Abstract
Elevated levels of reactive oxygen species can damage proteins. Sulfur-containing amino acid residues, cysteine and methionine, are particularly susceptible to such damage. Various enzymes evolved to protect proteins or repair oxidized residues, including methionine sulfoxide reductases MsrA and MsrB, which reduce methionine-S-sulfoxide (Met-SO), and methionine-R-sulfoxide (Met-RO) residues, respectively, back to methionine. Here, we show that MsrA and MsrB are involved in the regulation of mitochondrial function. Saccharomyces cerevisiae mutant cells lacking MsrA, MsrB or both proteins, had normal levels of mitochondria, but lower levels of cytochrome c and fewer respiration-competent mitochondria. The growth of single MsrA or MsrB mutants on respiratory carbon sources was inhibited, and that of the double mutant was severely compromised, indicating impairment of mitochondrial function. Although MsrA and MsrB are thought to have similar roles in oxidative protein repair each targeting a diastereomer of methionine sulfoxide, their deletion resulted in different phenotypes. GFP fusions of MsrA and MsrB showed different localization patterns and primarily localized to cytoplasm and mitochondria, respectively. This finding agreed with compartment-specific enrichment of MsrA and MsrB activities. These results show that oxidative stress contributes to mitochondrial dysfunction through oxidation of methionine residues in proteins located in different cellular compartments.
Elevated levels of reactive oxygen species (ROS) can cause damage to cellular components, including nucleic acids, proteins, and lipids (1–2). Aside their deleterious effects, ROS are also involved in regulation of numerous cellular processes, such as gene expression and cell growth. Therefore, cells need to tightly regulate the levels of these oxidants. Cysteine and methionine, the two sulfur-containing amino acid residues, are particularly susceptible to oxidation by ROS. However, several oxidized forms of cysteine (e.g., sulfenic acid, sulfinic acid and disulfide) and methionine (e.g., methionine sulfoxide) can be enzymatically reduced to the original residues, restoring protein activities (3–5). In addition, cyclic oxidation and reduction of cysteines and methionines in proteins is thought to be a mechanism that protects cells from the deleterious effects of oxidants (6–7).
Methionine sulfoxide has an asymmetric sulfur atom, which gives rise to two diastereomers. Methionine sulfoxide reductase A (MsrA) was the first enzyme identified in the methionine repair pathway (8), and it is stereospecific for methionine-S-sulfoxide (Met-SO). A second enzyme, methionine sulfoxide reductase B (MsrB), which is a selenium-containing protein in mammals and a cysteine-containing protein in most other organisms (9–11), was also identified, and it is specific for methionine-R-sulfoxide (Met-RO). Because both enzymes participate in the methionine sulfoxide reduction pathway, they are often expressed in a coordinated fashion and, in many bacteria, the genes for these two proteins are located within the same operon.
In addition, MsrA and MsrB have very similar phylogenetic profiles: their patterns of occurrence in completed genomes are essentially identical (9–12). This observation is particularly interesting since these two enzymes evolved from unrelated ancestral genes. On the other hand, MsrA and MsrB use alternate substrates and are not known to interact with each other. In this regard, it is surprising that these two enzymes are sometimes fused to form a single polypeptide. The PilB protein of Neisseria gonorrhoeae has a secreted form that is associated with the outher membrane and contains both MsrA and MsrB activities (13). What could then be the evolutionary advantage of linking these two proteins/activities? The requirement of Msrs for pathogenicity of certain microorganisms (14) suggests that some periplasmic proteins contain methionine residues that are oxidized and then repaired by Msrs. The pathogenecity of the bacteria containing periplasmic Msr systems may simply reflect the ability of these organisms to protect their electron transport chain components from the host defense response.
Oxidative stress is known to participate in programmed cell death. One early consequence of oxidative stress is the release of cytochrome c from mitochondria to cytosol. In animal cells, this process initiates a program that leads to cell death. However, the mechanisms by which the stress is sensed and causes mitochondrial dysfunction are not well understood. The cytochrome c has a conserved methionine that coordinates the heme iron and regulates the redox potential of this protein. This methionine has been described as target for oxidation by ROS (15–16) and the resulting methionine sulfoxide form of cytochrome c was shown to be impaired in electron transfer (15). In a recent study, mouse MsrA was shown to be important in cytochrome c repair and protection against cataract (17).
In this study, we found that Msrs regulate mitochondrial functions in yeast. Cells lacking MsrA and MsrB genes were respiratory-deficient due to lower levels of mitochondrial cytochrome c and had shorter lifespan. Mitochondrial function was particularly compromised in cells lacking both Msrs. However, these enzymes unequally affected mitochondrial function, with MsrA being primarily located in the cytosol and MsrB in mitochondria. This study illustrates how the cellular system for methionine sulfoxide reduction regulates mitochondrial function.
MATERIALS AND METHODS
Yeast growth and media
Isolation of yeast mutants lacking methionine sulfoxide reductases was previously described (9–18). These cells were grown in either YPD media (1% yeast extract, 2% peptone, 2% dextrose or other carbon sources) or supplemented YNB (Yeast Nitrogen Base). To analyze growth rates, 2% glycerol, 2% ethanol or 1% lactic acid were used as carbon sources. SDC liquid medium (0.18% yeast nitrogen base without amino acids, 0.5% ammonium sulfate, 0.14% NaH2PO4, 0.173% complete amino acid mix, 2% glucose; pH adjusted to 6.0 with NaOH) was used for chronological lifespan and IRC assays.
Localization of MsrA and MsrB
Open reading frames of S. cerevisiae MsrA and MsrB, without stop codons and with BamHI and HindIII cloning sites, were PCR amplified and cloned into the Sma1 site of pBS. GFP was subcloned from pEGFP-N2 into the HindIII-XhoI site of MsrA and MsrB constructs preserving the reading frames. A BamH1/XhoI fragment coding for Msr-GFP fusion was moved from pBS to p426 yeast vector containing a galactose-inducible promoter. Cells were transformed and initially grown in the presence of 2% glucose, then switched to growth on galactose to induce MsrA-GFP or MsrB-GFP expression. After 3 h, cells were pictured with a confocal fluorescent microscope.
Growth curves
Yeast mutant strains were grown in the presence of 2% glycerol, 2% ethanol or 1% lactic acid and the absorbance was measured at 600 nm at indicated time points.
Western blot analysis
Logarithmically growing cells (5 ml) were harvested and lysed in extraction buffer containing 1× protease inhibitor cocktail (Sigma). 10 µg of total protein from these cell extracts were fractionated on SDS-PAGE gels and electrotransferred onto PVDF membranes (Invitrogen). Membranes were incubated with anti-cytochrome c1 (Cyc1), anti-Tom40 and anti-phosphoglycerate kinase (PGK) antibodies. The blots were developed using an ECL detection system (Amersham).
Cytochrome c isolation
Cells were grown to stationary phase in YPD medium, shifted to YPG (3% glycerol) and incubated with gentle shaking. 10 g (wet weight) of WT cells and 10 g of double mutant cells were harvested and Cyc1 protein was extracted in parallel from these cells as described (19).
Analysis of mitochondria
Flow cytometry was used to monitor the levels of functional mitochondria in WT and indicated mutant strains. (20). Cells were grown to stationary phase in YPD medium, then shifted to YPG and kept for additional 12 h. Approximately 107 cells were harvested and stained with MitoTracker Red (Molecular Probes, Eugene, OR) as described by the manufacturer and analyzed with a flow cytometer.
Yeast aging and index of respiratory competence (IRC) assays
For replicative aging assays, cells were grown on fresh media for 2 days. For each strain, 35 daughter cells (starter mothers) were collected and lined up with a micromanipulator on agar plates. New buds (daughters) from these virgin cells were removed and discarded as they formed. This process continued until cells ceased dividing. Lifespan was determined as the total number of daughter cells that each mother cell generated. Chronological lifespan assays were done as described previously (21). Briefly, frozen stocks of mutant yeast strains were spread onto YPD plates and incubated at 30 °C for 2 days. Several colonies were chosen and incubated in 5 ml SDC medium overnight. These cultures were diluted to 50 ml SDC media and incubated at 30 °C with shaking at 250 rpm. After 3 days, cells were diluted to an A600 of 0.1–0.2, and used for viability assays wherein cells were spread onto YPD plates every 2 days, incubated at 30 °C, and colonies counted after 3 days.
IRC assays were done according to (21). Briefly, from the same cultures that were used for chronological aging assays, cells were diluted to an A600 of 0.1–0.2 and used for the IRC assay once in 2 days. Diluted cells were spread onto YPG plates, incubated at 30 °C, and colonies were counted after 3 days. The IRC was calculated as colony number that was observed from YPG plates divided by the number of colonies on YPD plates.
Overexpression of MsrA and MsrB in wild-type and Msr mutant cells
MsrA and MsrB genes were amplified from yeast genomic DNA and cloned into BamHI and XhoI restriction sites of p425 or p423 yeast expression vectors under GPD promoter. These constructs were transformed into the WT, Msr double (MsrAΔ/MsrBΔ), and Msr triple (MsrAΔ/MsrBΔ/fRMsrΔ) mutant cells using lithium acetate method. For co-expression, plasmids for MsrA and MsrB were used in transformations. Also 1 kb upstream of MsrA and MsrB along with ORF of each genes were cloned into Sac1 and BamHI restriction site of p425 which disrupt the GPD promoter for native expression of each gene under their own promoter.
Complementation assay
Logarithmically growing wild-type, double (MsrAΔ/MsrBΔ) and triple (MsrAΔ/MsrBΔ/fRMsrΔ) mutant cells, and the same cells expressing individual Msrs, were grown in YNB media, harvested and washed with distilled water. The cultures were diluted to an A600 of 0.5. The double mutants spotted on the appropriate YNB plates which contained glycerol for complementation of respiratory growth. Since double Msr mutant strain had residual Msr activity, due to presence of fRMsr (free methionine-R-sulfoxide reductase) specific for the reduction of free Met-RO (22), complementation assay for reductase activity was checked using the triple Msr mutant (MsrAΔ/MsrBΔ/fRMsrΔ). Appropriate YNB plates missing methionine but containing a racemic mixture of methionine sulfoxides (20 mg/L) for reductase activity was used for the spotting assay. Plates were then incubated at 30 °C for four days.
Assays of superoxide levels in yeast cells
To assays for mitochondrial superoxide, mitochondria-specific dye Mitosox (Invitrogen) was used as described previously (23). For cytoplasmic superoxide levels, dihydroethidium dye (Sigma), which detects superoxide radicals in cytosol regardless of their origin, was used (23). Briefly, cells were grown overnight in YPD, diluted to an A600 of 0.6, centrifuged and washed with water. Pellets were incubated in 5 ml of 2 mM H2O2 for 1 h, washed out of peroxide and incubated in YPD media. Dihydroethidium (DHet) was added to cultures to 160 µM for detection of cytosolic superoxide, and Mitosox was added to 5 µM to detect mitochondrial superoxide levels. Cultures were incubated for 45 min, then washed and resuspended in 2 ml 1× PBS. Fluorescence was measured with a flow cytometer for 10,000 cells for each strain.
Isolation of mitochondria and analysis of MsrA and MsrB activities in cellular compartments
The procedures were as previously described (24). Wild type, MsrAΔ, MsrBΔ, and MsrAΔ/MsrBΔ yeast strains were grown at 30 °C for 16 h and then collected by centrifugation. 70–80 g cells were suspended with 50 ml of 100 mM Tris, pH 9.4, in the presence of 10 mM DTT and incubated at 30 °C for 15 min with gentle shaking. After removing the supernatant by centrifugation at 3,500 rpm, cells were resuspended in 40 ml of 1.2 M sorbitol buffer containing 2.5 mg zymolyase 20T per gram of cells. Following incubation at 30 °C for 30 min with gentle shaking, cells were centrifuged at 5,000 rpm for 5 min and resuspended in 100 ml of breaking buffer (0.6 M sorbitol, 20 mM MES potassium, pH 6.0, for mitochondria or pH 7.4 for cytosolic fractions) containing 0.5 mM PMSF. These cells were then homogenized and centrifuged at 3,500 rpm for 5 min. After removing the supernatant, it was centrifuged again at 10,000 rpm for 10 min. The supernatant was collected as a cytosolic part and the pellet was resuspended in breaking buffer (pH 6.0) lacking PMSF. After repeating centrifugation and resuspension of this pellet to get pure mitochondria, this mitochondrial pellet was finally resuspended in breaking buffer (pH 7.4) by using a Teflon dounce and then stored at − 80 °C. Western blot analyses were performed to examine preparation of subcellular fractions using 10 µg of cytosolic or mitochondrial protein lysates and anti-Tom40 and anti-PGK antibodies.
MsrA and MsrB activities were measured in isolated mitochondria and cytosol of each strain using an HPLC procedure. Briefly, 200 µg protein was used in each assay. The reaction (100 µl) was carried out at 37 °C for 30 min in the presence of 20 mM DTT and either 200 µM dabsyl-methionine-S-sulfoxide (MsrA assay) or 200 µM dabsyl-methionine-R-sulfoxide (MsrB assay) were added to the reaction mixture. After stopping the reaction by adding 200 µl acetonitrile, the mixture was centrifuged at 4 °C for 15 min at 13,000 rpm and then supernatant (50 µl) was injected onto a C18 column (ZORBAX Eclipse XDB-C18) to quantify dabsyl-methionine.
RESULTS
Localization of MsrA and MsrB in yeast
To test the role of Msrs in mitochondrial function, we used a yeast model organism, S. cerevisiae, for which strains deficient in MsrA, MsrB or both enzymes are available (9). S. cerevisiae has single MsrA and MsrB genes, and these proteins stereospecifically reduce Met-SO and Met-RO, respectively. Interestingly, the specific activity of the yeast MsrA is at least 10 times higher than that of MsrB, and this difference is also reflected in the severity of phenotypes associated with deficiency of one or the other enzymes (9). To examine relevance of MsrA and MsrB to mitochondrial function, the localization of these enzymes was determined by expressing fusion proteins with GFP. Figure 1 shows fluorescent microscopy images of WT cells transformed with MsrA and MsrB expression constructs. MsrA showed diffuse distribution in the cytosol, whereas MsrB was located within defined subcellular compartments that resembled mitochondrial staining. Mitochondrial location of MsrB was further confirmed by co-localization with Mitotracker Red. In addition, it was noted that the MsrB protein sequence displays a predicted mitochondrial signal, whereas MsrA lacked such signal peptide. Thus, localization experiments found that MsrA was located primarily in the cytosol, whereas MsrB resided mainly in mitochondria.
FIGURE 1.
Localization of MsrA and MsrB in yeast cells. Confocal microscopy analysis of WT cells expressing C-terminal GFP fused to MsrA or MsrB. The MsrA signal (derived from the MsrA-GFP fusion) shows distribution throughout the cell, whereas the MsrB signal (from the MsrB-GFP fusion) shows a dotted pattern which overlaps with staining with MitoRed, a mitochondrial fluorescent dye.
MsrA and MsrB activities in cytosol and mitochondria
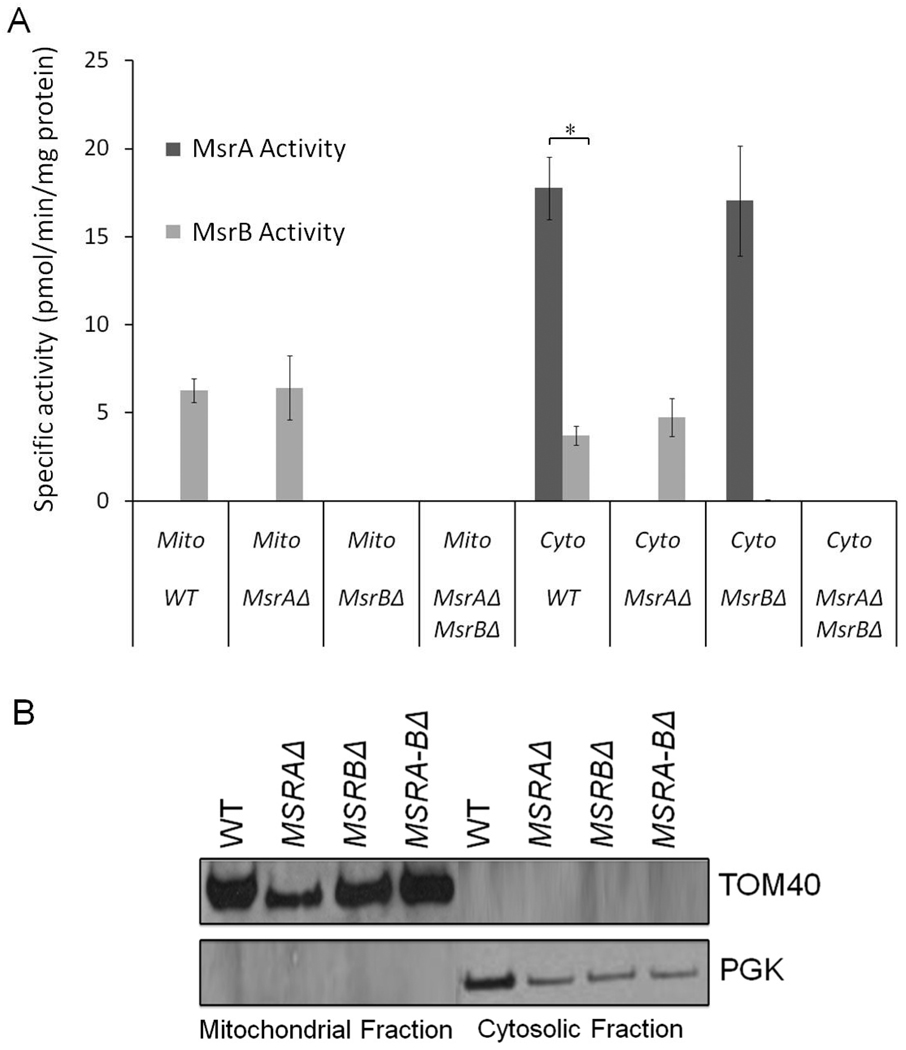
We further examined the occurrence of MsrA and MsrB by characterizing the ability of cytosolic and mitochondrial fractions to reduce dabsyl derivates of Met-RO and Met-SO. In wild type cells, mitochondria had higher MsrB activity than cytosol, whereas the cytosolic fraction was highly enriched for MsrA activity (Figure 2A). Single mutants lacked the corresponding activities and the double mutant showed no activity with either Met-SO or Met-RO (Figure 2A). Thus, MsrA is the main enzyme that reduces Met-SO residues in proteins and it is primarily located in the cytosol (p<0.0089), whereas MsrB is the main protein that repairs Met-RO residues and it is mostly located in mitochondria. The differential location of MsrA and MsrB activities was further verified by examining the purity of cytosolic and mitochondrial fractions by Western blot analysis (Figure 2B).
FIGURE 2.
Met-SO and Met-RO activities in preparations of yeast cytosol and mitochondria. (A) Cytosol and mitochondria were isolated from wild type and single and double Msr mutant strains and examined for ability to reduce Met-SO and Met-RO in an in vitro assay. Error bars show the standard error of the mean of three independent analyses. (B) Purity of mitochondrial and cytosolic fractions analyzed by Western blot analysis using anti-PGK and anti-Tom40 antibodies.
Being antioxidant repair proteins, Msrs have roles in decreasing the intracellular ROS as initially postulated by Levine et al. (6). In addition, it was shown that MsrA knockout mice had higher levels of ROS in lens cells (17) and that MsrA-overexpressing PC12 cells had lover levels of ROS (25). To assess the role of yeast Msrs in regulating ROS, superoxide levels in single and double Msr mutants were assayed in cytosol and mitochondria using fluorescent dyes and compared with those in control cells (Figure 3). We found that deletion of either Msr gene or deletion of both genes led to a significant increase in ROS levels in both compartments. This result shows that both MsrA and MsrB play protective roles against ROS in mitochondria and cytosol in S. cerevisiae.
FIGURE 3.
Superoxide levels in Msr mutants. Cytosolic and mitochondrial superoxide levels were assayed for WT and Msr mutant cells as described in Materials and Methods. Three independent analyses were performed for each strain.
Methionine sulfoxide reductases regulate respiration
To test whether mitochondrial function is impaired in cells lacking MsrA and/or MsrB, the deletion mutant cells were grown in the presence of respiratory substrates: lactic acid (Figure 4A), ethanol (Figure 4B), and glycerol (Figure 4C). In order to derive energy from these compounds, yeast cells need to utilize TCA cycle enzymes and electron transport system, which requires functional mitochondria. We found that the growth of MsrAΔ and MsrBΔ mutants was partially impaired and that of the MsrAΔ/MsrBΔ mutant was dramatically decreased in cells grown on these respiratory substrates. In lactate and ethanol media, the absence of MsrA retarded the growth more significantly than the absence of MsrB; thus, MsrA seems to be a more prominent regulator of mitochondrial function even if it is located mainly in the cytosol.
FIGURE 4.
MsrA and MsrB regulate growth of yeast cells on respiratory substrates. Indicated WT and mutant cells were diluted to A600 of 0.05 in glucose-free rich media containing 1% lactic acid (A), 2% ethanol (B), or 2% glycerol (C) and incubated at 30 °C. Growth rates were monitored based on A600 measurements. Error bars show the standard error of the mean. Three independent analyses were performed for each strain.
To better understand the finding of the slow growth of mutant cells on respiratory substrates, we determined Cyc1 levels in mutant and WT cells by Western blotting. As shown in Figure 5A, MsrAΔ and MsrBΔ single mutants had lower levels of cytochrome c, and the double mutant had extremely small amounts of Cyc1. In addition, we purified native cytochrome c, in parallel, from the same mass of WT and double mutant cells and analyzed proteins on an SDS-PAGE gel (Figure 5B). Little cytochrome c could be seen in the double mutant sample, whereas this protein was abundant in the sample derived from WT cells. To test whether the decreased level of Cyc1 in mutants was due to lower levels of mitochondria, lysates from mutant and WT cells were analyzed for a mitochondria-specific marker protein, Tom40 (Figure 5A). Interestingly, MsrAΔ and MsrBΔ single mutants and the double mutant showed the same levels of mitochondria as WT cells.
FIGURE 5.
Cytochrome c and functional mitochondria are lower in methionine sulfoxide reductase mutants. (A) Western blot analysis of cell extracts with polyclonal antibodies specific for cytochrome c1 and Tom40. 10 µg of protein was loaded in each lane, and PGK was used as a loading control. (B) SDS-PAGE analysis of native cytochrome c preparations isolated from WT and double mutant cells. Same amount of cells (10 g wet weight) was used for both strains, proteins purified in parallel using identical procedures, and 10 µl of each preparation was loaded on the gel. (C) WT and indicated mutant cells were stained with MitoRed, analyzed by flow cytometry, and relative ratios of signal densities of a representative assay were plotted.
We next utilized a flow cytometry approach to monitor the levels of functional mitochondria in cells. The MitoRed marker selectively accumulates within mitochondria in a membrane-potential dependent manner, which separates respiration-competent cells from respiration-deficient cells. Figure 5C shows the relative ratios of MitoRed stained cells in WT or mutant strains. As could be deduced from this experiment, respiration-competent mitochondria in the double mutant cells were at the level of 8%, and in MsrAΔ and MsrBΔ single mutants at the level of 17 % and 75 % of WT cells, respectively. Thus, the effect of MsrA deletion was stronger than that of MsrB deletion. The double mutant cells had few respiring mitochondria showing that Msr deficiency leads to non-functional mitochondria rather than to a decrease in the number of mitochondria.
Msrs activities and aging
It has previously been shown that calorie restriction extends replicative lifespan of yeast cells by promoting a metabolic shift towards increased mitochondrial respiration (26). If higher rate of respiration leads to longer lifespan, then respiratory substrates, such as lactic acid, would be expected to increase the lifespan as well. We carried out yeast aging assays and indeed found that all strains tested had 12–45% longer lifespan on lactic acid compared to that of the corresponding cells maintained on 2% glucose. However, when analyzed within each growth condition, single MsrAΔ and MsrAΔ/MsrBΔ double mutants had reduced lifespan compared to WT cells (Figure 6A). The lack of mitochondrial MsrB extended life span by 25% in cells grown on lactate compared to that on glucose (Figure 6B). Because of the growth defect on respiratory substrates, we could not carry out aging assays for MsrAΔ/MsrBΔ strain under lactic acid conditions. In addition, we assayed chronological aging for single and double mutant strains (Figure 6C). With the samples used for chronological aging assays, we further analyzed the index of respiratory competence (IRC), which reflects the fraction of cells that can grow on both fermentable (glucose) and non-fermentable (ethanol, glycerol or lactate) carbon sources (21). The data suggested that the MsrAΔ/MsrBΔ strain could not use non-fermentable respiratory sources, whereas MsrBΔ used them better than MsrAΔ (Figure 7). Thus, MsrA plays an important role in the mitochondrial function, and MsrB contributes to this function.
FIGURE 6.
Lifespan analysis of WT and Msr mutant cells. Replicative (A, B) and chronological lifespan (C) was determined for indicated WT and Msr mutant cells. For replicative lifespan assay, cells were grown on glucose (A) or lactate (B).
FIGURE 7.
Analysis of index of respiratory competence. An IRC assay shows how respiratory carbon sources support the growth of WT and Msr mutant cells. Error bars show the standard error of the mean. Three independent analyses were performed for each strain.
Double deletion of Msr genes causes irreversible damage to mitochondrial function
To test whether impaired mitochondrial function could be rescued by overexpression of Msrs, we performed a complementation assay. Both genes were expressed under the control of their own promoters by cloning a 1 kb upstream region of each gene along with the ORF. Figure 8A shows that overexpression of MsrA or MsrB did not normalize mitochondrial function of the double mutant grown on a respiratory substrate, since respiratory substrates require functional mitochondria. Furthermore, co-expression of each Msr gene in the same cells also did not result in cell growth (Figure 8B). This complementation assay was repeated using different types of respiratory substrates (glycerol and ethanol) and different vectors (with GPD and TEF promoters). The same constructs were utilized for cell growth assays in the presence of racemic Met-RO and Met-SO mixture to test whether Msr activity can be complemented by overexpression of Msr genes. Because WT and indicated mutant strains are methionine auxotrophs (9), cells could be supplemented with methionine sulfoxides to test the function of Msrs. As we discussed above (see Materials and Methods), we used a triple Msr mutant (MsrAΔ/MsrBΔ/fRMsrΔ) to assay the reductase activity. Figure 8C shows that respiratory deficiency of mutants could not be rescued by overexpressing Msr genes. However, the reductase activity could be restored by overexpressing each of the Msr genes in the triple mutant (Figure 8D).
FIGURE 8.
Complementation assay. The GPD promoter was disrupted in p425 yeast expression vector and MsrA and MsrB expressed under their own promoters in Msr mutants. Designations of yeast strains are as follows: ABΔ, cells missing MsrA and MsrB genes: ABFΔ, cells missing MsrA, MsrB and fRMsr genes. (A, B) Spotting assay using respiratory substrates. WT and double Msr mutant cells were analyzed as shown, together with mutant cells containing a vector (+v) or expressing MsrA (+pMsrA), MsrB (+pMsrB), or co-expressing MsrA and MsrB (+pMsrA-B). (C) The triple Msr mutant was also analyzed for growth on respiratory substrates and (D) following complementation of mitochondrial function of cells grown on Met-free YNB medium supplemented with 20 mg/liter Met-RO or Met-SO.
DISCUSSION
This study uncovered unexpectedly complex roles of MsrA and MsrB in regulating mitochondrial function in S. cerevisiae. Although these two oxidoreductases were expected to have complementary functions each repairing one-half of methionine sulfoxide residues in proteins, we observed differences in localization of these enzymes as well as differences in MsrA and MsrB activities in cytosol and mitochondria. MsrA, in the form of fusion with GFP, localized primarily to cytosol, whereas MsrB localized to mitochondria. Nevertheless, the absence of MsrA caused stronger phenotypes with regard to mitochondrial function.
In agreement with earlier observations in mice (27) and fruit flies (28), it was found that replicative lifespan of yeast cells could be modulated by Msrs (29). Deletion and overexpression of Msrs could decrease and increase lifespan, respectively, under aerobic conditions. Interestingly, under anaerobic conditions, lifespan was significantly reduced compared to that in the presence of oxygen, suggesting a role for ROS-independent components of aging. However, under anaerobiosis, yeast lifespan was not influenced by Msrs, likely because ROS levels were low and thus the enzymes that repair ROS damage were not needed (29). In contrast, a recent study showed that the shortened lifespan in MsrA knockout mice cannot be reproduced, even though the loss of MsrA made mice much more sensitive to oxidative stress (30).
The specific targets of Msrs are poorly understood. It is thought that MsrA and MsrB catalyze reduction of all accessible methionine sulfoxide residues, but this contrasts with the findings that reversible methionine sulfoxide reduction regulates activities of certain proteins, such as ion channels (31). In addition, one should expect that if MsrA and MsrB lack specificity for protein targets, susceptible methionine residues would have been counter-selected from occurring in proteins that reside in regions most susceptible to oxidative damage. Following the findings that Msrs may be required to protect electron transport system components in bacteria (14) and cytochromes c in mouse (17), we identified that mitochondrial function requires Msrs in yeast.
Mitochondria, being generators of ROS in eukaryotic cells, are also the targets of these reactive species. The steady state concentration of ROS in mitochondria may be higher than that in the rest of the cell (32). Mitochondrial DNA, due to its proximity to ROS generation sites and lack of protective histones, is exposed to ROS damage and, as a result, mtDNA accumulates mutations approximately 10 times faster than the nuclear DNA. In eukaryotes, studies established a clear link between mitochondrial function and aging (33). When mitochondria become non-functional, NADH is not processed by the electron transport chain, and instead is oxidized by cytosolic NADH oxidase, which produces higher levels of free radicals that eventually may lead to cell death (34).
Our experimental analysis showed that yeast cells completely lacking Msrs had decreased growth rates on lactic acid and other respiratory substrates in liquid media and that they generally displayed shorter lifespan. Further analysis of these mutant cells revealed that the levels of active respiring mitochondria were lower than those in WT cells and that cytochrome c levels were significantly lower in both single and double mutant strains even though they had normal levels of mitochondria. Indeed, overexpression of Msrs did not rescue mitochondrial function in double mutant cells, while the Msr function was restored under these conditions. An earlier DNA microarray analysis showed that the expression level of cytochrome c and other respiratory genes did not change in MsrAΔ, MsrBΔ and MsrAΔ/MsrBΔ mutants (29), suggesting that the loss of cytochrome c was not due to a transcriptional regulation in mutants. The absence of Msrs is expected to lead to ROS accumulation both in mitochondria and cytosol and the accumulation of ROS could lead to mitochondrial genome instability, accumulation of mutations and eventually impaired mitochondrial function.
Yeast cells growing on 2% glucose preferentially utilize glycolysis and repress the synthesis of mitochondrial proteins. As a result, free radicals may be produced at intermediate levels (26–35). However, when cells are grown on 0.5% glucose, repression of respiratory genes is released, whereas lifespan is increased (26–36). A similar effect may be seen in cells that grow on respiratory substrates, such as lactate. When lactic acid is used as a carbon source, most of the respiratory genes were moderately upregulated in wild type cells (data not shown). Deficiency in mitochondrial MsrB led to a dramatic activation of respiratory genes by lactic acid and this upregulation of respiration may be the underlying reason why MsrB mutants had a longer lifespan in lactic acid. However, in the cells overexpressing MsrB, respiratory genes were not upregulated when these cells were grown on lactic acid. Thus, the lack of MsrB led to a deficiency in the respiratory system and lactate-induced activation of the respiratory genes to meet the increased demand for energy production.
Since MsrB-GFP fusion protein was primarily localized to mitochondria, we also searched for MsrB interacting partners using yeast two-hybrid system and found that Uth1 interacts with MsrB (data not shown). The Uth1 protein, a member of the SUN family, was initially identified from a mutational screen for stress resistance and lifespan extension during starvation (37). Uth1 is a mitochondrial protein and is also known to reside in the outer mitochondrial membrane. It was shown to play a role in mitochondrial biogenesis and autophagy and yeast cell death induced by heterologous expression of the pro-apoptotic protein Bax (38–39) Interestingly, Uth1 deficiency was shown to decrease the level of cytochrome proteins by up to 25%; however, the mechanisms by which Uth1 regulates mitochondrial function are not clear. The functional significance of the observed Uth1/MsrB interaction needs further investigation.
In eukaryotes, cytochrome c release from mitochondria upon exposure to oxidative stress has been known for a long time, but the molecular mechanism of how oxidative stress initiates cytochrome c release is not fully understood. Our data suggest that, in the absence of Msrs, oxidation of cytochrome c and possibly other respiratory enzymes causes mitochondrial dysfunction, which leads to mitochondrial collapse, premature aging and cell death.
The functional differences between yeast MsrA and MsrB pose new questions. For example, it is not clear how the complementary methionine sulfoxide substrates are reduced in cytosol and mitochondria if each of these compartments has widely different levels of individual Msrs. One clue may come from the observation that, in yeast, MsrA is 10-fold more active than MsrB (29), which is consistent with the stronger phenotypes and changes in gene expression observed in MsrA deficiency compared to the deficiency of MsrB. Besides its involvement in oxidative protein repair, MsrA has a role in reducing free methionine sulfoxide in a process that supplies methionine to cells, whereas MsrB is only active with protein substrates. The need for targeting Msrs to different compartments is evident from the studies in mammals, which evolved separate MsrBs for cytosolic (MsrB1), mitochondria (MsrB2) and the endoplasmic reticulum (MsrB3) (40). Moreover, MsrB3 and MsrA are alternatively spliced and MsrA can produce both mitochondrial and cytosolic forms from the same transcript (41).
AKNOWLEDGEMENTS
We thank Drs. M. E. Dumont, F. Sherman and B. Polevoda for providing Cyc1 antibodies, and Drs. D. Pain and B. Musuvathi for Tom40 antibodies. We also thank Terri Fangman and Dr. You Zhou from the University of Nebraska Microscopy Core Facility and Danielle Shea from the Flow Cytometry Core Facility for expert assistance.
This study was supported by NIH grant AG021518 (to VNG).
Abbreviations
- MsrA
methionine sulfoxide reductase A
- MsrB
methionine sulfoxide reductase B
- Met-SO
methionine S-sulfoxide
- Met-RO
methionine R-sulfoxide
- ROS
reactive oxygen species
- YPLA
yeast rich media with lactic acid
REFERENCES
- 1.Stadtman ER. Protein oxidation and aging. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- 2.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 3.Brot N, Weissbach L, Werth J, Weissbach H. Enzymatic reduction of oxidized alpha-1-proteinase inhibitor restores biological activity. Proc. Natl. Acad. Sci. U.S.A. 1981;78:2155–2158. doi: 10.1073/pnas.78.12.7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friguet B. Oxidized protein degradation and repair in ageing and oxidative stress. FEBS Lett. 2006;580:2910–2916. doi: 10.1016/j.febslet.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 5.Mary J, Vougier S, Picot CR, Perichon M, Petropoulos I, Friguet B. Enzymatic reactions involved in the repair of oxidized proteins. Exp. Gerontol. 2004;8:1117–1123. doi: 10.1016/j.exger.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Levine RL, Mosoni L, Berlett BS, Stadtman ER. Methionine residues as endogenous antioxidants in proteins. Proc. Natl. Acad. Sci. U.S.A. 1996;93:15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stadtman ER, Levine RL. Protein oxidation. Ann. N Y Acad. Sci. 2000;899:191–208. doi: 10.1111/j.1749-6632.2000.tb06187.x. [DOI] [PubMed] [Google Scholar]
- 8.Brot N, Weissbach L, Werth J, Weissbach H. Enzymatic reduction of protein-bound methionine sulfoxide. Proc. Natl. Acad. Sci. U.S.A. 1980;78:2155–2158. doi: 10.1073/pnas.78.4.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kryukov GV, Kumar RA, Koc A, Sun Z, Gladyshev VN. Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase. Proc. Natl. Acad. Sci. U.S.A. 2002;99:4245–4250. doi: 10.1073/pnas.072603099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang W, Escribano J, Sarfazi M, Coca-Prados M. Identification, expression and chromosome localization of a human gene encoding a novel protein with similarity to the pilB family of transcriptional factors (pilin) and to bacterial peptide methionine sulfoxide reductases. Gene. 1999;233:233–240. doi: 10.1016/s0378-1119(99)00131-6. [DOI] [PubMed] [Google Scholar]
- 11.Grimaud R, Ezraty B, Mitchell JK, Lafitte D, Briand C, Derrick PJ, Barras F. Repair of oxidized proteins. Identification of a new methionine sulfoxide reductase. J. Biol.Chem. 2001;276:48915–48920. doi: 10.1074/jbc.M105509200. [DOI] [PubMed] [Google Scholar]
- 12.Ezraty B, Aussel L, Barras F. Methionine sulfoxide reductases in prokaryotes. Biochim. Biophys. Acta. 2005;1703:221–229. doi: 10.1016/j.bbapap.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 13.Skaar EP, Tobiason DM, Quick J, Judd RC, Weissbach H, Etienne F, Brot N, Seifert HS. The outer membrane localization of the Neisseria gonorrhoeae MsrA/B is involved in survival against reactive oxygen species. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10108–10113. doi: 10.1073/pnas.152334799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hassouni ME, Chambost JP, Expert D, Gijsegem VF, Barras F. The minimal gene set member msrA, encoding peptide methionine sulfoxide reductase, is a virulence determinant of the plant pathogen Erwinia chrysanthemi. Proc. Natl. Acad. Sci. U.S.A. 1999;96:887–892. doi: 10.1073/pnas.96.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feinberg BA, Bedore JE, Jr, Ferguson-Miller S. Methionine-80-sulfoxide cytochrome c: preparation, purification and electron-transfer capabilities. Biochim. Biophys. Acta. 1986;851:157–165. doi: 10.1016/0005-2728(86)90121-0. [DOI] [PubMed] [Google Scholar]
- 16.Ivanetich KM, Bradshaw JJ, Kaminsky LS. Methionine sulfoxide cytochrome c. Biochemistry. 1976;15:1144–1153. doi: 10.1021/bi00650a029. [DOI] [PubMed] [Google Scholar]
- 17.Marchetti M, Lee W, Cowell T, Wells T, Weissbach H, Kantorow M. Silencing of the methionine sulfoxide reductase A gene results in loss of mitochondrial membrane potential and increased ROS production in human lens cells. J. of Exp. Eye Research. 2006;83:1281–1286. doi: 10.1016/j.exer.2006.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le DT, Lee BC, Marino SM, Zhang Y, Fomenko DE, Kaya A, Hacioglu E, Kwak GH, Koc A, Kim HY, Gladyshev VN. Functional analysis of free methionine-R-sulfoxide reductase from Saccharomyces cerevisiae. J. Biol. Chem. 2009;284:4354–4364. doi: 10.1074/jbc.M805891200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komar-Panicucci S, Sherman F, McLendon G. Modulated growth of Saccharomyces cerevisiae by altering the driving force of the reactions of cytochrome c: Marcus' theory in vitro and in vivo. Biochemistry. 1996;35:4878–4885. doi: 10.1021/bi952771z. [DOI] [PubMed] [Google Scholar]
- 20.Skowronek P, Krummeck G, Haferkamp O, Rodel G. Flow cytometry as a tool to discriminate respiratory-competent and respiratory-deficient yeast cells. Curr. Genet. 1990;18:265–267. doi: 10.1007/BF00318391. [DOI] [PubMed] [Google Scholar]
- 21.Parrella E, Longo VD. The chronological life span of Saccharomyces cerevisiae to study mitochondrial dysfunction and disease. Methods. 2008;46:256–262. doi: 10.1016/j.ymeth.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Lin Z, Johnson LC, Weissbach H, Brot N, Lively MO, Lowther WT. Free methionine-(R)-sulfoxide reductase from Escherichia coli reveals a new GAF domain function. Proc. Natl. Acad. Sci. U. S. A. 2007;104:9597–9602. doi: 10.1073/pnas.0703774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffiths L, Swartzlander D, Meadows KL, Wilkinson KD, Corbett AH, Doetsch PW. Dynamic compartmentalization of base excision repair proteins in response to nuclear and mitochondrial oxidative stress. Mol. Cell. Biol. 2008;29:794–807. doi: 10.1128/MCB.01357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meisinger C, Sommer T, Pfanner N. Purification of Saccharomyces cerevisiae mitochondria devoid of microsomal and cytosolic contaminations. Anal. Biochem. 2000;287:339–342. doi: 10.1006/abio.2000.4868. [DOI] [PubMed] [Google Scholar]
- 25.Yermolaieva O, Xu R, Schinstock C, Brot N, Weissbach H, Heinemann SH, Hosh T. Methionine sulfoxide reductase A protects neuronal cells against brief hypoxia/reoxygenation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:1159–1164. doi: 10.1073/pnas.0308215100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 27.Moskovitz J, Bar-Noy S, Williams WM, Requena J, Berlett BS, Stadtman ER. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc. Natl. Acad. Sci. U.S.A. 2001;98:12920–12925. doi: 10.1073/pnas.231472998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruan H, Tang XD, Chen ML, Joiner ML, Sun G, Brot N, Weissbach H, Heinemann SH, Iverson L, Wu CF, Hoshi T. High-quality life extension by the enzyme peptide methionine sulfoxide reductase. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2748–2753. doi: 10.1073/pnas.032671199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koc A, Gasch AP, Rutherford JC, Kim HY, Gladyshev VN. Methionine sulfoxide reductase regulation of yeast lifespan reveals reactive oxygen species-dependent and - independent components of aging. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7999–8004. doi: 10.1073/pnas.0307929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salmon AB, Perez VI, Bokov A, Jernigan A, Kim G, Zhao H, Levine RL, Richardson A. Lack of methionine sulfoxide reductase A in mice increases sensitivity to oxidative stress but does not diminish life span. FASEB. 2009;10:3601–3608. doi: 10.1096/fj.08-127415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang XD, Daggett H, Hanner M, Garcia ML, McManus OB, Brot N, Weissbach H, Heinemann SH, Hoshi T. Oxidative regulation of large conductance calcium-activated potassium channels. J. Gen. Physiol. 2001;117:253–274. doi: 10.1085/jgp.117.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richter C, Park JW, Ames BN. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc. Natl. Acad. Sci. U.S.A. 1988;85:6465–6467. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh KK. Mitochondrial dysfunction is a common phenotype in aging and cancer. Ann. NY Acad. Sci. 2004;1019:260–264. doi: 10.1196/annals.1297.043. [DOI] [PubMed] [Google Scholar]
- 34.Wissing S, Ludovico P, Herker E, Buttner S, Engelhardt SM, Decker T, Link A, Proksch A, Rodrigues F, Corte-Real M, Frohlich KU, Manns J, Cande C, Sigrist SJ, Kroemer G, Madeo F. An AIF orthologue regulates apoptosis in yeast. J. Cell. Biol. 2004;166:969–974. doi: 10.1083/jcb.200404138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuller HJ. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 2003;43:139–160. doi: 10.1007/s00294-003-0381-8. [DOI] [PubMed] [Google Scholar]
- 36.Barros MH, Bandy B, Tahara EB, Kowaltowski AJ. Higher respiratory activity decreases mitochondrial reactive oxygen release and increases life span in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:49883–49888. doi: 10.1074/jbc.M408918200. [DOI] [PubMed] [Google Scholar]
- 37.Bandara PD, Flattery-O'Brien JA, Grant CM, Dawes IW. Involvement of the Saccharomyces cerevisiae UTH1 gene in the oxidative-stress response. Curr. Genet. 1998;34:259–268. doi: 10.1007/s002940050395. [DOI] [PubMed] [Google Scholar]
- 38.Kissova I, Deffieu M, Manon S, Camougrand N. Uth1p is involved in the autophagic degradation of mitochondria. J. Biol. Chem. 2004;279:39068–39074. doi: 10.1074/jbc.M406960200. [DOI] [PubMed] [Google Scholar]
- 39.Camougrand N, Grelaud-Coq A, Marza E, Priault M, Bessoule JJ, Manon S. The product of the UTH1 gene, required for Bax-induced cell death in yeast, is involved in the response to rapamycin. Mol. Microbiol. 2003;47:495–506. doi: 10.1046/j.1365-2958.2003.03311.x. [DOI] [PubMed] [Google Scholar]
- 40.Kim HY, Gladyshev VN. Methionine sulfoxide reduction in mammals: characterization of methionine-R-sulfoxide reductases. Mol. Biol. Cell. 2004;15:1055–1064. doi: 10.1091/mbc.E03-08-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim HY, Gladyshev VN. Alternative first exon splicing regulates subcellular distribution of methionine sulfoxide reductases. BMC Mol. Biol. 2006;7:11. doi: 10.1186/1471-2199-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]