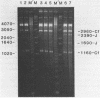
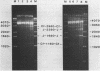
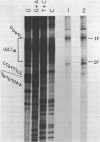
Abstract
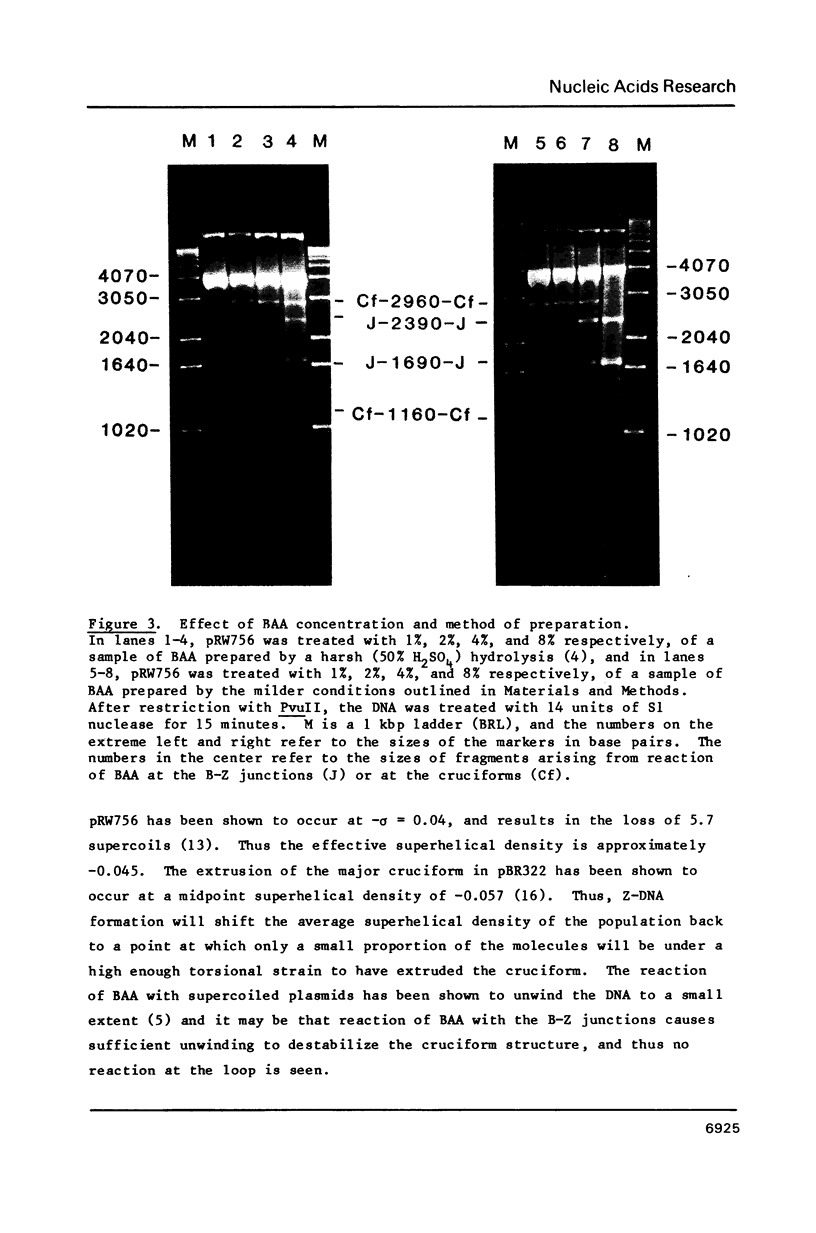
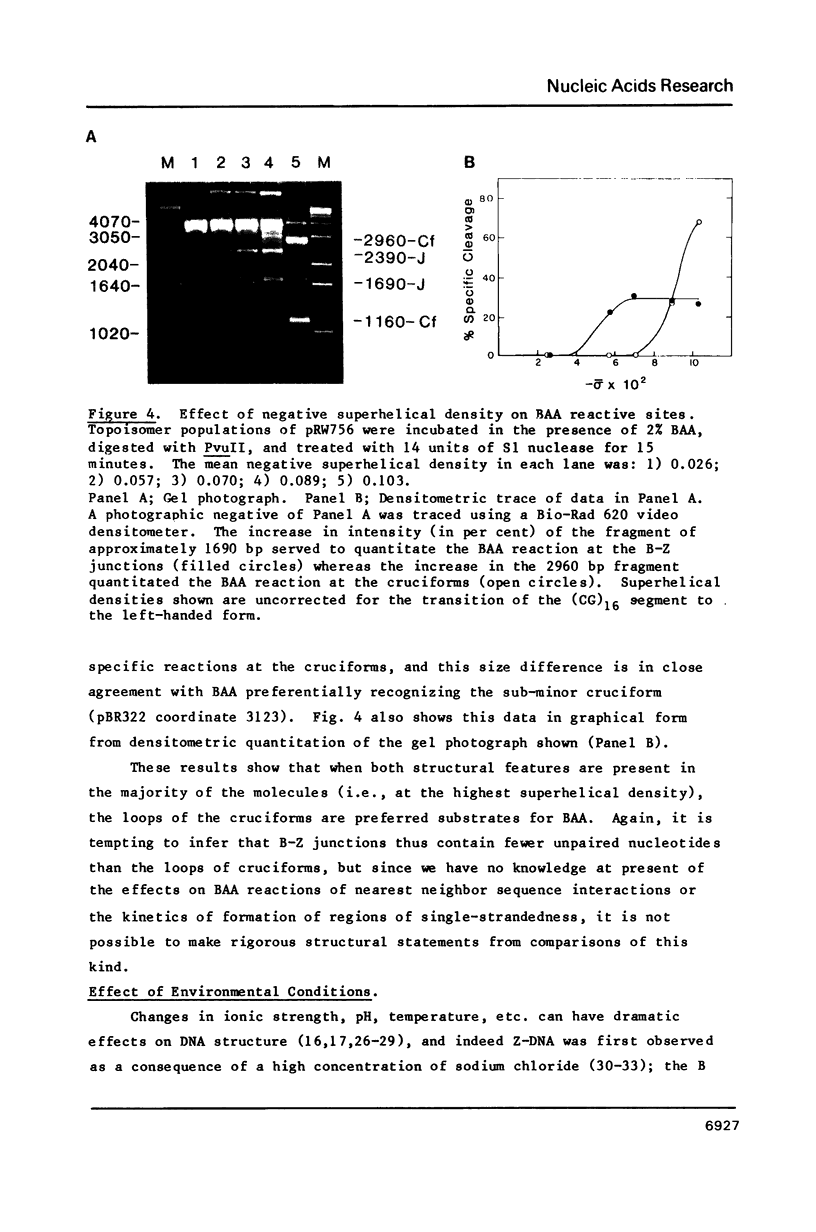
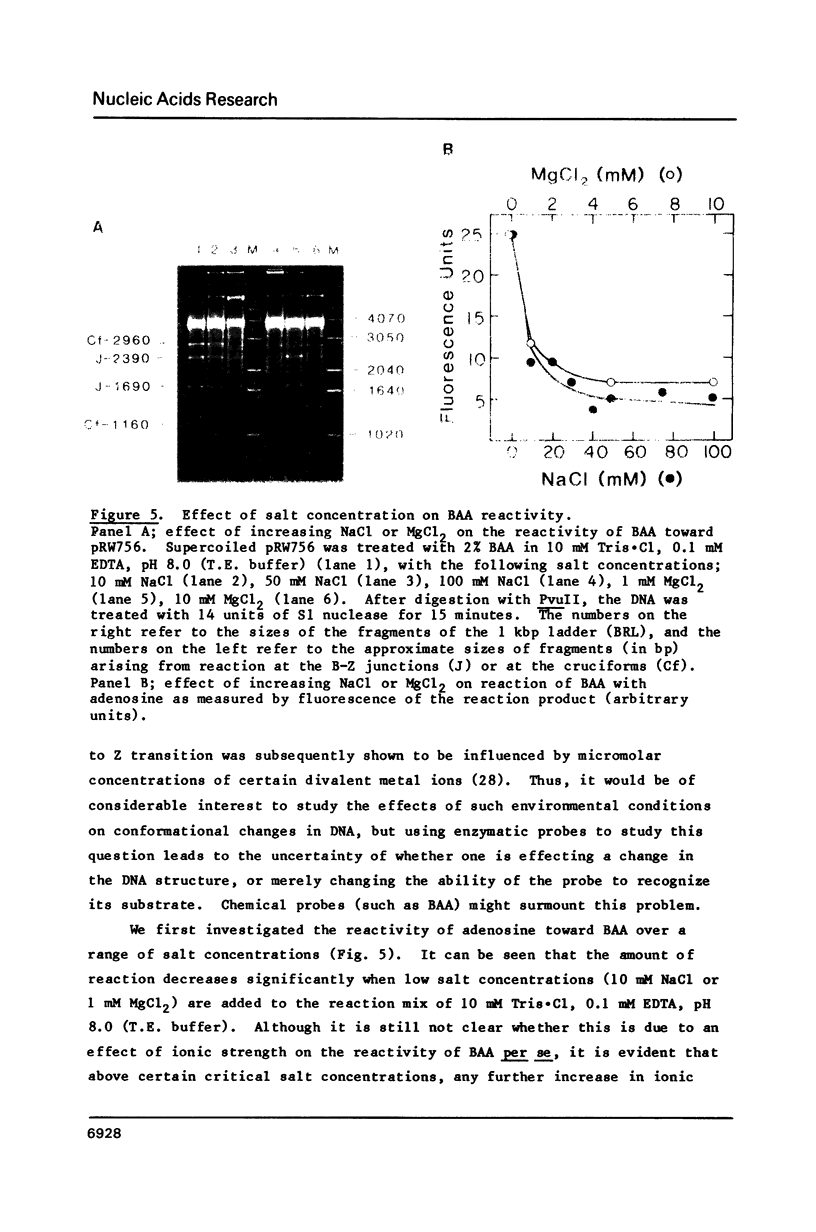
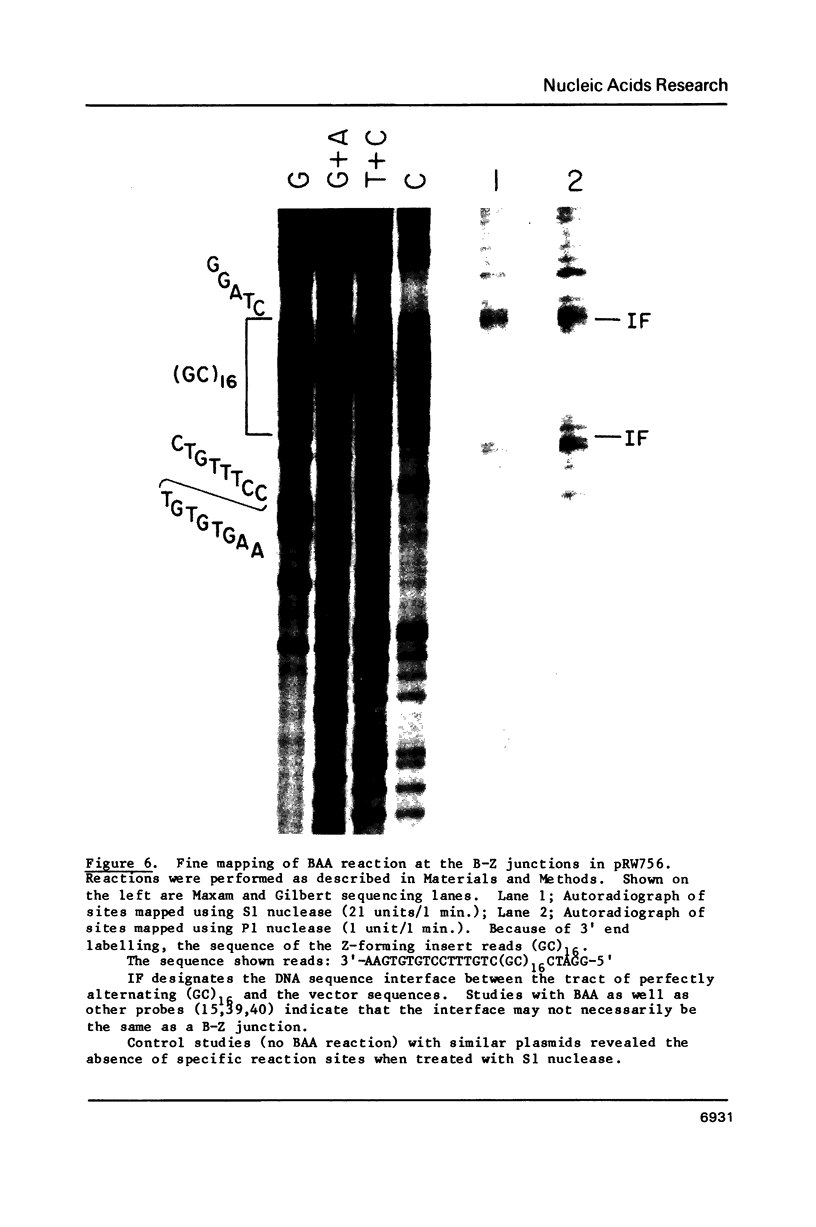
The reaction of bromoacetaldehyde (BAA) was investigated further with recombinant plasmids containing tracts of (CG)16, in pRW756, or (CA)32, in pRW777, which adopt left-handed Z-structures under the influence of negative supercoiling. The cruciform structures adopted by the inverted repeat sequences near the replication origins of the pBR322 vectors served as internal controls for the number of unpaired bases. The extent of reaction with the B-Z junctions and the cruciforms was dependent on the reaction and analysis conditions, the method of preparation of BAA, ionic conditions, and the amount of negative supercoiling. In contrast to the previous results of Kang and Wells, B-Z junctions in addition to cruciforms do react with BAA. However, more forcing conditions are required to detect this reaction since B-Z junctions appear to be less reactive than the single stranded loops of cruciforms. The site of reaction with DNA was readily mapped with high precision at the nucleotide level. Also, a simple method is described for determining the concentration of BAA as well as its intrinsic reactivity in a given ionic medium.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avigad G., Damle S. Fluorometric assay of adenine and its derivatives. Anal Biochem. 1972 Nov;50(1):321–323. doi: 10.1016/0003-2697(72)90511-8. [DOI] [PubMed] [Google Scholar]
- Barrio J. R., Secrist J. A., 3rd, Leonard N. J. Fluorescent adenosine and cytidine derivatives. Biochem Biophys Res Commun. 1972 Jan 31;46(2):597–604. doi: 10.1016/s0006-291x(72)80181-5. [DOI] [PubMed] [Google Scholar]
- Camilloni G., Della Seta F., Negri R., Grazia Ficca A., Di Mauro E. Structure of RNA polymerase II promoters. Conformational alterations and template properties of circularized Saccharomyces cerevisiae GAL1-GAL10 divergent promoters. EMBO J. 1986 Apr;5(4):763–771. doi: 10.1002/j.1460-2075.1986.tb04279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor C. R., Efstratiadis A. Possible structures of homopurine-homopyrimidine S1-hypersensitive sites. Nucleic Acids Res. 1984 Nov 12;12(21):8059–8072. doi: 10.1093/nar/12.21.8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan M., Dodgson J. B., Engel J. D. Analysis of the adult chicken beta-globin gene. Nucleotide sequence of the locus, microheterogeneity at the 5'-end of beta-globin mRNA, and aberrant nuclear RNA species. J Biol Chem. 1983 Mar 25;258(6):3983–3990. [PubMed] [Google Scholar]
- Drew H., Takano T., Tanaka S., Itakura K., Dickerson R. E. High-salt d(CpGpCpG), a left-handed Z' DNA double helix. Nature. 1980 Aug 7;286(5773):567–573. doi: 10.1038/286567a0. [DOI] [PubMed] [Google Scholar]
- Fowler R. F., Skinner D. M. Eukaryotic DNA diverges at a long and complex pyrimidine.purine tract that can adopt altered conformations. J Biol Chem. 1986 Jul 5;261(19):8994–9001. [PubMed] [Google Scholar]
- Galazka G., Palecek E., Wells R. D., Klysik J. Site-specific OsO4 modification of the B-Z junctions formed at the (dA-dC)32 region in supercoiled DNA. J Biol Chem. 1986 May 25;261(15):7093–7098. [PubMed] [Google Scholar]
- Haniford D. B., Pulleyblank D. E. Facile transition of poly[d(TG) x d(CA)] into a left-handed helix in physiological conditions. Nature. 1983 Apr 14;302(5909):632–634. doi: 10.1038/302632a0. [DOI] [PubMed] [Google Scholar]
- Haniford D. B., Pulleyblank D. E. Transition of a cloned d(AT)n-d(AT)n tract to a cruciform in vivo. Nucleic Acids Res. 1985 Jun 25;13(12):4343–4363. doi: 10.1093/nar/13.12.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htun H., Lund E., Dahlberg J. E. Human U1 RNA genes contain an unusually sensitive nuclease S1 cleavage site within the conserved 3' flanking region. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7288–7292. doi: 10.1073/pnas.81.23.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston B. H., Rich A. Chemical probes of DNA conformation: detection of Z-DNA at nucleotide resolution. Cell. 1985 Oct;42(3):713–724. doi: 10.1016/0092-8674(85)90268-5. [DOI] [PubMed] [Google Scholar]
- Kang D. S., Wells R. D. B-Z DNA junctions contain few, if any, nonpaired bases at physiological superhelical densities. J Biol Chem. 1985 Jun 25;260(12):7783–7790. [PubMed] [Google Scholar]
- Kilpatrick M. W., Klysik J., Singleton C. K., Zarling D. A., Jovin T. M., Hanau L. H., Erlanger B. F., Wells R. D. Intervening sequences in human fetal globin genes adopt left-handed Z helices. J Biol Chem. 1984 Jun 10;259(11):7268–7274. [PubMed] [Google Scholar]
- Kilpatrick M. W., Torri A., Kang D. S., Engler J. A., Wells R. D. Unusual DNA structures in the adenovirus genome. J Biol Chem. 1986 Aug 25;261(24):11350–11354. [PubMed] [Google Scholar]
- Kilpatrick M. W., Wei C. F., Gray H. B., Jr, Wells R. D. BAL 31 nuclease as a probe in concentrated salt for the B-Z DNA junction. Nucleic Acids Res. 1983 Jun 11;11(11):3811–3822. doi: 10.1093/nar/11.11.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klysik J., Stirdivant S. M., Singleton C. K., Zacharias W., Wells R. D. Effects of 5 cytosine methylation on the B-Z transition in DNA restriction fragments and recombinant plasmids. J Mol Biol. 1983 Jul 25;168(1):51–71. doi: 10.1016/s0022-2836(83)80322-2. [DOI] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T., Gelinas R., Weintraub H. Detection of an altered DNA conformation at specific sites in chromatin and supercoiled DNA. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4389–4393. doi: 10.1073/pnas.80.14.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T., Kohwi Y. Poly(dG)-poly(dC) sequences, under torsional stress, induce an altered DNA conformation upon neighboring DNA sequences. Cell. 1985 Nov;43(1):199–206. doi: 10.1016/0092-8674(85)90024-8. [DOI] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T., Manes T., Kohwi Y. Unusual conformational effect exerted by Z-DNA upon its neighboring sequences. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2223–2227. doi: 10.1073/pnas.84.8.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuśmierek J. T., Singer B. Chloroacetaldehyde-treated ribo- and deoxyribopolynucleotides. 1. Reaction products. Biochemistry. 1982 Oct 26;21(22):5717–5722. doi: 10.1021/bi00265a050. [DOI] [PubMed] [Google Scholar]
- Kłysik J., Stirdivant S. M., Larson J. E., Hart P. A., Wells R. D. Left-handed DNA in restriction fragments and a recombinant plasmid. Nature. 1981 Apr 23;290(5808):672–677. doi: 10.1038/290672a0. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. Structural perturbation in supercoiled DNA: hypersensitivity to modification by a single-strand-selective chemical reagent conferred by inverted repeat sequences. Nucleic Acids Res. 1983 May 25;11(10):3097–3112. doi: 10.1093/nar/11.10.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKeon C., Schmidt A., de Crombrugghe B. A sequence conserved in both the chicken and mouse alpha 2(I) collagen promoter contains sites sensitive to S1 nuclease. J Biol Chem. 1984 May 25;259(10):6636–6640. [PubMed] [Google Scholar]
- McLean M. J., Blaho J. A., Kilpatrick M. W., Wells R. D. Consecutive A X T pairs can adopt a left-handed DNA structure. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5884–5888. doi: 10.1073/pnas.83.16.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejedlý K., Kwinkowski M., Gałazka G., Kłysik J., Palecek E. Recognition of the structural distortions at the junctions between B and Z segments in negatively supercoiled DNA by osmium tetroxide. J Biomol Struct Dyn. 1985 Dec;3(3):467–478. doi: 10.1080/07391102.1985.10508435. [DOI] [PubMed] [Google Scholar]
- Nickol J. M., Felsenfeld G. DNA conformation at the 5' end of the chicken adult beta-globin gene. Cell. 1983 Dec;35(2 Pt 1):467–477. doi: 10.1016/0092-8674(83)90180-0. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Rich A. The sequence (dC-dA)n X (dG-dT)n forms left-handed Z-DNA in negatively supercoiled plasmids. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1821–1825. doi: 10.1073/pnas.80.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Transcriptional block caused by a negative supercoiling induced structural change in an alternating CG sequence. Cell. 1985 Jan;40(1):129–137. doi: 10.1016/0092-8674(85)90316-2. [DOI] [PubMed] [Google Scholar]
- Pohl F. M., Jovin T. M. Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly (dG-dC). J Mol Biol. 1972 Jun 28;67(3):375–396. doi: 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- Schon E., Evans T., Welsh J., Efstratiadis A. Conformation of promoter DNA: fine mapping of S1-hypersensitive sites. Cell. 1983 Dec;35(3 Pt 2):837–848. doi: 10.1016/0092-8674(83)90116-2. [DOI] [PubMed] [Google Scholar]
- Singleton C. K. Effects of salts, temperature, and stem length on supercoil-induced formation of cruciforms. J Biol Chem. 1983 Jun 25;258(12):7661–7668. [PubMed] [Google Scholar]
- Singleton C. K., Kilpatrick M. W., Wells R. D. S1 nuclease recognizes DNA conformational junctions between left-handed helical (dT-dG n. dC-dA)n and contiguous right-handed sequences. J Biol Chem. 1984 Feb 10;259(3):1963–1967. [PubMed] [Google Scholar]
- Singleton C. K., Kilpatrick M. W., Wells R. D. S1 nuclease recognizes DNA conformational junctions between left-handed helical (dT-dG n. dC-dA)n and contiguous right-handed sequences. J Biol Chem. 1984 Feb 10;259(3):1963–1967. [PubMed] [Google Scholar]
- Singleton C. K., Klysik J., Stirdivant S. M., Wells R. D. Left-handed Z-DNA is induced by supercoiling in physiological ionic conditions. Nature. 1982 Sep 23;299(5881):312–316. doi: 10.1038/299312a0. [DOI] [PubMed] [Google Scholar]
- Singleton C. K., Klysik J., Wells R. D. Conformational flexibility of junctions between contiguous B- and Z-DNAs in supercoiled plasmids. Proc Natl Acad Sci U S A. 1983 May;80(9):2447–2451. doi: 10.1073/pnas.80.9.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton C. K., Wells R. D. The facile generation of covalently closed, circular DNAs with defined negative superhelical densities. Anal Biochem. 1982 May 15;122(2):253–257. doi: 10.1016/0003-2697(82)90277-9. [DOI] [PubMed] [Google Scholar]
- Thomas T. J., Messner R. P. A left-handed (Z) conformation of poly(dA-dC).poly(dG-dT) induced by polyamines. Nucleic Acids Res. 1986 Aug 26;14(16):6721–6733. doi: 10.1093/nar/14.16.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wohlrab F., McLean M. J., Wells R. D. The segment inversion site of herpes simplex virus type 1 adopts a novel DNA structure. J Biol Chem. 1987 May 5;262(13):6407–6416. [PubMed] [Google Scholar]
- Zacharias W., Larson J. E., Klysik J., Stirdivant S. M., Wells R. D. Conditions which cause the right-handed to left-handed DNA conformational transitions. Evidence for several types of left-handed DNA structures in solution. J Biol Chem. 1982 Mar 25;257(6):2775–2782. [PubMed] [Google Scholar]
- Zimmerman S. B. The three-dimensional structure of DNA. Annu Rev Biochem. 1982;51:395–427. doi: 10.1146/annurev.bi.51.070182.002143. [DOI] [PubMed] [Google Scholar]