Abstract
Nanoscale surface features that mimic extracellular matrix are critical environmental cues for cell contact guidance and are vital in advanced medical devices in order to manipulate cell behaviors. Among them, nanogratings (line-and-space grating) are common platforms to study geometric effects on cell contact guidance, especially, cell alignment, but generally are one pattern height per platform. In this study, we developed a strategy to fabricate controlled substrates with a wide range of pattern shapes and surface chemistries and to separate surface chemistry and topography effects. As a demonstration of this strategy, six nanograting platforms on three materials were fabricated and applied to examine and differentiate the effects of surface topography and surface chemistry on cell contact guidance of murine preosteoblasts. All of the six platforms contained the same gradient in pattern height (0 nm to ≈ 350 nm). They were prepared using nanoimprint lithography and annealing for thermoplastic materials (low molecular weight polystyrene (PS) and polymethylmethacrylate (PMMA)) and photo-imprint for a thermoset material (a cross-linked dimethacrylate (DMA)). Each material contains two platforms that are only different in line-and-space pitches (420 nm or 800 nm). The DMA nanogratings had a reverse line-and-space profiles to those of the PS and PMMS nanogratings. Using these platforms, a full range of cell alignment, from randomly orientated to completely parallel to the grating direction was achieved. Results from focal adhesion assay and scanning electronic microscopy (SEM) indicated a change in cell-substrate contact from a non-composite state (full contact) to a composite state (partial contact between cell and substrate) as pattern height increased. These gradient platforms allowed for the separation of surface chemistry and surface topography to provide insight into the mechanisms responsible for cell contact guidance on nanopatterned surfaces.
Keywords: Surface topography, nanoimprint lithography, cell contact guidance, tissue engineering, extracellular matrix, focal adhesion
Introduction
Surface topography of extracellular matrix (ECM) is recognized as one of the most important environmental cues for cell contact guidance.1–3 Cells respond to surface features by adjusting their morphology and/or orientation as well as their gene expression and in some cases, ultimately their differentiation pathway.4–8 For cells that respond to surface topography, cell morphology, including cell alignment can be regarded as one of the early indicators for cell differentiation and gene expression.9–11 Because these cell responses are important in many scientific and technological areas – e.g., stem cell biology, regenerative medicine, and disease pathology – there have been significant efforts to understand the mechanisms responsible for cell contact guidance response to chemical signaling and topological cues, and eventually to freely manipulate the cell contact guidance.12–19
Recent advances in nanofabrication have led to the development of substrates that recapitulate many of the features presented in ECM.20–23 In addition to the biological factors present (i.e., cell type and growth conditions),24,25 cells have been shown to exhibit a range of responses that depend on not only the surface feature size and shape 26, 27 but also the organization (order/disorder) of the surface features.8,21 Gratings are common model platforms to study geometric effects on cell contact guidance due to the simplicity of their geometries, and in some cases the line-and-space variation of grating is simulated as ordered surface features.28 The key points for the success of regenerative medicine are to identify the ideal features that can manipulate or direct cell behaviors. A combinatorial grating platform that spans a range of a specific topographical parameter (e.g., pattern height) and can easily transfer to different materials will facilitate the search for these features.
In the current study, we developed a simple and versatile strategy to fabricate controlled surface topographies with a wide range of continuous pattern heights, different shapes, and various surface chemistries, enabling the separation of surface chemistry and topography effects. We then used these platforms to investigate the effects of grating structure (in terms of height, ridge and trough width, and surface chemistry) on cell alignment. As a demonstration, the alignment of murine preosteoblasts on grating platforms was examined. A full range of cell alignment, from randomly oriented to highlyparallel to the direction of the grating, was observed on a single platform. The competing effects between pattern height and width of trough are discussed particularly with respect to cell-substrate contact. Further, cell-substrate contact was evaluated using focal adhesion assays and scanning electronic microscopy (SEM).
1. Materials and Methods*
2.1. Gradient gratings prepared via nanoimprint lithography (NIL)
Poly(styrene) (PS, MW = 18.7 kg/mol, Polymer Laboratories) or polymethylmethacrylate (PMMA, MW = 19.0 kg/mol, Polymer Laboratories) dissolved in toluene (6.7 % by mass) was spun cast onto Si wafers at 209 rad/s (2000 rpm) and baked for 30 min at 170 °C under vacuum. The imprints (20 mm × 5 mm) were fabricated using a NX-2000 imprint tool with a mold consisting of parallel line-and-space gratings in silicon oxide (Λ = 420 nm or 800 nm, H ≈ 360 nm). Imprints were made in two steps: 10 s at 100 °C and 1.4 MPa followed by 3 min at 140 °C and 3.4 MPa. To form the gradient in pattern height, imprinted structures were annealed on a hot stage with a gradient of temperatures for a predefined period of time, depending on the polymer.29 Briefly, for PS gratings, the temperature gradient was 1 °C/mm between 102 °C and 122 °C, and the annealing times were 60 s and 120 s for the 420 nm and 800 nm gratings, respectively. For PMMA gratings, the temperature gradient was 1.2 °C/mm between 120 °C and 144 °C, and the annealing times were 7 min and 14 min for the 420 nm and 800 nm gratings, respectively.
2.2. Gradient gratings prepared via photopolymerization
Ethoxylated bisphenol-A dimethacrylate (DMA, degree of ethoxylation = 6, Esstech Inc.) was activated using the photoinitiator system of camphorquinone (0.2 % by mass, Aldrich) and ethyl 4-N, N-dimethylaminobenzoate (0.8 % by mass, Aldrich). Activated DMA (0.1 mL) was spread onto a methacryloyl propyl trimethoxysilane treated glass slide (20 mm × 25 mm). The silane treatment was used to enhance the adhesion of the DMA to the glass slide. A gradient PS platform was pressed directly into the DMA monomer, which was then cured using a Dentsply Triad 2000 visible light curing unit with a tungsten halogen light bulb (250W and 120V) for 2 min per side, and post-cured at 60 °C for 1 h. Immediately after post-curing, filtered air was used to rapidly cool the sample to room temperature. The PS substrate was removed, and residual PS on the DMA grating was removed via a toluene rinse. The DMA grating was then washed using 70 % ethanol and air dried.
2.3. Characterization of grating platforms
Atomic force microscopy (AFM, Dimension 3100, Vecco Instruments) was used to quantify the pattern height. The height measurements were calibrated using standard grating patterns. For 420 nm and 800 nm pitch patterns, 3 μm × 3 μm and 5 μm × 5 μm scans were performed, respectively. Pattern uniformity was confirmed by performing measurements on multiple spots across each sample. Height data is presented as the average of at least five such measurements. For visualization via electron microscopy, grating patterns were freeze-fractured and examined via field emission-SEM (FE-SEM, 15 kV, Hitachi S-4700-II, Pleasanton, CA). DMA gratings were sputter-coated with gold prior to imaging.
2.4. Cell seeding
Gradient patterns were sterilized in 70 % (volume fraction in water) ethanol for 20 min and placed in 6-well tissue culture polystyrene plates. MC3T3-E1 murine pre-osteoblasts (Riken Cell Bank, Hirosaka, Japan) were maintained in alpha minimum essential medium (αMEM) containing 10 % (volume fraction) fetal bovine serum, 5 mmol/L L-glutamine, and 60 mg/mL kanamycin sulfate. Cells were cultured in humidified incubators (5 % by volume CO2, 37 °C). Cells of passage 4–8 were seeded on the grating platforms at 1 × 104 cells/cm2. After 24 h, samples were fixed with 3.7 % (volume fraction in phosphate buffered saline, PBS) formaldehyde, permeabilized with 0.5 % (by volume in PBS) Triton X-100, blocked with 10 mg/mL bovine serum albumin in PBS, and rinsed with PBS.
2.5. Quantification of cell alignment
Cells on gratings were stained with 2 μg/mL 4′,6-diamidino-2-phenylindole (DAPI, nuclear stain), 1 μg/mL Alexa Fluor 488 C5 maleimide (sulfhydryl stain to label cell bodies), and 0.17 μmol/L Alexa Fluor 546 Phalloidin (F-actin stain) in PBS for 1 h. The entire gradient pattern was imaged using a Leica DMR 1200 upright microscope with a 10X objective and a Hamamatsu Orca ER digital camera (Vashaw Scientific, Inc.). A tile of 13 × 5 (L × W) images was collect for each pattern, and each image was 1.6 × 1.2 mm (L × W). The pattern height of the cell position in the images was determined by AFM before cell culture. Orientation angle of cells, defined as the angle between the major axis of the cell and the grating line direction, was determined using custom macros in ImagePro Plus. The population of aligned cells was calculated as the fraction of cells with orientation angle within 10 degrees of the grating line direction. Over 200 cells were characterized per grating location.
2.6. SEM of cellular extensions
Cells on grating platforms were rinsed with saline, fixed with 3.7 % volume fraction of formaldehyde, subjected to gradient alcohol dehydrations, rinsed with hexamethyldisilazane, and sputter coated with gold for imaging via FE-SEM.
2.7. Focal adhesions
Fixed cells on grating platforms were incubated with anti-murine monoclonal anti-vinculin (Sigma, 1:200 dilution) for 1 h and followed by Alexa Fluor 488 goat anti-mouse IgG (Invitrogen, 1:200 dilution) for 1 h. Cells were imaged using epifluorescence microscopy (Nikon ECLIPSE, TE300) and a 60X oil immersion objective. Fluorescence microscopy images of the focal adhesion patterns were transformed via Fast Fourier transform (FFT) into scattering images to determine the presence of periodicity.30 An Igro macro was used to take a circular average of the intensity of the pattern in the scattering images, and the repeat distance (d) was determined according to the scattering wave vector (q) value at the peak intensity using the relation d = 2π/q; q was calibrated using the diffraction peak from an 800 nm DMA grating platform. The DMA grating platforms were used as the grating controls because the PS grating pattern is not visible by light microscopy (PS substrates are on opaque silicon wafers).
2.8. Water contact angle measurement
The water contact angle measurements were carried out using sessile drop method with a Kruss G2 system on flat surfaces at room temperature. The volume of the deionized water droplet was ≈ 2 μL, and the images of the sessile droplet were taken immediately after the deposition on the substrate.
2. Results and discussion
The current study establishes strategies to fabricate line-and-space gratings as model substrates on variety of materials, enabling rapid assessment of the respective roles of surface topography and chemistry on cell alignment. The parameters of interest for guiding cell alignment are pattern height, width of the ridge and trough, and surface chemistry of the substrate material. To systematically study the contribution of each parameter, we fabricated grating platforms from three materials, each with a constant pitch (Λ = 420 nm or 800 nm) and a gradient in height (H = 0 nm to ≈ 330 nm) (Fig. 1a). Platforms made of PS and PMMA are nearly identical in surface topography (wider ridges and narrower troughs), but differ in surface chemistry. The average water contact angles on PS and PMMA surfaces are 90.4 ± 0.6 degrees and 67.8 ± 1.4 degrees, respectively. The third platform is based on a photopolymerized DMA that has similar surface chemistry as PMMA (average water contact angle of DMA is 69.4 ± 2.3 degrees), but with an inverse line and space geometries (narrower ridges and wider troughs) as compared to those of the PS and PMMA gratings.
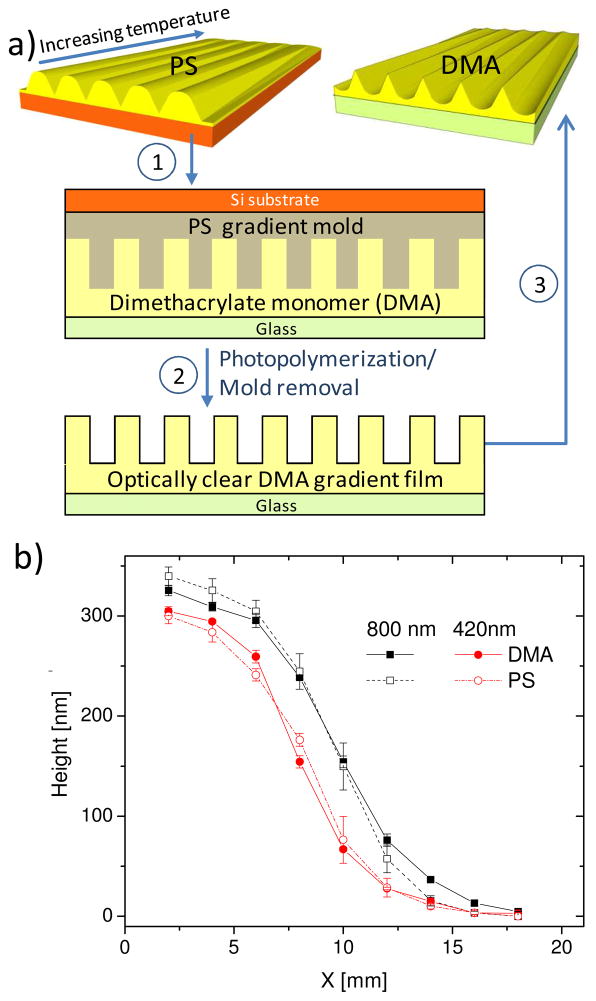
Figure 1.
(a) Fabrication of gradient platforms with inverse line and space profiles. PS gratings were produced by NIL followed by annealing on a temperature gradient. Some PS gratings were subsequently used as a mold to form DMA gratings with the inverted pattern structure through photopolymerization. (b) Pattern height as a function of position (x) for PS and DMA grating platforms determined by AFM for Λ = 420 nm and 800 nm. Lines are drawn to guide the reader’s eyes.
Two approaches were applied to fabricate all gradient grating and are amenable to fabricate a variety of grating platforms on different materials. First, PS and PMMA (thermoplastic polymers) gradient gratings were fabricated by thermal annealing of nanoimprinted polymer structures,29 a process driven by the Laplace pressure associated with the polymer line curvature at temperatures above the glass transition temperature (Tg). Thermal annealing results in a reduction in the pattern height to a predicted value through a “leveling” of the initial imprinted structure. Specifically, a gradient in H can be quantitatively controlled by the viscoelastic properties of the linear polymer, annealing time and temperature, initial H, and Λ.31,32 The gradient fabrication approach by thermal annealing is suitable for thermoplastic polymers but cannot be used for chemically cross-linked polymers. Second, we produced gradient patterns of inversed line-and-space structures using the gradient PS (or PMMA) platform as the replicating mold to generate chemically cross-linked DMA platforms with height gradients (Fig. 1a). We note that while a PMMA-like dimethacrylate was chosen for this study, dimethacrylates with strong hydrophobic characteristics (such as C12 dimethacrylate) or hydrophilic characteristics (such as polyethylene glycol dimethacrylate) can be readily used to generate structures of tunable surface properties. This pattern transfer process is also amenable to cross-linked polymers formed by other reaction mechanisms. Moreover, the DMA selected for this study is optically clear, chemically stable, and has good handling characteristics. These properties allow the DMA platform to be used for label-free in situ live cell imaging. In contrast, similar live cell imaging on a PS grating over an opaque silicon substrate is difficult without the use of fluorescent labeling. Additionally, the excellent chemical stability of DMA makes the platform amenable for processes involving alcohol or other solvents, such as the sequential cell dehydration during sample preparation for SEM.
Using this strategy, a range of pattern heights are fabricated on one platform, and the height can be precisely controlled via selection of materials and annealing conditions. In this study, the range of pattern height is 0 nm to 320 nm, which is ideal for cell contact guidance investigation. Further, these well defined surface topographies can be produced onto a large variety of materials, which enables the separation of surface chemistry and topography effects.
Precise control over surface topography was confirmed by FE-SEM and quantitative AFM analysis. AFM revealed a gradient in H for all gratings with the maximum H for the 420 nm pitch at ≈ 300 nm, and the maximum H for the 800 nm pitch at ≈ 330 nm. A comparison of the height profile of the PS mold and DMA replica gradient patterns shows excellent agreement, indicating high fidelity in pattern transfer (Fig. 1b). PMMA platforms had a height gradient equivalent to that of PS. From this point on, unless otherwise noted, high H will refer to a grating height of approximately 300 nm, while low H will refer to a grating height of less than 10 nm,.
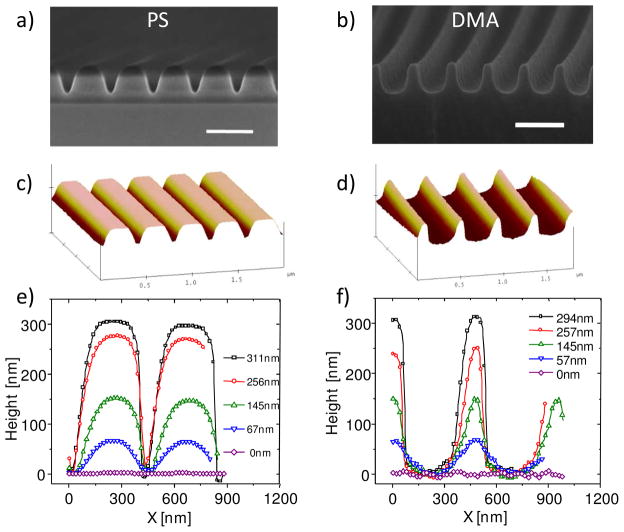
The pattern shapes were determined by analyzing the cross section of the grating platforms at different H using AFM and SEM (Fig. 2). As-imprinted PS gratings had the expected rectangular shape (not shown), but even a slight thermal annealing resulted in a slumping of the polymer and narrowing of the trough (Fig. 2a and c). As H decreased due to the higher annealing temperatures, the grating shape approached a sinusoidal form at low H (< 60 nm), before becoming a smooth surface (Fig. 2e). The PMMA grating had a nearly identical pattern profile as the PS grating. Note that the AFM profile of the grating at high H is convoluted between the real surface topography and the shape of the AFM tip, which would slightly distort the cross-sectional profile (appears artificially narrower in this case) near the bottom of the trough. In comparison, SEM images provide reliable pattern shapes even in the narrow troughs. SEM and AFM together confirmed that the DMA gratings had the inverse structure to that of the PS gratings, displaying wider troughs and narrower ridges, particularly at high H (Fig 2b and d). The differences in shape between PS/PMMA and DMA grating diminished at low H (≈ 60 nm) as both approached sinusoidal form and eventually became a flat surface (Fig. 2e and f).
Figure 2.
Cross-sectional shape profile of grating platforms at different H. (a) and (b) are SEM images for PS and DMA platforms, respectively, at H ≈ 300 nm. Scale bars = 500 nm; (c) and (d) are AFM images for PS and DMA platforms, respectively, at H ≈ 250 nm; (e) and (f) are AFM cross-sectional measurements for PS and DMA platforms, respectively, at 5 different positions.
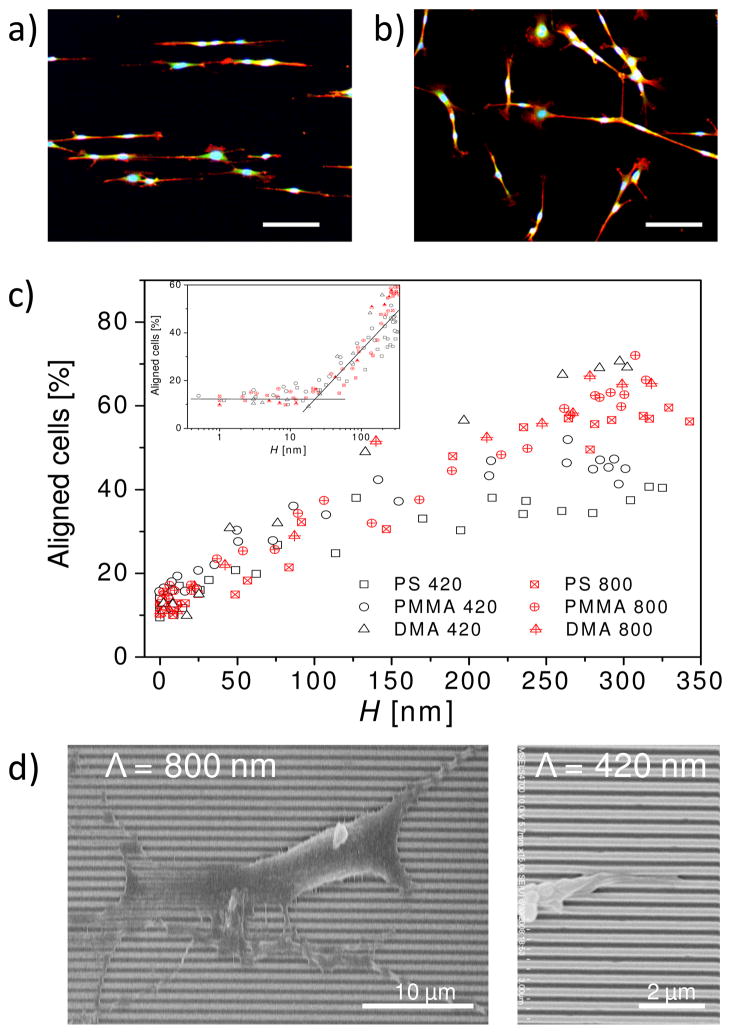
As a demonstration of utilizing these platforms, we examined cell alignment of murine pre-osteoblasts. MC3T3-E1 cells are a well-characterized cell line often used as a model cell for bone tissue engineering and regenerative medicine, applications that would benefit from the ability to adjust the cell contact guidance. On all of the gradient gratings, cells preferentially aligned along the grating line direction at high H (Fig. 3a), and the degree of alignment decreased with diminishing H. As expected, cell alignment was random at very low H (and on flat surfaces) (Fig. 3b). The population of aligned cells (those with the long axis of the cell body oriented within 10° of the grating direction) is ≈ 11 % on flat surfaces and low H (< 30 nm), consistent with the expected value for randomly orientated cells (fraction of cells in 9 equally distributed bins). The critical grating height, the height at which cells begin to sense substrate effects and align (≈ 30 nm), was similar for all gratings, irrespective of surface chemistry, pitch size and pattern shape (Fig. 3c inset). These results indicate the onset of cell alignment is predominately due to height, and less dependent on the surface chemistry and ridge/trough width. These results are consistent with a recent report indicating that the critical height for the onset of cell alignment is independent of surface chemistry and Λ on gratings fabricated by e-beam lithography.33 Our previous study has shown that initial cell spreading is driven by thermodynamics and that cell alignment occurs during initial contact with the grating platform, bearing a strong resemblance to contact angle anisotropy (the difference between the contact angles parallel and perpendicular to the grating direction). Our thermodynamic calculations identified energetic barriers that both a cell and a water droplet would encounter during spreading on a grating surface.29 The present results suggest that the free energy barrier across the grating patterns is sufficiently small below the critical height, such that cells are able to overcome the substrate effects.
Figure 3.
Cell alignment on grating platforms. Fluorescent images of cells on PS platforms taken at (a) high H (≈ 300 nm) and (b) low H (≈ 10 nm). Scale bars = 100 μm. Cells were elongated on the hydrophobic PS patterns. Cells on low H regions of the relatively more hydrophilic PMMA substrate were more spread (image not shown) (c) Percentage of aligned cells as a function of H on PS, PMMA, and DMA platforms (Λ = 420 nm and 800 nm). Inset plots the same data in a semi-log format to identify the critical height for cell alignment. (d) FE-SEM images of a representative cell and cell extension on DMA platforms illustrate that cell extensions follow the surface of the gratings and penetrate at least partially into the troughs.
The amount of cell alignment gradually increased above the critical H and eventually reached a maximum (Fig. 3c). While the exact values varied slightly, two general ranges of maxima were evident and appeared to highly depend on the pattern geometry (pitch and trough width). For PS and PMMA patterns (narrower troughs) with Λ = 420 nm, the maximum percentage of aligned cells was ≈ 40 % and was reached at H ≈ 200 nm. Cell alignment on all 800 nm pitch patterns (narrower and wider troughs) and the DMA 420 nm pitch pattern (wider troughs) approached a plateau in the range of 60 % to 70 % at a higher H (> 250 nm). While indicating that cell alignment is height dependent, these results clearly reveal that the trough dimensions also play a critical role in controlling the degree of cell alignment. For a given height, the main topography feature leading to the observed differences in cell alignment is the trough width. All platforms that reached the higher degree of alignment (all of the 800 nm and 420 nm DMA platforms) had wider troughs relative to the 420 nm PS and PMMA. At any given height, the 800 nm pitch platforms had wider troughs than the 420 nm pitch PS and PMMA platforms, simply due to the increased pitch size. The 420 nm DMA platforms had wider troughs than their PS/PMMA counterparts because the DMA platforms had the inverse line and space profiles of the PS and PMMA platforms. Moreover, the 420 nm DMA had a significantly narrower ridge as compared to 800 nm PS and PMMA patterns. The fact that 420 nm pitch DMA grating behaved nearly identical to the 800 nm pitch gratings indicates that cells are more sensitive to the width of the trough and less sensitive to the width of the ridge. These results suggest that cells can create conformal contacts with platforms when the trough is sufficiently wide, consistent with constraints in membrane curvature. We note that the analysis showed that the 800 nm pitch patterns and the 420 nm pitch DMA patterns reached a maximum degree of cell alignment of ≈ 70 % using our predefined criteria (± 10 ° to grating direction). It is clear from Fig. 3a that all cells are preferentially aligned in the grating direction (± 20 ° to grating direction).
The presence of a maximum cell alignment at high H indicates that physical limitations imposed by the surface topography affected cell alignment. In the current system, the height at which cells could no longer “sense” further increases in pattern depth suggests a lack of cell-substrate contact in the deeper troughs. This phenomenon is similar to the wetting of a simple water droplet on gratings where a transition from the non-composite state (full contact between droplet and grating) to the composite state (partial contact between droplet and grating mostly near the ridge surfaces). 30,31 For cell spreading on grating surfaces, the analog for the non-composite state is full contact between cells and undulating grating surfaces. Above a certain threshold, defined by the pattern height and the width of the trough, cells no longer have full contact with the entire grating surface due to restrictions in membrane curvature. Once the transition to partial contact occurs, there can be no increases in cell alignment.
SEM images of cells on gradient gratings were used to gain visual confirmation of the cell-substrate contact or lack thereof. As noted previously, only the DMA platform withstood the processing for SEM visualization. Cell extensions were shown to cross several DMA grating lines, and portions of cell extensions appear to have at least partially penetrated into the troughs (Fig. 3d). Cell extensions on the 420 nm pitch DMA pattern appear to have significant cell-substrate contact both in and out of the trough at high H, consistent with the increased alignment exhibited on these DMA gratings. These results are in agreement with our explanation for the cell-substrate contact to induce cell alignment.
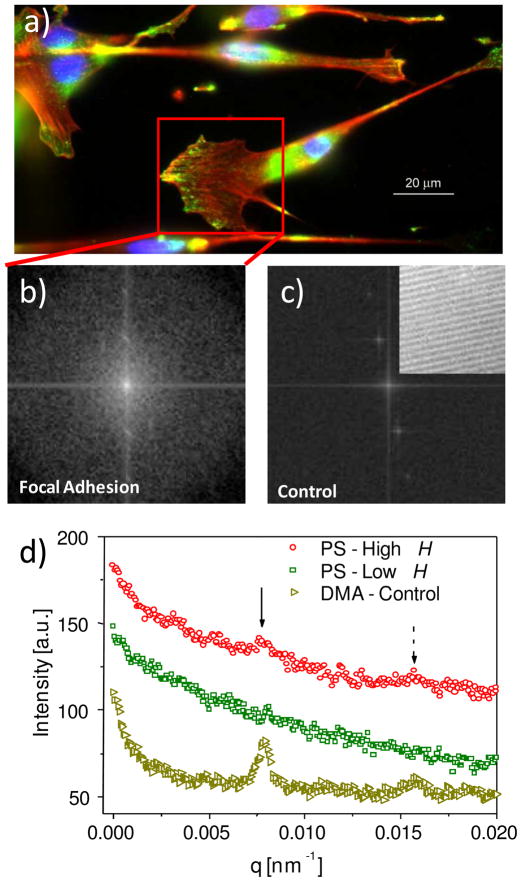
The transition from full cell-substrate contact to partial cell-substrate contact at higher H was further supported by the periodicity or striations in focal adhesion complexes correlated with the grating direction (Fig. 4a). FFT of regions containing these cellular extensions (Fig. 4b) followed by conversion to an intensity versus wave vector (q) plot revealed distinct peaks comparable to those exhibited by the patterns alone (Fig. 4c), indicating a strong periodicity in focal adhesions with a repeat distance equal to the pitch size of the pattern (Fig. 4d). FFT is a quantitative, efficient method to identify periodic features within an image. An image is transformed into its discrete frequency domain representation, where the repeating patterns in the spectrum are identifiable. Similar image processing and data analysis methods have been applied to images collected using small-angle neutron scattering and small-angle X-ray scattering, but have rarely been reported on fluorescent microscopy images. In this study, the strong periodicity further substantiates cell contact on the ridges along the grating direction and is consistent with incomplete contact between cells and bottom of the trough. Focal adhesion complexes appear to be uniformly distributed at lower H. Accordingly, FFT analysis showed no distinct peaks in the intensity profile at low H, consistent with the physical picture of complete cell-substrate contact and the formation of focal adhesion complexes on both ridges and troughs.
Figure 4.
(a) Fluorescent images of focal adhesion complexes for cells on PS grating (H ≈ 300 nm). The nuclei are in blue, the actin is in red, and the focal adhesion is in green. A box is drawn to enclose a region with a high concentration of focal adhesion complexes. The green channel in the box was subsequently analyzed via FFT. (b) Image generated by FFT of the boxed region. (c) Image by FFT of a control grating at high H without cells (DMA, real space image provided as inset). (d) Intensity versus q plot for focal adhesion complexes on PS gratings at high and low H, and on the control gratings without cells. Solid and dashed arrows correspond to the first and second order diffraction peaks, respectively.
Finally, we note that topography has a far greater effect on cell alignment than surface chemistry for materials examined. The grating platforms did not receive pretreatment prior to cell seeding and the differences in surface chemistry presented in this study should not have greatly altered protein adsorption. For PS and PMMA gratings, where the gradient patterns had very similar topography but different surface chemistries (and wetting characteristics), cell alignment was nearly identical, indicating that surface chemistry contributed minimally here. Likewise, when comparing PMMA and DMA gratings which had similar surface chemistry but different surface topography, a pronounced difference in cell alignment was clearly evident and primarily due to the inversed line-and-space geometry at high H.
3. Conclusion
In summary, NIL and a versatile imprint method were applied to generate six (three materials and two pitch sizes) correlated grating platforms designed to separate the effects of surface chemistry and topography on cell alignment. The grating platforms contained the same gradient in pattern height but differed in the ridge and trough profiles. Using our imprint method, the gradient in pattern height was successfully transferred from PS platforms, which contained wider ridges and narrower troughs, to photopolymerized DMA platforms to produce an inversed line and space geometry with wider troughs and narrower ridges. These platforms were adapted to evaluate the competing effects of surface chemistry and surface topography on cell contact guidance in terms of the orientation of murine preosteoblasts cells. Surface chemistry had little effect on cell alignment, whereas topography had significant effects in terms of both height and trough width. Cells oriented randomly at low pattern heights (H < 30 nm) and aligned parallel to the grating direction at high H, which agrees well with results in literature. Cell alignment on the grating was a function of pattern height when the width of the ridge and trough were consistent. At high H, wider troughs induced greater cell alignment, likely due to increased contact of the cell with the trough surfaces. Focal adhesion assays and SEM studies indicated that cells could cross the grating lines, even at high H. The greater cell alignment at high H and wider troughs is likely due to the smaller energy cost for cells to stay on the ridges rather than cross over the lines. Further, a full range of cell alignment, from randomly oriented to almost perfectly parallel to the grating direction, was achieved on a single platform. Thus, this platform is a vital step toward being able to evaluate the effects of important parameters on cell contact guidance, ultimately to understand and control cell orientation.
Acknowledgments
This work is partially funded by the National Institute of Dental and Craniofacial Research (NIDCR) through an Interagency Agreement (Y1-DE-7005-01) with NIST and by the NIST Office of Microelectronic Programs. Ding acknowledges partial funding support from the National Science Foundation under Grant No. CMMI-0928067. We acknowledge the nanofabrication laboratory of the Center for Nanoscale Science and Technology (CNST) at NIST for providing facilities for the nanoimprint process, and acknowledge the use of the NIST Combinatorial Methods Center equipment. The authors would like to thank Drs. Nathan Gallant, Xiaohua Zhang, and Carl Simon for their advice and technical recommendations.
Footnotes
Official contribution of the National Institute of Standards and Technology; not subject to copyright in the United States
Certain equipment, instruments or materials are identified in this paper to adequately specify the experimental details. Such identification does not imply recommendation by the National Institute of Standards and Technology, nor does it imply the materials are necessary the best available for the purpose.
References
- 1.Stevens MM, George JH. Science. 2005;310(5751):1135–1138. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- 2.Cukierman E, Pankov R, Stevens DR, Yamada KM. Science. 2001;294(5547):1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 3.Langer R, Vacanti JP. Science. 1993;260(5110):920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 4.Oh S, Brammer KS, Li YSJ, Teng D, Engler AJ, Chien S, Jin S. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(7):2130–2135. doi: 10.1073/pnas.0813200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park J, Bauer S, von der Mark K, Schmuki P. Nano Lett. 2007;7(6):1686–1691. doi: 10.1021/nl070678d. [DOI] [PubMed] [Google Scholar]
- 6.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Science. 1997;276(5317):1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 7.Curtis A, Wilkinson C. Biomaterials. 1997;18(24):1573–1583. doi: 10.1016/s0142-9612(97)00144-0. [DOI] [PubMed] [Google Scholar]
- 8.Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, Wilkinson CDW, Oreffo ROC. Nat Mater. 2007;6(12):997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 9.Kurpinski K, Chu J, Hashi C, Li S. Proc Natl Acad Sci U S A. 2006;103(44):16095–16100. doi: 10.1073/pnas.0604182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teixeira AI, McKie GA, Foley JD, Berticsc PJ, Nealey PF, Murphy CJ. Biomaterials. 2006;27(21):3945–3954. doi: 10.1016/j.biomaterials.2006.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yim EKF, Pang SW, Leong KW. Exp Cell Res. 2007;313(9):1820–1829. doi: 10.1016/j.yexcr.2007.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendonca G, Mendonca DBS, Aragao FJL, Cooper LF. Biomaterials. 2008;29(28):3822–3835. doi: 10.1016/j.biomaterials.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 13.Baksh D, Song L, Tuan RSJ. Cell Mol Med. 2004;8(3):301–316. doi: 10.1111/j.1582-4934.2004.tb00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen CJ, Ou YC, Liao SL, Chen WY, Chen SY, Wu CW, Wang CC, Wang WY, Huang YS, Hsu SH. Experimental Neurology. 2007;204(1):443–453. doi: 10.1016/j.expneurol.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Saha K, Pollock JF, Schaffer DV, Healy KE. Curr Opin Chem Biol. 2007;11(4):381–387. doi: 10.1016/j.cbpa.2007.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bettinger CJ, Orrick B, Misra A, Langer R, Borenstein JT. Biomaterials. 2006;27(12):2558–2565. doi: 10.1016/j.biomaterials.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 17.Choi CH, Hagvall SH, Wu BM, Dunn JCY, Beygui RE, Kim CJ. Biomaterials. 2007;28(9):1672–1679. doi: 10.1016/j.biomaterials.2006.11.031. [DOI] [PubMed] [Google Scholar]
- 18.Dalby MJ, Hart A, Yarwood SJ. Biomaterials. 2008;29(3):282–289. doi: 10.1016/j.biomaterials.2007.09.030. [DOI] [PubMed] [Google Scholar]
- 19.Gerecht S, Bettinger CJ, Zhang Z, Borenstein JT, Vuniak-Novakovic G, Langer R. Biomaterials. 2007;28(28):4068–4077. doi: 10.1016/j.biomaterials.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curtis ASG, Wilkinson CDJ. Biomater Sci-Polym Ed. 1998;9(12):1313–1329. doi: 10.1163/156856298x00415. [DOI] [PubMed] [Google Scholar]
- 21.Bettinger CJ, Langer R, Borenstein JT. Angew Chem-Int Edit. 2009;48(30):5406–5415. doi: 10.1002/anie.200805179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chou SY, Krauss PR, Renstrom PJ. Science. 1996;272(5258):85–87. [Google Scholar]
- 23.Guo LJ. Adv Mater. 2007;19(4):495–513. [Google Scholar]
- 24.Lu X, Leng Y. Journal of Biomedical Materials Research Part B-Applied Biomaterials. 2009;90B(1):438–445. doi: 10.1002/jbm.b.31304. [DOI] [PubMed] [Google Scholar]
- 25.Rajnicek AM, Britland S, McCaig CDJ. Cell Sci. 1997;110:2905–2913. doi: 10.1242/jcs.110.23.2905. [DOI] [PubMed] [Google Scholar]
- 26.Lim JY, Donahue HJ. Tissue Eng. 2007;13(8):1879–1891. doi: 10.1089/ten.2006.0154. [DOI] [PubMed] [Google Scholar]
- 27.Flemming RG, Murphy CJ, Abrams GA, Goodman SL, Nealey PF. Biomaterials. 1999;20(6):573–588. doi: 10.1016/s0142-9612(98)00209-9. [DOI] [PubMed] [Google Scholar]
- 28.Li W, Fang GP, Lij YF, Qiao GJJ. Phys Chem B. 2008;112(24):7234–7243. doi: 10.1021/jp712019y. [DOI] [PubMed] [Google Scholar]
- 29.Ding Y, Sun J, Ro H, Wang Z, Zhou J, Lin NJ, Lin-Gibson S, Cicerone MT, Soles CL. Thermodynamic underpinnings of cell alignment on controlled topography. Adv Mater. 2010 doi: 10.1002/adma.201001757. [DOI] [PubMed] [Google Scholar]
- 30.Zhang XH, Berry BC, Yager KG, Kim S, Jones RL, Satija S, Pickel DL, Douglas JF, Karim A. Acs Nano. 2008;2(11):2331–2341. doi: 10.1021/nn800643x. [DOI] [PubMed] [Google Scholar]
- 31.Ding YF, Ro HW, Douglas JF, Jones RL, Hine DR, Karim A, Soles CL. Adv Mater. 2007;19(10):1377–1380. [Google Scholar]
- 32.Ding YF, Ro HW, Germer TA, Douglas JF, Okerberg BC, Karim A, Soles CL. Acs Nano. 2007;1(2):84–92. doi: 10.1021/nn700014p. [DOI] [PubMed] [Google Scholar]
- 33.Loesberg WA, te Riet J, van Delft FCMJ, Schon P, Figdor CG, Speller S, van Loon JJWA, Walboomers XF, Jansen JA. Biomaterials. 2007;28(27):3944–3951. doi: 10.1016/j.biomaterials.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 34.Zhao Y, Lu QH, Li M, Li X. Langmuir. 2007;23(11):6212–6217. doi: 10.1021/la0702077. [DOI] [PubMed] [Google Scholar]