Abstract
Alternative polyadenylation (APA) of mRNAs has emerged as an important mechanism for post-transcriptional gene regulation in higher eukaryotes. Although microarrays have recently been used to characterize APA globally, they have a number of serious limitations that prevents comprehensive and highly quantitative analysis. To better characterize APA and its regulation, we have developed a deep sequencing-based method called Poly(A) Site Sequencing (PAS-Seq) for quantitatively profiling RNA polyadenylation at the transcriptome level. PAS-Seq not only accurately and comprehensively identifies poly(A) junctions in mRNAs and noncoding RNAs, but also provides quantitative information on the relative abundance of polyadenylated RNAs. PAS-Seq analyses of human and mouse transcriptomes showed that 40%–50% of all expressed genes produce alternatively polyadenylated mRNAs. Furthermore, our study detected evolutionarily conserved polyadenylation of histone mRNAs and revealed novel features of mitochondrial RNA polyadenylation. Finally, PAS-Seq analyses of mouse embryonic stem (ES) cells, neural stem/progenitor (NSP) cells, and neurons not only identified more poly(A) sites than what was found in the entire mouse EST database, but also detected significant changes in the global APA profile that lead to lengthening of 3′ untranslated regions (UTR) in many mRNAs during stem cell differentiation. Together, our PAS-Seq analyses revealed a complex landscape of RNA polyadenylation in mammalian cells and the dynamic regulation of APA during stem cell differentiation.
Keywords: alternative polyadenylation, genomics, polyadenylation, sequencing
INTRODUCTION
Cleavage/polyadenylation of pre-mRNAs is not only a nearly universal step of eukaryotic gene expression, but also a versatile mechanism for post-transcriptional gene regulation (Colgan and Manley 1997; Zhao et al. 1999). It is estimated that over half of human and >30% of mouse genes produce alternatively polyadenylated mRNAs that encode different protein isoforms and/or have distinct 3′ UTRs (Tian et al. 2005). Several recent studies have reported widespread APA regulation in the immune and neural systems (Flavell et al. 2008; Sandberg et al. 2008) during development (Ji et al. 2009), oncogenesis (Mayr and Bartel 2009), and the generation of induced pluripotent stem (iPS) cells (Ji and Tian 2009). Interestingly, the global APA profile of a cell seems to be tightly associated with its proliferation and differentiation status. For example, proximal poly(A) sites tend to be used in proliferating or undifferentiated cells, leading to the production of mRNAs with shorter 3′ UTRs, whereas distal poly(A) sites are favored in differentiated cells, resulting in mRNAs with longer 3′ UTRs (Sandberg et al. 2008; Ji and Tian 2009; Mayr and Bartel 2009). In keeping with this trend, progressive lengthening of mRNA 3′ UTRs through APA has been observed during mouse embryonic development (Ji et al. 2009). By contrast, 3′ UTR shortening was reported during Caenorhabditis elegans development (Mangone et al. 2010). Nonetheless, it is becoming increasingly clear that APA is much more prevalent than previously anticipated and that APA regulation plays important roles in a variety of physiological and pathological processes.
APA can impact gene expression through multiple mechanisms (Lutz 2008; Millevoi and Vagner 2009; Neilson and Sandberg 2010). First, alternatively polyadenylated mRNAs can code for different protein isoforms with distinct physiological properties. For example, a switch from the proximal to the distal poly(A) site in the IgM transcripts upon differentiation of primary B cells leads to a switch in their protein products from the secreted form to the membrane-bound form of IgM (Takagaki et al. 1996). Thus, like alternative splicing, APA contributes to the expansion of protein diversity in higher eukaryotes. Secondly, APA generates mRNAs with distinct 3′ UTRs. Many sequence elements found in extended 3′ UTRs, such as AU-rich and GU-rich elements and microRNA target sites, have a largely negative influence on mRNA stability and/or translation efficiency. As a result, mRNAs with shorter 3′ UTRs tend to express higher levels of proteins (Sandberg et al. 2008; Mayr and Bartel 2009). Indeed it has been shown that the shortening of 3′ UTRs through APA can lead to the activation of oncogenes in cancer cells (Mayr and Bartel 2009). Thirdly, as mRNA cleavage/polyadenylation is intimately linked to other steps of gene expression (Hirose and Manley 2000; Bentley 2005; Moore and Proudfoot 2009), the choice of alternative poly(A) sites can influence other gene expression steps. For example, APA and alternative splicing seem to be coordinately regulated in a tissue-specific manner (Wang et al. 2008). Finally, it has recently been reported in plant that APA of anti-sense RNAs can differentially silence the expression of a sense gene at least in part through modulating histone modifications (Hornyik et al. 2010; Liu et al. 2010). Therefore, it is critical to understand how APA is regulated temporally and spatially.
Despite recent progress, the APA field has been hampered by the lack of a reliable method for profiling RNA polyadenylation quantitatively at the transcriptome level. Previous global APA studies have used microarray analyses (Flavell et al. 2008; Sandberg et al. 2008; Ji et al. 2009; Ji and Tian 2009), which have several serious limitations. First, microarray-based APA analyses are limited by the availability and the design of the microarrays (for example, variable coverage of the 3′ UTRs and downstream regions by different microarray platforms). Secondly, microarray data cannot directly identify poly(A) sites. Instead, databases of known poly(A) sites were used as a reference for locating poly(A) junctions. Thirdly, quantification of APA isoforms using microarrays requires calculating the difference in average signal intensities between probes that detect common regions shared by all isoforms and those that detect the extended regions only found in the longer isoforms. When more than two APA isoforms are present, the quantification for each isoform becomes difficult and unreliable. As a result, most previous studies have been limited to genes that produce only two APA isoforms (Sandberg et al. 2008; Ji and Tian 2009). To overcome these limitations, we have developed PAS-Seq, a deep sequencing-based method for quantitative and global analysis of RNA polyadenylation, and have applied this method to study RNA polyadenylation in a variety of human and mouse cells.
RESULTS
PAS-Seq
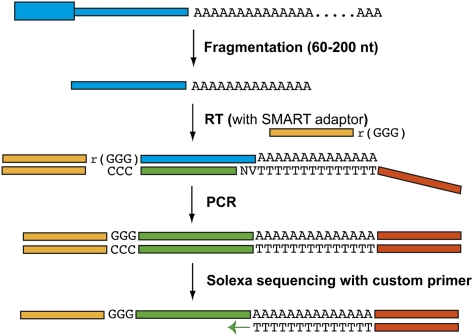
The PAS-Seq procedure is outlined in Figure 1. Poly(A)+ RNAs are fragmented to ∼60–200 nucleotide pieces and reverse transcription (RT) is carried out using the MML-V SMART RT system (Zhu et al. 2001). A double nucleotide anchored oligo(dT) primer was used to ensure that RT starts at the poly(A) junctions. When RT reaches the 5′ end of the RNA template, MML-V reverse transcriptase adds a few untemplated deoxycytodines to the 3′ ends of the cDNAs through its terminal transferase activity. Included in the RT reaction is a SMART adaptor (a hybrid oligonucleotide that has three guanine ribonucleotides linked to the 5′ end of a DNA linker) that can anneal to the untemplated deoxycytodines. The MML-V reverse transcriptase then switches the template and extends the cDNA until the 5′ end of the SMART adaptor. Thus, a single RT reaction generates cDNAs with linkers attached on both ends, eliminating the end-repair, A-tailing, and linker ligation steps required for conventional mRNA-seq. After synthesis of the second strand, cDNAs are size-selected and amplified with a low-cycle PCR to prepare the library. Single-end sequencing (40 nucleotides) was carried out using the Illumina Genome Analyzer II platform. A custom primer (standard Illumina sequencing primer with a oligo(dT) extension at the 3′ end) was used for sequencing to ensure that the sequencing reads start at or near the poly(A) junction (Fig. 1). T's were removed from all PAS-Seq reads before mapping. Reads that have six consecutive A's or seven or more A's in the 10 nt window downstream from the poly(A) junction were likely due to internal priming (Beaudoing et al. 2000; Tian et al. 2005), and therefore were excluded from further analyses. PAS-Seq reads in close vicinity were clustered together and the average locations were assigned as the poly(A) sites (see Materials and Methods for details on the computational pipeline).
FIGURE 1.
PAS-Seq. The procedure of PAS-Seq is described in detail in text. The blue boxes represent RNAs and orange, yellow, and green boxes represent linkers and cDNAs. Two degenerate nucleotides are present in the anchored oligo(dT) primer; “V” represents A/C/G and “N” represents A/T/C/G.
PAS-Seq analyses of a human transcriptome
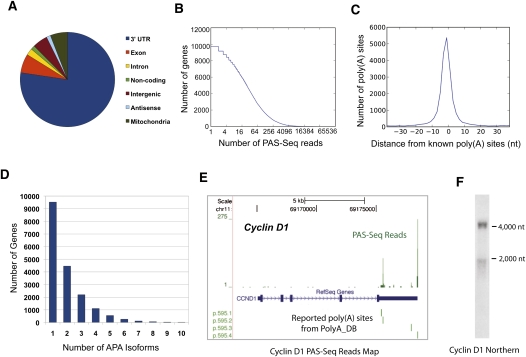
We first analyzed the transcriptome of a human cervical cancer cell line (HeLa) using the PAS-Seq method. From a single sequencing run, ∼1.9 million reads were obtained that could be mapped to unique locations in the human genome (estimated mapping error rate: 0.5%. See Materials and Methods for details). Out of these, ∼0.4 million reads were likely due to internal priming and therefore removed. Of the remaining reads, ∼80% mapped to the 3′ UTR of annotated genes while the rest mapped to exons (excluding 3′ UTRs), introns, noncoding RNA genes, intergenic regions, potential antisense transcripts, and the mitochondria genome (Fig. 2A). In total, our PAS-Seq reads covered 11,992 genes, including ∼300 noncoding RNA genes. A recent mRNA-seq study of the HeLa transcriptome detected mRNAs of 11,246 genes (Morin et al. 2008). Therefore, our PAS-Seq analysis provided a similar coverage level of the transcriptome as in mRNA-seq.
FIGURE 2.
PAS-Seq analyses of HeLa cell transcriptome. (A) A pie chart showing the distribution of HeLa PAS-Seq reads mapping result. “Exon” represents exonic regions excluding 3′ UTRs. (B) The depth of PAS-Seq analyses of HeLa transcriptome. Y-axis: the number of genes. X-axis: the number of reads detected in log2 scale. (C) Comparison between PAS-Seq-identified vs. known poly(A) sites. Y-axis: the number of poly(A) sites. X-axis: the distance between PAS-Seq-identified and the nearest known poly(A) sites. (D) A bar graph showing the number of genes with different number of poly(A) sites detected and at least 2 PAS-Seq reads are required per poly(A) site. Each number “n” on the x-axis means n or more APA isoforms were detected. (E) PAS-Seq mapping results for cyclin D1 transcripts. The green bars above “RefSeq Gene” are the PAS-Seq reads shown in BED format. The green lines below “RefSeq” mark the known poly(A) sites found in polyA_DB. (F) Northern blot for cyclin D1. Size markers are labeled.
As shown in Figure 2B, the number of reads detected per gene varied widely and reached up to 38,906 for the RPS14 gene, which encodes a ribosomal protein. On average, 96 reads were obtained per gene. Poly(A) junctions identified by PAS-Seq reads that are in close vicinity of one another were clustered and the average location was assigned as the poly(A) sites. To determine how well the poly(A) sites identified by PAS-Seq match known poly(A) sites supported by EST data, we plotted the distance between the poly(A) sites identified by PAS-Seq reads to the closest known poly(A) sites (within 40 nt) found in the polyA_DB2 database (Lee et al. 2007). The distribution of the PAS-Seq-derived poly(A) sites showed a sharp peak centered on known poly(A) sites (Fig. 2C; distance: 0.05 ± 9.4 nt), demonstrating that PAS-Seq accurately identifies poly(A) sites.
In order to obtain an estimate of the false discovery rate (FDR) for PAS-Seq identified poly(A) sites, we took a motif-based approach. Previous studies have shown that ∼92% of human poly(A) sites have the AAUAAA hexamer or one of 11 other close variants within the 40 nucleotide upstream region (Beaudoing et al. 2000; Tian et al. 2005). We found that, when a 2 reads/poly(A) site threshold is used, 82% of PAS-Seq identified poly(A) sites in HeLa cells have at least one of the 12 hexamers within a 40 nt upstream region. If a 10 reads/poly(A) sites threshold is used, the percentage becomes 91%. Based on these data, we estimate that 12.2% of PAS-Seq identified poly(A) sites are false positive when a 2 reads/poly(A) site threshold is used and 1.2% with a 10 reads/poly(A) site threshold (see Materials and Methods for details).
Our PAS-Seq analyses showed that ∼47% of genes that are expressed in HeLa cells produce alternatively polyadenylated mRNAs (Fig. 2D) and, on average, 1.9 APA isoforms were detected per gene. Ten or more poly(A) sites were used in the mRNAs of 19 genes (Fig. 2D). An example of APA identified by PAS-Seq is shown in Figure 2E. PAS-Seq reads mapped to two major sites in the 3′ UTR of the protooncogene cyclin D1 (CCND1) mRNAs that are over 2000 nt apart from each other, and both matched known poly(A) sites. Furthermore, a significantly greater number of reads mapped to the distal poly(A) site than to the proximal one, as is reflected by the relative sizes of the corresponding peaks. Our Northern analysis of HeLa poly(A)+ RNAs indeed detected the two APA isoforms of cyclin D1 mRNAs and showed that the relative abundance of these two isoforms mirrors the number of PAS-Seq reads obtained for the respective poly(A) sites (Fig. 2F; Supplemental Fig. S1F). Northern analyses of five other targets further supported the notion that PAS-Seq correctly and quantitatively identified major and minor APA isoforms (Supplemental Fig. S1A–F). We conclude that PAS-Seq faithfully provides quantitative information on the relative abundance of APA mRNA isoforms.
Polyadenylation and APA of histone mRNAs
The mRNAs of the metazoan replication-dependent histone genes are the only known eukaryotic mRNAs that are not polyadenylated (Marzluff et al. 2008). The 3′ ends of these mRNAs are formed by an endonucleolytic cleavage between a highly conserved stem–loop secondary structure and a downstream element (HDE). The 3′ processing of histone mRNAs requires a number of unique factors as well as several factors that are components of the mRNA 3′ processing (cleavage/polyadenylation) machinery. When normal 3′-end formation of histone mRNAs is inhibited by either mutation or knockdown of essential histone 3′ processing factors, sequences downstream from the normal cleavage site can be recognized by the mRNA 3′ processing machinery to produce polyadenylated mRNAs (Sullivan et al. 2001; Godfrey et al. 2006; Wagner et al. 2007). However, these polyadenylated histone mRNAs are generally considered as “misprocessed” products formed under nonphysiological conditions (Godfrey et al. 2006).
Interestingly, our PAS-seq analyses of both human (HeLa) and mouse cells (embryonic stem cell [ES], neural stem/progenitor cells [NSP], and neurons) detected polyadenylated mRNAs for over 20 replication-dependent histone genes that encode all four core histones (human data presented in Supplemental Table S1). Polyadenylation takes place mostly at or near the normal cleavage sites. The polyadenylation signal hexamer AAUAAA or its close variants were found upstream of some but not all poly(A) sites, indicating that polyadenylation of histone mRNAs is mediated by the canonical (AAUAAA-dependent) or a noncanonical (AAUAAA-independent) mechanism. Furthermore, APA was detected for multiple histone genes and some alternative poly(A) sites are evolutionarily conserved (Supplemental Fig. S2A,B).
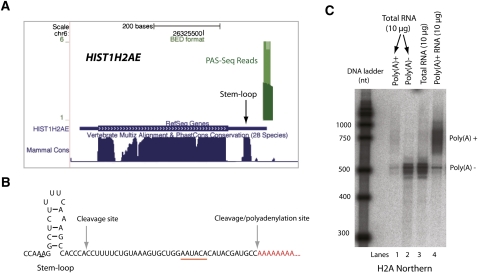
To experimentally verify the existence of polyadenylated mRNAs of the replication-dependent histone genes and to determine what percentage of their mRNAs is polyadenylated, we investigated H2A mRNAs in more detail using several approaches. Our PAS-Seq analyses detected polyadenylation in transcripts of four replication dependent H2A genes (Supplemental Table S1; PAS-Seq reads for HIST1H2AE transcripts shown in Fig. 3A). First, we mapped the exact poly(A) site by 3′-RACE to 36 nt downstream from the stem–loop-dependent cleavage site (Fig. 3B), consistent with our PAS-seq data (Fig. 3A). A potential poly(A) signal hexamer (AAUACA) was found within 20 nt upstream of the poly(A) site. Secondly, we carried out Northern analyses of H2A mRNAs to quantitatively compare the polyA− and polyA+ forms (Fig. 3C). When HeLa total RNAs were probed, a prominent band of ∼500 nt and a defused band of ∼750 nt were observed (lane 3). Northern analyses of polyA− and polyA+ RNAs demonstrated that the prominent ∼500 nt species corresponded to unpolyadenylated H2A mRNAs (lane 2), whereas the ∼750 nt band represented polyadenylated H2A mRNAs (lane 1), which were more prominent when higher amounts of polyA+ RNAs were probed (lane 4). Quantification of the Northern data showed that ∼4.3% of the H2A mRNAs are polyadenylated. Similar ratios between the unpolyadenylated vs. polyadenylated H2A mRNAs were observed using an RNase-protection assay (RPA) (Supplemental Fig. S3). Therefore, our data provided evidence that a portion of the mRNAs of replication-dependent histone genes is polyadenylated in mammalian cells even when the histone mRNA 3′-end processing machinery is functional.
FIGURE 3.
Polyadenylated histone mRNAs. (A) PAS-Seq mapping results for HIST1H2AE. The green bar above the RefSeq Gene represents PAS-Seq reads. The sequence conservation levels among mammals are shown below the RefSeq Gene. The position of the stem–loop sequence is marked. (B) A diagram of HIST1H2AE 3′-UTR stem–loop structure, normal cleavage site, and cleavage/polyadenylation site detected by 3′-RACE. (C) Northern blot of H2A. Lanes 1 and 2 are polyA+ and polyA− RNAs isolated from 10 μg of HeLa total RNA.
A polyadenylation map for the human and mouse mitochondria transcriptomes
Mitochondrial RNA polyadenylation remains poorly characterized. In plant mitochondria and bacteria, RNA polyadenylation is transient and primarily functions in targeting RNAs for degradation (Slomovic et al. 2005; Nagaike et al. 2008). In human mitochondria, both transiently polyadenylated truncated RNA species and stable poly(A) tails at the 3′ ends of transcripts have been reported (Nagaike et al. 2008). However, RNA polyadenylation has not been comprehensively characterized in mammalian mitochondria and it is not clear whether distinct classes of RNAs (mRNA, tRNAs, and rRNAs) have different polyadenylation patterns.
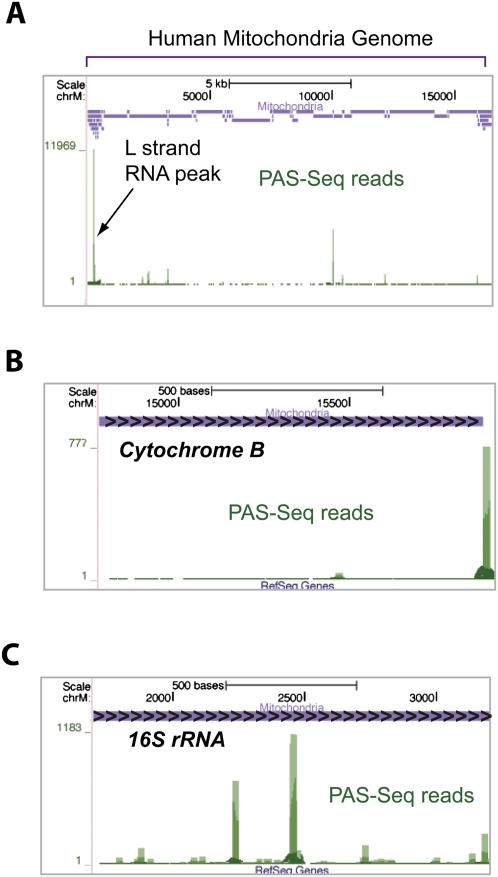
Our PAS-seq analyses generated the first comprehensive RNA polyadenylation map for the human and mouse mitochondrial transcriptomes (Fig. 4A) and three prominent features have emerged from this map. First, a high level of polyadenylated RNA species was detected for light (L) strand transcripts in the D-loop region (Fig. 4A). In fact, ∼12,000 reads mapped to this region, significantly higher than any other region in the entire human mitochondrial genome. Polyadenylated RNAs in this region were first observed almost 30 yr ago and were further confirmed by EST data (Ojala et al. 1981; Slomovic et al. 2005). Our data suggest that these polyadenylated RNA species are highly abundant, but their functions remain unknown. Second, PAS-Seq read peaks were detected at the 3′ ends for mRNAs of seven out of the 13 human mitochondrial protein-coding genes (an example, cytochrome B gene, is shown in Fig. 4B and Supplemental Fig. S4A), suggesting that at least some mitochondrial mRNAs have stable polyadenylated 3′ ends. Unlike cellular mRNAs, however, many PAS-Seq reads mapped to internal sites of mRNAs of most mitochondrial protein-coding genes (nine out of 13 in human), in some cases covering essentially the entire transcript (Fig. 4B). These observations suggest that the internally polyadenylated truncated mRNAs are widespread and may represent transient degradation intermediates. Third, internally polyadenylated mitochondrial rRNAs (12S and 16S) were also detected. But distinct from the pattern observed in mitochondrial mRNAs (Fig. 4B), much higher levels of internal polyadenylation of rRNAs take place at discrete locations (Fig. 4C; Supplemental Fig. S4B). These RNAs could represent prematurely terminated transcripts or stalled degradation intermediates. Since polyadenylation facilitates degradation of RNA secondary structures in bacteria (Blum et al. 1999), we examined mitochondrial rRNAs for stable secondary structures, but found no apparent correlation between secondary structure-forming sites and internal polyadenylation sites (Supplemental Fig. S4C). These highly abundant polyadenylated truncated rRNAs remain to be characterized.
FIGURE 4.
A human mitochondria RNA polyadenylation map. (A) PAS-Seq mapping results on the entire mitochondrial genome. A custom track (purple) shows the positions of all human mitochondrial genes. The green bars represent PAS-Seq reads and the peak corresponding to the polyadenylated L strand transcripts is marked. (B) PAS-Seq mapping results on the cytochrome B gene. Overlapping PAS-Seq reads appear as a continuous line covering the majority of the gene. (C) PAS-Seq mapping results on the 16S rRNA gene.
PAS-Seq analyses of mouse ES cells, NSP cells, and neurons
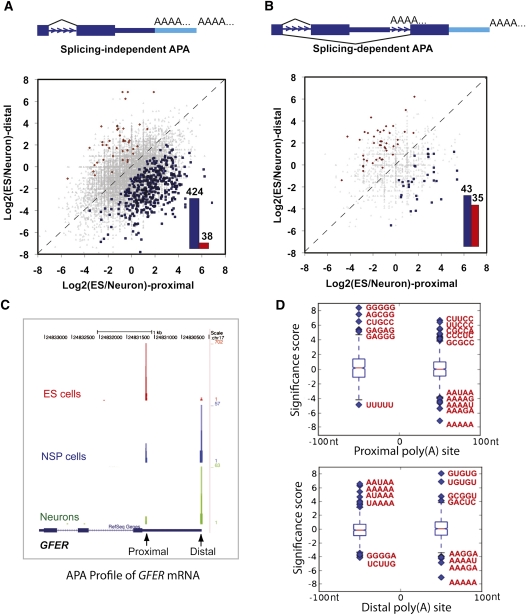
We used stem cell differentiation as a model system to study the dynamic changes of APA. To this end, we carried out PAS-Seq analyses of the pluripotent mouse ES cells, partially differentiated NSP cells, and terminally differentiated neurons. Similar read coverage and depth were obtained for all three cell types (Supplemental Fig. S5). Alternatively polyadenylated mRNAs were detected for 38%–47% of expressed genes in each cell type (Supplemental Fig. S5E), comparable to the levels detected in HeLa cells (Fig. 2D). When data from all three mouse cell types are combined, APA was detected for 52% of expressed mouse genes, a significantly higher percentage than the previous estimate (32%) based on the entire mouse EST database (Tian et al. 2005). Next, pairwise comparisons of APA profiles were carried out and proximal-distal poly(A) site pairs showing statistically significant changes were identified using Fisher Exact Test (P < 0.05; see Materials and Methods for details). When comparing mouse ES cells and neurons, we identified >13,000 splicing-independent APA events (both poly(A) sites in the same terminal exon), but only ∼1200 splicing-dependent ones (alternative poly(A) sites located in different exons or introns), suggesting that the great majority of APA events are independent of splicing. For splicing-independent APA events, the differentiation of ES cells into neurons is accompanied by significantly more proximal-to-distal switch events than distal-to-proximal switch events (Fig. 5A, cf. blue and red bars). This bias was also observed in the ES vs. NSP and NSP vs. neuron comparisons (Supplemental Fig. S6), suggesting that 3′ UTR lengthening takes place during stem cell differentiation. This was in sharp contrast with splicing-dependent APA events, which did not show a systematic shift to either the proximal or distal poly(A) sites (Fig. 5B; Supplemental Fig. S6C,D, cf. blue and red bars). An example of APA changes during stem cell differentiation is shown in Figure 5C. The proximal poly(A) site of the GFER mRNA, which encodes a growth factor, was predominantly used in ES cells while the distal site was favored in neurons. An intermediate APA profile was observed in NSP cells. This type of switch-like APA changes was observed for the mRNAs of many other genes. These results suggest that APA is coordinated in a regulated fashion during stem cell differentiation and the regulation of APA is largely mediated by splicing-independent mechanisms.
FIGURE 5.
Dynamic APA landscape during stem cell differentiation. (A) Splicing-independent APA changes between ES and neurons. Y-axis: ratio of PAS-Seq reads counts between ES and neurons for the distal poly(A) sites in log2 scale. X-axis: ratio of PAS-Seq reads counts between ES and neurons for the proximal poly(A) sites in log2 scale. APA events with statistically significant change (P < 0.05 by Fisher Exact Test) are colored in red (distal sites preferentially used in ES) or blue (proximal sites preferentially used in ES). Bar graph shows the number of red or blue data points in the scatter plot. (B) Splicing-dependent APA changes between ES and neurons. (C) PAS-Seq reads mapping results for the GFER gene in ES, NSP, and neurons. Proximal and distal sites are marked. (D) Box plots showing over- and underrepresented motifs at the regulated proximal and distal poly(A) sites. The sequences of the most over- and underrepresented motifs (Z score >5) are shown in red.
Since we observed a striking shift in the splicing-independent APA profile during stem cell differentiation, we investigated the underlying mechanisms by taking the proximal-distal poly(A) site pairs that showed statistically significant changes between the ES cells and neurons and carried out sequence motif analysis to identify over- or underrepresented 5-mers around these poly(A) sites (from −100 nucleotides to +100 nucleotides of the poly(A) sites). Interestingly, AAUAAA-like sequences and GU-rich downstream elements were found to be significantly overrepresented at the distal poly(A) sites (Z score >5; Fig. 5D, bottom panel), suggesting that the regulated distal poly(A) sites tend to be strong canonical poly(A) sites. By contrast, G-rich and C/U-rich elements were overrepresented around the proximal poly(A) sites (Z score >5; Fig. 5D, top panel). These data suggest that the regulated proximal and distal poly(A) sites have distinct characteristics, which may provide the molecular basis for differential poly(A) site selection during stem cell differentiation.
To understand the functional significance of the APA regulation in stem cell differentiation, we performed gene ontology (GO) analyses of the genes whose transcripts displayed significantly different APA profiles between ES cells and neurons. We found that genes with neuronal functions are highly enriched (Supplemental Fig. S7), indicating that APA is involved in establishing a neuron-specific gene expression program.
DISCUSSION
We have developed a high throughput sequencing-based method called PAS-Seq for quantitative characterization of RNA polyadenylation at the transcriptome-wide level. PAS-Seq is a simple method that not only accurately identifies poly(A) junctions, but also provides quantitative information on the relative abundance of polyadenylated mRNAs. Compared to microarrays, PAS-Seq offers several advantages in APA analyses. First, PAS-Seq can be used to analyze polyadenylation in any organism whose genome has been sequenced and is not limited by the availability or the design of microarrays. Second, PAS-Seq directly identifies poly(A) sites and is well suited for discovering novel polyadenylation events. Third, PAS-Seq provides quantitative information on APA isoforms. Unlike microarray-based methods, PAS-Seq detects each APA isoform independently and, therefore, the quantification is not compromised by the total number of APA isoforms. As mentioned earlier, microarray-based APA analyses have been limited to genes that have only two APA isoforms (Sandberg et al. 2008; Ji and Tian 2009). We found that ∼2000 genes produce three or more APA isoforms in all the human and mouse cells examined (Fig. 2D; Supplemental Fig. S5E), and these mRNAs would be difficult to study using microarray-based methods. Thus, PAS-Seq permits much more comprehensive and truly transcriptome-wide analysis of RNA polyadenylation.
It is estimated that up to 12% of the ESTs are generated from internal priming of the oligo(dT) primer during the RT step (Nam et al. 2002). Such mispriming can also occur in PAS-Seq. We have taken two steps to minimize the FDR. First, we have used a double nucleotide-anchored oligo(dT) primer for RT, which has been shown to reduce internal priming by up to threefold compared to the regular oligo(dT) primer that has been used to generate most of the 3′ ESTs (Nam et al. 2002). Second, we have applied a stringent filtering step during data analysis to remove PAS-Seq reads that are likely due to internal priming (see Materials and Methods for details). Given these additional measures, we believe that the FDR of PAS-Seq analysis is comparable to or better than that of the EST data.
Our PAS-Seq analyses detected APA in 52% of mouse genes, significantly higher than the previous estimate based on the entire mouse EST database and similar to the levels of APA detected for humans (Tian et al. 2005). Our study also highlighted dynamic changes in APA profiles during stem cell differentiation. By analyzing over 13,000 splicing-independent APA events and ∼1200 splicing-dependent events, we found that the majority of APA changes during stem cell differentiation are splicing-independent and are from proximal to distal poly(A) sites, leading to the lengthening of 3′-UTR as differentiation progresses (Fig. 5; Supplemental Fig. S6). These results are consistent with recent microarray-based studies. For example, Sandberg et al. (2008) reported that proliferating cells tend to express mRNAs with shortened 3′ UTRs by selecting a proximal poly(A) site (Sandberg et al. 2008). Similarly, shortening of 3′ UTR through APA during the generation of iPS from different cell types has been reported (Ji and Tian 2009). Lastly, in cancer cells proximal poly(A) sites are preferentially used in transcripts of proto-oncogenes (Mayr and Bartel 2009). Together with these studies, our results suggest that APA profile strongly correlates with cell proliferation and differentiation status and APA regulation may be involved in establishing and/or maintaining cell identities.
How is APA regulated during stem cell differentiation? During B cell activation and generation of iPS cells, it has been proposed that APA regulation is mediated by a change in the overall activities of the cleavage/polyadenylation machinery (Takagaki et al. 1996; Ji et al. 2009). Our analysis demonstrated that proximal poly(A) sites preferentially used in ES cells tend to have G-rich and C/U-rich sequences while the distal poly(A) sites favored in differentiated neurons have strong features of canonical poly(A) sites, including AAUAAA and G/U-rich element (Fig. 5). Based on these observations, we propose an alternative mechanism for APA regulation during stem cell differentiation. We propose that certain regulatory factor(s) highly expressed in ES cells specifically bind to G-rich and C/U-rich sequences around the proximal poly(A) sites and stimulate polyadenylation at these sites. For example, hnRNP H and related proteins have been shown to regulate polyadenylation by binding to G-rich auxiliary elements (Arhin et al. 2002). Differentiated cells may express lower levels of such regulatory factors and, as a result, canonical distal poly(A) sites are preferentially used as they are recognized by the cleavage/polyadenylation machinery with higher affinity. The above two models are not mutually exclusive.
Our PAS-Seq, 3′-RACE, Northern, and RPA analyses provided evidence that a portion of mRNAs for replication-dependent histone genes is polyadenylated in mammalian cells. A recent study of C. elegans mRNA 3′ UTRs also detected polyadenylated histone mRNAs (Mangone et al. 2010). Together these results suggest that polyadenylation is an evolutionarily conserved alternative mechanism for histone mRNA 3′ end formation. What might be the functional significance of polyadenylation of histone mRNAs? First, polyadenylation may be a fail-safe mechanism for histone mRNA 3′ processing. Like other steps of gene expression, 3′ end formation of histone transcripts is not 100% efficient and if the SLBP-mediated histone mRNA 3′ processing fails, polyadenylation ensures that the mRNA is produced and transcription is terminated. Since most replication-dependent histone genes are found in tight clusters (Marzluff et al. 2002), polyadenylation may help to prevent transcription read-through and disruption of gene expression at downstream sequences when normal 3′-end formation fails. Second, although the majority of histone synthesis occurs during S phase, basal synthesis of histones persists throughout the cell cycle (Wu and Bonner 1981). As nonpolyadenylated histone mRNAs are degraded at the end of S phase (Marzluff et al. 2008), the polyadenylated histone mRNAs may be responsible for the basal synthesis of histones. A comparison of histone mRNA polyadenylation profiles between mouse ES cells (a mixture of cells in different stages of the cell cycle) and post-mitotic neurons revealed noticeable differences (Supplemental Fig. S2C), supporting the notion that histone mRNA polyadenylation may be regulated by the cell cycle.
In summary, we have developed a high throughput sequencing-based method for transcriptome-wide quantitative analysis of RNA polyadenylation. Using this method, we have demonstrated that RNA polyadenylation is more complex than previously thought and APA is widespread, dynamic, and tightly regulated during stem cell differentiation. Global analyses of mRNA polyadenylation using methods like PAS-Seq, coupled with detailed biochemical and structural studies of mRNA cleavage/polyadenylation factors and regulators, will be key to our future effort in deciphering the “poly(A) code,” the rules by which poly(A) sites are defined and chosen under different biological conditions.
MATERIALS AND METHODS
Cell culture
HeLa cells were grown in DMEM plus 10% fetal bovine serum or FetalPlex (Gemini). Mouse C57BL/6NTac ES cells were a kind gift of Thomas Fielder (see Acknowledgments). Mouse fetal-derived neural stem/progenitor cells (NSPCs) were isolated from cerebral cortical regions of wild-type CD1 mice at embryonic day 12.5 (E12.5), using established protocols with the addition of a trypsinization step to increase cell survival (Flanagan et al. 2008). Neurons were derived from the same set of E12.5 mouse cortices as NSPCs, using conditions described previously (Flanagan et al. 2002).
PAS-Seq
Total RNAs were isolated using the Trizol reagent (Invitrogen) and poly(A)+ RNAs were purified using the mRNA purification kit (Invitrogen). 0.5–1 μg of poly(A)+ RNA was fragmented using the RNA Fragmentation Buffer (Ambion) and then precipitated. Reverse transcription (RT) was performed as follows: 22 μl RNA is incubated with 2 μl HITS-3′ (12μM) at 65°C for 5 min and then on ice for 5 min. Then 8 μl of 5X buffer, 2 μl 0.1M DTT, 2 μl HITS-5′ (a SMART oligo), 2 μl RNaseOUT, and 2 μl Superscript III were added. The mixture is incubated at 50°C for 30 min and then 42°C for 30 min. cDNAs were purified using the PCR Cleanup kit (Qiagen) and the second strand cDNAs were synthesized by a three cycle PCR (98°C 10 sec, 60°C 30 sec, 72°C 30 sec) using the Phusion polymerase (New England Biolabs) and PE 1.0 and PE 2.0 primers. The PCR reaction was run on a 2% agarose gel and the 200–300 bp band was excised. Gel-extracted DNAs were further amplified by a 15 cycle PCR (98°C 10 sec, 65°C 30 sec, 72°C 30 sec) using the Phusion polymerase and PE 1.0 and PE 2.0 primers. The PCR reaction was purified using the PCR Cleanup kit before sequencing using the Illumina Genome Analyzer using a custom sequencing primer (PAS-Seq). Sequences of the oligos are listed in Table 1.
TABLE 1.
Oligos for PAS-Seq
Bioinformatic methods
Read filtering and mapping
PAS-Seq reads were retrieved from GERALD (Generation of Recursive Analysis Linked by Dependency). These reads were filtered by trimming T's off the beginning of reads and removing reads that have a string of 12 or more T's. The remaining reads were mapped to the human (hg18) and mouse genomes (mm9), allowing up to two mismatches using Bowtie with the settings “bowtie -m 1–best -p 4.” Reads that have six or more consecutive A's or seven or more A's in the 10 nt immediately downstream from the poly(A) junction (where the 5′ end of the read maps) are removed as they are likely due to internal priming.
Estimation of mismapping rate
To estimate the probability of a sequence from our reads mapping to the incorrect position in the genome, we used the alignments from our reads mapping to the (hg 18) reference genome. First, the sequencing error rate was estimated using a scheme employed by Wang et al. (2008), resulting in an estimated error rate of 0.52 mismatches/read, a rate that results in mostly zero or one incorrect bases out of 40. We then corrected all mismatches to match the reference genome, resulting in ∼1M perfectly matching reads. Next we generated a set of mock reads, introducing errors at a frequency of 0.52 errors/read, which were then mapped to the genome and using Bowtie, allowing for up to two mismatches. The fraction of mock reads mapping to a unique genomic location was 98.2%. Of those uniquely mapping reads, 99.5% were correctly remapped to their original location. Therefore the mismapping rate is estimated to be 0.5%.
Clustering of PAS-Seq reads and identification of poly(A) sites
Due to the intrinsic heterogeneity in cleavage/polyadenylation, for any poly(A) signal, most of the corresponding cleavage sites cluster in a 24 nt window, but the distance between the most upstream and the most downstream cleavage sites could reach up to 30–40 nt (Pauws et al. 2001; Tian et al. 2005). Therefore, PAS-Seq reads within 40 nt windows are pooled and the weighted average of the genomic coordinates of all the reads in a cluster is assigned as the poly(A) site. If multiple poly(A) sites are found within 40 nt of one another, a second round of clustering is performed and a new poly(A) site is assigned.
Estimation of false discovery rate
FDR is calculated using the following formula: (Pt-Pp)/(Pt-Pf). Pt: percentage of true poly(A) sites that have one of the 12 hexamers within 40 nt upstream; Pf: percentage of random 40 mers that have one of the 12 hexamers within 40 nt upstream; Pp: percentage of PAS-Seq identified poly(A) sites that have one of the 12 hexamers within 40 nt upstream. Pt is set at 92% based on previous studies (Beaudoing et al. 2000; Tian et al. 2005). Pf is the percentage of random 40 mers that have at least one of the 12 hexamers. We randomly picked 50,000 exons from the human genome and randomly selected 40 mers from these exons (equivalent to 50,000 40 mers) and calculated Pf to be 9.9%. Pp is 82% with a 2 reads/poly(A) site threshold, and 91% with a 10 reads/poly(A) site threshold. Therefore, the FDR is 12.2% with a 2 reads/poly(A) site threshold, and 1.2% with a 10 reads/poly(A) site threshold.
Making a unique 3′ UTR database
A set of unique 3′ UTRs for both mouse and humans was made using the following strategy: First a list of all 3′ UTRs was extracted from the Known Genes database of the UCSC table browser (Karolchik et al. 2009). The 3′ UTRs from each isoform in this database were grouped together by stop codon. The isoform with the longest 3′ UTR in each group of common stop codons was kept and the rest discarded. For each 3′ UTR in the new set, if a stop codon occurs in the last exon, the beginning of the 3′ UTR is equal to the position of the stop codon. If the stop codon does not occur in the last exon, the beginning of the 3′ UTR is equal to the 3′ splice site of the last exon. 30 nts were added to the 3′ ends of all UTRs in the new set to ensure that reads mapping to the ends of the transcript are captured. A final filtering step was performed to check for overlap in the database. If any overlapping segments occurred between 3′ UTRs in the database, both 3′ UTRs were discarded.
Mapping reads to the genome
For both human and mouse, the Known Genes database was used to map our reads to the genome. We classified the genome into seven groups and mapped the poly(A) clusters to these groups using the following hierarchy: 3′ UTRs, exonic regions (excluding the 3′ UTRs), noncoding genes, introns, mitochondrial regions, antisense regions, intergenic regions. Reads belonging to a region higher up in the hierarchy cannot belong to a region further down in the hierarchy.
Comparison of APA profiles between different samples
Poly(A) sites in two samples that are within 24 nt of each other are considered the same poly(A) site. For splicing-independent APA, the ratio of PAS-Seq read counts for one poly(A) site between two samples is compared to the ratio of another poly(A) site found in the same 3′ UTR. The statistical significance of the difference is estimated using a Fisher's exact test, and the poly(A) site pairs with significant P-values corresponding to a false discovery rate cutoff of 5% (Benjamini-Hochberg) were considered as significantly different between the two samples. For splicing-dependent APA, poly(A) sites from different 3′ UTRs of the same gene are compared and the P-values are calculated the same way as described above.
Sequence motif analyses
To identify potential cis-elements associated with APA regulation, the significantly regulated APA events between ES cells and neurons were split into two groups, those events that are shifted toward the proximal site in ES cells and those events that shift toward the distal site in ES cells. For each of these groups the sequences +/−100 nucleotides in both the proximal and distal poly(A) site were downloaded. The background model was made by taking the frequency of all possible 5-mers in the 100 nt upstream or downstream from all poly(A) sites detected in both cell lines. For both groups, we compared the background model to the frequency of 5-mers in four different regions: upstream of the proximal poly(A) site; downstream from the proximal poly(A) site; upstream of the distal poly(A) site; and downstream from the distal poly(A) site using a Z-score.
GO analysis
Among transcripts that displayed significantly different APA profiles between ES cells and neurons, we searched for significantly overrepresented Biological Process GO terms against a background model of all alternatively polyadenylation transcripts found in both ES cells and neurons using GeneMerge (Castillo-Davis and Hartl 2003). P-values were conservatively corrected for multiple testing as provided by GeneMerge and recorded below a cutoff of 0.1.
Accession codes
All sequence read data have been submitted to the Short Read Archive section of the GEO database at NCBI under accession number GSE25450.
Northern blotting
2.5 μg of polyA+ RNA was glyoxylated, separated on a 1.2% agarose gel, and then transferred to BrightStar TM-Nylon membrane (Ambion). The membrane was hybridized with radio-labeled DNA probe (1.5–2 × 106 cpm/mL) for overnight at 55°C, washed, and scanned using a phosphorimager. For DNA probe preparation, specific PCR products of each target were labeled by randomly priming in the presence of 32P-dCTP (Table 2).
TABLE 2.
Oligos for preparing Northern probes
3′ RACE
2.5 μg of total RNA was reverse transcribed with a primer that consists of Oligo(dT) and adaptor sequences. Amplification was performed using a primer containing part of the adaptor and a primer specific to each target. A second amplification was then carried out using nested primers for 30 cycles. The PCR products were resolved on a 1.2% agarose gel, and the DNA band with expected size was extracted from the gel and sequenced (Table 3).
TABLE 3.
Oligos for 3′ RACE
RNase protection assay (RPA)
A HIST1H2AE PCR product encompassing the coding and 3′ UTR region was cloned into pBluescript KS II+ vector. Antisense RNA synthesized using T7 polymerase in the presence of 32P-UTP and was used as probe. Approximately fivefold molar excess of probe was hybridized with 10 μg of total HeLa RNA at 45°C and digested with RNase A/T1 mixture. The protected fragments were resolved on a 5% polyacrylamide-8M urea gel and scanned using a phosphorimager.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Thomas Fielder at the UCI Transgenic Mouse Facility for providing the mouse ES cells, Kip Bodi and the Tufts University Core Facility for help with sequencing, and Drs. Jim Manley, Bert Semler, Marian Waterman, Xiaohui Xie, and Inna and Ruslan Aphasizhev for technical advice and comments on the manuscript. This study was supported by a NIH T15LM07443 (P.J.S.), the California Institute for Regenerative Medicine RT1-01074 and NIH AG23583 (L.A.F.), NIH RO1GM62287 and R21CA149548 (K.J.H.), NIH R01GM090056, a seed grant from American Cancer Society (ACS/IRG–98-279-07), and start-up funds from UC Irvine (Y.S.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2581711.
REFERENCES
- Arhin GK, Boots M, Bagga PS, Milcarek C, Wilusz J 2002. Downstream sequence elements with different affinities for the hnRNP H/H′ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res 30: 1842–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D 2000. Patterns of variant polyadenylation signal usage in human genes. Genome Res 10: 1001–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL 2005. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 17: 251–256 [DOI] [PubMed] [Google Scholar]
- Blum E, Carpousis AJ, Higgins CF 1999. Polyadenylation promotes degradation of 3′-structured RNA by the Escherichia coli mRNA degradosome in vitro. J Biol Chem 274: 4009–4016 [DOI] [PubMed] [Google Scholar]
- Castillo-Davis CI, Hartl DL 2003. GeneMerge–post-genomic analysis, data mining, and hypothesis testing. Bioinformatics 19: 891–892 [DOI] [PubMed] [Google Scholar]
- Colgan DF, Manley JL 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev 11: 2755–2766 [DOI] [PubMed] [Google Scholar]
- Flanagan LA, Ju YE, Marg B, Osterfield M, Janmey PA 2002. Neurite branching on deformable substrates. Neuroreport 13: 2411–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan LA, Lu J, Wang L, Marchenko SA, Jeon NL, Lee AP, Monuki ES 2008. Unique dielectric properties distinguish stem cells and their differentiated progeny. Stem Cells 26: 656–665 [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME 2008. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron 60: 1022–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey AC, Kupsco JM, Burch BD, Zimmerman RM, Dominski Z, Marzluff WF, Duronio RJ 2006. U7 snRNA mutations in Drosophila block histone pre-mRNA processing and disrupt oogenesis. RNA 12: 396–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Manley JL 2000. RNA polymerase II and the integration of nuclear events. Genes Dev 14: 1415–1429 [PubMed] [Google Scholar]
- Hornyik C, Terzi LC, Simpson GG 2010. The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev Cell 18: 203–213 [DOI] [PubMed] [Google Scholar]
- Ji Z, Tian B 2009. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS ONE 4: e8419 doi: 10.1371/journal.pone.0008419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B 2009. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci 106: 7028–7033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Kent WJ 2009. The UCSC Genome Browser. Curr Protoc Bioinformatics Chapter 1: Unit 1.4 doi: 10.1002/0471250953.bi0104s28 [DOI] [PubMed] [Google Scholar]
- Lee JY, Yeh I, Park JY, Tian B 2007. PolyA_DB 2: mRNA polyadenylation sites in vertebrate genes. Nucleic Acids Res 35: D165–D168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Marquardt S, Lister C, Swiezewski S, Dean C 2010. Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327: 94–97 [DOI] [PubMed] [Google Scholar]
- Lutz CS 2008. Alternative polyadenylation: a twist on mRNA 3′ end formation. ACS Chem Biol 3: 609–617 [DOI] [PubMed] [Google Scholar]
- Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, et al. 2010. The landscape of C. elegans 3′UTRs. Science 329: 432–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ 2002. The human and mouse replication-dependent histone genes. Genomics 80: 487–498 [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 9: 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Vagner S 2009. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res 38: 2757–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136: 688–700 [DOI] [PubMed] [Google Scholar]
- Morin R, Bainbridge M, Fejes A, Hirst M, Krzywinski M, Pugh T, McDonald H, Varhol R, Jones S, Marra M 2008. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques 45: 81–94 [DOI] [PubMed] [Google Scholar]
- Nagaike T, Suzuki T, Ueda T 2008Polyadenylation in mammalian mitochondria: insights from recent studies. Biochim Biophys Acta 1779 266–269 [DOI] [PubMed] [Google Scholar]
- Nam DK, Lee S, Zhou G, Cao X, Wang C, Clark T, Chen J, Rowley JD, Wang SM 2002. Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. Proc Natl Acad Sci 99: 6152–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson JR, Sandberg R 2010. Heterogeneity in mammalian RNA 3′ end formation. Exp Cell Res 316: 1357–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala D, Montoya J, Attardi G 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature 290: 470–474 [DOI] [PubMed] [Google Scholar]
- Pauws E, van Kampen AH, van de Graaf SA, de Vijlder JJ, Ris-Stalpers C 2001. Heterogeneity in polyadenylation cleavage sites in mammalian mRNA sequences: implications for SAGE analysis. Nucleic Acids Res 29: 1690–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic S, Laufer D, Geiger D, Schuster G 2005. Polyadenylation and degradation of human mitochondrial RNA: the prokaryotic past leaves its mark. Mol Cell Biol 25: 6427–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan E, Santiago C, Parker ED, Dominski Z, Yang X, Lanzotti DJ, Ingledue TC, Marzluff WF, Duronio RJ 2001. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev 15: 173–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL 1996. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87: 941–952 [DOI] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res 33: 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Burch BD, Godfrey AC, Salzler HR, Duronio RJ, Marzluff WF 2007. A genome-wide RNA interference screen reveals that variant histones are necessary for replication-dependent histone pre-mRNA processing. Mol Cell 28: 692–699 [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RS, Bonner WM 1981. Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell 27: 321–330 [DOI] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YY, Machleder EM, Chenchik A, Li R, Siebert PD 2001. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques 30: 892–897 [DOI] [PubMed] [Google Scholar]