are slightly more distant with regard to NO-Cys may play a role other than a direct acid-base motif-dependent Cys activation.
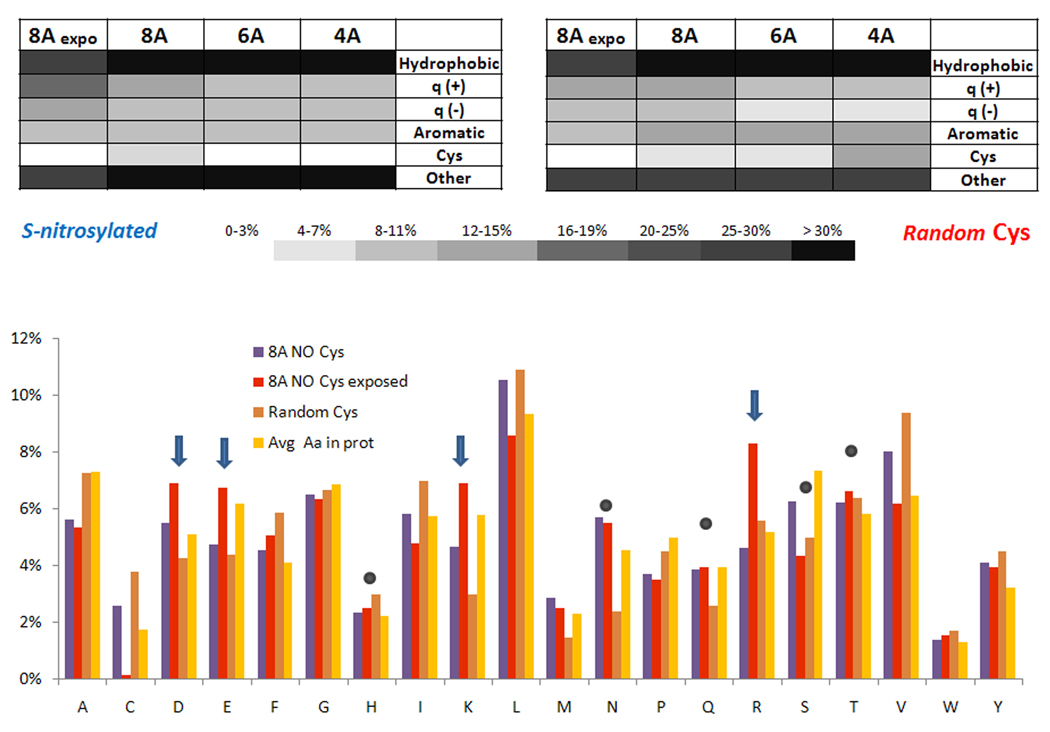
Analysis of the exposed 8 Å region
In the following text, we refer to the region composed of exposed amino acids within 8 Å from NO-Cys sulfur atoms as the exposed 8 Å region. To define this region, we considered exposed residues with at least one of their atoms accessible to the solvent (area greater than 1.0 Å2, a permissive cut-off value, which should not exclude a priori less polar residues). Figure 4 shows the comparison of amino acid composition of the exposed 8 Å region and the whole 8 Å region for NO-Cys and control proteins. An increase in both positively and negatively charged residues (Arg, Lys, Glu and Asp; but not His) clearly characterized the exposed regions of NO-Cys sites (Figure 4B). At the same time, other polar but non-charged residues were not over-represented in the NO-Cys exposed region. The complete list of amino acids composing the exposed 8 Å region is given in Supplementary Information, Table S2.
Figure 4. Amino acid composition of structural regions around NO-Cys sites.
A. NO-Cys sites (left) and a control set of randomly chosen Cys residues (right) were analyzed for hydrophobic, positively and negatively charged, aromatic, Cys and other residues. The data are shown for 4, 6, and 8 Å in comparison with the interface region exposed to solvent located within 8 Å from NO-Cys (see text for further details). B. Distribution of NO-Cys residues within 8 Å and solvent-exposed 8 Å regions, compared to two control Cys sets: a set of randomly chosen proteins (Random Cys) and relative abundance of amino acids in proteins, retrieved from http://www.russell.embl-heidelberg.de/aas/. Blue arrows point to charged residues that show increased representation in the exposed 8 Å region (particularly in respect to the complete 8 Å region, which also accounts for non-exposed residues), while filled gray circles indicate polar residues (including His) which show unchanged occurrence.
Charged residues often play important roles in proteins. Besides participation in active sites (e.g., acid-base catalysis) and metal binding, they usually exert strong influence on the electrostatic potential distribution of proteins. This feature is often crucial for protein-protein interactions, one of the main reasons why the distribution of the electrostatic potential on a protein molecular surface or solvent-accessible area (e.g., the Connelly surface) is a common analysis in structural biology. Among the charged residues responsible for this function, His is used less frequently (in part because of its higher hydrophobicity), instead more often playing a role in chemical plasticity due to its imidazole ring (metal binding, π-stacking interaction, etc.). As seen in Figure 4, the unchanged occurrence of His and the higher representation of all other charged residues in regions proximal to NO-Cys support the idea that the latter amino acids may be involved in protein-protein interactions that occur in proximity of NO-Cys sites. Interestingly, when the exposed 8 Å region was considered, the average conservation of all residues involved increased to 69.5 +/−16%, which was higher than the conservation of NO-Cys itself (Table S2), again suggesting an important role for this region. These results support the idea that the acid-base motif, found in proximity to molecular surface, could play a role in protein-protein interactions, possibly extending the previous findings for TRX1 and caspase 3 (i.e., exposed acid-base motif being in proximity but not too close to NO-Cys) to a larger set of proteins.
Deviating NO-Cys: NO-Cys lacking the acid-base motif in the exposed 8 Å region
As discussed above, some NO-Cys sites in our dataset lacked the acid-base motif, including GSTp (Cys 48), GSTt (Cys 50), Ser/Thr phosphatase (Cys 228), malate dehydrogenase (Cys 137), GAPDH (Cys 150), PRX2 (Cys 121), and tubulin beta (Cys 12). Additionally, histone deacetylase with its Cys 262 had the acid-base motif with 8 Å, but its only basic residue (His 282) was not exposed. Thus, 8 out of 70 NO-Cys did not have a solvent-exposed acid-base motif. Searching for common features among these Cys, we found that while they were exposed, the output values were highly variable: pKa ranged from 2 (PRX2) to 11 (tubulin beta), conservation ranged from less than 1% (GSTt) to more than 95% (Ser-Thr phosphatase, GAPDH, PRX2, tubulin beta), and proximity to other Cys thiols also varied significantly. We further discuss each of these proteins in detail.
Direct reactivity with NO
When considering potential mechanisms of S-nitrosylation, besides the case-specific protein-protein interaction, the most obvious alternatives are (i) direct S-nitrosylation; and (ii) trans-nitrosylation via amino-acid derivatives, such as S-nitrosoglutathione (GSNO). We first tested the possibility of direct NO/Cys reaction employing the chemical-physical features thought to be necessary for an effective reaction [18,19], i.e., that the NO/Cys reaction may take place in small and slightly hydrophobic pockets (i.e., clusters of spatially related amino acids, defining a partially solvent accessible region), with concomitant presence of a basic residue with its functional atom(s) in proximity to the modifiable Cys sulfur atom.
We employed a simple algorithm, which analyzed the NO-Cys sites in the dataset in the following steps: (i) analysis of protein pockets with Castp (http://sts-fw.bioengr.uic.edu/castp/); (ii) analysis of NO-Cys belonging to one or more pockets; (iii) analysis for occurrence of basic residues in the same pocket (if there was more than one potential pocket, each was evaluated separately) at a distance less than 8 Å from the sulfur atom; and (iv) calculation of the hydrophobicity index for the pocket according to the Kyte-Doolittle scale. These assumptions were meant to detect NO-Cys sites characterized by direct nitrosylation. We tested all proteins in our dataset, and the positives included GSTp, malate dehydrogenase, GSTm, IKKB, vesicle-fusing ATPase, myosin heavy chain 9, Stress-70 protein, and MMP9 (Table 1). Interestingly, two proteins, GSTp and malate dehydrogenase, were those with deviating NO-Cys (absence of the exposed acid-base motif). Therefore, for direct S-nitrosylation, the presence of both acid and basic residues near the NO-Cys does not appear to be a necessary feature.
Table 1.
Properties of NO-Cys sites relevant for direct S-nitrosylation
| Protein | Cys | Pocket Composition |
Pocket KD score |
Basic residue |
Charged atom-sulfur distance (in Å) |
|---|---|---|---|---|---|
| Glutathione S-transferase Pi | 48 | LTCK^Y | 0.1 | Lys 55 (NZ) | 6.3 |
| Glutathione S-transferase Mu 1 | 173 | LLH^CL | 2.1 | His 166 (NE2) | 6.9 |
| inhibitor of nuclear factor kappa-B kinase subunit beta # | 179 | LR^CTY | 0.1 | Arg 144 (NH2) | 5 |
| Vesicle-fusing ATPase | 91 | FK^QCL | 0.3 | Lys 89 (NZ) | 6.2 |
| Malate dehydrogenase, cytoplasmic | 154 | ANCLTH^G | 0.5 | His (NE2) | 7.9 |
| Myosin heavy chain 9 # | 91 | YYCVH^IITAI CK^NTVLIIVC | 1.7 | His 770 (NE2) | 5.6 |
| Stress-70 protein # | 487 | CANK^L | 0.1 | Lys 485 (NZ) | 5.5 |
| MMP9 | 99 | CVH^AH | 0.4 | His 401 (NE2) | 3.8 |
| Glutathione S-transferase Theta * | 50 | CVLTEK^ | 0.7 | Lys 149 (NZ) | 4.9 |
basic residue of the pocket whose charged atom is located close to NO-Cys
homology model is characterized by less than 50% identity with the template
modeled quaternary structure (i.e., modeled dimer)
Pocket KD score refers to an average Kyte-Doolittle score for amino acids that form pockets. In the Basic residue column, the basic amino acid belonging to the pocket and its charged atom are specified. The actual distance between the NO-Cys sulfur atom and this charged atom is shown in the adjacent column (Charged atom-sulfur distance).
Docking analysis: NO-Cys and GSNO
To computationally investigate trans-nitrosylation via GSNO, we carried out docking calculations for all NO-Cys-containing proteins in our dataset. In a recent study, influences exerted by the NO group on Cys residues were investigated [32] and new force field parameters specific for this modification determined, providing a robust starting point for a variety of structure-based investigations of NO modification sites. We implemented these parameters for NO-Cys in docking calculations; in particular, we transferred the QM-theory level partial charges in the modified site of interest (i.e., Cys-NO side chain of GSNO, as detailed in Experimental Procedures). In our analysis, we required the binding positions to be characterized by an overall favorable energy (i.e., energy < −1 kcal/mol) and with a distance between the sulfur atoms or between the nitrogen of the NO substituent lower than 3.0 Å.
First, we discuss the data for the models lacking the acid-base motif in the exposed 8 Å region, i.e., GADPH, tubulin beta, histone deacetylase, PRX2, GSTp, GSTt, Ser/Thr phosphatase and malate dehydrogenase. These proteins, except GSTs and malate dehydrogenase, showed good affinity for GSNO with more than one binding position (the best are shown in Figure 6 and further information is provided in Table 2). In the case of malate dehydrogenase and GSTp, GSNO docking did not yield reasonable structural models, suggesting that GSNO is unlikely to serve as the nitrosylating agent. However, these proteins were found as positive candidates for the direct NO reactivity test described above.
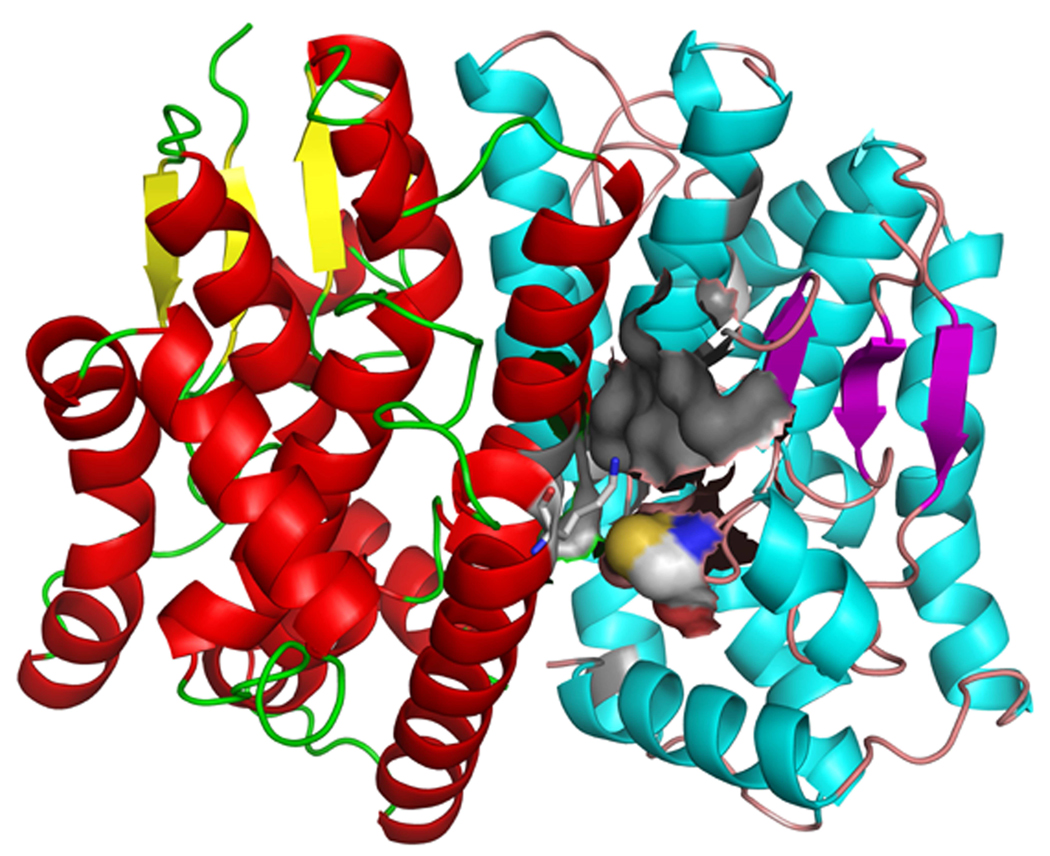
Figure 6. GSTt dimerization interface region.
Molecular model of the dimerization complex for rat GSTt. The two monomers are shown in cartoon representation in different color schemes. Cys 50 of one monomer is shown in spacefill, and the nearby Lys 149 (belongs to the other monomer) is shown in sticks representation. The grey area depicts (in semi-transparent molecular surface visualization) a hydrophobic pocket described in detail in the text.
Table 2.
Proteins with predicted reactivity toward GSNO
| protein | id | Cys position |
Distance, S-S |
Distance, S-N |
Energy (Kcal/mol) |
|---|---|---|---|---|---|
| Glyceraldehyde 3-phosphate dehydrogenase | P04797 | 150 | 2.7 | 4.1 | −4.7 |
| Histone deacetylase 2 # | Q92769 | 262 | 2.5 | 3.9 | −4.9 |
| Tubulin beta | P69897 | 12 | 2.4 | 2.5 | −3.3 |
| Peroxiredoxin 2 | Q949U7 | 121 | 1.9 | 2.6 | −1.5 |
| OxyR | Q1R3U8 | 113 | 2.9 | 3.5 | −4.9 |
| S-adenosylmethionine synthetase, isoform type-1 | P13444 | 121 | 3.8 | 2.4 | −4.5 |
| Creatine kinase | P09605 | 283 | 2.4 | 4.1 | −6.9 |
| ISG15 ubiquitin-like modifier | Q4FJR9 | 76 | 3.6 | 3.8 | −3.4 |
| Calpain 2, large subunit | Q07009 | 301 | 3.7 | 2.7 | −3.7 |
| Tubulin alpha | Q6P9V9 | 347 | 4 | 2.5 | −4.1 |
| Myosin heavy chain 9# | P35579 | 91 | 2.2 | 2.5 | −3.1 |
| Thioredoxin 1 | P10599 | 73 | 2.5 | 4 | −3.9 |
| Mitogen-activated protein kinase | Q99683 | 116 | 4.6 | 3 | −4.8 |
| Serine/threonine protein phosphatase 2B | Q99683 | 256 * | 2.4 | 2.8 | −5.4 |
| Sodium/potassium-transporting ATPase | P06687 | 42 * | 3 | 4.8 | −4.1 |
homology model employed is characterized by less than 50% identity with the template
the reported candidate Cys is not the previously reported NO-Cys residue (i.e., not in our NO-Cys dataset, Table S1).
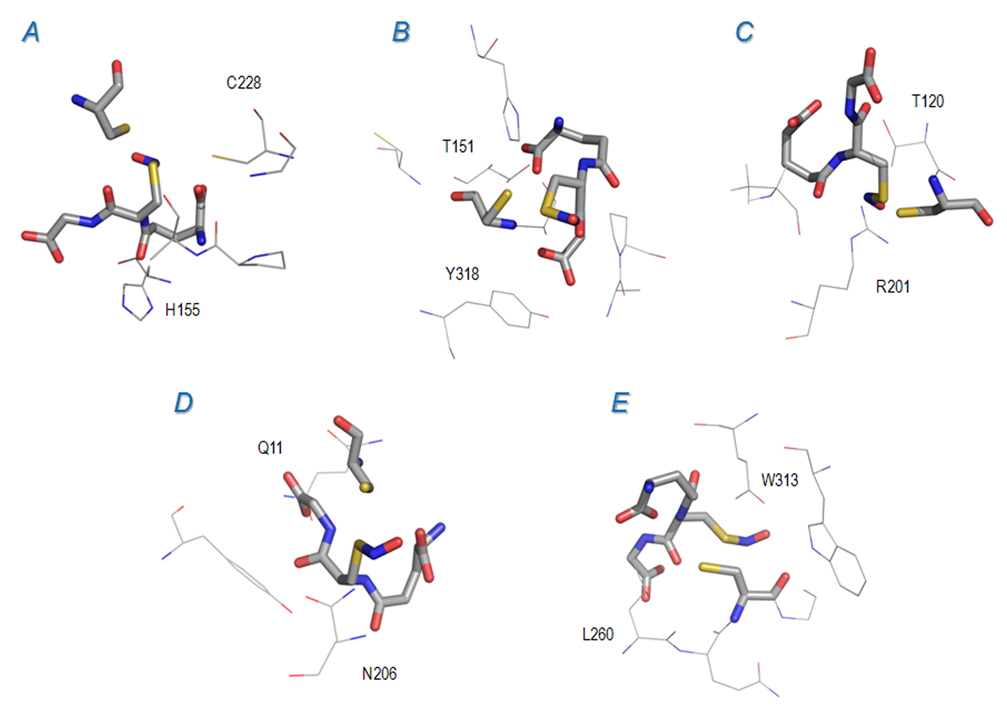
An interesting case was the analysis of Ser/Thr phosphatase: docking GSNO to the reported NO-Cys site (Cys 228) did not yield a reliable model. Instead, a good affinity was found between GSNO and a nearby Cys 256 (4 Å from Cys 228), as shown in Table 2 and Figure 5A. In the best model (with the energy of −5.4 kcal/mol, Figure 5A), the GSNO sulfur atom was within 2.4 Å of the Cys 256 sulfur (and 2.8 Å from the NO nitrogen), suggesting that Cys 256 (rather than 228) was a good NO-Cys candidate, via interaction with GSNO. Thus, it is possible that Cys 256 may act as a donor in the trans-nitrosylation reaction between Cys 228 (the reported final acceptor) and Cys 256. This possibility raises an important question: could Cys residues in the same protein be engaged in trans-nitrosylation? If so, the low occurrence (Figure 2 and Figure 4) of nearby Cys (especially within 6 Å) could be explained by the need to avoid promiscuous and uncontrolled NO transfer within proteins whose functionality is affected by NO-Cys modification. It would be important to address this question in further direct experiments. In this regard, Ser/Thr phosphatase may be an excellent model protein for such study.
Figure 5. Docking calculations for proteins in the NO-Cys dataset.
S-nitrosylated glutathione (GSNO) docking models for proteins in the NO-Cys dataset lacking the solvent accessible 8 Å acid-base motif. GSNO and Cys residues are depicted in sticks. In each panel, some nearby residues (i.e., located close to GSNO) are labeled for reference. A. Serine/threonine phosphatase (Cys 228, in wireframe and labeled, and Cys 256); B. GAPDH (Cys 150); C. PRX2 (Cys 121); D. Tubulin beta (Cys 12); E. Histone deacetylase (Cys 262). Relevant thermodynamic and geometric parameters are given in Table 2.
As to the other NO-Cys-containing proteins in the dataset, we found that only few showed good affinity toward GSNO. The other positive probes (Table 2) included OxyR (Cys 113), MAT (Cys 121), creatine kinase (Cys 283), calpain 2 (Cys 301), tubulin alpha (Cys 347), myosin heavy chain 9 (Cys 91), JNK1 (Cys 116), TRX1 (Cys 73), and Isg15 (Cys 76). Additionally, sodium/potassium-transporting ATPase showed a reactive Cys 42, which was not previously reported to be S-nitrosylated, but it was close to the known NO-Cys site (Cys 49, with the S-S distance of 4.3 Å from Cys 42), similar to what was described for Ser/Thr phosphatase.
We found good affinity of TRX1 for GSNO with preferential binding in the exposed region between Cys 32 (catalytic Cys) and Cys 73. Indeed, in our calculations (based on the reduced form of TRX1), only the latter was found in a position such that its sulfur atom could react with the NO group of the substrate (Figure S6). These observations are in line with previous experimental studies [33–35]. We did not find good candidates for the Cys 69-GSNO complex (Figure S6), notwithstanding Cys 69 is also a potential target of S-nitrosylation via GSNO [35]. Finally, our docking data indicated that the acid-base motif did not correlate with the affinity toward GSNO: 5 out of 8 “deviating” NO-Cys sites (i.e., without the acid-base motif) were predicted as reactive toward GSNO (Table 1 and Figure 5). Thus, similarly to what was found for direct reactivity with NO, GSNO affinity also did not require the presence of both basic and charged residues in proximity of the modifiable Cys.
Overall, 7 out of 8 proteins lacking the exposed distant acid-base motif in proximity of NO-Cys were found to be good candidates for reaction with GSNO (5 proteins) or NO (the other 2 proteins). Altogether, our results support the hypothesis that the acid-base motif may play an indirect role in the NO/Cys reaction: its exposure, positioning (with functional groups distal to NO-Cys) and lack of control with GSNO and NO reactivity, point directly to a role in defining the molecular surface near NO-Cys (e.g., electrostatic potential and ionic interactions).
However, while the role of the acid could be explained in this way, the role of basic residues may be more complex. Hypothetically, their presence can be linked to thiolate stabilization, protein-protein interactions, or both. Additionally, it has to be considered that both acidic and basic residues may contribute, together with the other nearby residues (e.g,. Ser and Thr), to interactions (electrostatic, H-bond) with GSNO. However, this contribution varies case by case, and from our docking analysis appears not to be a generalizable feature of NO-Cys reactivity with GSNO.
Effects of protein-protein interactions on Cys reactivity
These considerations do not explain the case of the single remaining deviating protein that lacked the exposed acid-base motif and did not show reactivity with GSNO or NO): GSTt. Cytosolic GSTs, a family of multifunctional enzymes, naturally form homodimers [36]. The importance of their subunit interface region is well understood and fundamental to the function of these proteins [37]. Two types of interaction emerged as critical for GSTs dimerization: hydrophobic contacts (particularly in the Alpha, Mu and Pi classes) and electrostatic interactions driven by class-specific patterns of charged amino acids exposed in the contact region [38]. We modeled a rat GSTt dimer based on the crystal structure of the human protein (1LJR which could not be directly used due to significant differences with the rat protein, including Cys50Ser mutation). In the dimer structure, Cys 50 was well exposed (>10 Å2) and found in proximity to the dimerization interface (Figure 6). Moreover, its reactivity was significantly enhanced, with a calculated pKa of 6.9 for the dimeric form of the protein (8.2 for the monomeric form).
We tested the GSTt dimer model for (i) direct S-nitrosylation; and (ii) GSNO docking assay. Its NO-Cys (Cys 50) was predicted with the sulfur atom in a well defined pocket (Cys 50, Val 63, Leu 64, Thr 65 of one monomer, and Glu 97, Lys 149 of the other monomer), which was slightly hydrophobic (0.4 is the overall Kyte-Doolittle score). Moreover, the basic residue had its charged atom at 4.8 Å from the sulfur (Figure 6, Lys depicted in sticks representation). Thus, Cys 50 was a potential candidate for direct S-nitrosylation (Table 2).
As to the docking assay, GSNO showed affinity for Cys 50 of the dimeric GSTt, in clear contrast with the zero affinity found for the monomeric protein. However, no strictly reactive positions were found for the first 10 ranked docking models (Figure S7). In turn, GSH consistently docked, in both monomeric and dimeric forms of GSTt, close to its natural binding site (near Gln 12, Figure S7). So, in the case of this protein, the NO modification appears to exert the effect toward GSH properties, leading to an increased affinity of Cys 50 toward GSNO. For comparison purposes, we conducted the same docking analysis for the modeled dimer of GSTp, and the results were clearly different: neither GSH nor GSNO arrived closer than 10 Å to Cys 48. Instead, both substrates docked in the natural GSH-binding pocket.
Thus, dimerized GSTt showed clear enhancement in Cys 50 reactivity (for all tested parameters), transforming a previously apparently inert Cys into a reactive residue. In particular, Cys 50 in the dimer appeared to form a suitable site for direct S-nitrosylation. This could be taken as a general case of how protein-protein interactions could change the character of Cys in an interface region, enhancing its reactivity. This switch in Cys reactivity upon interaction with another protein may be crucial in the interprotein NO transfer process, and also could explain why many reported NO-Cys sites eluded efficient predictions based on standard reactivity parameters (e.g., exposure, pKa).
Topology and relative mobility of NO-Cys
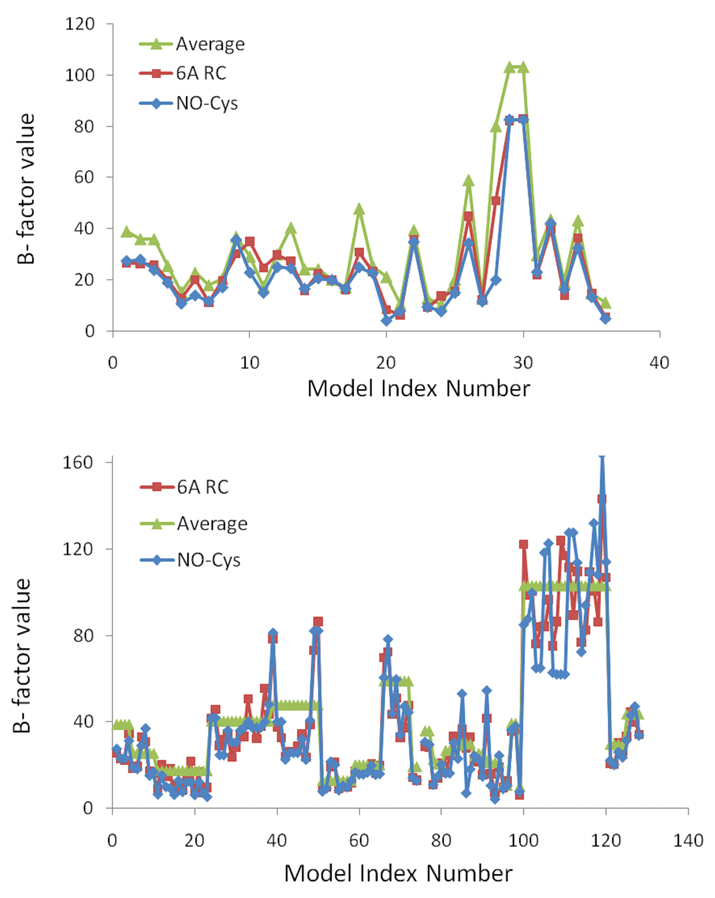
Finally, notable observations were obtained from the analysis of B factors (Figure 7). In a clear pattern, shared by all modified Cys residues, nitrosylated sites were located in portions of proteins characterized by lower mobility than an average value (i.e., the B factor value relative to the modified Cys was lower than the average B factor value for the whole protein) for the same protein (Figure 7A).
Figure 7. B factor analysis of NO-Cys sites.
The upper panel shows calculated B factor values of NO-Cys sites (i.e., dataset protein, Table S1), and the lower panel the B-factor values for all other Cys in the same proteins. NO-Cys alone is shown in blue, the average value for amino acids in proximity (within 6 Å, shown as 6A RC) to the Cys in red, and the average B factor value for the protein in green. On the X axis, Model Index Number refers to the NO-Cys ordered as in Table S1 (upper panel), or to all Cys of the same proteins ordered in the N-terminal to C-terminal direction: thus for each protein in Table S1, all Cys (excluding the NO-Cys itself) are listed in N-terminal to C-terminal order in the graph.
However, increased mobility characterized these proteins in the progression from NO-Cys to proximal (6 Å) region and then to the whole protein. Thus, a clear trend was revealed by our analysis wherein both the modified Cys and, to a lower extent, its proximally located residues showed lower mobility (as revealed by experimentally derived indicators - the B-factors) when compared to other regions in the same proteins (Figure 7). Importantly, this pattern was absent in the control set of proteins (Figure 7B). These data suggest that S-nitrosylation occurs on structured Cys residues, wherein the local effect of NO modification could influence at a greater extent the regions distant from the modified site.
Effects of Cys nitrosylation on electrostatic potential distribution
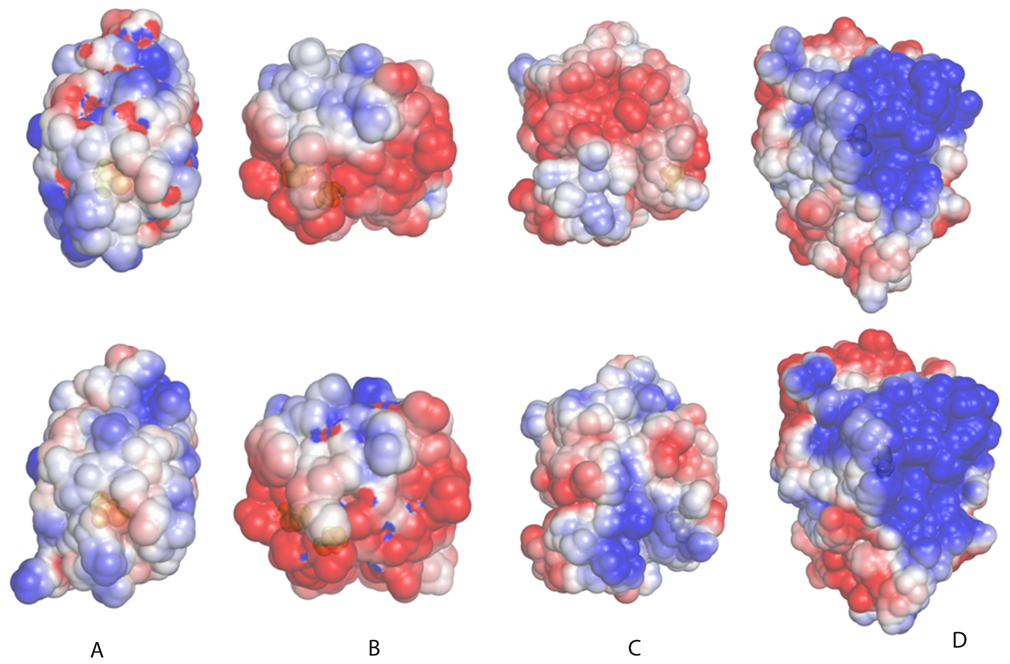
We analyzed available crystallographic structures of proteins containing NO-Cys, including human Trx1 (pdb 2HXK), Blackfin tuna myoglobin (2NRM), human hemoglobin (1BUW) and human PTP1b (3EU0), and compared them with the corresponding non-modified structures (see Table 3, Fig. 8 for details on pdb codes). Upon modification, these proteins show moderate structural rearrangements. Superimposing NO-sylated and non-modified proteins, the all atom root mean square deviation (rmsd) varied from 0.8 Å for Trx1, to 1.3 Å for hemoglobin, with 1.1 Å for myoglobin and 0.9 Å for PTP1b (Table 3). Interestingly, the average displacement differed for various sets of amino acids, if grouped by their physico-chemical properties (i.e., basic, acidic, polar not charged, and apolar residues, as defined by DeepView 4.0). Charged residues clearly showed larger displacement than other categories of amino acids (Table 3). In three proteins (myoglobin, TRX1 and hemoglobin), basic residues were the most displaced, whereas acidic residues were the most displaced in PTP1b. A common trend was apparent wherein following S-nitrosylation, the structural rearrangement affected charged residues on a greater extent. Of particular interest was the observation that many highly displaced charged residues were exposed (present in the molecular surface), and were located far (more than 8 Å) from the Cys modification site. For example, upon nitrosylation of Cys 10 of myoglobin, a loop (Lys 73 to His 78) that was laterally placed with respect to the N-terminal α-helix containing the NO-Cys showed complete repositioning. This movement corresponded to a rmsd of 4.4 Å, which was significantly higher than the average for the whole protein (Table 3) and also higher than the displacement for the NO-Cys region itself (rmsd of 1.6 Å, the α-helical region spanning Asp 4 and Ala 15). This region contained three positively charged residues (Lys 73, Lys 75 and His 78), each located at more than 15 Å from the modification site. As consequence of the movement of exposed charged residues, the molecular surface was locally rearranged, and this led to a change in its electrostatic properties (i.e., when charged residues moved in the surface, both the surface and relative positions of charged atoms were changed, thus affecting the surface electrostatic potential distribution), as shown in Fig. 8. At least in three cases (myoglobin, TRX1 and hemoglobin), a marked redistribution of the electrostatic potential was observed. In the case of PTP1B, which was curiously the protein showing the lowest displacement of basic residues (Table 3), the redistribution following S-nitrosylation of its catalytic Cys was smaller though still detectable. Given the low number of available paired experimental structures with and without NO-Cys, it may be difficult to draw general conclusions. However, from the analysis of this limited protein dataset, it seems that an important effect of S-nitrosylation is to modify the electrostatic properties of molecular surfaces by triggering the movement of its exposed charged residues. This hypothesis fits well with sophisticated theoretical calculations on NO-Cys in proteins, showing that nitrosylation causes substantial charge redistribution in Cys side chain atoms [32]. Intriguingly, in this scenario an additional role for acidic and basic residues might be to enhance the electrostatic perturbation introduced by S-nitrosylation through a mechanism wherein the message propagates from a receiver (Cys) to peripheral parts of the protein.
Table 3.
Average displacement of amino acids following S-nitrosylation
| Basic residues (r.m.s.d.) |
Acidic residues (r.m.s.d.) |
Polar residues (r.m.s.d.) |
Apolar residues (r.m.s.d.) |
All residues (r.m.s.d.) |
|
|---|---|---|---|---|---|
| Myoglobin | 2.1 Å | 1.2 Å | 1.3 Å | 1.1 Å | 1.4 Å |
| Trx1 | 1.3 Å | 1.1 Å | 0.8 Å | 0.5 Å | 0.8 Å |
| Hemoglobin | 1.9 Å | 1.7 Å | 1.3 Å | 1 Å | 1.3 Å |
| PTP1b | 1.1 Å | 1.5 Å | 0.8 Å | 0.5 Å | 0.9 Å |
The structures of human Trx1, with NO-Cys (1ERU) and without this modification (2HXK), Blackfin tuna myoglobin (2NRM and 1MYT), human hemoglobin (1BUW and 1A3N) and human PTP1B (3EU0 and 2F6F) were superimposed with DeepView 4.0. The root mean square deviation (rmsd) was calculated for all atoms of proteins, and separately, for all atoms belonging to each different set of amino acids grouped on the basis of their chemical similarities as defined by DeepView (i.e. basic, acidic, polar not charged and apolar residues). Values reported refer to the rmsd expressed in Å, and reflects different extent of displacement for each group of amino acids following S-nitrosylation event.
Figure 8. Effect of S-nitrosylation on electrostatic potential distribution.
Electrostatic potential distribution plotted on the solvent accessible surface is shown for four proteins, for which crystal structures are available for both S-nitrosylated and non-modified forms. In the upper row, non-modified proteins are shown, and in the lower row the corresponding structures with NO-Cys. A) Blackfin tuna myoglobin; B) human TRX1; C) human hemoglobin (chain B); D) human PTP1b. PDB codes for these proteins are (first number corresponds to NO-Cys-containing protein, and second to non-modified protein): myoglobin (2NRM and 1MYT), TRX1 (1ERU, 2HXK), hemoglobin (1BUW and 1A3N), and PTP1B (3EU0 and 2F6F). The potentials range from −2.5 kT per proton charge (red) to +2.5 kT per proton charge (blue). Proteins are oriented with the modified Cys (shown in space fill, colored in yellow, below the semi-transparent protein surface) oriented toward the reader.
Discussion
Bioinformatics approaches have been applied to examine different types of protein modifications (phosphorylation, acetylation, etc.), with variable success in term of the ability to employ common patterns for predictive purposes. These studies have helped identifying some of the common features responsible for specificity of these modifications, as well as providing the basis for rules and patterns implemented in predictive algorithms (e.g., various web accessible services at Expasy, http://www.expasy.ch/). However, modifications of Cys residues still represent a big challenge. In this regard, NO-Cys provides an important model for the analysis of Cys modification as a whole and is important in itself considering the role of NO-Cys modifications in biology. In this work, we applied several theoretical approaches to address sequence- and structure-based features that may be behind the specificity of Cys nitrosylation.
Our analyses revealed that known NO-Cys sites in proteins form a heterogeneous set. We found that the features of NO-Cys sites widely discussed in the literature, including (i) the acid-base motif flanking NO-Cys sites in sequence and/or structure; (ii) higher than average hydrophobic content; (iii) lower pKa; and (iv) higher exposure of Cys sulfur atoms, did not distinguish Cys residues known to be modified with NO from random Cys residues in proteins. However, further analysis revealed that occurrence of a charged residue (usually a basic amino acid) in close proximity (6 Å) to NO-Cys and another oppositely charged within a larger region (up to 8 Å from NO-Cys) better describe NO-Cys sites. In addition, the residues in the modified acid-base motif often point outward with respect to the NO-Cys sulfur atom, and they are exposed and located in conserved regions of solvent accessible surfaces in proteins.
Interestingly, these features also apply to the NO-Cys sites containing flanking acidic residues (Figure S4), quite frequently (but not invariantly) found nearby modifiable Cys (Figure 2). During preparation of this manuscript, a study was published showing that human DPR-1 possesses a single NO-Cys site, Cys 644, which leads to a series of neurotoxic events in the cell, linked to the development of Alzheimer’s disease [31]. The authors modeled the structure of human DPR-1 and showed that the flanking acidic and the closest basic residues were present in the solvent accessible surface surrounding the modifiable site, rather than pointing to Cys 644 itself. This observation is consistent with our extensive bioinformatics analysis and the model of S-nitrosylation.
Altogether, our results suggest a hypothesis that NO-Cys sites are characterized by exposed acid-base motifs serving a function different than activation of Cys for reaction with NO or stabilization of NO-Cys. We propose that the finding for TRX1/caspase 3 (i.e., an exposed acid-base motif nearby the NO-Cys, necessary for protein-protein interaction and NO-transfer) can be extended to many other NO-Cys sites. Specific S-nitrosylation involving protein-protein interactions has been assessed in various physiologically relevant cases [39, 40]. Addressing how these interactions may affect NO-Cys formation (or vice versa) is potentially crucial in unraveling the specificity for this type of redox signaling. It appears that protein-protein interaction-based trans-nitrosylation may better explain the observations that apparently inert Cys residues are specifically and selectively nitrosylated. Some NO-Cys sites have high pKa and are not exposed while other Cys in the same protein, not known to be S-nitrosylated, show a higher reactivity.
An explanation could be that other proteins (or other NO-Cys sites within the same protein) specifically interact with the Cys in question, enhancing its reactivity and specifically “turning on” the residue for interaction with NO or NO donors. We discussed one such case (rat GSTt) wherein protein-protein interaction seems to activate one of its Cys residues. Here, the modifiable Cys (Cys 50) is inert in the monomer, but in the dimer its reactivity is enhanced. Combined with the analyses of suitable features for direct NO reactivity, such as occurrence of protein pockets containing modifiable Cys residues, our data show that the heterogeneity of NO-Cys sites is difficult to rationalize if only direct reactivity with NO is accounted for or, more generally, if a single mechanism is considered. On the other hand, interactions between some proteins, such as TRX1/caspase 3, or with agents, such as GSNO, could better explain the high variation in amino acid composition and other parameters. In this scenario, the specificity is driven by protein-protein interactions or by affinity of proteins towards GSNO. We presented the first attempt to date to systematically address this issue, analyzing all reliable NO-Cys sites for reactivity toward NO and GSNO, and found several potential candidates for both. Additionally, analyzing four available paired cases of NO-Cys-containing and non-modified structures, we found that the link between S-nitrosylation, displacement of charged residues and protein-protein interactions could be further extended, as NO modification leads to a redistribution of charged residues in the molecular surface, inducing a shift in its electrostatic properties and thus affecting specificity of interaction. These observations suggest that S-nitrosylation is linked to protein-protein interactions in two ways: first, interaction between two proteins, one of which is S-nitrosylated, may promote NO transfer to the other protein, as previously discussed; second, following S-nitrosylation, a change in the spectrum of binding partners for the modified protein occur (e.g., via redistribution of the electrostatic potential), possibly affecting protein function.
Finally, we propose that for future bioinformatics analyses aiming to identify common features of NO-Cys sites and use them for predictive purposes, separate datasets should be built and analyzed. For instance, candidate NO-Cys sites could be classified based on (i) NO donor compounds (e.g., GSNO, NO or Cys-NO), (ii) concentrations of these compounds, (iii) function of the NO-Cys in stress and/or signaling responses, and (iv) groups of organisms in which this regulation occurs (e.g., prokaryotes, lower eukaryotes, plants, vertebrates). In addition, as trans-nitrosylation may further complicate the analyses, biochemical pathways to which a protein is involved may prove of fundamental importance: interacting proteins may promote NO transfer from one Cys to another. This way, it might be possible to identify common features of various NO-Cys classes, each characteristically related to the type of NO-sylating agents and physiological conditions in which the modifications occur. This possibility, even if intriguing, will require additional experimental data before extensive bioinformatics analysis could adequately address the issue.
Experimental Procedures
Sequence analysis
A set of NO-Cys containing proteins was collected by manually curated literature searches and selecting proteins reported to be nitrosylated in more than one study. Sequence alignments were prepared with PSI-BLAST against the NCBI non-redundant protein database with the following search parameters: expectation value 1e-4, expectation value for multipass model 1e-3, and maximal number of output sequences 1,000. Cys conservation for proteins was determined using an in-house Python-script by parsing the PSI-BLAST output. For the structural profile analysis and multiple sequence alignments, ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html) standalone version 2.0.3 was employed. Hydrophobicity was analyzed with in-house Python programs, implementing the standard Kyte-Doolittle scale [41].
Molecular modeling
Models were built via Swiss Model (http://swissmodel.expasy.org/). VegaZZ 2.2.0 molecular modeling package was used to check for missing residues, and for minimization runs (with CHARMM22 force field) to fix planarity issues, edit multiple side chain conformations, and adjust for incorrect geometries. These operations were needed for subsequent structure-based analysis, particularly for docking and protein pocket calculations.
For modeling of human DPR-1, the mGENTHREADER (http://bioinf.cs.ucl.ac.uk/psipred/psiform.html) method was employed. The alignment with bacterial dynamin-like protein BDLP (pdb code 2j69) was chosen, and human DPR-1 was then modeled with HOMER (http://protein.cribi.unipd.it/Homer/).
Calculations of pKa values for dataset proteins were done with PropKa implementation in VegaZZ. Calculations of accessible surface area were performed with a standalone program, Surface 4.0, downloaded from http://www.pharmacy.umich.edu/tsodikovlab/. The same program was used to assess the topology of cavities in proteins of our dataset, while the server Castp (http://stsfw.bioengr.uic.edu/castp/) was employed for protein pocket detection. Electrostatic potential calculations were made with the standalone program APBS (http://apbs.sourceforge.net/); partial charges and atomic radii needed for the APBS input file (pqr files) were calculated with VegaZZ.
Bioinformatics approaches to structure-based analyses of the dataset
All calculations were made with in-house Python programs (version 2.5, http://www.python.org/download/releases/2.5/). To define the sites of N-nitrosylation, we calculated the distance between the sulfur atom of NO-Cys and any other atom in the protein. Residues containing at least one atom within the cut-off distance were kept, defining the distance-dependent NO-Cys site. To assess the location of charged residues relative to the Cys sulfur atom, the distance between the sulfur atom (S) and the functional group atoms of charged residues (GrF) were calculated and compared with the distance between the S atom and the charged residue approximated center of mass (CM). Thus, the S to GrF distance was compared with the S to CM distance: when the first was lower, the charged residue was thought to point toward the sulfur atom (and potentially, directly contributing to chemical activation of the SG atom).
For the B factor analysis of crystallographic structures, we extracted B values for each amino acid of interest. When a protein from the dataset was characterized by only an NMR structure (in our case, the only such protein was myeloid differentiation primary response protein, pdb code 2js7), the average root mean square deviation (rmsd) for each atom and each model of the ensemble was calculated (resulting in an average rmsd for each atom). These values were then normalized to 100, leading to a B-factor like scale, and were then employed as substitute for the B factor to calculate protein-specific mobility of NO-Cys as compared to other protein regions.
Computational docking
VegaZZ 2.2.0 was employed to prepare substrate models (GSNO and GSH). The SP4 force field and the AMMP-mom method were employed to assign charges to GSH and minimize the structures. GSNO has also been treated with SP4 force field, and assignment of initial charges was done with AMMP-mom. Then, we manually implemented the charge for the NO-Cys portion of the substrate following the charge distribution as reported in a recently published study [32]. It provided parameters for NO-Cys: at a more sophisticated (compared to force field SP4) level, the authors found charge redistribution on Cys residue upon NO addition, which mainly involved its side chain atoms. We implemented the findings of this study in our analysis (Supplementary information, Figure S1). GSNO was then exported to ArgusLab for docking calculations. The substrate was analyzed in the intercalation site using a docking box of 20×20×20 Å centered on the candidate modifiable Cys residues. We employed the GAdock docking engine, a genetic algorithm search technique implemented in ArgusLab. To set up the docking parameters, calculations were performed with the following values: population size 250, max generations 10000, mutation rate 0.02, grid resolution 0.15 Å, flexible ligand mode (other parameters were kept with default values). The interaction energy values were calculated as the energy of the complex minus the energy of the ligand, minus the energy of protein: ΔEinter = E(complex) – (E(ligand) + E(protein)).
Supplementary Material
Acknowledgments
We would like to thank Dr. Jonathan Stamler for discussion and helpful insights.
This study was supported by NIH GM065204 (to VNG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Murad F. Cyclic guanosine monophosphate as a mediator of vasodilation. J. Clin. Invest. 1986;78:1–5. doi: 10.1172/JCI112536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolotina VM, Najibi S, Palacino JJ, Pagano PJ, Cohen RA. Nitric oxide directly activates calcium-dependent potassium channels in vascular smooth muscle. Nature. 1994;368:850–853. doi: 10.1038/368850a0. [DOI] [PubMed] [Google Scholar]
- 3.Stamler JS, Lamas S, Fang FC. Nitrosylation. the prototypic redox-based signaling mechanism. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- 4.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat. Cell. Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 5.Gaston B, Singel D, Doctor A, Stamler JS. S-nitrosothiol signaling in respiratory biology. Am. J. Respir. Crit. Care. Med. 2006;173:1186–1193. doi: 10.1164/rccm.200510-1584PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hare JM, Stamler JS. NO/redox disequilibrium in the failing heart and cardiovascular system. J. Clin. Invest. 2005;115:509–517. doi: 10.1172/JCI200524459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mannick JB, Schonhoff CM. Analysis of protein S-nitrosylation Chapter 14. Curr. Protoc. Protein Sci. 2006 doi: 10.1002/0471140864.ps1406s46. Unit 14.6. [DOI] [PubMed] [Google Scholar]
- 8.Han P, Zhou X, Huang B, Zhang X, Chen C. On-gel fluorescent visualization and the site identification of S-nitrosylated proteins. Anal. Biochem. 2008;377:150–155. doi: 10.1016/j.ab.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 9.Mannick JB, Schonhoff CM. Measurement of protein S-nitrosylation during cell signaling. Methods Enzymol. 2008;440:231–242. doi: 10.1016/S0076-6879(07)00814-2. [DOI] [PubMed] [Google Scholar]
- 10.Palmer LA, Gaston B. S-nitrosothiol assays that avoid the use of iodine. Methods Enzymol. 2008;440:157–176. doi: 10.1016/S0076-6879(07)00809-9. [DOI] [PubMed] [Google Scholar]
- 11.Gao C, Guo H, Wei J, Mi Z, Wai PY, Kuo PC. Identification of S-nitrosylated proteins in endotoxin-stimulated RAW264.7 murine macrophages. Nitric Oxide. 2005;12:121–126. doi: 10.1016/j.niox.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat. Cell. Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 13.Torta F, Usuelli V, Malgaroli A, Bachi A. Proteomic analysis of protein S-nitrosylation. Proteomics. 2008;8:4484–4494. doi: 10.1002/pmic.200800089. [DOI] [PubMed] [Google Scholar]
- 14.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci. STKE. 2001;2001:PL1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 15.Kuncewicz T, Sheta EA, Goldknopf IL, Kone BC. Proteomic analysis of S-nitrosylated proteins in mesangial cells. Mol. Cell. Proteomics. 2003;2:156–163. doi: 10.1074/mcp.M300003-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Martínez-Ruiz A, Lamas S. Detection and proteomic identification of S-nitrosylated proteins in endothelial cells. Arch. Biochem. Biophys. 2004;423:192–199. doi: 10.1016/j.abb.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 17.Rhee KY, Erdjument-Bromage H, Tempst P, Nathan CF. S-nitroso proteome of Mycobacterium tuberculosis: Enzymes of intermediary metabolism and antioxidant defense. Proc. Natl. Acad. Sci. USA. 2005;102:467–472. doi: 10.1073/pnas.0406133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc. Natl. Acad. Sci. USA. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greco TM, Hodara R, Parastatidis I, Heijnen HF, Dennehy MK, Liebler DC, Ischiropoulos H. Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proc. Natl. Acad. Sci. USA. 2006;103:7420–7425. doi: 10.1073/pnas.0600729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stamler JS, Jia L, Eu JP, McMahon TJ, Demchenko IT, Bonaventura J, Gernert K, Piantadosi CA. Blood flow regulation by S-nitrosohemoglobin in the physiological oxygen gradient. Science. 1997;276:2034–2037. doi: 10.1126/science.276.5321.2034. [DOI] [PubMed] [Google Scholar]
- 21.Sun J, Xin C, Eu JP, Stamler JS, Meissner G. Cysteine-3635 is responsible for skeletal muscle ryanodine receptor modulation by NO. Proc. Natl. Acad. Sci. USA. 2001;98:11158–11162. doi: 10.1073/pnas.201289098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nedospasov A, Rafikov R, Beda N, Nudler E. An autocatalytic mechanism of protein nitrosylation. Proc. Natl. Acad. Sci. USA. 2000;97:13543–13548. doi: 10.1073/pnas.250398197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell. Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 24.Derakhshan B, Hao G, Gross SS. Balancing reactivity against selectivity: the evolution of protein S-nitrosylation as an effector of cell signaling by nitric oxide. Cardiovasc. Res. 2007;75:210–219. doi: 10.1016/j.cardiores.2007.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asada K, Kurokawa J, Furukawa T. Redox- and calmodulin-dependent S-nitrosylation of the KCNQ1 channel. J. Biol. Chem. 2009;284:6014–6020. doi: 10.1074/jbc.M807158200. [DOI] [PubMed] [Google Scholar]
- 26.Cammer SA, Hoffman BT, Speir JA, Canady MA, Nelson MR, Knutson S, Gallina M, Baxter SM, Fetrow JS. Structure-based active site profiles for genome analysis and functional family subclassification. J. Mol. Biol. 2003;334:387–401. doi: 10.1016/j.jmb.2003.09.062. [DOI] [PubMed] [Google Scholar]
- 27.Marino SM, Gladyshev VN. A structure-based approach for detection of thiol oxidoreductases and their catalytic redox-active cysteine residues. PLoS Comput Biol. 2009;5:e1000383. doi: 10.1371/journal.pcbi.1000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell DA, Morton SU, Fernhoff NB, Marletta MA. Thioredoxin is required for S-nitrosation of procaspase-3 and the inhibition of apoptosis in Jurkat cells. Proc. Natl. Acad. Sci. USA. 2007;104:11609–11614. doi: 10.1073/pnas.0704898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benhar M, Forrester MT, Hess DT, Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmgren A. Biochemistry. SNO removal. Science. 2008;320:1019–1020. doi: 10.1126/science.1159246. [DOI] [PubMed] [Google Scholar]
- 31.Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, Lipton SA. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han S. Force field parameters for S-nitrosocysteine and molecular dynamics simulations of S-nitrosated thioredoxin. Biochem. Biophys. Res. Commun. 2008;377:612–616. doi: 10.1016/j.bbrc.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 33.Sumbayev VV. S-nitrosylation of thioredoxin mediates activation of apoptosis signal-regulating kinase 1. Arch. Biochem. Biophys. 2003;415:133–136. doi: 10.1016/s0003-9861(03)00199-1. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Liu T, Wu C, Li H. A strategy for direct identification of protein S-nitrosylation sites by quadrupole time-of-flight mass spectrometry. J. Am. Soc. Mass. Spectrom. 2008;19:1353–1360. doi: 10.1016/j.jasms.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hashemy SI, Holmgren A. Regulation of the catalytic activity and structure of human thioredoxin 1 via oxidation and S-nitrosylation of cysteine residues. J. Biol. Chem. 2008;283:21890–21898. doi: 10.1074/jbc.M801047200. [DOI] [PubMed] [Google Scholar]
- 36.Luo JK, Hornby JA, Wallace LA, Chen J, Armstrong RN, Dirr HW. Impact of domain interchange on conformational stability and equilibrium folding of chimeric class micro glutathione transferases. Protein Sci. 2002;11:2208–2217. doi: 10.1110/ps.0208002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hornby JA, Luo JK, Stevens JM, Wallace LA, Kaplan W, Armstrong RN, Dirr HW. Equilibrium folding of dimeric class mu glutathione transferases involves a stable monomeric intermediate. Biochemistry. 2000;39:12336–12344. doi: 10.1021/bi000176d. [DOI] [PubMed] [Google Scholar]
- 38.Piromjitpong J, Wongsantichon J, Ketterman AJ. Differences in the subunit interface residues of alternatively spliced glutathione transferases affects catalytic and structural functions. Biochem. J. 2007;401:635–644. doi: 10.1042/BJ20060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park HS, Yu JW, Cho JH, Kim MS, Huh SH, Ryoo K, Choi EJ. Inhibition of apoptosis signal-regulating kinase 1 by nitric oxide through a thiol redox mechanism. J. Biol. Chem. 2004;279:7584–7590. doi: 10.1074/jbc.M304183200. [DOI] [PubMed] [Google Scholar]
- 40.Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, Ferris CD, Hayward SD, Snyder SH, Sawa A. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell. Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 41.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.