Abstract
Glycosylation is a very common modification of protein and lipid, and most glycosylation reactions occur in the Golgi. Although the transfer of initial sugar(s) to glycoproteins or glycolipids occurs in the ER or on the ER membrane, the subsequent addition of the many different sugars that make up a mature glycan is accomplished in the Golgi. Golgi membranes are studded with glycosyltransferases, glycosidases, and nucleotide sugar transporters arrayed in a generally ordered manner from the cis-Golgi to the trans-Golgi network (TGN), such that each activity is able to act on specific substrate(s) generated earlier in the pathway. The spectrum of glycosyltransferases and other activities that effect glycosylation may vary with cell type, and thus the final complement of glycans on glycoconjugates is variable. In addition, glycan synthesis is affected by Golgi pH, the integrity of Golgi peripheral membrane proteins, growth factor signaling, Golgi membrane dynamics, and cellular stress. Knowledge of Golgi glycosylation has fostered the development of assays to identify mechanisms of intracellular vesicular trafficking and facilitated glycosylation engineering of recombinant glycoproteins.
Many sugars are added to proteins and lipids in the Golgi. The glycosyltransferases and other activities responsible are concentrated in successive Golgi compartments such that each acts on a substrate generated earlier in the pathway.
The Golgi is home to a multitude of glycosyltransferases (GTs), glycosidases, and nucleotide sugar transporters that function together to complete the synthesis of glycans from founding sugars covalently attached to protein or lipid in the endoplasmic reticulum (ER) (Fig. 1, sugars shaded in green). Thus, glycoproteins, glycosphingolipids (GSLs), proteoglycans, and glycophosphatidylinositol (GPI) anchors acquire their final sugar complement during passage through the Golgi. Most glycoproteins and proteoglycans are either secreted from the cell, or span the plasma membrane with their glycans becoming the molecular frontier of the cell (Fig. 1). GSLs and GPI-anchored proteins also reside in the plasma membrane, the latter being confined to the outer leaflet of the lipid bilayer. The forest of glycans at the cell surface is often called the glycocalyx and can be visualized by electron microscopy after staining for sugars.
Figure 1.
Glycans that mature in the Golgi. The diagram depicts simple N- and O-glycans attached to glycoproteins, proteoglycans, glycosphingolipids, and a GPI anchor in the plasma membrane. Rather rare O-glycans are found attached to EGF-like repeats (EGF; pink) or thrombospondin repeats (TSR; gray) with a particular consensus sequence. The WxxW motif in a TSR is C-mannosylated. Core regions boxed in teal are sugars added in the ER. The remaining sugars in each class of glycan are added during passage through the cis-, medial-, and trans-Golgi network (TGN) compartments of the Golgi. Abbreviations are: Man, mannose; Gal, galactose; Glc, glucose; GlcNAc, N-acetylglucosamine; GlcNH2, Glucosamine; GlcA, glucuronic acid; IdoA, iduronic acid; GalNAc, N-acetylgalactosamine; Xyl, xylose; Fuc, Fucose; Sia, sialic acid; 3S, 3-O-sulfated; 6S, 6-O-sulfated, PO4−, phosphate. (Modified from Figure 1.6 in Essentials of glycobiology, with permission from Varki and Sharon 2009.)
Glycosylation is the most common posttranslational modification of proteins. Mature glycans at any one glycosylation site may be as simple as a single sugar, or as complex as a polymer of more than 200 sugars, potentially modified with phosphate, sulfate, acetate, or phosphorylcholine. Most importantly, glycans are often branched. For example, a complex N-glycan (Fig. 1) may have up to six branches or antennae, and each antenna may contain many repeating disaccharide units. This article will describe the nature of resident Golgi GTs and other activities involved in Golgi glycosylation from entry into the cis-Golgi through passage to the trans-Golgi network (TGN). The focus is on mammalian Golgi glycosylation but comparisons with yeast, Caenorhabditis elegans, and Drosophila are made where appropriate.
GOLGI GLYCOSYLTRANSFERASES
General Characteristics
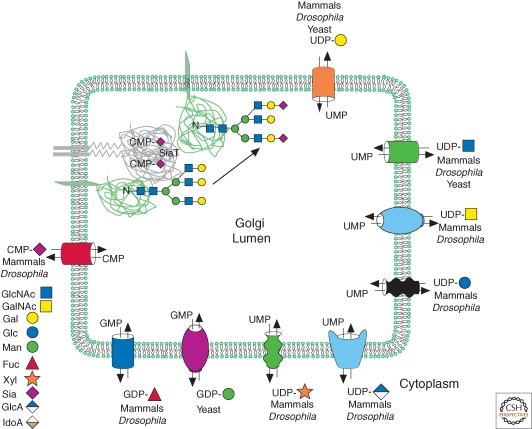
In mammals, there are more than 250 GTs that reside in the Golgi where they catalyze the transfer of one sugar to another sugar on a glycan acceptor, generally covalently attached to protein or lipid. GTs are grouped into families of related catalytic or sequence activity in the Carbohydrate Active Enzymes database (CAZy) (Cantarel et al. 2009). There are, for example, 20 mammalian sialyltransferases (Takashima 2008). By contrast, Drosophila has a single sialyltransferase (Koles et al. 2004), and yeast have none. Most GTs transfer a single sugar to an acceptor but there are a few GTs that catalyze the transfer of two different sugars, typically to generate a polymer of repeating units, as in proteoglycans (Fig. 1). The Golgi GT is a type II transmembrane protein with a short amino-terminal cytoplasmic tail in the cytosol, a transmembrane domain, a stalk-like stem region, and a globular catalytic domain in the Golgi lumen (Fig. 2). Some GTs contain two functional domains. For example, several polypeptide GalNAcTs have a lectin domain that binds to GalNAc and a catalytic domain that transfers GalNAc (Hassan et al. 2000; Fritz et al. 2006); a family of N-deacetylase/N-sulfotransferases are important in heparan sulfate biosynthesis (Aikawa et al. 2001); and the putative glycosyltransferase Large has two glycosyltransferase domains (Longman et al. 2003; Aguilan et al. 2009). GTs are often glycosylated by other GTs or, in some cases, by autocatalytic transferase activity. The glycans on some GTs are required for their correct folding during synthesis, and these GTs often lack catalytic activity when generated in bacteria. Interestingly, many GTs are cleaved in their stem region by proteases in the Golgi and secreted from the cell (Shifley and Cole 2008; Kitazume et al. 2009). This may be a mechanism to regulate their activity in the Golgi, or to allow them to act at the cell surface or in the extracellular environment. The latter would require nucleotide sugars to be available outside the cell. Intriguingly, nucleotide sugar transporters of the Golgi have been found to facilitate nucleotide sugar release from the cell (Sesma et al. 2009).
Figure 2.
Glycosyltransferases and nucleotide sugar transporters of the Golgi. The diagram depicts a variety of nucleotide sugar transporters that transfer a nucleotide sugar from the cytoplasm into the Golgi lumen in exchange for a nucleotide monophosphate generated by hydrolysis of the nucleotide diphosphate released after transfer of the sugar to an acceptor. A typical type II transmembrane glycosyltransferase (a sialyltransferase; gray) with its short cytoplasmic tail, transmembrane domain, extended stem region, and globular catalytic domain is shown binding CMP-Sia from which it transfers Sia to Gal on a complex N-glycan on a glycoprotein (green). Abbreviations are defined in the legend to Figure 1. (Modified from Figure 4.4 in Essentials of glycobiology, with permission from Freeze and Elbein 2009.)
General GT Reaction
The general reaction catalyzed by a Golgi GT is shown for a sialyltransferase (SiaT) in Figure 2. The sugar transferred by each GT comes from a high energy nucleotide sugar which is synthesized in the cytoplasm (or nucleus in the case of CMP-Sia), and must be imported into the Golgi lumen. This is achieved by nucleotide sugar transporters (Berninsone and Hirschberg 2000), families of multitransmembrane transporters (Cantarel et al. 2009) that reside throughout the compartments of the Golgi (Fig. 2). Importantly, mammals do not have a GDP-Man transporter but use Dolichol-P-Man as the donor for the transfer of core Man residues in the ER (Fig. 1, green shaded Man residues). Nucleotide sugar transport is driven by reverse transport of the monophosphate nucleotide generated by Golgi resident pyrophosphorylases that hydrolyse the released nucleotide diphosphate following sugar transfer (Berninsone and Hirschberg 2000). The addition of each sugar creates the substrate or acceptor of the next GT. Many glycosyltransferases require a metal ion to optimize catalysis. In the case of N-glycans linked via Asn to protein, the acceptor substrate is often a specifically branched glycan with a particular sugar composition (Stanley et al. 2009). Each sugar within a glycan may be substituted with another sugar at any carbon with a free hydroxyl group, and thus branched antennae are a feature of many glycans (Fig. 1). The specificity of each GT is defined not only by the sugar it transfers but also by the sugar transferred to and the glycosidic linkage formed. In fact, sequence comparisons group GTs based on the glycosidic linkage catalysed rather than the sugar transferred (Oriol et al. 1999).
Targeting GTs to the Golgi
Each Golgi GT transfers a sugar to a specific acceptor generated by preceding GTs, and must act at a particular stage in the glycosylation pathway. Thus, Golgi GTs must be appropriately localized in the cis-, medial-, trans-Golgi, or the TGN. This does not mean that a GT needs to be strictly confined to one Golgi compartment. In fact, Golgi GTs cycle from the Golgi to the ER, and at any time, a small proportion of those GTs tested have been found in the ER (Rhee et al. 2005). Nevertheless, an appropriate proportion of the enzyme population must be in the right place for a sufficient amount of time to act on glycoconjugates carrying the appropriate substrate. The factors that localize Golgi GTs to a particular compartment of the Golgi include sequence motifs located in the cytosolic domain, the transmembrane domain and/or the stem region, sometimes leading to oligomerization as shown for the sialyltransferase in Figure 2 (Colley et al. 1992; Fenteany and Colley 2005). Enzymes that act sequentially in a glycosylation pathway may also be found in a complex. For example, GlcNAcT-I, the GT that initiates the synthesis of complex N-glycans, interacts with α-mannosidase II, which removes two Man residues, and GlcNAcT-II which acts next in the pathway, by a mechanism termed kin recognition (Nilsson et al. 1993b, 1994, 1996; Opat et al. 2000). Golgi GTs generally have a shorter transmembrane domain than glycoproteins that span the plasma membrane, and this is proposed to promote retention of GTs in the Golgi (Munro 1995). In some cases, a Golgi GT associates with another protein, which is important for Golgi localization or transferase activity. For example, T-synthase must be synthesized together with the chaperone COSMC to acquire activity, and the complex constitutes the active enzyme in the Golgi (Ju and Cummings 2002; Aryal et al. 2010). The Drosophila protein pipe is similar to mammalian enzymes that modify glycosaminoglycans, and requires another protein termed “windbeutel” to localize to the Golgi (Sen et al. 2000). A glycoprotein inhibitor of complex and hybrid N-glycan synthesis (GnT1IP) forms a complex with GlcNAcT-I, α-mannosidase IIX and/or GlcNAcT-III, but inhibits the enzyme activity of only GlcNAcT-I (Huang and Stanley 2010). An extensive discussion of the mechanisms of Golgi resident protein retention and compartmental localization is presented in Banfield 2011.
Sugars Added and Removed in the Golgi
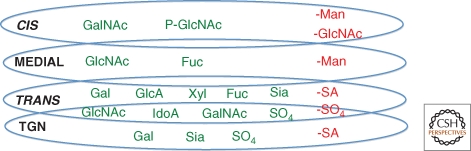
The diagram in Figure 3 depicts which sugars are added by glycosyltransferases, may be removed by glycosidases, or modified by epimerases and sulfotransferases in the different Golgi compartments. The compartments are shown as overlapping to denote the anterograde and retrograde vesicular trafficking of Golgi GTs known to occur. Because of the dynamic and competitive nature of Golgi glycan synthesis, glycans on a mature glycoconjugate are a heterogeneous collection, even at a single glycosylation site. An immature glycan may be a substrate for many different GTs that compete, and a glycoconjugate may transit a Golgi compartment too quickly to be acted on by all GTs capable of using it as a substrate. Thus, a purified glycoprotein population will include many glycoforms and these may vary in functional activity. Nevertheless, in this sea of heterogeneity, there often exists a strategically placed glycan whose precise structure is the key to a biological activity of the molecule, as in the case of the P-selectin glycoprotein ligand (PSGL-1) (Somers et al. 2000). In PSGL-1, an amino-terminal core 2 O-glycan with a sialylated Lewis X glycan determinant and adjacent sulfated tyrosine(s) are required for the binding of PSGL-1 by P-selectin (Xia et al. 2003). The related L-selectin binds ligands with a sialylated Lewis X determinant that is sulfated (Hiraoka et al. 1999, 2004).
Figure 3.
Compartmentalization of sugar addition and removal in the Golgi. Although Golgi membranes are dynamically recycling, compartments corresponding to cis-, medial-, and trans-Golgi and the TGN are identified by resident proteins that mostly localize to a particular compartment (Nilsson et al. 1993a; Rabouille et al. 1995). The diagram shows in which Golgi compartment particular sugars are added or generated (green) or may be removed (red). Compartments are shown overlapping to signify that distinctions are not precise. Sugars added in the ER or ERGIC compartments before the cis-Golgi are boxed in teal in Figure 1. Abbreviations are defined in the legend to Figure 1.
Sulfation of specific OH and NH2 groups on sugars is a very important feature of proteoglycans (Selleck 2000; Fig. 1). Phosphorylation is required on high mannose N-glycans of lysosomal hydrolases for them to be recognized by mannose-6-phosphate receptors and routed to lysosomes (Kornfeld 1990; Fig. 1). Therefore, the glycans that emerge from the TGN may be quite simple or extremely complex with several branches that contain linear polymeric chains of sugars modified by sulfate, phosphate, or potentially other moieties. For example, in C. elegans some glycans contain phosphorylcholine (Cipollo et al. 2005). In additon, a glycoprotein often carries glycans of many different types. Thus, GPI-anchored proteins usually possess N- and/or O-glycans, glycoproteins may carry one or more glycosaminoglycan chains, proteoglycans may carry N- and O-glycans, and molecules with EGF-like (EGF) or thrombospondin (TSR) repeats may carry many different classes of glycan (Fig. 1). For example, Notch1 has 36 EGF repeats, some of which may carry O-fucose-, O-glucose- and O-GlcNAc glycans, and an N-glycan (Stanley and Okajima 2010).
PROTEIN AND LIPID GLYCOSYLATION IN THE GOLGI
N-Glycosylation
Glycoproteins that are N-glycosylated arrive in the cis-Golgi carrying N-glycans added cotranslationally in the ER (Kelleher and Gilmore 2006) to a proportion of their Asn-X (not Pro)-Ser/Thr (rarely Cys) sites (Zielinska et al. 2010). The consensus motif for N-glycosylation is necessary but not sufficient for N-glycosylation to occur. The N-glycans on glycoproteins in the cis-Golgi are of the high mannose type and usually contain eight or nine mannose (Man) residues (Stanley et al. 2009). These may remain unchanged during passage through the Golgi and be present on cell surface or secreted glycoproteins (Fig. 1). However, N-glycans are often processed in the Golgi, initially in the cis-Golgi by a set of α-mannosidases that remove Man residues to generate the Man5GlcNAc2Asn intermediate which is the substrate of the medial Golgi GT GlcNAcT-I. The transfer of GlcNAc to Man5GlcNAc2Asn by GlcNAcT-I initiates the synthesis of hybrid and complex N-glycans. Hybrid N-glycans keep the five Man residues and extend the arm that received GlcNAc by adding Gal and sialic acid and/or other sugars. To become complex, N-glycans lose the terminal two of the five Man residues and acquire a second GlcNAc to form a biantennary, complex N-glycan (Fig. 1). This may be further branched up to six times and each branch may be elongated by the addition of different sugars including Gal, GlcNAc, GalNAc, Fuc, Sia, and disaccharide units (Stanley et al. 2009). Even a simple CHO cell has complex N-glycans that may carry 60 or more sugar residues (North et al. 2010).
Drosophila glycoproteins carry mainly high mannose N-glycans sensitive to cleavage by endoglycosidase H (Endo H), but may also carry complex N-glycans (Aoki et al. 2007; Koles et al. 2007). The complex N-glycans have few sugars. Some neural glycoproteins have N-glycans with terminal sialic acid, and other glycoproteins contain glucuronic acid in antennae and fucose in the core region of N-glycans (Aoki and Tiemeyer 2010; Rendic et al. 2010). C. elegans has high mannose and complex N-glycans with some specialized features such as the incorporation of phosphorylcholine on GlcNAc (Cipollo et al. 2005). Yeast glycoproteins enter the cis-Golgi with mainly Man8GlcNAc2Asn at N-glycan sites, the same as Drosophila and mammals. However, these N-glycans quickly acquire additional Man residues and ultimately form yeast mannan with long chains of Man that may be substituted with phosphate (Munro 2001).
O-Glycosylation
The initiation of O-glycosylation also occurs in the ER for most O-glycans and consists of the addition of only a single sugar residue to Ser or Thr. The most abundant types of O-glycosylation are represented by the O-glycans termed mucins initiated by GalNAc-Ser/Thr and the glycosaminoglycan (GAG) chains on proteoglycans initiated by Xyl-Ser (Fig. 1). Mucin O-glycans are extended in the Golgi by the addition of Gal, GlcNAc, sialic acid and fucose to form linear or branched O-GalNAc glycans (Brockhausen et al. 2009)(Fig. 1). There are more than 15 polypeptide GalNAc transferases (ppGalNAcT) in mammals and several in Drosophila (Zhang and Ten Hagen 2010), but none in yeast. Much work has been devoted to determining consensus sites for O-GalNAc addition to Ser or Thr and criteria are beginning to appear (Caragea et al. 2007; Gerken et al. 2008).
GAG chains attached to Ser have a common core of four sugars (Fig. 1, green shaded) completed in the early Golgi and extended by disaccharide units to form long linear polymers to generate heparan sulfate and chondroitin sulfate (Fig. 1). Characteristic of GAGs is the modification of their sugars by sulfate at specific positions. These modifications occur in blocks that generate discrete regions of binding specificity and have been referred to in heparan sulfate and heparin as the “HS code” (Bulow et al. 2008). Sulfotransferases and sulfamidases of the Golgi may dynamically regulate such a code. Drosophila and C. elegans synthesize GAGs and proteoglycans, but yeast do not. The consensus site for addition of xylose and initiation of a GAG chain is a-a-a-a-G-S-G-a-a/G-a (“a” representing Asp or Glu) (Roch et al. 2010).
The EGF-like repeats in the extracellular domain of Notch and other vertebrate proteins are modified by O-fucose and O-glucose glycans (Rampal et al. 2007; Stanley and Okajima 2010), as well as O-GlcNAc (Matsuura et al. 2008; Fig. 1). The first sugar in each case is added in the ER and requires a correctly folded EGF-like repeat that contains a specific consensus site (Panin et al. 2002; Takeuchi and Haltiwanger 2010; T. Okajima, personal communication). O-fucose and O-glucose glycans are known to be extended in the Golgi of mammalian cells (Fig. 1), and there is the potential for O-GlcNAc to acquire Gal, GlcNAc, sialic acid, and fucose. Drosophila also expresses these O-glycans on EGF-like repeats of Notch and other proteins. However, O-fucose glycans in Drosophila may have a glucuronic acid attached to the O-fucose (Aoki et al. 2008), and there is no evidence for the addition of Gal or sialic acid to the GlcNAc (Xu et al. 2007). C. elegans has protein O-fucosyltransferase 1 (Loriol et al. 2006), but no close homologue of Fringe, the transferase that adds GlcNAc to Fucose-O-EGF (Bruckner et al. 2000; Moloney et al. 2000). The O-glucose glycans on EGF repeats appear to have emerged with Notch signaling in the metazoa (Acar et al. 2008; Sethi et al. 2010).
Thrombospondins and other proteins with TSP repeats have consensus sites for two unusual glycans (Fig. 1). The addition of C-linked Man to tryptophan is thought to occur in the ER (Doucey et al. 1998) and no further modification in the Golgi has been observed to date (Wang et al. 2009). The O-fucose glycan of TSP repeats is also initiated in the ER, but it is extended in the Golgi with a single Glc (Kozma et al. 2006).
Yeast are not known to generate any of the O-glycans described above. However, yeast synthesize O-glycans initiated with O-mannose in the ER and extended with Man in the Golgi. Mammals and Drosophila also make O-mannose glycans, initiated from Dol-P-Man in the ER and extended in mammals in the Golgi (Fig. 1). O-mannose glycans are rare and concentrated in brain and other tissues that express α-dystroglycan. They are required on α-dystroglycan for it to functionally bind to the extracellular matrix, and disruptions of O-mannose glycan synthesis lead to muscular dystrophies (Moore and Hewitt 2009; Nakamura et al. 2010).
Glycosylation of Lipids
In mammals, the first sugar to be added to ceramide (Cer) is added in the ER. Glc-Cer may not be further modified during passage through the Golgi, may have one or two sugars added (Fig. 1), or may be extensively modified by a battery of GTs localized in different compartments of the Golgi (Schnaar et al. 2009). GSL may carry up to eight sugars attached to Glc-Cer. Another abundant glycolipid in certain tissues is sulfatide (Fig. 1) (SO4-Gal-Cer).
GPI-Anchors
The synthesis and transfer of a GPI anchor occurs in the ER and the mature ER structure is shown in Figure 1 (teal shaded). A phosphatidylethanolamine is added in the Golgi in mammals (Fujita and Kinoshita 2010). In addition, in the Golgi the Man residues of the anchor may be substituted with additional Man residues or GlcNAc, which can then be extended with Gal and sialic acid (Ferguson et al. 2009). Novel Golgi GTs responsible for generating mature GPI anchors are still being uncovered (Izquierdo et al. 2009). The GPI-anchored protein is finally located in the outer leaflet of the plasma membrane (Fig. 1).
REGULATION AND USES OF GOLGI GLYCOSYLATION
Regulation of Golgi Glycosylation
Glycosylation in the Golgi is controlled by the spectrum of GTs, nucleotide sugar synthases, and transporters expressed by a cell, as well as factors that affect the lumenal Golgi environment, and the structure and organization of Golgi membranes. Changes of Golgi pH have long been known to alter glycosylation, and an anion channel has now been identified that regulates Golgi pH (Maeda et al. 2008; Maeda and Kinoshita 2010). Cells lacking this ion channel transport glycoproteins slowly through the secretory pathway, and the glycosylation of both glycoproteins and glycolipids is truncated in these cells. Interestingly, passage through the Golgi is also slowed if the CMP-Sia or GDP-fucose transporters or both are knocked down using RNAi, or by CMP-Sia transporter mutation in Lec2 CHO cells (Xu et al. 2010). In cells with reduced nucleotide sugar transport, glycoproteins with truncated glycans accumulate in the Golgi rather than proceeding to the plasma membrane or to secretion. In addition, ER stress pathways are activated in cells in which nucleotide sugar transport is inhibited (Xu et al. 2010). In Drosophila cells in culture, separate knockdown of two different ppGalNAcTs causes a slowdown in secretion and an alteration in Golgi organization, as well as reduced transfer of GalNAc to mucin glycoproteins (Zhang and Ten Hagen 2010). Thus, Golgi resident proteins appear to contribute to the overall integrity and function of the Golgi.
Other factors in the Golgi lumen that are important for glycosylation are the nucleotide sugar pyrophosphorylases that hydrolyse released nucleotide diphosphates, as their number and activity will affect nucleotide sugar import (Berninsone and Hirschberg 2000; Fig. 2). Chaperones important for certain glycosyltransferases to leave the ER may also function in the Golgi. For example, COSMC is necessary for T-synthase to move along the secretory pathway and to be active in the Golgi (Wang et al. 2010). In another example, the putative glycosyltansferase Large must physically associate with α-dystroglycan to be functional in modifying this substrate in the Golgi (Kanagawa et al. 2004). Recently, inhibitors of glycosyltransferase activities have been discovered. One of these inhibits the activity of GlcNAcT-I, the transferase that initiates the synthesis of complex and hybrid N-glycans (Huang and Stanley 2010). It is a testis-specific glycoprotein that binds to GlcNAcT-I as well as some other medial Golgi enzymes, but appears to inactivate only GlcNAcT-I. It is termed GlcNAcT-I Inhibitory Protein (GnT1IP) and its expression is tightly regulated during spermatogenesis. Another example is a class of proteins of the transmembrane BAX inhibitor motif-containing (TMBIM) family that inhibit Gb3 synthase that generates the glycolipid Gb3, the receptor of Shiga toxin (Yamaji et al. 2010). Yet another mechanism of glycosylation regulation is to change the Golgi content of a glycosyltransferase by relocation. This has been recently described for several ppGalNAcTs that transfer GalNAc to Ser or Thr and initiate mucin glycan synthesis (Gill et al. 2010). Growth factor stimulation of Src tyrosine kinase activity, or injection of activated Src kinase, causes a large proportion of all ppGalNAcTs tested to shift from the Golgi to the ER, which results in an increase in total GalNAc addition to proteins. This is a COP1-mediated process and is dependent on the Arf1 GTPase. Medial Golgi enzymes tested, or the Golgi tethering protein giantin, were not redistributed by Src activation.
Mutations in proteins of the conserved oligomeric Golgi (COG) complex that provides a scaffold important for Golgi membrane structure and tethering of retrograde vesicles, also cause alterations in glycosylation (Smith and Lupashin 2008). The first member of the eight subunits of the COG complex was discovered in a CHO mutant ldlC that has defects in N- and O-glycan and glycolipid synthesis (Podos et al. 1994). Several COG subunits have now been shown to be mutated and to give rise to glycosylation defects in patients with congenital diseases of glycosylation (Zeevaert et al. 2008; Foulquier 2009; Lubbehusen et al. 2010). The mechanism by which COG defects alter multiple glycosylation pathways appears to be caused by partial relocation and degradation of Golgi glycosyltransferases and other glycosylation activities when COG is dysfunctional (see Freeze and Elbein 2009).
Golgi Glycosylation and the Cell Biologist
The fact that glycosyltransferase and processing glycosidase reactions are spaced along Golgi membrane compartments means that glycans are gradually built up before exit from the TGN, and may be used as markers of passage through the Golgi. N-glycans in particular have been used extensively to monitor whether a glycoprotein traffics through the Golgi. The general rationale is shown in Figure 4. N-glycans of glycoproteins in the cis-Golgi are all high mannose and are susceptible to removal by cleavage using Endo H or peptide N-glycosidase (N-glycanase). Complex N-glycans with only three Man residues in the core, are resistant to removal by Endo H, but remain sensitive to N-glycanase. Because N-glycans usually contribute substantially to the molecular weight of a glycoprotein, simple comparisons by gel electrophoresis are used to detect glycosidase sensitivity. An important caveat however, is that only resistance to Endo H can be interpreted as evidence of passage through the Golgi. This is because a glycoprotein may fold in such a way that high mannose N-glycans never become complex, even though it passes through the Golgi. The same is true for hybrid N-glycans that contain five core Man residues and never become Endo H resistant.
Figure 4.
Golgi glycans as tags. Glycoproteins in the ER, ERGIC, and cis-Golgi have high mannose N-glycans that are all susceptible to release by Endo H. Processing α-mannosidases generate the Man5GlcNAc2Asn N-glycan shown above in the medial-Golgi to which GlcNAcT-I transfers a GlcNAc on the left terminal Man residue. Hybrid N-glycans result from the extension of this GlcNAc but the Man5GlcNAc2Asn core is not further processed and remains sensitive to Endo H. Complex N-glycans are formed by the removal of two Man residues following the action of GlcNAcT-I and the addition of a second GlcNAc by GlcNAcT-II. Further extension gives rise to the biantennary complex N-glycan shown in the diagram. Complex N-glycans are resistant to Endo H. Both high mannose and complex N-glycans are sensitive to removal by N-glycanase, whose action generates an Asp in place of Asn, a change that may be used to identify N-glycosylation sites by mass spectrometry.
Other glycosidases may also be used to detect exposure to a particular Golgi compartment. The presence of sialic acid residues on glycoproteins exposed to the trans-Golgi or TGN may be ascertained by sialidase treatment. Gal residues added in the trans-Golgi can be removed by treatment with a galactosidase and GlcNAc residues added in the medial Golgi can be removed using hexosaminidases. Monitoring sensitivity by gel electrophoresis is feasible provided at least two to three sugars are removed or sugars are specifically labeled.
Experimental approaches that have been very productive and still have much potential are the use of glycosylation mutants in intracellular trafficking assays. This approach was pioneered by James Rothman using the CHO mutant 15B (or Lec1) in mammalian cells (Fries and Rothman 1980). These mutants lack GlcNAcT-I activity in the medial Golgi, and thus by mixing Golgi membranes from mutant with membranes from wild type in vitro, glycoprotein transfer between them could be determined by the acquisition of the product of GlcNAcT-I activity only present in wild type Golgi membranes. This approach was widely adopted and several elegant trafficking assays were designed using CHO mutants with different glycosylation defects (Brandli 1991). Although these assays have now been superseded to a large extent by permeabilized cell and knockdown strategies, there are a battery of glycosylation mutants in CHO cells (Patnaik and Stanley 2006), BHK21 cells (Stojanovic et al. 1984), MDCK cells (Brandli et al. 1988), and HEK293T cells(Reeves et al. 2002; Crispin et al. 2009) that could be helpful. Mutant lines with a well-characterized defect in glycan biosynthesis have many applications such as determining functions for glycans in cell-cell and cell-pathogen interactions, and generating recombinant molecules with tailored glycans for functional studies and glycosylation engineering (Stanley 1992). For example, Lec1 CHO cells with inactive GlcNAcT-I (Chen and Stanley 2003), have an intact N-glycan pathway in the ER where N-glycans are important for folding and quality control (Maattanen et al. 2010), through the cis-Golgi and to the medial-Golgi, but glycoproteins made in Lec1 cells never acquire complex or hybrid N-glycans because their synthesis must be initiated by GlcNAcT-I. These high mannose N-glycans may be used to affinity purify a glycoprotein using lectins, to identify mannose binding proteins, to determine if complex N-glycans are necessary for function, and to generate correctly folded glycoproteins with only a single GlcNAc at most (if not all) N-glycan sites by Endo H treatment. The latter has been particularly helpful in generating crystals for structural studies (Bouyain et al. 2005).
CONCLUDING REMARKS
Golgi glycosylation is a complex and highly dynamic process that is essential for the production of fully functional glycoproteins, glycolipids, proteoglycans, and GPI-anchored proteins, and for the timely transport of membrane and secreted proteins. The enzymes, transporters, chaperones, and inhibitors that regulate glycosylation must function together in a crowded environment that is sensitive to perturbations in pH, Golgi peripheral membrane structure and the content of Golgi membrane proteins. Understanding each glycosylation step in all the different pathways, and the consequences of blocking glycosylation at any point, is important for the generation of tools and strategies in cell biology and Golgi research, and for understanding pathogenic mechanisms of Golgi glycosylation malfunction at the cellular and organismal level.
ACKNOWLEDGMENTS
The author thanks Yuko Tashima for critical comments on the manuscript and NCI grants RO1 30645 and 36434 for support.
Footnotes
Editors: Graham Warren and James Rothman
Additional Perspectives on The Golgi available at www.cshperspectives.org
REFERENCES
- Acar M, Jafar-Nejad H, Takeuchi H, Rajan A, Ibrani D, Rana NA, Pan H, Haltiwanger RS, Bellen HJ 2008. Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell 132: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilan JT, Sundaram S, Nieves E, Stanley P 2009. Mutational and functional analysis of Large in a novel CHO glycosylation mutant. Glycobiology 19: 971–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aikawa J, Grobe K, Tsujimoto M, Esko JD 2001. Multiple isozymes of heparan sulfate/heparin GlcNAc N-deacetylase/GlcN N-sulfotransferase. Structure and activity of the fourth member, NDST4. J Biol Chem 276: 5876–5882 [DOI] [PubMed] [Google Scholar]
- Aoki K, Tiemeyer M 2010. The glycomics of glycan glucuronylation in Drosophila melanogaster. Methods Enzymol 480: 297–321 [DOI] [PubMed] [Google Scholar]
- Aoki K, Perlman M, Lim JM, Cantu R, Wells L, Tiemeyer M 2007. Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. J Biol Chem 282: 9127–9142 [DOI] [PubMed] [Google Scholar]
- Aoki K, Porterfield M, Lee SS, Dong B, Nguyen K, McGlamry KH, Tiemeyer M 2008. The diversity of O-linked glycans expressed during Drosophila melanogaster development reflects stage- and tissue-specific requirements for cell signaling. J Biol Chem 283: 30385–30400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryal RP, Ju T, Cummings RD 2010. The endoplasmic reticulum chaperone Cosmc directly promotes in vitro folding of T-synthase. J Biol Chem 285: 2456–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfield DK 2011. Retention mechanisms within the Golgi. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a005264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berninsone PM, Hirschberg CB 2000. Nucleotide sugar transporters of the Golgi apparatus. Curr Opin Struct Biol 10: 542–547 [DOI] [PubMed] [Google Scholar]
- Bouyain S, Longo PA, Li S, Ferguson KM, Leahy DJ 2005. The extracellular region of ErbB4 adopts a tethered conformation in the absence of ligand. Proc Natl Acad Sci 102: 15024–15029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandli AW 1991. Mammalian glycosylation mutants as tools for the analysis and reconstitution of protein transport. Biochem J 276: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandli AW, Hansson GC, Rodriguez-Boulan E, Simons K 1988. A polarized epithelial cell mutant deficient in translocation of UDP- galactose into the Golgi complex. J Biol Chem 263: 16283–16290 [PubMed] [Google Scholar]
- Brockhausen I, Schachter H, Stanley P 2009. O-GalNAc Glycans. In Essentials of glycobiology (ed. Varki A, et al. ), pp. 115–128 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- Bruckner K, Perez L, Clausen H, Cohen S 2000. Glycosyltransferase activity of Fringe modulates Notch-δ interactions. Nature 406: 411–415 [DOI] [PubMed] [Google Scholar]
- Bulow HE, Tjoe N, Townley RA, Didiano D, van Kuppevelt TH, Hobert O 2008. Extracellular sugar modifications provide instructive and cell-specific information for axon-guidance choices. Curr Biol 18: 1978–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res 37: D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caragea C, Sinapov J, Silvescu A, Dobbs D, Honavar V 2007. Glycosylation site prediction using ensembles of Support Vector Machine classifiers. BMC Bioinformatics 8: 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Stanley P 2003. Five Lec1 CHO cell mutants have distinct Mgat1 gene mutations that encode truncated N-acetylglucosaminyltransferase I. Glycobiology 13: 43–50 [DOI] [PubMed] [Google Scholar]
- Cipollo JF, Awad AM, Costello CE, Hirschberg CB 2005. N-Glycans of Caenorhabditis elegans are specific to developmental stages. J Biol Chem 280: 26063–26072 [DOI] [PubMed] [Google Scholar]
- Colley KJ, Lee EU, Paulson JC 1992. The signal anchor and stem regions of the b-galactoside a2,6-sialyltransferase may each act to localize the enzyme to the Golgi apparatus. J Biol Chem 267: 7784–7793 [PubMed] [Google Scholar]
- Crispin M, Chang VT, Harvey DJ, Dwek RA, Evans EJ, Stuart DI, Jones EY, Lord JM, Spooner RA, Davis SJ 2009. A human embryonic kidney 293T cell line mutated at the Golgi α-mannosidase II locus. J Biol Chem 284: 21684–21695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucey MA, Hess D, Cacan R, Hofsteenge J 1998. Protein C-mannosylation is enzyme-catalysed and uses dolichyl-phosphate-mannose as a precursor. Mol Biol Cell 9: 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenteany FH, Colley KJ 2005. Multiple signals are required for α2,6-sialyltransferase (ST6Gal I) oligomerization and Golgi localization. J Biol Chem 280: 5423–5429 [DOI] [PubMed] [Google Scholar]
- Ferguson MAJ, Kinoshita T, Hart GW 2009. Glycosylphosphatidylinositol anchors. In Essentials of glycobiology (ed. Varki A, et al. ), pp. 143–162 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- Foulquier F 2009. COG defects, birth and rise! Biochim Biophys Acta 1792: 896–902 [DOI] [PubMed] [Google Scholar]
- Freeze HH, Elbein AD 2009. Glycosylation precursors. In Essentials of glycobiology (ed. Varki A, et al. ), pp. 47–62 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Fries E, Rothman JE 1980. Transport of vesicular stomatitis virus glycoprotein in a cell-free extract. Proc Natl Acad Sci 77: 3870–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz TA, Raman J, Tabak LA 2006. Dynamic association between the catalytic and lectin domains of human UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferase-2. J Biol Chem 281: 8613–8619 [DOI] [PubMed] [Google Scholar]
- Fujita M, Kinoshita T 2010. Structural remodeling of GPI anchors during biosynthesis and after attachment to proteins. FEBS Lett 584: 1670–1677 [DOI] [PubMed] [Google Scholar]
- Gerken TA, Ten Hagen KG, Jamison O 2008. Conservation of peptide acceptor preferences between Drosophila and mammalian polypeptide-GalNAc transferase ortholog pairs. Glycobiology 18: 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill DJ, Chia J, Senewiratne J, Bard F 2010. Regulation of O-glycosylation through Golgi-to-ER relocation of initiation enzymes. J Cell Biol 189: 843–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan H, Reis CA, Bennett EP, Mirgorodskaya E, Roepstorff P, Hollingsworth MA, Burchell J, Taylor-Papadimitriou J, Clausen H 2000. The lectin domain of UDP-N-acetyl-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase-T4 directs its glycopeptide specificities. J Biol Chem 275: 38197–38205 [DOI] [PubMed] [Google Scholar]
- Hiraoka N, Kawashima H, Petryniak B, Nakayama J, Mitoma J, Marth JD, Lowe JB, Fukuda M 2004. Core 2 branching β1,6-N-acetylglucosaminyltransferase and high endothelial venule-restricted sulfotransferase collaboratively control lymphocyte homing. J Biol Chem 279: 3058–3067 [DOI] [PubMed] [Google Scholar]
- Hiraoka N, Petryniak B, Nakayama J, Tsuboi S, Suzuki M, Yeh JC, Izawa D, Tanaka T, Miyasaka M, Lowe JB, et al. 1999. A novel, high endothelial venule-specific sulfotransferase expresses 6-sulfo sialyl Lewis(x), an L-selectin ligand displayed by CD34. Immunity 11: 79–89 [DOI] [PubMed] [Google Scholar]
- Huang H-H, Stanley P 2010. Testis-specific regulation of complex and hybrid glycan synthesis. J Cell Biol 190: 893–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo L, Nakanishi M, Mehlert A, Machray G, Barton GJ, Ferguson MA 2009. Identification of a glycosylphosphatidylinositol anchor-modifying β1-3 N-acetylglucosaminyl transferase in Trypanosoma brucei. Mol Microbiol 71: 478–491 [DOI] [PubMed] [Google Scholar]
- Ju T, Cummings RD 2002. A unique molecular chaperone Cosmc required for activity of the mammalian core 1 β 3-galactosyltransferase. Proc Natl Acad Sci 99: 16613–16618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagawa M, Saito F, Kunz S, Yoshida-Moriguchi T, Barresi R, Kobayashi YM, Muschler J, Dumanski JP, Michele DE, Oldstone MB, et al. 2004. Molecular recognition by LARGE is essential for expression of functional dystroglycan. Cell 117: 953–964 [DOI] [PubMed] [Google Scholar]
- Kelleher DJ, Gilmore R 2006. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16: 47R–62R [DOI] [PubMed] [Google Scholar]
- Kitazume S, Oka R, Ogawa K, Futakawa S, Hagiwara Y, Takikawa H, Kato M, Kasahara A, Miyoshi E, Taniguchi N, et al. 2009. Molecular insights into β-galactoside α2,6-sialyltransferase secretion in vivo. Glycobiology 19: 479–487 [DOI] [PubMed] [Google Scholar]
- Koles K, Irvine KD, Panin VM 2004. Functional characterization of Drosophila sialyltransferase. J Biol Chem 279: 4346–4357 [DOI] [PubMed] [Google Scholar]
- Koles K, Lim JM, Aoki K, Porterfield M, Tiemeyer M, Wells L, Panin V 2007. Identification of N-glycosylated proteins from the central nervous system of Drosophila melanogaster. Glycobiology 17: 1388–1403 [DOI] [PubMed] [Google Scholar]
- Kornfeld S 1990. Lysosomal enzyme targeting. BiochemSocTrans 18: 367–374 [DOI] [PubMed] [Google Scholar]
- Kozma K, Keusch JJ, Hegemann B, Luther KB, Klein D, Hess D, Haltiwanger RS, Hofsteenge J 2006. Identification and characterization of a β1,3-glucosyltransferase that synthesizes the Glc-β1,3-Fuc disaccharide on thrombospondin type 1 repeats. J Biol Chem 281: 36742–36751 [DOI] [PubMed] [Google Scholar]
- Longman C, Brockington M, Torelli S, Jimenez-Mallebrera C, Kennedy C, Khalil N, Feng L, Saran RK, Voit T, Merlini L, et al. 2003. Mutations in the human LARGE gene cause MDC1D, a novel form of congenital muscular dystrophy with severe mental retardation and abnormal glycosylation of α-dystroglycan. Hum Mol Genet 12: 2853–2861 [DOI] [PubMed] [Google Scholar]
- Loriol C, Dupuy F, Rampal R, Dlugosz MA, Haltiwanger RS, Maftah A, Germot A 2006. Molecular evolution of protein O-fucosyltransferase genes and splice variants. Glycobiology 16: 736–747 [DOI] [PubMed] [Google Scholar]
- Lubbehusen J, Thiel C, Rind N, Ungar D, Prinsen BH, de Koning TJ, van Hasselt PM, Korner C 2010. Fatal outcome due to deficiency of subunit 6 of the conserved oligomeric Golgi complex leading to a new type of congenital disorders of glycosylation. Hum Mol Genet 19: 3623–3633 [DOI] [PubMed] [Google Scholar]
- Maattanen P, Gehring K, Bergeron JJ, Thomas DY 2010. Protein quality control in the ER: the recognition of misfolded proteins. Semin Cell Dev Biol 21: 500–511 [DOI] [PubMed] [Google Scholar]
- Maeda Y, Kinoshita T 2010. The acidic environment of the Golgi is critical for glycosylation and transport. Methods Enzymol 480: 495–510 [DOI] [PubMed] [Google Scholar]
- Maeda Y, Ide T, Koike M, Uchiyama Y, Kinoshita T 2008. GPHR is a novel anion channel critical for acidification and functions of the Golgi apparatus. Nat Cell Biol 10: 1135–1145 [DOI] [PubMed] [Google Scholar]
- Matsuura A, Ito M, Sakaidani Y, Kondo T, Murakami K, Furukawa K, Nadano D, Matsuda T, Okajima T 2008. O-linked N-acetylglucosamine is present on the extracellular domain of notch receptors. J Biol Chem 283: 35486–35495 [DOI] [PubMed] [Google Scholar]
- Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine KD, Haltiwanger RS, et al. 2000. Fringe is a glycosyltransferase that modifies Notch. Nature 406: 369–375 [DOI] [PubMed] [Google Scholar]
- Moore CJ, Hewitt JE 2009. Dystroglycan glycosylation and muscular dystrophy. Glycoconj J 26: 349–357 [DOI] [PubMed] [Google Scholar]
- Munro S 1995. An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J 14: 4695–4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S 2001. What can yeast tell us about N-linked glycosylation in the Golgi apparatus? FEBS Lett 498: 223–227 [DOI] [PubMed] [Google Scholar]
- Nakamura N, Stalnaker SH, Lyalin D, Lavrova O, Wells L, Panin VM 2010. Drosophila Dystroglycan is a target of O-mannosyltransferase activity of two protein O-mannosyltransferases, Rotated Abdomen and Twisted. Glycobiology 20: 381–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Hoe MH, Slusarewicz P, Rabouille C, Watson R, Hunte F, Watzele G, Berger EG, Warren G 1994. Kin recognition between medial Golgi enzymes in HeLa cells. EMBO J 13: 562–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Pypaert M, Hoe MH, Slusarewicz P, Berger EG, Warren G 1993a. Overlapping distribution of two glycosyltransferases in the Golgi apparatus of HeLa cells. J Cell Biol 120: 5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Rabouille C, Hui N, Watson R, Warren G 1996. The role of the membrane-spanning domain and stalk region of N-acetylglucosaminyltransferase I in retention, kin recognition and structural maintenance of the Golgi apparatus in HeLa cells. J Cell Sci 109: 1975–1989 [DOI] [PubMed] [Google Scholar]
- Nilsson T, Slusarewicz P, Hoe MH, Warren G 1993b. Kin recognition. A model for the retention of Golgi enzymes. FEBS Letters 330: 1–4 [DOI] [PubMed] [Google Scholar]
- North SJ, Huang HH, Sundaram S, Jang-Lee J, Etienne AT, Trollope A, Chalabi S, Dell A, Stanley P, Haslam SM 2010. Glycomics profiling of Chinese hamster ovary cell glycosylation mutants reveals N-glycans of a novel size and complexity. J Biol Chem 285: 5759–5775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opat AS, Houghton F, Gleeson PA 2000. Medial Golgi but Not Late Golgi Glycosyltransferases Exist as High Molecular Weight Complexes. Role of luminal domain in complex formation and localization. J Biol Chem 275: 11836–11845 [DOI] [PubMed] [Google Scholar]
- Oriol R, Mollicone R, Cailleau A, Balanzino L, Breton C 1999. Divergent evolution of fucosyltransferase genes from vertebrates, invertebrates, and bacteria. Glycobiology 9: 323–334 [DOI] [PubMed] [Google Scholar]
- Panin VM, Shao L, Lei L, Moloney DJ, Irvine KD, Haltiwanger RS 2002. Notch ligands are substrates for protein O-fucosyltransferase-1 and Fringe. J Biol Chem 277: 29945–29952 [DOI] [PubMed] [Google Scholar]
- Patnaik SK, Stanley P 2006. Lectin-resistant CHO glycosylation mutants. Methods Enzymol 416: 159–182 [DOI] [PubMed] [Google Scholar]
- Podos SD, Reddy P, Ashkenas J, Krieger M 1994. LDLC encodes a brefeldin A-sensitive, peripheral Golgi protein required for normal Golgi function. J Cell Biol 127: 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C, Hui N, Hunte F, Kieckbusch R, Berger EG, Warren G, Nilsson T 1995. Mapping the distribution of Golgi enzymes involved in the construction of complex oligosaccharides. J Cell Sci 108: 1617–1627 [DOI] [PubMed] [Google Scholar]
- Rampal R, Luther KB, Haltiwanger RS 2007. Notch signaling in normal and disease States: possible therapies related to glycosylation. Curr Mol Med 7: 427–445 [DOI] [PubMed] [Google Scholar]
- Reeves PJ, Callewaert N, Contreras R, Khorana HG 2002. Structure and function in rhodopsin: High-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci 99: 13419–13424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendic D, Sharrow M, Katoh T, Overcarsh B, Nguyen K, Kapurch J, Aoki K, Wilson IB, Tiemeyer M 2010. Neural-specific {α}3-fucosylation of N-linked glycans in the Drosophila embryo requires Fucosyltransferase A and influences developmental signaling associated with O-glycosylation. Glycobiology 20: 1353–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SW, Starr T, Forsten-Williams K, Storrie B 2005. The steady-state distribution of glycosyltransferases between the Golgi apparatus and the endoplasmic reticulum is approximately 90:10. Traffic 6: 978–990 [DOI] [PubMed] [Google Scholar]
- Roch C, Kuhn J, Kleesiek K, Gotting C 2010. Differences in gene expression of human xylosyltransferases and determination of acceptor specificities for various proteoglycans. Biochem Biophys Res Commun 391: 685–691 [DOI] [PubMed] [Google Scholar]
- Schnaar RL, Suzuki A, Stanley P 2009. Glycosphingolipids. In Essentials of glycobiology (ed. Varki A, et al. ), pp. 129–142 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Selleck SB 2000. Proteoglycans and pattern formation: Sugar biochemistry meets developmental genetics. Trends Genet 16: 206–212 [DOI] [PubMed] [Google Scholar]
- Sen J, Goltz JS, Konsolaki M, Schupbach T, Stein D 2000. Windbeutel is required for function and correct subcellular localization of the Drosophila patterning protein Pipe. Development 127: 5541–5550 [DOI] [PubMed] [Google Scholar]
- Sesma JI, Esther CR Jr, Kreda SM, Jones L, O’Neal W, Nishihara S, Nicholas RA, Lazarowski ER 2009. Endoplasmic reticulum/golgi nucleotide sugar transporters contribute to the cellular release of UDP-sugar signaling molecules. J Biol Chem 284: 12572–12583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi MK, Buettner FF, Krylov VB, Takeuchi H, Nifantiev NE, Haltiwanger RS, Gerardy-Schahn R, Bakker H 2010. Identification of glycosyltransferase 8 family members as xylosyltransferases acting on O-glucosylated notch epidermal growth factor repeats. J Biol Chem 285: 1582–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifley ET, Cole SE 2008. Lunatic fringe protein processing by proprotein convertases may contribute to the short protein half-life in the segmentation clock. Biochim Biophys Acta 1783: 2384–2390 [DOI] [PubMed] [Google Scholar]
- Smith RD, Lupashin VV 2008. Role of the conserved oligomeric Golgi (COG) complex in protein glycosylation. Carbohydr Res 343: 2024–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers WS, Tang J, Shaw GD, Camphausen RT 2000. Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLe(X) and PSGL-1. Cell 103: 467–479 [DOI] [PubMed] [Google Scholar]
- Stanley P 1992. Glycosylation engineering. Glycobiology 2: 99–107 [DOI] [PubMed] [Google Scholar]
- Stanley P, Okajima T 2010. Roles of glycosylation in Notch signaling. Curr Top Dev Biol 92: 131–164 [DOI] [PubMed] [Google Scholar]
- Stanley P, Schachter H, Taniguchi N 2009. N-Glycans. In Essentials of glycobiology (ed. Varki A, et al. ), pp. 101–114 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Stojanovic D, Vischer P, Hughes RC 1984. Glycosyl transferases of baby hamster kidney cells and ricin-resistant mutants. O-glycan biosynthesis. EurJBiochem 138: 551–562 [DOI] [PubMed] [Google Scholar]
- Takashima S 2008. Characterization of mouse sialyltransferase genes: Their evolution and diversity. Biosci Biotechnol Biochem 72: 1155–1167 [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Haltiwanger RS 2010. Role of glycosylation of Notch in development. Semin Cell Dev Biol 21: 638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Sharon N 2009. Historical Background. In Essentials of glycobiology (ed. Varki A, et al. ), pp. 1–22 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Wang Y, Ju T, Ding X, Xia B, Wang W, Xia L, He M, Cummings RD 2010. Cosmc is an essential chaperone for correct protein O-glycosylation. Proc Natl Acad Sci 107: 9228–9233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LW, Leonhard-Melief C, Haltiwanger RS, Apte SS 2009. Post-translational modification of thrombospondin type-1 repeats in ADAMTS-like 1/punctin-1 by C-mannosylation of tryptophan. J Biol Chem 284: 30004–30015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L, Ramachandran V, McDaniel JM, Nguyen KN, Cummings RD, McEver RP 2003. N-terminal residues in murine P-selectin glycoprotein ligand-1 required for binding to murine P-selectin. Blood 101: 552–559 [DOI] [PubMed] [Google Scholar]
- Xu A, Haines N, Dlugosz M, Rana NA, Takeuchi H, Haltiwanger RS, Irvine KD 2007. In vitro reconstitution of the modulation of Drosophila Notch-ligand binding by Fringe. J Biol Chem 282: 35153–35162 [DOI] [PubMed] [Google Scholar]
- Xu YX, Liu L, Caffaro CE, Hirschberg CB 2010. Inhibition of Golgi apparatus glycosylation causes endoplasmic reticulum stress and decreased protein synthesis. J Biol Chem 285: 24600–24608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji T, Nishikawa K, Hanada K 2010. Transmembrane BAX inhibitor motif containing (TMBIM) family proteins perturb a trans-Golgi network enzyme, Gb3 synthase, and reduce Gb3 biosynthesis. J Biol Chem 285: 35505–35518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaert R, Foulquier F, Jaeken J, Matthijs G 2008. Deficiencies in subunits of the Conserved Oligomeric Golgi (COG) complex define a novel group of Congenital Disorders of Glycosylation. Mol Genet Metab 93: 15–21 [DOI] [PubMed] [Google Scholar]
- Zhang L, Ten Hagen KG 2010. Dissecting the biological role of mucin-type O-glycosylation using RNA interference in Drosophila cell culture. J Biol Chem 285: 34477–34484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinska DF, Gnad F, Wisniewski JR, Mann M 2010. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141: 897–907 [DOI] [PubMed] [Google Scholar]