Abstract
Rcl1 is an essential nucleolar protein required for U3 snoRNA-guided pre-rRNA processing at sites flanking the 18S rRNA sequence. A potential catalytic role for Rcl1 during pre-rRNA cleavage has been suggested based on its primary structure similarity to RNA 3′-terminal phosphate cyclase (Rtc) enzymes, which perform nucleotidyl transfer and phosphoryl transfer reactions at RNA ends. Here, we report the 2.6 Å crystal structure of a biologically active yeast Rcl1, which illuminates its modular 4-domain architecture and overall homology with RNA cyclases while revealing numerous local differences that account for why Rtcs possess metal-dependent adenylyltransferase activity and Rcls do not. A conserved oxyanion-binding site in Rcl1 was highlighted for possible catalytic or RNA-binding functions. However, the benign effects of mutations in and around the anion site on Rcl1 activity in vivo militate against such a role.
Keywords: rRNA processing, RNA cyclase
INTRODUCTION
The RNA components of the eukaryal ribosome are formed by an elaborate series of processing steps, whereby a primary transcript containing the 18S, 5.8S, and 25/28S rRNA sequences and external and internal spacers undergoes multiple nucleolytic cleavages to generate the mature rRNA ends (for review, see Henras et al. 2008). Most of the processing steps occur in the nucleus in the context of a preribosomal particle composed of intrinsic ribosomal proteins and a multitude of extrinsic ribosome maturation factors. The nuclear pre-rRNA cleavages flanking the 18S rRNA are guided by base pairing of the 5′ external spacer to the U3 small nuclear (sno) RNA (Beltrame and Tollervey 1995). U3 snoRNA has been isolated from budding yeast as a large ribonucleoprotein (RNP) complex—with dozens of associated proteins—called the small subunit (SSU) processosome (Dragon et al. 2002).
Although the immediate catalyst of the U3-guided endonucleolytic cleavages is not known, speculation has focused on the Rcl1 protein, which is essential for viability in budding yeast, localized in the nucleolus, loosely associated with U3 snoRNP, and required for pre-rRNA processing at the A0, A1, and A2 sites flanking the 18S rRNA sequence (Billy et al. 2000). Saccharomyces cerevisiae Rcl1 (a 367-aa polypeptide) is stably associated with its binding partner Bms1 (a 1183-aa polypeptide). Bms1, too, is essential, nucleolar, and required for pre-rRNA processing at the A0, A1, and A2 sites (Gelperin et al. 2001; Wegierski et al. 2001). Bms1, per se, binds U3 snoRNA in vitro; it also binds directly to Rcl1 and mediates the association of Rcl1 with pre-ribosomes (Karbstein et al. 2005). Bsm1 is a GTPase and the formation of a U3-Bms1–Rcl1 complex is thought to be regulated by guanine nucleotide binding (Karbstein and Doudna 2006).
The notion that Rcl1 might be acting catalytically during 18S rRNA processing stems from its primary structure similarity to the RNA 3′-terminal phosphate cyclase (Rtc) enzymes that catalyze de novo conversion of RNA 3′-monophosphate ends to 2′,3′-cyclic phosphates (Genschik et al. 1997; Billy et al. 2000). Cyclization entails a series of three nucleotidyl transfer reactions (Filipowicz et al. 1985; Reinberg et al. 1985). In the first step, Rtc reacts with ATP and a divalent cation to form a covalent Rtc-AMP intermediate and liberate PPi. Adenylate is linked via a phosphoamide bond to a histidine Nε atom (Billy et al. 1999; Tanaka et al. 2010). In the second step, the adenylate is transferred from Rtc-AMP to the RNA 3′-phosphate terminus to form an activated phosphoanhydride intermediate, RNA(3′)pp(5′)A. In the third step, the terminal ribose 2′-OH attacks the 3′-phosphate of RNA(3′)pp(5′)A to generate an RNA 2′,3′-cyclic phosphate product and release AMP. Yeast Rcl1 was surveyed thoroughly for cyclase activity and found to have none, prompting the suggestion that it might instead mediate a different type of phosphoryl transfer reaction: hydrolysis of an RNA phosphodiester (Billy et al. 2000).
Rtc enzymes are distributed widely among bacterial, archaeal, and eukaryal taxa (Genschik et al. 1997), though their biological functions and physiological substrates are uncharted. (It is noteworthy that budding yeast have no active RNA cyclase in their proteomes.) In contrast, the Rcl1 (“RNA cyclase-like”) clade is specific to eukarya. The structures and mechanisms of phosphoryl transfer by bacterial and human Rtc enzymes have been illuminated by X-ray crystallography and structure-guided mutagenesis (Billy et al. 1999; Palm et al. 2000; Tanaka and Shuman 2009; Tanaka et al. 2010). However, no structural information is available for a eukaryal Rcl1 protein.
Here, we report the 2.6 Å crystal structure of Kluyveromyces lactis Rcl1, a bona fide ortholog of S. cerevisiae Rcl1.
RESULTS AND DISCUSSION
Kluyveromyces lactis Rcl1
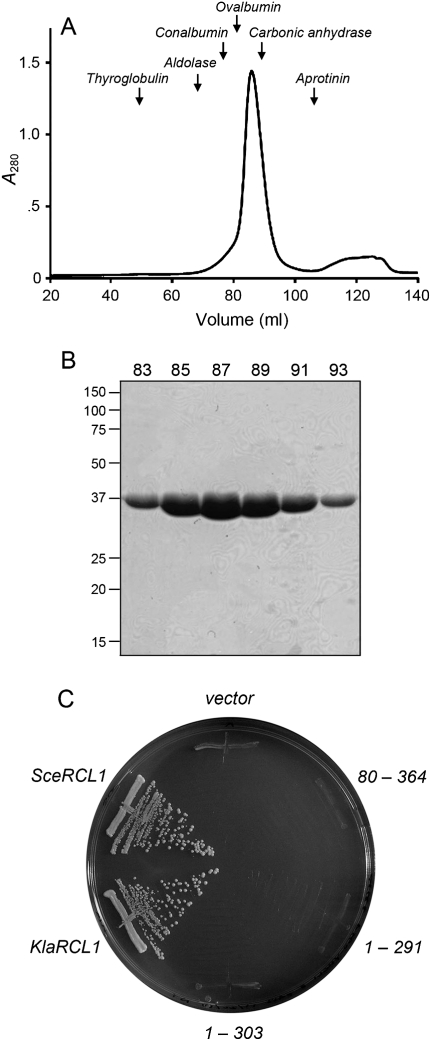
An alignment of the amino acid sequences of human Rcl1 and four fungal homologs highlights 155 positions of side-chain identity/similarity distributed across the entire length (363–372 aa) of the Rcl1 polypeptides (Fig. 1A). To cast the net broadly for crystallization candidates, we produced in bacteria His10- or His10-Smt3-tagged versions of human, Schizosacccharomyces pombe, S. cerevisiae, and K. lactis Rcl1. Human Rcl1 was intractably insoluble; S. pombe Rcl1 was poorly soluble. In contrast, K. lactis and S. cerevisiae Rcl1 were soluble and could be isolated from bacterial extracts by Ni-agarose chromatography in high yield: 30 and 40 mg per liter of bacterial culture, respectively. K. lactis Rcl1 behaved as a single component during gel-filtration chromatography (Fig. 2A,B); its elution profile relative to those of marker proteins of known size was consistent with Rcl1 being a monomer in solution (Fig. 2A).
FIGURE 1.
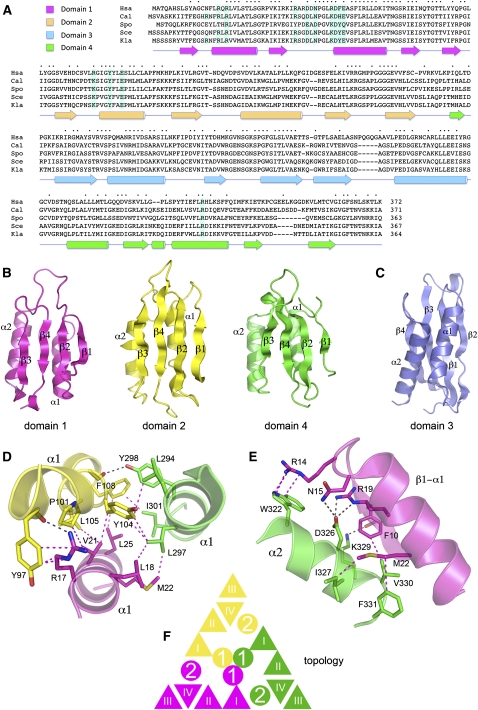
Rcl1 structure and domain organization. (A) The Rcl1 clade. The amino acid sequences of the Rcl1 proteins of Homo sapiens (Hsa), Candida albicans (Cal), Schizosaccharomyces pombe (Spo), Saccharomyces cerevisiae (Sce), and Kluyveromyces lactis (Kla) are aligned. Positions of amino acid side-chain identity/similarity are denoted by • above the sequences. Gaps in the alignment are indicated by –. The secondary structure elements of the KlaRcl1 fold (strands depicted as arrows and helices as cylinders) are shown below the sequence and are colored according to domain modules as specified at top, left (domain 1 in magenta, domain 2 in yellow, domain 3 in blue, and domain 4 in green). The conserved KlaRcl1 residues that were subjected to mutagenesis are highlighted in shaded boxes. (B) Rcl1 domain modules 1, 2, and 4 have a shared fold composed of a four-stranded β sheet plus two α helices arrayed with the same topology. (C) Rcl1 domain 3 has a distinctive folding topology. The secondary structure elements are labeled. (D) A three-helix bundle of the α1 helices of domains 1, 2, and 4 comprises the inner layer of the tri-radial domain architecture. Amino acid side chains and main-chain atoms engaged in cross-domain contacts are rendered as stick models. Hydrogen bonds are indicated by black dashed lines; van der Waals contacts are denoted by magenta dashed lines. (E) The α2 helices of domains 1, 2, and 4 comprise the middle tri-radial layer. The figure depicts the cross-domain contacts of the domain 4 α2 helix with the adjacent β1–α1 segment of domain 1. (F) Rcl1 domains 1, 2, and 4 are arrayed radially in three layers. The figure shows a cartoon diagram of the domain topologies. β strands are represented as triangles and α helices as circles.
FIGURE 2.
Physical and genetic characterization of KlaRcl1. (A) Gel filtration. SeMet-KlaRcl1 (17.5 mg in 3.5 mL) was analyzed by Sephadex 200 gel filtration. The protein elution profile was monitored continuously by UV absorbance. A280 is plotted as a function of elution volume. The column was calibrated by tracking the elution profiles of marker proteins of known native size. The peak positions of the markers thyglobulin (669 kDa), aldolase (158 kDa), conalbumin (75 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa), and aprotinin (6.5 kDa) are indicated by arrows. KlaRcl1 (a 39-kDa polypeptide) chromatographed as a single component eluting between ovalbumin and carbonic anhydrase. (B) Aliquots (5 μL) of the indicated Sephadex 200 column fractions were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The positions and sizes (kDa) of marker proteins are indicated on the left. (C) Complementation of S. cerevisiae rcl1Δ. Plasmid shuffle was performed with CEN HIS3 plasmids bearing wild-type SceRCL1, wild-type KlaRCL1, or the indicated KlaRCL1 deletion mutants. The empty CEN HIS3 plasmid vector served as a negative control. Single His+ transformants were streaked on agar medium containing 0.75 mg/mL of FOA; the plate was photographed after incubation for 3 d at 30°C.
To query whether K. lactis Rcl1 is functional in vivo, we established a plasmid shuffle assay for complementation of a lethal rcl1Δ mutation in S. cerevisiae (Fig. 2C). A haploid S. cerevisiae strain in which the chromosomal RCL1 gene is disrupted depends for its growth on a wild-type S. cerevisiae RCL1 gene (SceRCL1) carried on a CEN URA3 plasmid. This rcl1Δ strain was unable to grow on agar medium containing FOA (5-fluoroorotic acid), which selects against retention of the URA3 SceRCL1 plasmid. Transformation of rcl1Δ cells with a CEN HIS3 plasmid containing another copy of SceRCL1 allowed them to form FOA-resistant colonies, whereas transformation with an empty CEN vector did not (Fig. 2C). We cloned the K. lactis RCL1 gene (KlaRCL1) into a CEN HIS3 plasmid under the control of the S. cerevisiae TPI1 promoter. The salient finding was that transformation with the KlaRCL1 plasmid permitted growth of rcl1Δ cells on FOA (Fig. 2C). Individual FOA-resistant KlaRCL1 strains were tested for growth on rich medium (YPD agar) at 18°C, 25°C, 30°C, and 37°C in parallel with isogenic SceRCL1 strains; both grew equally well, as gauged by colony size (Supplemental Fig. S3; data not shown). Thus, we conclude that KlaRcl1 is a functional ortholog of SceRcl1.
Crystallization of KlaRcl1
Crystallization trials were conducted in parallel with SceRcl1 and KlaRcl1 and the latter emerged as the winner. Crystals of KlaRcl1 were grown by vapor diffusion against a precipitant solution containing PEG-3350 and tartrate. The crystals belonged to orthorhombic space group P212121 and contained four protomers in the asymmetric unit. SAD phases from crystals of SeMet-substituted KlaRcl1 were exploited to derive an initial model, which was then refined using the diffraction data from native crystals. The refined model at 2.6 Å resolution (R = 20.4%; Rfree = 25.9%) contained amino acids 5–356 of protomers A and D, 5–358 of protomer B, and 6–356 of protomer C (Supplemental Table S1). Our discussion of the KlaRcl1 structure will focus on protomer A.
Overview of the Rcl1 structure
The tertiary structure of KlaRcl1 is shown in Figure 3A; the secondary structure elements are displayed below the amino acid sequence in Figure 1A. Rcl1 consists of four domain modules: domain 1 (colored magenta), domain 2 (yellow), domain 3 (blue), and domain 4 (green). Domains 1, 2, and 4 are homologous to one other (Fig. 1B) and arranged in a radial pattern with pseudo-threefold symmetry (Figs. 1F, 3A). Domains 1, 2, and 4 are composed of a four-stranded β sheet with topology β1↑2↑4↓3↑ (Fig. 1B). Two parallel α helices, connecting β1 and β2 and β2 and β3, respectively, pack against the back face of the β sheet (Fig. 1B). Pairwise superpositions of these three domains in DALI (Holm et al. 2008) highlighted triplication of a core fold, with modest conservation of primary structure. To wit: (1) domains 1 and 2 superimposed with 2.8 Å RMSD at 65 Cα positions, of which 18 are identical or similar side chains; (2) domains 2 and 4 superimposed with 2.9 Å RMSD at 66 Cα positions (19 identical or similar side chains); (3) domains 1 and 4 superimposed with 2.9 Å RMSD at 71 Cα positions (16 identical or similar side chains).
FIGURE 3.
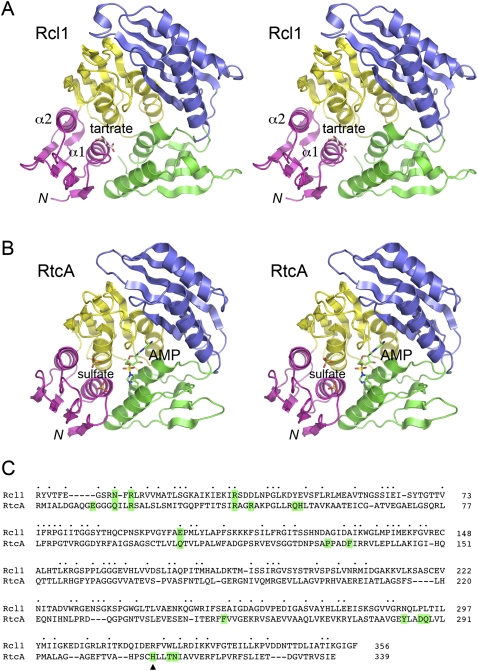
Structure of Rcl1 and comparison to RtcA. (A) Stereo view of the A protomer of KlaRcl1. The four component domains are colored as follows: N-terminal domain 1 in magenta, domain 2 in yellow, domain 3 in blue, and domain 4 in green. A tartrate anion bound to domain 1 is depicted as a stick model. (B) Stereo view of the A protomer of E. coli RtcA-AMP (PDB ID 3KGD) superimposed on KlaRcl1 and offset vertically. The AMP bound covalently to His309 is depicted as a stick model with green carbons. Two sulfate anions bound to domain 1 are depicted as stick models. (C) The folds of Rcl1 and RtcA-AMP were compared in DALI; the resulting primary structure alignment is shown. Positions of amino acid side-chain identity/similarity are denoted by • above the sequences. Gaps in the alignment are indicated by –. The His309 site of covalent AMP attachment to RtcA is indicated by ▴. The RtcA residues that interact with AMP or the sulfate anions or are imputed to bind the metal cofactor are highlighted in green boxes (as are any conserved equivalents in Rcl1).
The domains are arrayed radially in three layers (Figs. 1F, 3A). The innermost layer is composed of the α1 helices arranged as a parallel 3-helix bundle (Figs. 1D, 3A) stabilized by: (1) a network of interdomain van der Waals contacts among aliphatic and aromatic side chains; (2) side-chain to main-chain carbonyl hydrogen-bond donation (by Arg17 and Tyr298); and (3) an Arg17–Tyr97 π-cation interaction (Fig. 1D). The middle layer consists of the α2 helices; the β sheets comprise the surface layer (Figs. 1F, 3A). Each α2 helix makes a network of cross-domain contacts with the β1–α1 elements of the adjacent domain that stabilizes the radial architecture. These are illustrated in Figure 1E for the domain 1–domain 4 interface and include: (1) van der Waals contacts between hydrophobic side chains; (2) H-bond donation from Lys329 to a main-chain carbonyl; (3) H-bond donation from Asn15 and Arg19 to Asp326; and (4) an Arg14–Trp322 π-cation stack.
Rcl1 domain 3 is a distinctive fold inserted en bloc between the β1 strand and α1 helix of domain 4 (Fig. 1A). Domain 3 consists of a four-stranded β sheet plus two α helices (Fig. 1C), but the folding topology of the domain 3 sheet (β2↑1↑3↓4↑) is completely different from those in domains 1, 2, and 4 (Fig. 1B,C). When the sheets are co-oriented so that the directionality of the four strands are the same, it becomes apparent that the two helices of the domain threefold are located on the anterior face of the sheet (Fig. 1C) rather than on the posterior face as in domains 1, 2, and 4 (Fig. 1B). Domain 3 forms a “lid” over domain 2 via multiple interdomain contacts between residues in domains 3 and 2 and domains 3 and 4 (Supplemental Fig. S1). These include salt bridges at the base (Glu280–Arg289) and tip (Lys154–Glu232) of the lid and an orthogonal Trp229–Arg155–Tyr195 π-cation stack (Supplemental Fig. S1).
Inspection of the Rcl1 crystal lattice reveals a modest interface between protomers. Within the asymmetric unit, protomers B and D are related by a 180° rotation that aligns their respective domain 4 β3 strands to form a continuous cross-protomer 8-stranded β sheet. Protomer A forms the same continuous β sheet via alignment of its domain 4 with that of protomer C in the adjacent asymmetric unit (Supplemental Fig. S2). The A–C and B–D protomer interfaces bury ∼450 Å2 of surface area/protomer, with an average shape complementarity of 0.55 (Lee and Richards 1971). Interactions between bona fide protein oligomers typically bury >900 Å2 of surface area/protomer with shape complementarity values of >0.70 (Lawrence and Colman 1993). Thus, we surmise that the observed crystal packing interactions are not indicative of a physiological dimer interface, consistent with our observation that KlaRcl1 behaves as a monomer in solution, as gauged by gel filtration.
Comparison of Rcl1 and RtcA
The tertiary structure of Rcl1 is homologous to that of E. coli RNA 3′ cyclase RtcA (Fig. 3B). Rcl1 and RtcA superimpose in DALI with a RMSD of 2.5 Å at 328 Cα positions (Z score 34.0). A structure-based alignment of their amino acid sequences highlights 104 positions of side-chain identity or similarity (Fig. 3C). Whereas the shared four-domain architecture is apparently the defining feature of the Rtc/Rcl superfamily, the new structure of KlaRcl1 explains why the Rcl1 proteins have no detectable RNA cyclase or auto-adenylylation activity (Billy et al. 2000).
First, Rcl1 lacks the His309 nucleophile of RtcA (denoted by the arrowhead in Fig. 3C) that forms the covalent bond to the AMP phosphate (Billy et al. 1999; Tanaka et al. 2010). Instead, Rcl1 has an arginine (Arg319) at the equivalent position in domain 4 (Figs. 3C, 4A). Although a protein arginine side chain might, in principle, serve as a nucleophile for covalent phosphoryl or nucleotidyl transfer (Besant et al. 2009), we are not aware of any enzymatic nucleotidyl transfer reaction to which such a mechanism naturally applies. Arginine is an unlikely nucleophile given that its predicted pKa of 12.3 (Pace et al. 2009) strongly disfavors the deprotonated state required for attack on a phosphoester or phosphoanhydride substrate. Polynucleotide ligases and RNA-capping enzymes characteristically use lysines as nucleophiles for covalent NMP transfer from NTPs to RNA or DNA ends (Shuman and Lima 2004). However, RNA cyclases are stringently adapted to histidine-based catalysis, insofar as replacing the active-site histidine of human RNA 3′-phosphate cyclase with lysine-abolished RNA cyclization and Rtc auto-adenylylation (Tanaka and Shuman 2009).
FIGURE 4.
Anion binding site of Rcl1. (A) Stereo view of the tartrate complex in the A promoter of Rcl1. The folds of the four domains are colored as in Figures 1 and 3. Selected amino acid side chains and main-chain atoms and the tartrate anion bound to domain 1 are rendered as stick models. Hydrogen bonds are indicated by dashed lines. (B) Surface model of KlaRcl1 highlighting a deep cavity wherein the tartrate anion (stick model) sits at the left margin, surrounded by surface protein nitrogen atoms (blue).
Second, Rcl1 proteins lack the conserved glutamate found in the domain β1–α1 loop of the Rtc enzymes (Tanaka et al. 2010). This acidic residue (corresponding to Glu14 in E. coli and human Rtc) is essential for cyclization and is proposed to coordinate the divalent cation cofactor for the adenylyltransferase steps (Tanaka and Shuman 2009).
The Rcl1/RtcA structure superposition also informs us that the constituents of domains 2, 3, and 4 of RtcA that line the active site and contact the adenosine moiety of AMP are not conserved in Rcl1. These include RtcA side chains Asp287, Gln288, and Asn313 that coordinate the ribose hydroxyls (these correspond to Pro293, Leu294, and Leu323 in Rcl1, respectively) and Rcl1 residues Pro131, Phe135, Phe251, and Tyr284 that cage the adenine base (these correspond to Ala127, Ala131, Glu257, and Asn290 in Rcl1, respectively) (Fig. 3C). In sum, these distinctive features of the Rcl1 versus Rtc clades signify that Rcl1 proteins are unlikely to bind adenine nucleotides (at least not at the site exploited by cyclases) and, for the several reasons cited above, unable catalyze a covalent nucleotidyl transfer reaction.
An oxyanion-binding site in Rcl1
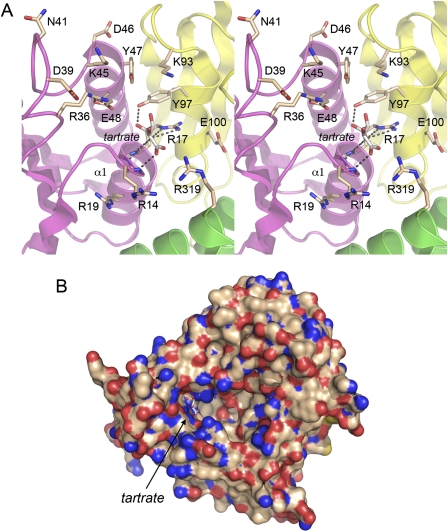
The Rcl1 crystal structure reveals a tartrate anion overlying the proximal end of the α1 helix of domain 1 (Fig. 3A). It is remarkable that a sulfate anion occupies the analogous site over the α1 helix in the crystal structure of RtcA-adenylate (Fig. 3B). A citrate anion is present at this site in the RtcA apoenzyme structure (Palm et al. 2000). In the case of RtcA, it is proposed that the sulfate anion bound to α1 mimics the terminal 3′-phosphate of the RNA substrate for cyclization (Tanaka et al. 2010). [A second neighboring sulfate anion (Fig. 3B) is thought to mimic the penultimate internucleotide phosphodiester of the RNA substrate.] The putative “3′-terminal” sulfate anion in RNA cyclase is coordinated by two arginines that are essential for cyclization activity and for the formation of a stable binary complex with 3′-phosphate terminated RNA (Tanaka and Shuman 2009; Tanaka et al. 2010). In light of these findings and the speculation that Rcl1 might be the RNA endonuclease responsible for 18S rRNA processing, we were attracted to the idea that the Rcl1 anion-binding site demarcated by tartrate could represent: (1) an endonuclease active site; or (2) an RNA-binding site.
The tartrate anion in Rcl1 occupies the lateral edge of a deep pocket in the protein surface, where it is surrounded by multiple protein nitrogen atoms (Fig. 4B). The fine anatomy of the Rcl1 anion-binding site is depicted in Figure 4A. The distal β1–α1 loop and the proximal segment of the α1 helix form a P-loop-like oxyanion hole, wherein the tartrate oxygens receive hydrogen bonds from the Asn15, Phe16, and Arg17 main-chain amide nitrogens, the Tyr97 side-chain hydroxyl, and two Arg17 side-chain guanidinium nitrogens (Fig. 4A). Other hydrogen bond donors in the vicinity of the tartrate oxygens (4.0–5.7 Å away) are Arg14, Arg36, Tyr47, Lys93, and Arg319 (Fig. 4A). Arg17 and Arg36 are conserved in RtcA (Fig. 3C).
Rcl1 mutagenesis
If the anion site in Rcl1 is functionally relevant, then we would expect mutations in its amino acid constituents to impact Rcl1 activity in vivo. Casting the net widely, we introduced single and clustered alanine substitutions at the 14 KlaRcl1 side chains depicted in Figure 4A (and highlighted in shaded boxes in Fig. 1A). The mutated KlaRCL1 alleles on CEN HIS3 plasmids were tested by plasmid shuffle for rcl1Δ complementation, and all were found to support colony formation during FOA selection for loss of the URA3 SceRCL1 plasmid (data not shown). Individual FOA-resistant rcl1Δ KlaRCL1-Ala isolates were then tested for growth on YPD agar at 18°C, 25°C, 30°C, and 37°C. All of the KlaRCL1-Ala strains grew as well as the isogenic wild-type strain at all temperatures tested (Supplemental Fig. S3; data not shown). Even the compound mutations R14A-R17A-R19A, K45A-Y47A-E48A, and K93A-Y97A had no apparent effect on yeast growth. We conclude that the crystallographically defined oxyanion-binding site and several surrounding side chains from domains 1, 2, and 4 are not crucial for Rcl1 activity in vivo.
Taking a more radical approach to mutagenesis, we queried the effects of deleting the N- and C-terminal domains of KlaRcl1, using the structure to guide the choice of deletion boundaries. We found that KlaRCL1-(80-364) fails to complement rcl1Δ (Fig. 2C), signifying that domain 1 cannot be deleted. KlaRCL1-(1-303) and KlaRCL1-(1-291) also failed to complement rcl1Δ (Fig. 2C). We infer that the radial domain architecture is necessary for Rcl1 activity.
CONCLUSIONS AND IMPLICATIONS
The first structure of a biologically active eukaryal Rcl protein highlights its overall homology with Rtc RNA cyclases (as anticipated), while underscoring numerous local differences that account for why Rtcs possess metal-dependent adenylyltransferase activity and Rcls do not. The identification of an oxyanion-binding site within domain 1 of Rcl1 raised the prospects of possible catalytic and RNA-binding functions of this site during Rcl1-dependent cleavage of preribosomal RNA. However, the benign effects of multiple mutations in and around the anion site argue against such a role. We infer that if it is the case that Rcl1 is an endonuclease, then none of the mutated residues are essential constituents of the active site. Similarly, if Rcl1 binds the pre-RNA substrate, then the tartrate-binding side chains and their neighbors are apparently not required to engage the RNA effectively enough to support yeast growth. In light of these results, it is worth re-evaluating the supposition that Rcl1 is the 18S rRNA-processing endoribonuclease. The attractiveness of this idea stems from Rcl1's link to cyclases, but that link is now attenuated with respect to catalysis.
MATERIALS AND METHODS
Rtc1 crystallization and structure determination
Crystals of KlaRcl1 were grown by the hanging-drop vapor-diffusion method at room temperature. Rcl1 (8 mg/mL in 10 mM Tris-HCl at pH 7.4, 100 mM NaCl, 2 mM DTT, 1 mM EDTA, 5% glycerol; purified as described in the Supplemental Methods) was mixed with an equal volume (2 μL) of the reservoir buffer containing 0.1 M disodium tartrate and 12.5% (w/v) PEG-3350. Crystals grew to their full size (100 × 50 × 10 μm) overnight. Single crystals were harvested, cryoprotected by sequential transfer to 0.1 M disodium tartrate 15% PEG-3350 solutions containing 10% and 20% (v/v) glycerol, and then flash-frozen in liquid nitrogen. X-ray diffraction data were collected at the National Synchrotron Light Source beamline X25, equipped with a ADXV-Q315 CCD detector. The KlaRcl1 crystals belonged to spacegroup P212121 with unit cell dimensions consistent with four to six protomers per asymmetric unit, assuming a solvent content of 40%–60% and a protein partial-specific volume of 1.23 Å3/Da. Diffraction data were collected for two crystals of native Rcl1 (grown from the same crystallization drop and cryoprotected under identical conditions); each data set comprised 185 sequential 1° oscillations. Diffraction data were collected for two crystals of SeMet-substituted Rcl1 (also from a single crystallization drop and cryoprotected in parallel); each SeMet data set comprised 365 sequential 1° oscillations. The single crystal diffraction data were processed in HKL2000 and then merged to yield the native and SeMet data sets described in Supplemental Table S1.
SeMet phases were obtained via single-wavelength anomalous diffraction (SAD) techniques as implemented in PHENIX (Adams et al. 2002). The combined two-crystal SeMet data set was analyzed in PHENIX HYSS using the automatically chosen resolution cutoff of 4.3 Å, which identified 40 anomalous scattering sites with fourfold NCS. Initial experimental phases from PHASER were improved in RESOLVE with the aid of fourfold NCS averaging. Iterative automated model building and refinement against the SeMet data were carried out using PHENIX AutoBuild. This initial model contained 1190 amino acids in four Rcl1 protomers. Following refinement and automated rebuilding of this model against the higher resolution native data, the model was extended and rebuilt by hand in COOT (Emsley and Cowtan 2004), during which NCS restraints were removed from protein segments where electron density varied from protomer to protomer, and solvent molecules were added. Continuous electron density allowed modeling of residues 6–356 in each Rcl1 protomer. Protomer B contains an additional two residues at its C terminus and protomers A, C, and D have one additional residue at their N-termini. Electron density consistent with a tartrate anion was located near the N terminus of helix 1 of domain 1 in each protomer. Additional tartrate, glycerol, and PEG molecules were modeled in accordance with difference Fourier maps and neighboring atomic features. The final Rcl1 model refined to R/Rfree values of 0.204/0.259 with no large difference Fourier peaks and excellent geometry apart from a single glycine Ramachandran outlier. Statistics for experimental phasing, model refinement, and model quality are found in Supplemental Table S1.
Plasmids for expression of SceRcl1 and KlaRcl1 in yeast
A 1.1-kbp DNA fragment containing the S. cerevisiae RCL1 (SceRCL1) ORF plus 300 bp of upstream (5′) and downstream (3′) chromosomal DNA was amplified by PCR from total yeast genomic DNA using primers that introduced an EcoRI site at the 5′ end and a SacI site at the 3′ end. This cassette was then modified by overlap extension PCR reactions with primers designed to introduce a NdeI site at the ATG start codon and a BamHI site immediately downstream of the stop codon. The modified cassette was inserted into yeast expression vector pSE360 (CEN URA3) to yield pSE360-SceRCL1. After sequencing the insert, the cassette was excised with EcoRI and SacI and transferred to vector pRS413 (CEN HIS3) to yield pRS413-SceRCL1.
The KlaRCL1 ORF was excised from pET28b-His10Smt3-KlaRCL1 with BamHI and XhoI, then inserted between the BamHI and XhoI sites of vector pRS413-TPI (CEN HIS3) to yield pRS413-TPI-KlaRCL1, in which expression of KlaRcl1 is driven by the constitutive yeast TPI1 promoter. Missense mutations were introduced into the KlaRCL1 ORF via the two-stage PCR overlap extension method. The mutated PCR products were digested with BamHI and XhoI and inserted into BamHI/XhoI-cut pRS413-TPI-KlaRCL1 in lieu of the wild-type gene. Deleted versions of KlaRCL1 were constructed by PCR amplification with primers that introduced a new start codon (plus a flanking BamHI site) or a new stop codon (plus a flanking XhoI site) at the desired positions in the ORF. The mutated KlaRCL1 genes were inserted into pRS413-TPI. The KlaRCL1 plasmid inserts were sequenced completely to confirm that no unwanted coding changes were introduced during amplification and cloning.
Plasmid shuffle assay for rcl1Δ complementation
An S. cerevisiae RCL1 rcl1::kanMX heterozygous diploid strain (purchased from Open Biosystems; clone ID 21701) was transformed with pSE360-SceRCL1. The resulting Ura+ diploid was sporulated and tetrads were dissected. We thereby recovered viable rcl1::kanMX haploids that were resistant to G418 and unable to grow on medium containing 0.75 mg/mL FOA (5-fluoroorotic acid), a drug that selects against the URA3 SceRCL1 plasmid. The rcl1Δ strain (Mat α ura3 his3 leu2 rcl1::kanMX pSE360-yRCL1 [CEN URA3]) was used to test plasmid-borne alleles of SceRCL1 and KlaRCL1 for rcl1Δ complementation by plasmid shuffle as follows. rcl1Δ pSE360-SceRCL1 cells were transformed with pR413 (CEN HIS3) plasmids bearing wild-type or mutant alleles of RCL1. Transformants were selected on His− agar medium. Individual colonies were patched on His− agar medium, and cells from each isolate were then streaked on agar containing 0.75 mg/mL of FOA. The plates were incubated at 18°C, 25°C, 30°C, and 37°C. In cases where the KlaRCL1 plasmid supported growth on FOA, individual rcl1Δ KlaRCL1 isolates were tested for growth on YPD agar medium at 18°C, 25°C, 30°C, and 37°C.
Accession number
The coordinates for the refined model of KlaRcl1 have been deposited in the RCSB protein structure database (PDB ID code 3PQV).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank the NSLS staff members Rick Jackimowicz and Alexei Soares for their assistance with data collection. The work was supported by NIH grant GM46330. S.S. is an American Cancer Society Research Professor.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2571811.
REFERENCES
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Beltrame M, Tollervey D 1995. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J 14: 4350–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besant PG, Atttwood PV, Piggott MJ 2009. Focus on phosphoarginine and phospholysine. Curr Protein Pept Sci 10: 536–550 [DOI] [PubMed] [Google Scholar]
- Billy E, Hess D, Hofsteenge J, Filipowicz W 1999. Characterization of the adenylation site in the RNA 3′-terminal phosphate cyclase from Escherichia coli. J Biol Chem 274: 34955–34960 [DOI] [PubMed] [Google Scholar]
- Billy E, Wegierski T, Nasr F, Filipowicz W 2000. Rcl1p, the yeast protein similar to the RNA 3′-phosphate cyclase, associates with U3 snoRNP and is required for 18S rRNA biogenesis. EMBO J 19: 2115–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon F, Gallagher JEG, Compagnone-Post PA, Mitchell BM, Porwacher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417: 967–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Strugala K, Konarska M, Shatkin AJ 1985. Cyclization of RNA 3′-terminal phosphate by cyclase from HeLa cells proceeds via formation of N(3′)pp(5′)A activated intermediate. Proc Natl Acad Sci 8: 1316–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperin D, Horton L, Beckman J, Hensold J, Lemmon SK 2001. Bms1p, a novel GTP-binding protein, and the related Tsr1p are required to distinct steps of 40S ribosome biogenesis in yeast. RNA 7: 1268–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P, Billy E, Swianiewicz M, Filipowicz W 1997. The human RNA 3′-terminal phosphate cyclase is a member of a new family of protein conserved in eukarya, bacteria and archaea. EMBO J 10: 2955–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Doudet J, Gérus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 65: 2334–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Rosenstrom P, Schenkel A 2008. Searching protein structure databases with DaliLite v.3. Bioinformatics 24: 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbstein K, Doudna JA 2006. GTP-dependent formation of a ribonucleoprotein subcomplex required for ribosome biogenesis. J Mol Biol 365: 423–443 [DOI] [PubMed] [Google Scholar]
- Karbstein K, Jonas S, Doudna JA 2005. An essential GTPase promotes assembly of preribosomal RNA processing complexes. Mol Cell 20: 633–643 [DOI] [PubMed] [Google Scholar]
- Lawrence MC, Colman PM 1993. Shape complementarity at protein/protein interfaces. J Mol Biol 234: 946–950 [DOI] [PubMed] [Google Scholar]
- Lee B, Richards F 1971. The interpretation of protein structures: estimation of static accessibility. J Mol Biol 55: 379–400 [DOI] [PubMed] [Google Scholar]
- Pace CN, Grimsley GR, Scholtz JM 2009. Protein ionizable groups: pK values and their contribution of protein stability and solubility. J Biol Chem 284: 13285–13289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm GJ, Billy E, Filipowicz W, Wlodawer A 2000. Crystal structure of RNA 3′-terminal phoshate cyclase, a ubiquitous enzyme with unusual topology. Structure 8: 12–23 [DOI] [PubMed] [Google Scholar]
- Reinberg D, Arenas J, Hurwitz J 1985. The enzymatic conversion of 3′-phosphate terminated RNA chains to 2′,3′-cyclic phosphate derivatives. J Biol Chem 260: 6068–6097 [PubMed] [Google Scholar]
- Shuman S, Lima CD 2004. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr Opin Struct Biol 14: 757–764 [DOI] [PubMed] [Google Scholar]
- Tanaka N, Shuman S 2009. Structure-activity relationships in human RNA 3′-phosphate cyclase. RNA 15: 1865–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Smith P, Shuman S 2010. Structure of the RNA 3′-phosphate cyclase–adenylate intermediate illuminates nucleotide specificity and covalent nucleotidyl transfer. Structure 18: 449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegierski T, Billy E, Nasr F, Filipowicz W 2001. Bms1p, a G-domain-containing protein, associates with Rcl1p and is required for 18S rRNA biogenesis in yeast. RNA 7: 1254–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]