Abstract
Genes involved in host-pathogen interactions are expected to be evolving under complex coevolutionary dynamics, including positive directional and/or frequency dependent selection. Empirical work has largely focused on the evolution of immune genes at the level of the protein sequence. We examine components of genetic variance for transcript abundance of defense genes in D. melanogaster and D. simulans using a diallel and a round robin breeding design, respectively, and infer modes of evolution from patterns of segregating genetic variation. Defense genes in D. melanogaster are overrepresented relative to non-defense genes among genes with evidence of significant additive variance for expression. Directional selection is expected to deplete additive genetic variance, whereas frequency dependent selection is expected to maintain additive variance. However, relaxed selection (reduced or no purifying selection) is an alternative interpretation of significant additive variation. Of the three classes of defense genes, the recognition and effector classes show an excess of genes with significant additive variance; whereas signaling genes, in contrast, are overrepresented for dominance variance. Analysis of protein coding sequences revealed no evidence for an association between additive or dominance variation in expression and directional selection. Both balancing selection driven by host-pathogen coevolution and relaxed selection for expression of uninduced defense genes are viable interpretations of these data.
Keywords: Red Queen, host-pathogen coevolution, diallel, microarray, gene expression, relaxed selection
Introduction
Rapid evolution of defense genes might be expected, given the obvious propensity of defense genes to coevolutionary dynamics (see Lazzaro and Little 2009), but precisely what evolutionary force is driving this pattern? The empirical literature of host-pathogen coevolution is rife with discussions of negative frequency dependent selection including such taxonomically diverse, classic host-pathogen systems such as flax and its rust Melampsora (Burdon and Thrall 2000); Daphnia and the bacterium Pasteuria (Carius et al. 2001); and the New Zealand snail Potamopyrgus and its trematode parasite Microphallus (Dybdahl and Lively 1998). While details of the mechanism (e.g. gene for gene vs. matching alleles models) differ depending on the system in question, most would agree that the Red Queen hypothesis, which posits that in order for hosts and pathogens to stay abreast of one another they must each run (that is, coevolve) as fast as they can, reigns (Dybdahl and Storfer 2003; Lazzaro and Little 2009). Although the original formulation of the Red Queen includes directional selection (Van Valen 1973; Van Valen 1976, 1977), the theory is often also modeled as negative frequency dependent selection, such that an allele is fit because it is rare; once it increases in frequency, its fitness decreases (Thompson 2005).
There is a rich literature on immune gene evolution in Drosophila (for a recent review, see Lazzaro 2008, also Obbard et al. 2009), documenting heterogeneous patterns of evolution for this group of genes as inferred from coding sequence evolution. Purifying, positive, and relaxed selection can be inferred by assessment of ratios of nonsynonymous to synonymous substitution rates, using a single protein coding sequence from each of two species. Other methods such as the HKA test and the McDonald-Kreitman test have also been applied to sequence data of defense genes, primarily with respect to coding sequences (but see Begun et al. 2007). The dN/dS approach has been elegantly applied to explore rates of evolution with respect to the distinct mechanistic roles played by different classes of immunity genes (Sackton et al. 2007). They divided genes into three functional categories: recognition (genes which detect the presence of a pathogen), signaling (genes internal to immune pathways which are involved in signal transduction), and effector (genes which interact directly with a pathogen to inhibit growth, etc.). Genes in both the recognition and effector classes might in particular be expected to be subject to coevolutionary dynamics, while genes in the signaling class might have been expected to be more conserved and evolve more slowly. Sackton et al. (2007) detected an excess of positive selection in the recognition class relative to non-immune single copy ortholog genes throughout the genome, while such an excess was not detected in either of the other two functional classes (although a subset of the signaling genes, those involved in signal modulation, also showed an excess of positive selection relative to other signaling genes).
Particular selective regimes are expected to result in particular patterns of components of genetic variance. For example, we expect directional selection-- both positive and purifying-- to reduce the amount of additive genetic variance, but have no effect on the other components of genetic variance. Similarly, frequency dependent selection is expected to maintain additive genetic variance, as is relaxed selection (Lynch and Walsh 1998). The effects of relaxed or frequency dependent selection, or of conversion of epistatic variance, on the other components of variance are less clear. Thus, inference on mode of selection on expression may be drawn from examining the components of genetic variance for expression.
Amino acid substitution rate, but not silent substitution rate, is positively correlated with expression divergence in Drosophila (Nuzhdin et al. 2004; Lemos et al. 2005; Bedford and Hartl 2009). Selection for gene expression might be expected to be correlated with selection at the level of the protein, at least for positive directional selection. If the association between substitution rate and expression divergence is caused by similar types of selection acting on both the protein and the transcript abundance, then partitioning of genetic variance of the transcript abundance of a gene might be expected on average to reflect the mode of selection at the level of the protein. However, this need not be true for every gene.
Given the sub-categorization of immune genes into functional groups (recognition, signaling, effector), what are the appropriate conditions for measuring expression of immune genes? For recognition and signaling genes, constitutive expression may be quite informative. The first response of a fly to a pathogen is likely to be important in preventing infection; and by definition, this first response occurs in an uninduced state. Indeed, expression (and translation of) immune genes in an uninduced state may well underlie the oft-discussed “cost of resistance” in the absence of challenge. Of course, some positive feedback may occur subsequent to exposure to a pathogen, which might affect expression of recognition and signaling genes. It is less clear that effector gene expression is equivalent in a resting state to an induced state (i.e. subsequent to exposure to a particular pathogen). However, it is possible that an effective response to a pathogen would require some non-negligible expression of effectors even in the absence of pathogen challenge. For example, constitutive expression of antimicrobial peptides can confer increased survival to challenge (reviewed in Vallet-Gely et al. 2008).
We consider expression data (i.e. transcript abundance) instead of protein data to investigate the evolution of genes involved in the immune response in two species of fruit fly, Drosophila melanogaster and D. simulans, using classical quantitative genetic breeding designs (the diallel and the round robin) to infer additive and dominance genetic variance for gene expression. Because genes involved in defense are expected to be rapidly evolving due to coevolution between host and pathogen, one might expect defense genes to have particular patterns of additive genetic variation distinct from other categories of genes. Positive selection should result in a paucity of additive variance, while frequency dependent or relaxed selection should result in an excess of additive variance. To quantify additive and dominance variance, we tested defense genes for GCA (general combining ability) and SCA (specific combining ability). GCA is largely attributable to additive variance, while SCA is largely attributable to dominance variance (Lynch and Walsh 1998). We estimated GCA for 388 defense genes in males of Drosophila simulans, using a round robin design (Wayne et al. 2004), and estimated GCA and SCA for females (598 defense genes) and males (609 defense genes) of D. melanogaster using a diallel design (Wayne et al. 2007). We compare 843 genes associated with host defenses with 8784 genes not expected to be involved in host-pathogen interactions, and conclude that defense genes are more likely to have significant additive genetic variance than non-defense genes. These results are consistent with either relaxed selection or balancing selection for the evolution of expression of defense genes.
Materials and Methods
D. melanogaster diallel
D. melanogaster originally captured in Wolfskill orchard (Winters, CA) were subjected to > 20 generations of full sibling inbreeding (Nuzhdin et al. 1999). Nine of these lines were used as parents in a full diallel design (all pairwise combinations, excluding homozygous parents; 72 F1 progeny). Three day old virgin adults were collected (Wayne et al. 2007). RNA was extracted and hybridized to a custom Agilent chip AMADID 012798 (McIntyre et al. 2006). Background-corrected spot intensities were log transformed. Additional details of the experimental design are given in Wayne et al. (2007).
Infection status of lines/evidence for non-induction of defense genes
Our inferences are based on the assumption that the variation between lines is genetic, rather than due to environmental variation confounded with line such as differential pathogen infection status. The block structure we used, with sub-blocks run at different times and in different incubators (described in detail in Wayne et al. 2007), makes it highly unlikely that horizontally transferred, episodic infections would be confounded with line (i.e., contribute to our estimate of genetic variance).
Our previous study (Wayne et al. 2007) would have identified most vertically transmitted infections. Maternally transmitted infections, which are effectively non-genetic maternal effects, would have been identified in our original analysis as RGCA (reciprocal general combining ability, see Lynch & Walsh 1998). RGCA was rare and almost entirely restricted to males (69 genes in the genome vs. 2 in females), and thus is inconsistent with a non-genetic maternal effect, which would have affected both sexes equally. Biparentally inherited infections in some lines and not others could potentially cause genetic variation for expression of defense genes; however, sigma virus is the only well known example of a biparentally transmitted pathogen, and the lines are negative for sigma.
D. simulans round robin
The parental stocks of D. simulans were obtained from flies caught in Wolfskill orchard, and were full sib mated for 25–29 generations to create nearly homozygous stocks (Nuzhdin et al. 2004). Ten parental stocks were crossed together in a round robin design and three-day old virgin adult males were collected (Wayne et al. 2004). Affymetrix Drosophila Genechips™ 1.0 were used for the hybridizations. Data were quantified using Affymetrix MAS5 software. Average difference values for each chip were normalized to the chip median and then log transformed.
Comparison of power between round robin and diallel design
We simulated 1,000 round robin datasets from the diallel data. Both the round robin and the diallel can be used to detect GCA, the general combining ability of a genotype. GCA is largely attributable to additive variance, though epistasis also contributes to GCA ( ; Lynch and Walsh 1998). Perhaps unsurprisingly, the diallel had much greater power than the round robin, in particular greater sensitivity (see Supplementary Table 1). Interestingly, there was greater (albeit low) power to detect GCA in males than in females using the round robin. This may reflect the more complex genetic architecture in females than in males (Wayne et al. 2007), such that GCA tends to be a smaller proportion of the variance in females than in males. Point estimates for GCA and SCA, merged with the functional category classifications, are presented in Supplementary Table 2.
Comparison between Affymetrix® and Agilent® platforms
Expression level for D. simulans was measured on an Affymetrix® platform while the D. melanogaster data were obtained on a custom Agilent ® platform. To compare results across the two platforms, all individual probe sequences were compared against FlyBase (www.flybase.org) 5.1 transcripts using BLAST (Altschul et al. 1990). FBTR numbers were linked to FBGN numbers and then FBGN numbers used to identify gene names (Supplementary Table 3). If prior annotation existed for probes which were missing annotation from FlyBase 5.1 (www.affymetrix.com; McIntyre et al. 2006)) then that annotation was used and the probe retained. Data from the two platforms were linked via the gene name. There were a total of 11,310 genes in common across the two platforms out of 15,182 genes in FlyBase 5.1 for a total coverage of 72%. The numbers of genes present for each of the experiments out of the 11,310 genes in common are given in Supplementary Table 3.
Multiple probes per gene
A number of genes that were present on both chips had more than one probe for that gene. For genes with multiple probes, if the FDR corrected P values for the test of GCA across such probes disagreed in terms of significance, then these genes were discarded, leaving a total of 11,042 common symbols with congruent GCA estimates (Supplementary Table 3).
Defining genes involved in Drosophila defense and immunity
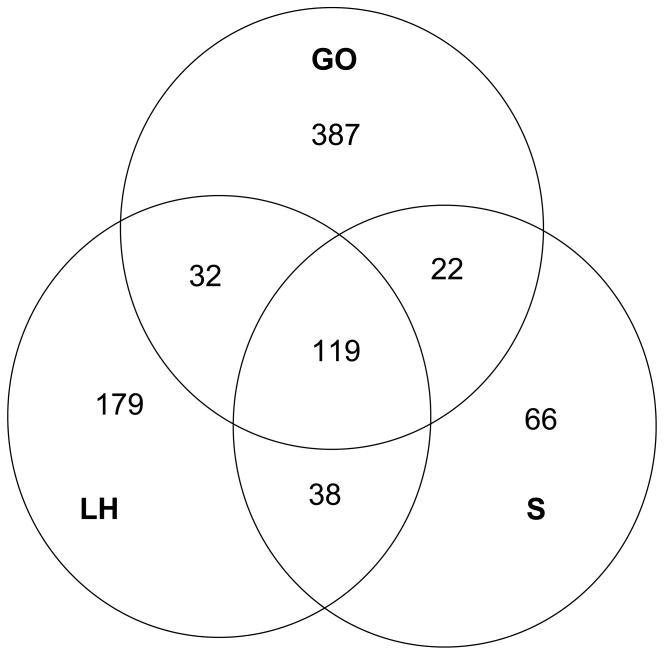
Genes present on both arrays were annotated as having a role in defense by searching the Gene Ontology (GO) database using the key words “defense”, “virus”, “antioxidant”, “antimicrobial”, “antibacterial”, “immune” and “antiviral” in the categories “Biological function”, “Cellular process” and “Molecular function”. 560 genes were identified as “defense” by this search (Figure 1). Recent papers (Lemaitre and Hoffmann 2007; Sackton et al. 2007) identified an additional 283 genes involved in defense, for a total of 843 genes (see Figure 1).
Figure 1.
Identification of genes involved in defense. There are a total of 843 genes identified as being involved in defense through three different criteria: gene ontology (GO), definition by Lemaitre & Hoffman 2007 (LH), or definition by Sackton et. al. 2007 (S). The relationship among these lists is depicted above.
Both Lemaître & Hoffman (2007) and Sackton et al. (2007) indicate functional categories for immunity genes included in their lists. In the four categories given by Sackton et al. (2007), 28 recognition, 106 signaling, 50 effectors, and 3 “other immune” were present in our data. For the Lemaître & Hoffman categories, 38 microbial recognition and phagocytosis genes; 53 signalling genes; 21 antimicrobial peptide genes; 138 microarray induced by infection genes; 16 haematopoiesis and cellular response genes; 8 coagulation genes; 6 antiviral defense genes; 10 melanization genes; and 11 miscellaneous/other were present in our data.
Testing for significance of variance components
For each of the probes detected in the diallel design (Table 1) the mixed effects linear model: ydijk = μ + dyed + GCAij + SCAij + RGCAij + RSCAij + εdijk was fit, where ydijk is the estimated gene expression of the kth replicate for the dth dye for the cross between parents i and j; μ is the overall mean; GCAij is a matrix of indicator variables for the parents; SCAij is the specific combining ability (SCA) of the cross between parent i and j; RGCAij is the reciprocal general combining ability and RSCAij is the reciprocal specific combining ability and εdijk is the error term. The GCA of a genotype is its average expression as a parent in hybrid combination with all other genotypes, and is expressed as the deviation from the overall mean. GCA approximates the additive variance, although it also includes epistatic terms: (Lynch and Walsh 1998). Similarly, SCA also includes epistatic terms as well as dominance terms: . The GCA component of variance were evaluated by use of an F ratio test statistic MSGCA/MSSCA (Lynch and Walsh 1998); for SCA, we used the F ratio MSSCA/MSerror (Wayne et al. 2007). Thus, genes significant for GCA or for SCA are those genes where the variance component is significantly greater than zero. The terms RGCA and RSCA are not considered further here, as they are present only in the diallel with D. melanogaster, not in the round robin with D. simulans.
Table 1. Defense genes are associated with additive genetic variation.
The number of genes detected (of 11,042 genes compared; Supplementary Table 1) for each experiment, and proportion significant for GCA. The number of defense genes detected (of 658; supplementary Figure 1); and the number and percent of defense genes with GCA. A Chi-Square test of association between the detection of GCA and the designation of a gene as being involved in defense was performed. P values were calculated exactly (Edgington 1995).
| D. sim. ♂ | D. mel. ♂ | D. mel. ♀ | D sim. ♂ + D. mel. ♂ | D. mel. ♂ + ♀ | |
|---|---|---|---|---|---|
| Detected | 6236 | 10046 | 9886 | 5691 | 9627 |
| Significant for GCA (%) | 459 (7.36) | 1595 (15.88) | 674 (6.82) | 66 (1.16) | 554 (5.75) |
| Defense genes detected | 388 | 609 | 598 | 360 | 584 |
| Defense genes with GCA (%) | 32 (8.23) | 170 (27.91) | 91 (15.22) | 9 (2.50) | 81 (13.87) |
| χ2 | 0.48 | 70.34 | 70.69 | 6.24 | 31.67 |
| P value | 0.55 | 4.8E-15 ** | 1.00E-13 ** | 0.02 * | 5.12E-07 ** |
For the D. simulans round robin data, the mRNA expression of each gene was analyzed using the reduced form of the model given above: yijk = μ + GCAij + εijk where yijk is the estimated gene expression for the kth replicate for the cross between parents i and j, μ is the overall mean and GCAij is a matrix of indicator variables for the parents and εijk is the error term. In this case, GCA effects were tested for significance with the F ratio MSGCA/MSε.
To account for multiple tests, balance type I and type II error, and balance the sensitivity of the round robin design compared to the diallel (Supplementary Table 1), an FDR level 0.1 was used (Benjamini and Hochberg 1995; Verhoeven et al. 2005). However, inferences were checked at other FDR levels (0.2 and 0.05) and were consistent regardless of the specific level chosen.
D. melanogaster round robin simulations
Since the two species were evaluated using different mating designs, we tested whether the mating design choice introduced a bias in the testing for GCA using simulation. A round robin design of 9 crosses was simulated by selecting at random from among the 72 diallel crosses. There are as many round robin designs as unique orders of 9 parents. One thousand randomly generated sets of round robin designs from 9 parents were selected. For the 1,000 simulated round robins we fit the model: yijk = μ + dk + GCAij + εijk for males and females separately. As dye effects are fixed, total phenotypic variance and proportion of variance explained by GCA were estimated and tested for in the same manner as the D. simulans data (MSGCA/MSε).
McNemar tests were performed to examine whether the diallel detects significantly more GCA than the round robin design in D. melanogaster. We found these tests to be significant most of the time; indicating that there is greater power in the diallel than in the round robin.
Sequence alignments, McDonald-Kreitman tests, NI, and DoS
Significant genomic resources now exist for many species and many isolates of Drosophila. However, these sequences are not all of the same quality or completeness. In addition, nomenclature varies with accession, such that the same gene may be found under a variety of non-overlapping names. To find all reported partial or complete sequences for the defense genes of interest, the coding sequence for each gene was blasted against all nucleotide sequences in GenBank (release 166.0) labeled D. simulans or D. melanogaster. BLAST hits were “stacked” still aligned to the complete CDS, and any missing bases were replaced by Ns to preserve the correct reading frame. We then pared down the set of matching sequences to those representing independent sequences for defense genes by removing any aligned sequence annotated as a whole chromosome, whole genome, chromosome region, or genomic contig. We also removed sequences labeled “draft” or “sequence in progress”.
To test competing hypotheses for the evolution of immunity genes significant for GCA and/or SCA, we used the McDonald-Kreitman (MK, McDonald and Kreitman 1991) test as well as two modifications of the MK test, the neutrality index (NI, Rand and Kann 1996) and the Direction of Selection (DoS, Stoletzki and Eyre-Walker 2010). All three compare patterns of within and between species variation for silent and replacement variation, and require sequence from at least two species. Therefore, genes without at least one D. simulans sequence were excluded, while genes meeting this criterion were carried forward. Next, a minimum sequence quality was enforced by requiring that sequences from both D. melanogaster and D. simulans have at least one run of 100 bp uninterrupted by Ns; again, genes meeting this criterion were carried forward while genes which did not were excluded. Our goal was to maximize the number of codons, rather than the number of individuals, to minimize the possibility of oversampling rare, deleterious segregating mutations biasing MK test results (Charlesworth and Eyre-Walker 2008). Therefore, for genes with multiple alternative splice products, the product with the maximum number of codons was selected for further analysis. After identification of the region with the longest D. simulans allele, D. melanogaster and D. simulans alleles with Ns were excluded from the analysis.
If evolution is proceeding neutrally, we expect the ratio of silent to replacement variation to be similar both within and between species (McDonald and Kreitman 1992). Two by two contingency tables were scrutinized to determine the pattern of deviation from the neutral expectation, and thus to infer specific selective scenarios. All genes with two or more cell values less than four were excluded due to ambiguity in interpretation of such sparse contingency tables. An excess of replacements between species is consistent with an interpretation of positive selection. MK tests were evaluated using the program DnaSP (Rozas et al. 2003), using the Williams’ adjusted G test as the test statistic.
The neutrality index (NI; Rand and Kann 1996) is an odds ratio of the table used for the MK test [NI = Pn/Ps)/(Dn/Ds)] and may be used to infer what departure from neutrality is causing a violation of the MK test. For example, under neutrality, Pn Ps should equal Dn Ds. In the case of NI > 1, an excess of amino acid polymorphisms is frequently interpreted as slightly deleterious mutations, while NI < 1 indicates an excess of fixations of amino acids, as might be expected under positive directional selection. However, NI has been criticized as biased, particularly when cell counts are small; accordingly we also considered the “direction of selection”, DoS (Stoletzki and Eyre-Walker 2010): DoS = DoS = Dn/(Dn + Ds) − Pn/(Pn + Ps). The DoS statistic is a difference of proportions, rather than a ratio of ratios. Similar to NI, DoS is positive in the case of adaptive evolution (directional selection), zero in the case of neutrality, and negative in the case of segregating slightly deleterious mutations.
Obbard et al. 09 also performed MK tests and calculated the components for NI and for DoS for 417 genes, which they classified as “immune” (99 genes) or “control” (318). Of the 417 genes in their paper, 334 were also present in our study, including 98 genes previously classified as “immunity”. Of these 98, we had classified 85 as defense. Interestingly, there is no ‘bias’ between our classifications, in that there are 14 genes Obbard and colleagues classify as “immune” that we do not classify as defense; and 13 genes classified by Obbard as control that we do classify as defense. The trends with respect to significant GCA for genes classified as immune relative to genes not involved in immunity are qualitatively similar between their classification “immune” and our classification “defense”. We also use the data of Obbard et al. 09 to calculate NI and DoS for the set of 98 genes (see Supplementary Table 4).
Results
Significant GCA (i.e., the component of variance for GCA was significantly different from zero; see methods for more detail) was disproportionately associated with defense genes in D. melanogaster relative to non-defense genes for both sexes (Table 1). This was not the case for D. simulans, although the trend in the D. simulans data mirrors that of D. melanogaster. The most parsimonious explanation for the absence of a statistically significant overrepresentation of defense genes with significant GCA in D. simulans is the lower power of the round robin design (see materials and methods).
Following Sackton et al (2007), we analyzed defense genes separately by functional category. Regardless of sex or species, the proportion of defense genes that have significant GCA is larger than the proportion of non-defense genes that have significant GCA. A greater proportion of genes from the recognition and effector classes have significant GCA in D. melanogaster males than in D. melanogaster females (43.18% and 28.74%, respectively; see Table 2). Again, we see that significant genes are less common in males of D. simulans than in D. melanogaster, and again the most straightforward interpretation is the lower power of the round robin design to detect GCA relative to the diallel design, rather than some biologically interesting difference between species.
Table 2. Additive genetic variation is associated with specific categories of defense genes.
The number and proportion of genes detected as well as those significant for GCA in functional categories of defense genes (Lemaitre & Hoffman 2007, Sackton et al. 2007). Chi-squared tests were used to evaluate whether specific classes were enriched for significant GCA relative to non-defense genes; P-values were calculated exactly (Edgington 1995).
| D. sim. ♂ | D. mel. ♂ | D. mel. ♀ | D sim. ♂ + D. mel. ♂ | D. mel. ♂ + D. mel. ♀ | |
|---|---|---|---|---|---|
| Signaling | |||||
| Detected | 76 | 106 | 103 | 71 | 101 |
| Significant for GCA (%) | 4 (5.26%) | 6 (5.66%) | 2 (1.94%) | 0 (0.00%) | 2 (1.98%) |
| χ2 | 0.49 | 8.37 | 3.90 | 0.84 | 2.68 |
| P value | 0.53 | 0.005** | 0.05* | 0.64 | 0.13 |
| Recognition | |||||
| Detected | 26 | 37 | 35 | 22 | 35 |
| Significant for GCA (%) | 4 (15.38%) | 18 (48.65%) | 8 (22.86%) | 3 (13.64%) | 7 (20.00%) |
| χ2 | 2.46 | 29.86 | 14.22 | 29.99 | 13.14 |
| P value | 0.12 | 3.16 × 10−06** | 0.002 | 0.002 | 0.0033 |
| Effector | |||||
| Detected | 33 | 51 | 52 | 31 | 51 |
| Significant for GCA (%) | 4 (12.12%) | 20 (39.22%) | 17 (32.69%) | 1 (3.23) | 14 (27.45%) |
| χ2 | 1.10 | 20.9 | 55.09 | 1.16 | 44.5 |
| P value | 0.30 | 4.92 × 10−05** | 2.79 × 10−08** | 0.3041 | 6.62 × 10−07** |
| Recognition/effector | |||||
| Detected | 59 | 88 | 87 | 53 | 86 |
| Significant for GCA (%) | 8 (13.56%) | 38 (43.18%) | 25 (28.74%) | 4 (7.69%) | 21 (24.42%) |
| χ2 | 3.356 | 49.55 | 66.37 | 19.04 | 55.74 |
| P value | 0.08* | 9.28 × 10−10** | 3.44 × 10−10** | 0.0032** | 1.06 × 10−08** |
Genes in the signaling class, which might be expected a priori to be more evolutionarily conserved than recognition or effector genes, are underrepresented for significant GCA relative to total genes (5.66% vs. 15.88% in D. melanogaster males; 1.94% vs. 6.82% in D. melanogaster females; see Table 2). Interestingly, signaling genes are also the only functional category of defense genes where there is significant overrepresentation of SCA relative to non-defense genes in the genome (Table 3).
Table 3. Defense genes in signalling pathways are associated with dominance variance in D. melanogaster females.
The number and proportion of genes significant for specific combining ability (SCA) among genes detected in D. melanogaster females was tested for association with all defense genes using a chi-squared test. Defense genes as a category were defined as presence on any of three lists: GO ontology, Lemaitre & Hoffman (2007) or Sackton et al. (2007). In addition, the pooled categories of “recognition” and “signaling” defense genes from the three lists were tested against total defense genes. P values were calculated exactly (Edgington 1995).
| D. melanogaster genes | D. melanogaster ♀ defense genes | Pooled recognition/effectors/phagocytosis/AMP’s | Pooled signaling | |
|---|---|---|---|---|
| Detected | 9886 | 598 | 86 | 103 |
| Significant SCA (%) | 2221 (22.47%) | 60 (21.43%) | 18 (20.93%) | 33 (32.04%) |
| χ2 | - | 2.10 | 0.12 | 5.48 |
| P value | - | 0.18 | 0.80 | 0.02 * |
Because we were curious about possible concordance of expression evolution and protein evolution, we utilized the large dataset of Obbard et al. 2009 to examine relationships between expression evolution and protein evolution, testing for an association between positive directional selection at the level of the protein and significant GCA for expression. We also tested for an association between positive directional selection in the protein and the presence of significant SCA for expression. We did not find any evidence of an association between positive directional selection, regardless of the indicator used (MK, NI, DoS), and components of genetic variance for expression (GCA or SCA; see Supplementary Tables 5 and 6, respectively).
We compiled sequence data for McDonald-Kreitman (MK) tests for 12 defense genes which were significant for GCA (9) or SCA (3), but were not included in Obbard et al. 09 data and had sufficiently long, high quality sequences in two species to be used for MK tests, NI, and DoS (see Materials and Methods for details; see Supplementary Table 7). Given that we had only a single sequence from D. simulans for half the samples, and the maximum number of sequences from D. simulans was 4 for those genes where more than one long, high quality sequence was available, we did not calculate NI or DoS for this species. Because a dozen genes is a very small sample, and because data are sparse (i.e. one or more individual cell counts per gene less than five), these results should be interpreted with caution.
Of the 12 genes analyzed, 7 showed a signature of positive directional selection by MK analysis (excess of nonsynonymous fixed differences), though only one, mys, is significant (P = 0.036, G with Williams’ correction). mys has significant SCA, but not significant GCA. Interestingly, mys had an NI for D. melanogaster of zero (consistent with neutrality) but a positive DoS (0.32, consistent with positive directional selection). Five genes had an NI for D. melanogaster greater than 1 (GCA: CG10912, CG15784, CG17575, hsf; SCA: fst), consistent with positive directional selection. Five genes in addition to mys had a positive DoS for D. melanogaster (GCA: cat, CG10680, CG18067, gstd1, jon65aiv), again consistent with positive directional selection. Lack of concordance between NI and DoS is discussed elsewhere in detail (Stoletzki and Eyre-Walker 2010), but is most easily explained here due to sparse data, including zero values in some cells.
Discussion
We find evidence that genes significant for GCA for expression are overrepresented among genes identified as playing a role in immunity and host defense relative to other genes. In particular, significant GCA, which is expected to be caused predominantly by additive genetic variation, is disproportionately associated with defense genes in the recognition and effector classes— interestingly, precisely those classes of defense genes which are most transparently expected to be targets of coevolution. An excess of GCA in recognition and effector classes would be consistent with two selective scenarios: relaxed or balancing selection. However, we can not distinguish between these two scenarios; and moreover, neither is intuitively pleasing. In contrast to recognition and effector genes, defense genes classified as signaling genes which also have significant GCA are underrepresented relative to non-defense genes. Also, significant SCA, which is expected to be caused predominantly be dominance variation, is disproportionately associated only with signaling genes, not with recognition or effector genes. These results are discussed in turn below.
Relaxed selection and defense gene expression evolution
We would not expect the resting level of expression of defense genes in the absence of pathogen challenge to be under relaxed selection. First, any response to pathogens requires some low level of expression of at least the recognition genes. Second, there is sometimes a cost of resistance in the absence of pathogen challenge (for example, Fritz and Simms 1992), suggesting simultaneously that in these cases, expression of defense genes in the absence of induction is non-negligible; and that expression of defense genes in the absence of pathogens should be minimized. Indeed, by this reasoning, one might expect that expression of defense genes would be under stabilizing selection rather than relaxed selection. Gene expression is thought to be largely controlled by stabilizing selection (including Rifkin et al. 2003; Denver et al. 2005; Lemos et al. 2005; Rifkin et al. 2005; Gilad et al. 2006; Whitehead and Crawford 2006; Bedford and Hartl 2009; but see also Khaitovich et al. 2004). However, though we do not test directly for stabilizing selection, the evolution of expression for defense genes seems to be exceptional relative to other genes in the genome, suggesting that expression of defense genes is not subject to stabilizing selection.
Balancing selection and defense gene expression evolution
While it is easy to imagine proteins evolving under balancing selection, it is more difficult to envision a mechanism for gene expression itself to be under balancing selection. However, one might conjecture that, on average, the mutational target size for expression (including both cis and trans acting factors) of a given gene is likely to be greater than that for functional changes in the protein sequence of the same gene. If so, once a beneficial protein variant arose, a concomitant modifier of gene expression might be selected for. Selection for modifiers of expression might be analogous to arguments on the evolution of dominance (see Fisher 1928, but see also Wright 1934; Charlesworth 1979; Kacser and Burns 1981; Orr 1991). However, evolution of modifiers of transcription arising in response to particular linked protein variants may not be so unlikely: evidence for coevolution between cis and trans acting factors for the expression of particular genes has been reported previously (Landry et al. 2005; McManus et al. 2010). Perhaps coevolution between cis and/or trans acting factors and the protein alleles whose expression they govern is not such a great leap.
Concerted evolution of protein and gene expression
If evolutionary forces acting at the level of the protein act similarly at the level of expression (Nuzhdin et al. 2004), then we may be able to make meaningful inferences about modes of selection by combining data on the genetic components of variance for expression (i.e. significance tests for GCA and SCA) with data on patterns of protein evolution. Accordingly, we compiled coding sequence data for defense genes and inferred presence or absence of positive selection using the frameworks of McDonald-Kreitman (MK), the neutrality index NI, and the direction of selection (DoS). We were able to compile such data for 346 genes total (334 from Obbard et al. 09, and 12 additional genes, which were analyzed seperately). We failed to detect any evidence for an association between significant GCA and directional selection, or between significant SCA and directional selection, for defense genes overall or for any of the three functional categories, regardless of which analytical framework to detect selection was employed.
Previous observations that cis acting sites are more likely to be associated with additive variance while trans acting sites are more likely to be associated with dominance variance (Lemos et al. 2008; McManus et al. 2010) suggest a different explanation for patterns of genetic variation for expression: linkage. Linkage might drive concerted evolution of proteins and their expression, because cis acting sites are likely to associated with additive variance. Thus, a paucity of additive variation for expression could signal the presence of selective sweeps, driven by directional selection in a protein but also removing additive variation for expression by homogenizing nearby cis-acting sequences. However, the lack of evidence for an association, negative or positive, between significant GCA and evidence for positive selection suggests that linkage is not a general explanation for excess significant GCA in defense genes.
Alternatively, if multiple protein variants are segregating in the population as might be expected under balancing selection, then we might expect to see additive variance for expression due to linked, effectively neutral cis acting variants. We are unable to test directly for balancing selection given available data, so this hypothesis remains untested.
The presence of significant SCA (along with the absence of significant GCA) in signaling genes is particularly intriguing. One might expect signaling genes to be highly conserved, given the observation of orthologs throughout the Drosophila group (Clark et al. 2007). However, the protein sequences of these genes actually seem to be rapidly evolving (Lazzaro et al. 2004; Lazzaro et al. 2006; Sackton et al. 2007). The signaling cascade is often the target of pathogen strategies in mammals (Finlay and McFadden 2006; Frank and Schmid-Hempel 2008; Maizels 2009), though examples in invertebrates are less well known (but see Vallet-Gely et al. 2008). A pathogen, rather than evading detection or defending itself against host weaponry, could subvert deployment of said weaponry simply by disrupting host communication (B. Lazzaro, personal communication). Interestingly, an excess of positive selection on the protein coding sequences of signaling genes has previously been observed (Obbard et al. 09). Our result of fewer signaling genes with significant GCA (Table 2), i.e. less additive variance than expected, would be consistent with directional selection for expression for signaling genes. However, there is no evidence in the signaling genes for a negative association between significant GCA for expression and detection of directional selection at the level of the protein. This observation is again inconsistent with the theory that linkage between nearby cis acting sites and coding sequences drive concerted evolution between protein and expression.
While depletion of additive variance could inflate the relative contribution of dominance variance, the absolute magnitude of SCA, i.e. whether or not it is different from zero, should be unaffected by lower additive variance. Thus, it would be unreasonable to expect an association between significant SCA for a gene’s expression and the detection of positive selection on the protein product of said gene; and indeed, no such association is not observed.
SCA includes epistasis as well as dominance variance (Lynch and Walsh 1998). The signaling genes are the only set of immune genes where genes significant for SCA was overrepresented relative to the genome at large. Interestingly, others have noted that the signaling genes seem to have more epistasis than recognition or effector genes (Lazzaro et al. 2004; but see Lazzaro et al. 2006). However, while mutations in interacting sites of individual proteins of the signaling cascade may be responsible for epistasis at the protein level, it is difficult to understand how protein interaction per se might translate into epistasis at the level of mRNA abundance. In other words, the fact that protein A interacts with protein B does not mean that the transcript for A interacts with the transcript for B; nor does the fact that protein A has more interactions with other proteins than protein C necessarily suggest that the transcript for A will also have more interactions than the transcript for C. Alternatively, or perhaps complementarily, signaling genes may have dominance variance because of antagonistic pleiotropy (Charlesworth and Hughes 2000): signaling pathways may well be extremely pleiotropic, as multiple recognition pathways use the same set of signaling pathways. However, as noted previously, trans acting factors are less likely to additive and more likely to have dominance variation (Lemos et al. 2008; McManus et al. 2010). Perhaps the architecture of expression of signaling genes is constructed from relatively more trans-acting factors than variation in cis-acting factors; or, perhaps genetic variation is disproportionately associated with trans- rather than cis-acting factors for these genes.
What then may we conclude about the evolution of expression of defense genes? First, expression of defense genes is clearly evolving differently from that of the genome at large. Second, both balancing or relaxed selection are consistent with the data. We are unable to explicitly test and therefore to exclude either of these hypotheses. Additional data including a detailed understanding of the genetic architecture of gene expression for defense genes, perhaps gene by gene, will be required to further refine our understanding of processes governing defense gene expression evolution.
Supplementary Material
Acknowledgments
Special thanks to L. M. Bono for the generation of the round robin simulation datasets and B. Walts for sequence alignments, as well as to B. Lazzaro for input on signaling cascades and pathogen tactics. Additional thanks to the anonymous reviewers and P. J. Wittkopp, as well as to B. M. Bolker, J. J. Bull, C. F. Baer, R. Heinemann, and T. Keller for helpful comments on the manuscript. This work was supported by NIH grant GM077618 to LMM, NIH grant GM076051 to SVN, and NIH grant GM083192 to MLW.
References
- Agresti A. Categorical Data Analysis. John H. Wiley & Sons; 2002. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. BASIC LOCAL ALIGNMENT SEARCH TOOL. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bedford T, Hartl DL. Optimization of gene expression by natural selection. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1133–1138. doi: 10.1073/pnas.0812009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Holloway AK, Stevens K, Hillier LW, Poh YP, Hahn MW, Nista PM, Jones CD, Kern AD, Dewey CN, Pachter L, Myers E, Langley CH. Population genomics: Whole-genome analysis of polymorphism and divergence in Drosophila simulans. Plos Biology. 2007;5:2534–2559. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- Burdon JJ, Thrall PH. Coevolution at multiple spatial scales: Linum marginale-Melampsora lini - from the individual to the species. Evolutionary Ecology. 2000;14:261–281. [Google Scholar]
- Carius HJ, Little TJ, Ebert D. Genetic variation in a host-parasite association: Potential for coevolution and frequency-dependent selection. Evolution. 2001;55:1136–1145. doi: 10.1111/j.0014-3820.2001.tb00633.x. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. Evidence against Fisher’s theory of dominance. Nature. 1979;278:848–849. [Google Scholar]
- Charlesworth B, Hughes KA. The maintenance of genetic variation in life-history traits. In: Singh RS, Krimbas CB, editors. Evolutionary Genetics: from Molecules to Morphology. Cambridge University Press; Cambridge, UK: 2000. pp. 369–392 . [Google Scholar]
- Charlesworth J, Eyre-Walker A. The McDonald-Kreitman test and slightly deleterious mutations. Molecular Biology and Evolution. 2008;25:1007–1015. doi: 10.1093/molbev/msn005. [DOI] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, Pollard DA, Sackton TB, Larracuente AM, Singh ND, Abad JP, Abt DN, Adryan B, Aguade M, Akashi H, Anderson WW, Aquadro CF, Ardell DH, Arguello R, Artieri CG, Barbash DA, Barker D, Barsanti P, Batterham P, Batzoglou S, Begun D, Bhutkar A, Blanco E, Bosak SA, Bradley RK, Brand AD, Brent MR, Brooks AN, Brown RH, Butlin RK, Caggese C, Calvi BR, de Carvalho AB, Caspi A, Castrezana S, Celniker SE, Chang JL, Chapple C, Chatterji S, Chinwalla A, Civetta A, Clifton SW, Comeron JM, Costello JC, Coyne JA, Daub J, David RG, Delcher AL, Delehaunty K, Do CB, Ebling H, Edwards K, Eickbush T, Evans JD, Filipski A, Findeiss S, Freyhult E, Fulton L, Fulton R, Garcia ACL, Gardiner A, Garfield DA, Garvin BE, Gibson G, Gilbert D, Gnerre S, Godfrey J, Good R, Gotea V, Gravely B, Greenberg AJ, Griffiths-Jones S, Gross S, Guigo R, Gustafson EA, Haerty W, Hahn MW, Halligan DL, Halpern AL, Halter GM, Han MV, Heger A, Hillier L, Hinrichs AS, Holmes I, Hoskins RA, Hubisz MJ, Hultmark D, Huntley MA, Jaffe DB, Jagadeeshan S, Jeck WR, Johnson J, Jones CD, Jordan WC, Karpen GH, Kataoka E, Keightley PD, Kheradpour P, Kirkness EF, Koerich LB, Kristiansen K, Kudrna D, Kulathinal RJ, Kumar S, Kwok R, Lander E, Langley CH, Lapoint R, Lazzaro BP, Lee SJ, Levesque L, Li RQ, Lin CF, Lin MF, Lindblad-Toh K, Llopart A, Long MY, Low L, Lozovsky E, Lu J, Luo MH, Machado CA, Makalowski W, Marzo M, Matsuda M, Matzkin L, McAllister B, McBride CS, McKernan B, McKernan K, Mendez-Lago M, Minx P, Mollenhauer MU, Montooth K, Mount SM, Mu X, Myers E, Negre B, Newfeld S, Nielsen R, Noor MAF, O’Grady P, Pachter L, Papaceit M, Parisi MJ, Parisi M, Parts L, Pedersen JS, Pesole G, Phillippy AM, Ponting CP, Pop M, Porcelli D, Powell JR, Prohaska S, Pruitt K, Puig M, Quesneville H, Ram KR, Rand D, Rasmussen MD, Reed LK, Reenan R, Reily A, Remington KA, Rieger TT, Ritchie MG, Robin C, Rogers YH, Rohde C, Rozas J, Rubenfield MJ, Ruiz A, Russo S, Salzberg SL, Sanchez-Gracia A, Saranga DJ, Sato H, Schaeffer SW, Schatz MC, Schlenke T, Schwartz R, Segarra C, Singh RS, Sirot L, Sirota M, Sisneros NB, Smith CD, Smith TF, Spieth J, Stage DE, Stark A, Stephan W, Strausberg RL, Strempel S, Sturgill D, Sutton G, Sutton GG, Tao W, Teichmann S, Tobari YN, Tomimura Y, Tsolas JM, Valente VLS, Venter E, Venter JC, Vicario S, Vieira FG, Vilella AJ, Villasante A, Walenz B, Wang J, Wasserman M, Watts T, Wilson D, Wilson RK, Wing RA, Wolfner MF, Wong A, Wong GKS, Wu CI, Wu G, Yamamoto D, Yang HP, Yang SP, Yorke JA, Yoshida K, Zdobnov E, Zhang PL, Zhang Y, Zimin AV, Baldwin J, Abdouelleil A, Abdulkadir J, Abebe A, Abera B, Abreu J, Acer SC, Aftuck L, Alexander A, An P, Anderson E, Anderson S, Arachi H, Azer M, Bachantsang P, Barry A, Bayul T, Berlin A, Bessette D, Bloom T, Blye J, Boguslavskiy L, Bonnet C, Boukhgalter B, Bourzgui I, Brown A, Cahill P, Channer S, Cheshatsang Y, Chuda L, Citroen M, Collymore A, Cooke P, Costello M, D’Aco K, Daza R, De Haan G, DeGray S, DeMaso C, Dhargay N, Dooley K, Dooley E, Doricent M, Dorje P, Dorjee K, Dupes A, Elong R, Falk J, Farina A, Faro S, Ferguson D, Fisher S, Foley CD, Franke A, Friedrich D, Gadbois L, Gearin G, Gearin CR, Giannoukos G, Goode T, Graham J, Grandbois E, Grewal S, Gyaltsen K, Hafez N, Hagos B, Hall J, Henson C, Hollinger A, Honan T, Huard MD, Hughes L, Hurhula B, Husby ME, Kamat A, Kanga B, Kashin S, Khazanovich D, Kisner P, Lance K, Lara M, Lee W, Lennon N, Letendre F, LeVine R, Lipovsky A, Liu XH, Liu JL, Liu ST, Lokyitsang T, Lokyitsang Y, Lubonja R, Lui A, MacDonald P, Magnisalis V, Maru K, Matthews C, McCusker W, McDonough S, Mehta T, Meldrim J, Meneus L, Mihai O, Mihalev A, Mihova T, Mittelman R, Mlenga V, Montmayeur A, Mulrain L, Navidi A, Naylor J, Negash T, Nguyen T, Nguyen N, Nicol R, Norbu C, Norbu N, Novod N, O’Neill B, Osman S, Markiewicz E, Oyono OL, Patti C, Phunkhang P, Pierre F, Priest M, Raghuraman S, Rege F, Reyes R, Rise C, Rogov P, Ross K, Ryan E, Settipalli S, Shea T, Sherpa N, Shi L, Shih D, Sparrow T, Spaulding J, Stalker J, Stange-Thomann N, Stavropoulos S, Stone C, Strader C, Tesfaye S, Thomson T, Thoulutsang Y, Thoulutsang D, Topham K, Topping I, Tsamla T, Vassiliev H, Vo A, Wangchuk T, Wangdi T, Weiand M, Wilkinson J, Wilson A, Yadav S, Young G, Yu Q, Zembek L, Zhong D, Zimmer A, Zwirko Z, Alvarez P, Brockman W, Butler J, Chin C, Grabherr M, Kleber M, Mauceli E, MacCallum I I. Drosophila 12 Genomes Consor; Broad Inst Genome Sequencing; Broad, and A. Whole Genome. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Denver DR, Morris K, Streelman JT, Kim SK, Lynch M, Thomas WK. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nature Genet. 2005;37:544–548. doi: 10.1038/ng1554. [DOI] [PubMed] [Google Scholar]
- Dybdahl MF, Lively CM. Host-parasite coevolution: Evidence for rare advantage and time-lagged selection in a natural population. Evolution. 1998;52:1057–1066. doi: 10.1111/j.1558-5646.1998.tb01833.x. [DOI] [PubMed] [Google Scholar]
- Dybdahl MF, Storfer A. Parasite local adaptation: Red Queen versus Suicide King. Trends in Ecology & Evolution. 2003;18:523–530. [Google Scholar]
- Edgington E. Randomization Tests. Marcel Dekker; New York City: 1995. [Google Scholar]
- Finlay BB, McFadden G. Anti-immunology: Evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124:767–782. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The possible modification of the response of the wild type to recurrent mutations. Am Nat. 1928;62:115–126. [Google Scholar]
- Frank SA, Schmid-Hempel P. Mechanisms of pathogenesis and the evolution of parasite virulence. Journal of Evolutionary Biology. 2008;21:396–404. doi: 10.1111/j.1420-9101.2007.01480.x. [DOI] [PubMed] [Google Scholar]
- Fritz Robert S, Simms Ellen Louise. Plant resistance to herbivores and pathogens: ecology, evolution, and genetics. Chicago: University of Chicago Press; 1992. [Google Scholar]
- Gilad Y, Oshlack A, Rifkin SA. Natural selection on gene expression. Trends Genet. 2006;22:456–461. doi: 10.1016/j.tig.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Kacser H, Burns JA. The molecular basis of dominance. Genetics. 1981;97:639–666. doi: 10.1093/genetics/97.3-4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Weiss G, Lachmann M, Hellmann I, Enard W, Muetzel B, Wirkner U, Ansorge W, Paabo S. A neutral model of transcriptome evolution. Plos Biology. 2004;2:682–689. doi: 10.1371/journal.pbio.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Wittkopp PJ, Taubes CH, Ranz JM, Clark AG, Hartl DL. Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics. 2005;171:1813–1822. doi: 10.1534/genetics.105.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro BP. Natural selection on the Drosophila antimicrobial immune system. Current Opinion in Microbiology. 2008;11:284–289. doi: 10.1016/j.mib.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro BP, Little TJ. Immunity in a variable world. Philosophical Transactions of the Royal Society B-Biological Sciences. 2009;364:15–26. doi: 10.1098/rstb.2008.0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro BP, Sackton TB, Clark AG. Genetic variation in Drosophila melanogaster resistance to infection: A comparison across bacteria. Genetics. 2006;174:1539–1554. doi: 10.1534/genetics.105.054593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro BP, Sceurman BK, Clark AG. Genetic basis of natural variation in D-melanogaster antibacterial immunity. Science. 2004;303:1873–1876. doi: 10.1126/science.1092447. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annual Review Of Immunology. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Lemos B, Araripe LO, Fontanillas P, Hartl DL. Dominance and the evolutionary accumulation of cis- and trans-effects on gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14471–14476. doi: 10.1073/pnas.0805160105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Bettencourt BR, Meiklejohn CD, Hartl DL. Evolution of proteins and gene expression levels are coupled in Drosophila and are independently associated with mRNA abundance, protein length, and number of protein-protein interactions. Molecular Biology and Evolution. 2005;22:1345–1354. doi: 10.1093/molbev/msi122. [DOI] [PubMed] [Google Scholar]
- Lynch M, Walsh J. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Inc; Sunderland, MA: 1998. [Google Scholar]
- Maizels RM. Parasite immunomodulation and polymorphisms of the immune system. Journal of Biology. 2009;8:62. doi: 10.1186/jbiol166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- McIntyre LM, Bono LM, Genissel A, Westerman R, Junk D, Telonis-Scott M, Harshman L, Wayne ML, Kopp A, Nuzhdin SV. Sex-specific expression of alternative transcripts in Drosophila. Genome Biology. 2006;7 doi: 10.1186/gb-2006-7-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, Coolon JD, Duff MO, Eipper-Mains J, Graveley BR, Wittkopp PJ. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Research. 2010;20:816–825. doi: 10.1101/gr.102491.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin SV, Keightley PD, Pasyukova EG, Morozova EA. Mapping quantitative trait loci affecting sternopleural bristle number in Drosophila melanogaster using changes of marker allele frequencies in divergently selected lines. Genet Res. 1999;72:79–91. doi: 10.1017/s001667239800336x. [DOI] [PubMed] [Google Scholar]
- Nuzhdin SV, Wayne ML, Harmon KL, McIntyre LM. Common pattern of evolution of gene expression level and protein sequence in Drosophila. Molecular Biology and Evolution. 2004;21:1308–1317. doi: 10.1093/molbev/msh128. [DOI] [PubMed] [Google Scholar]
- Obbard DJ, Welch JJ, Kim KW, Jiggins FM. Quantifying Adaptive Evolution in the Drosophila Immune System. Plos Genetics. 2009:5. doi: 10.1371/journal.pgen.1000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. A Test of Fisher’s Theory of Dominance. Proc Natl Acad Sci U S A. 1991;88:11413–11415. doi: 10.1073/pnas.88.24.11413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch J, Zhang Z, Baines JF. The Influence of Demography and Weak Selection on the McDonald-Kreitman Test: An Empirical Study in Drosophila. Molecular Biology and Evolution. 2009;26:691–698. doi: 10.1093/molbev/msn297. [DOI] [PubMed] [Google Scholar]
- Rand DM, Kann A. Polymorphism in Mitochondrial DNA: Contrasts Among Genes from Drosophila, Mice, and Humans. Molecular Biology and Evolution. 1996;13:735–748. doi: 10.1093/oxfordjournals.molbev.a025634. [DOI] [PubMed] [Google Scholar]
- Rifkin SA, Houle D, Kim J, White KP. A mutation accumulation assay reveals a broad capacity for rapid evolution of gene expression. Nature. 2005;438:220–223. doi: 10.1038/nature04114. [DOI] [PubMed] [Google Scholar]
- Rifkin SA, Kim J, White KP. Evolution of gene expression in the Drosophila melanogaster subgroup. Nature Genet. 2003;33:138–144. doi: 10.1038/ng1086. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Sackton TB, Lazzaro BP, Schlenke TA, Evans JD, Hultmark D, Clark AG. Dynamic evolution of the innate immune system in Drosophila. Nature Genetics. 2007;39:1461–1468. doi: 10.1038/ng.2007.60. [DOI] [PubMed] [Google Scholar]
- Stoletzki N, EyreWalker A. Estimation of the Neutrality Index. Molecular Biology and Evolution. 2010 doi: 10.1093/molbev/msq249.. first published online September 13, 2010. [DOI] [PubMed] [Google Scholar]
- Thompson JN. The Geographic Mosaic of Coevolution. University of Chicago Press; Chicago: 2005. [Google Scholar]
- Vallet-Gely I, Lemaitre B, Boccard F. Bacterial strategies to overcome insect defenses. Nature Reviews Microbiology. 2008;6:302–313. doi: 10.1038/nrmicro1870. [DOI] [PubMed] [Google Scholar]
- Van Valen L. A new evolutionary law. Evolutionary Theory. 1973;1:1–30. [Google Scholar]
- Van Valen L. Red Queen lives. Nature. 1976;260:575–575. [Google Scholar]
- Van Valen L. Red Queen. American Naturalist. 1977;111:809–810. [Google Scholar]
- Verhoeven KJF, Simonsen KL, McIntyre LM. Implementing false discovery rate control: increasing your power. Oikos. 2005;108:643–647. [Google Scholar]
- Wayne ML, Pan YJ, Nuzhdin SV, McIntyre LM. Additivity and transacting effects on gene expression in male Drosophila simulans. Genetics. 2004;168:1413–1420. doi: 10.1534/genetics.104.030973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne ML, Telonis-Scott M, Bono LM, Harshman L, Kopp A, Nuzhdin SV, McIntyre LM. Simpler mode of inheritance of transcriptional variation in male Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:18577–18582. doi: 10.1073/pnas.0705441104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A, Crawford DL. Neutral and adaptive variation in gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:5425–5430. doi: 10.1073/pnas.0507648103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Physiological and evolutionary theories of dominance. Am Nat. 1934;68:24–53. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.