Abstract
Bacterial cells are frequently exposed to dramatic fluctuations in their environment, which cause perturbation in protein homeostasis and lead to protein misfolding. Bacteria have therefore evolved powerful quality control networks consisting of chaperones and proteases that cooperate to monitor the folding states of proteins and to remove misfolded conformers through either refolding or degradation. The levels of the quality control components are adjusted to the folding state of the cellular proteome through the induction of compartment specific stress responses. In addition, the activities of several quality control components are directly controlled by these stresses, allowing for fast activation. Severe stress can, however, overcome the protective function of the proteostasis network leading to the formation of protein aggregates, which are sequestered at the cell poles. Protein aggregates are either solubilized by AAA+ chaperones or eliminated through cell division, allowing for the generation of damage-free daughter cells.
Bacteria combat stresses that cause proteins to misfold by up-regulating chaperones and proteases that remove them. Protein aggregates formed under severe stress are solubilized by AAA+ chaperones or lost at cell division.
PROTEIN MISFOLDING AND QUALITY CONTROL SYSTEMS
Although the general principles governing protein folding are similar in all organisms, there are a number of important differences in the folding environments of bacteria and eukaryotic cells. For example, the rate of polypeptide elongation during protein synthesis is significantly faster in bacteria (20 amino acids/sec) compared to eukaryotes (4 amino acids/sec). This difference has an intrinsic impact on cotranslational protein folding. In addition, although some bacteria can form biofilms, many bacteria can flourish unicellularly. As a result, they are more directly exposed to such environmental stresses (e.g., radiation, extreme temperatures or oxidative stress) that could interfere with protein folding, than are multicellular organisms.
To ensure protein homeostasis, bacteria have evolved sophisticated quality control systems consisting primarily of chaperones and proteases that exert multiple activities, which can be roughly divided into the following categories: (1) de novo folding of newly synthesized proteins, (2) preventing aggregation of unfolded proteins, (3) removing of terminally misfolded proteins by degradation, and (4) resolubilizing protein aggregates for subsequent refolding or degradation. In this article, we describe the prokaryotic cytosolic proteostasis network with a focus on Escherichia coli. However, because of the conserved nature of the proteostatic network, many of the principles derived from studies of E. coli can be applied to other bacteria.
RIBOSOME-BOUND TRIGGER FACTOR WELCOMES NASCENT POLYPEPTIDE CHAINS
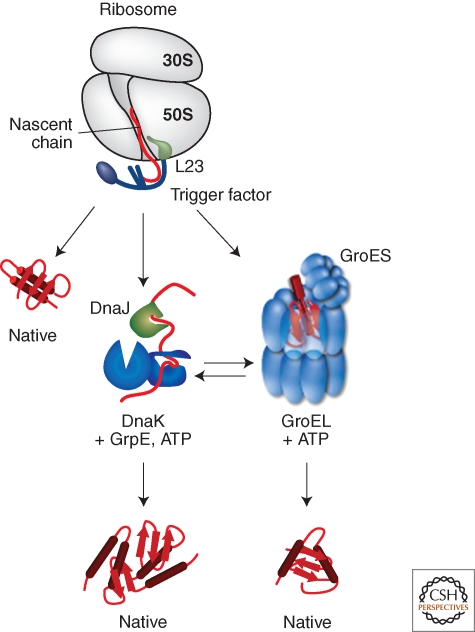
Nascent polypeptides emerge from the polypeptide exit tunnel of the ribosome in a more-or-less unfolded conformation. Consequently, nascent proteins could populate partially folded, aggregation-prone states that require the cotranslational assistance of chaperones to fold correctly. The ribosome-associated chaperone trigger factor (TF) interacts with nascent proteins to promote efficient de novo protein folding (Fig. 1) (Hartl and Hayer-Hartl 2009; Hesterkamp et al. 1996; Kramer et al. 2009; Maier et al. 2005; Valent et al. 1995).
Figure 1.
Interplay of chaperone system during de novo protein folding in E. coli. Nascent polypeptides initially interact with ribosome-bound Trigger Factor (TF), which binds to the ribosomal protein L23. On release from TF, newly synthesized proteins either fold spontaneously (roughly estimated two thirds of cytosolic proteins under physiological growth conditions) or require further folding-assistance by downstream chaperones, namely the Hsp70 chaperone DnaK, which acts together with its co-chaperone DnaJ and the nucleotide exchange factor GrpE, and/or the Hsp60 chaperone GroEL with its co-chaperone GroES. The ATP-dependent DnaK- and GroEL-machineries may act co- and/or posttranslationally.
TF consists of three domains, which are arranged in a long elongated structure. The amino-terminal domain of TF is necessary and sufficient to direct ribosome binding through ribosomal protein L23, which is located near the polypeptide exit channel and allows TF to efficiently interact with nascent polypeptides (Ferbitz et al. 2004; Kramer et al. 2002). Although distant in the amino acid sequence, the amino-terminal domain of TF is structurally adjacent to its carboxy-terminal domain. The carboxy-terminal domain together with the amino-terminal domain forms a cavity whose surface has both hydrophobic and hydrophilic characteristics, potentially enabling TF to promiscuously interact with a variety of substrates (Ferbitz et al. 2004; Lakshmipathy et al. 2007; Martinez-Hackert and Hendrickson 2009; Merz et al 2008). The middle domain of TF is located distal to the amino-terminal ribosome-binding domain and has peptidyl-prolyl cis/trans isomerase (PPIase) activity.
Mutations that inactivate the gene encoding TF cause no obvious phenotype, but they are synthetically lethal when combined with mutations that inactivate the Hsp70 chaperone DnaK. A fragment of TF consisting of a fusion between the amino- and carboxy-terminal domains is sufficient to complement this phenotype, suggesting that the chaperone activity of TF resides in these domains and that the PPIase domain is dispensable for function (Genevaux et al. 2004; Kramer et al. 2004a; Kramer et al. 2004b).
Two distinct mechanisms have been proposed for how TF promotes de novo protein folding: (1) the aqueous cavity formed by the amino- and carboxy-terminal domains promotes cotranslational folding by providing a protective folding space of restricted size. Indeed, the cavity formed by these domains is large enough to fit small folded domains (Hoffmann et al. 2006; Martinez-Hackert and Hendrickson 2009; Merz et al. 2008). (2) TF binds to unfolded nascent polypeptides to delay folding until sufficient information (encoded in more carboxy-terminal residues) is present to allow stable folding of the newly synthesized protein (Agashe et al. 2004). These mechanisms need not be mutually exclusive. For example, partially folded nascent substrates could bind within the protected cavity of TF (i.e., mechanism [1]), but binding could restrict the conformational freedom of the bound substrate thereby delaying folding until enough of the protein is synthesized (i.e., mechanism [2]). Such a hybrid mechanism might also enable TF to act on multidomain proteins by promoting folding of individual domains like beads on a string. Finally, such activity mode would be consistent with recent experiments that suggest TF can hold ribosomal proteins (and perhaps components of other large macromolecular complexes) in a partially folded form that is activated for assembly into the ribosome (Martinez-Hackert and Hendrickson 2009).
COOPERATION OF CHAPERONE SYSTEMS IN PROMOTING PROTEIN FOLDING
Two other chaperone systems that assist folding of newly synthesized proteins are the DnaK and GroE chaperone systems. Depletion of DnaK in strains lacking TF results in massive protein aggregation especially of large-sized proteins and components of protein complexes, which suggests that DnaK plays an important role in de novo protein folding, and indeed, about 5%–18% of nascent proteins have been found to associate with DnaK (Fig. 1) (Deuerling et al. 1999; Teter et al. 1999). In addition, the synthetic lethal growth defect of E. coli cells lacking DnaK and TF can be partially suppressed by overproduction of the GroE chaperone system or SecB, which is normally involved in protein translocation (Ullers et al. 2004; Vorderwulbecke et al. 2004).
DnaK has two functional domains, an amino-terminal nucleotide-binding (NBD) and a carboxy-terminal substrate-binding domain (SBD) that binds substrate segments in a cleft (Flaherty et al. 1990; Zhu et al. 1996). The nucleotide bound by the NBD controls whether the helical lid and the ß-sheet core of the SBD are in an open conformation (ATP-bound state), which allows fast substrate binding and release, or in a closed conformation (ADP-bound state), which results in locking of the substrate chain in the SBD. The DnaK ATPase cycle is controlled by the co-chaperones DnaJ, which targets misfolded substrates to DnaK and concomitantly stimulates ATP hydrolysis, and GrpE, which promotes ADP dissociation and substrate release (Mayer and Bukau 2005). DnaK acts by binding to short hydrophobic peptide segments of substrates and prevents aggregation of aggregation-prone protein conformers (Rüdiger et al. 1997). DnaK binding could also lead to local structural rearrangements in bound substrates, potentially allowing energetically trapped folding intermediates to re-enter a new folding round. In agreement with such activity, DnaK can reactivate some aggregated proteins and causes structural rearrangements in bound substrates (Ben-Zvi et al. 2004; Rodriguez et al. 2008; Skowyra et al. 1990).
The GroE system, which consists of GroEL and its co-chaperone GroES, is the only chaperone system that is known to be essential for viability in E. coli under all growth conditions, and depletion of GroEL results in the aggregation of a multitude of proteins (Chapman et al. 2006; Fayet et al. 1989). This system interacts with 10%–15% of newly synthesized proteins among which are 85 stringent GroEL substrates that cannot use TF or the DnaK system for productive folding (Fig. 1) (Ewalt et al. 1997; Houry et al. 1999; Kerner et al. 2005). In addition, many of these obligate GroEL substrates are essential for viability, which provides an explanation as to why the GroE system is essential (Kerner et al. 2005). Proteins with (αβ)8 TIM-barrel domains are enriched among the stringent GroEL substrates, suggesting a particular role of GroEL in the folding of this protein superfamily (Kerner et al. 2005).
The GroE system promotes folding by an entirely different mechanism compared to either TF or the DnaK system. GroEL oligomerizes to form a two-chambered barrel-like structure, and an oligomeric ring of the co-chaperone GroES caps the end of the GroEL cylinder (Xu et al. 1997). Nonnative polypeptides are captured by binding to hydrophobic substrate binding sites on an open ring of GroEL (Farr et al. 2000). Binding of ATP and GroES induces major conformational changes within GroEL that simultaneously hide the hydrophobic binding sites and create an expanded, closed cavity, which traps the substrate polypeptide in a hydrophilic chamber for protein folding (Horwich et al. 2009). However, the specific mechanism of GroEL chaperone activity is still under debate. GroEL has been proposed to act as a passive Anfinsen cage, which promotes protein folding by sequestering substrates in an isolated environment (Apetri and Horwich 2008; Tyagi et al. 2009). Alternatively, GroEL might induce substrate unfolding prior to encapsulation, which could facilitate productive folding by enabling a new round of folding for trapped, nonnative folding intermediates (Lin et al. 2008; Sharma et al. 2008; Tang et al. 2006).
Although not directly involved in de novo protein folding in the cytoplasm, the dedicated secretion chaperone SecB effectively illustrates how E. coli copes with the challenge to protein homeostasis posed by posttranslational translocation. Most of the proteins in extracytoplasmic compartments are translocated across the cytoplasmic membrane, which requires that proteins be in an unfolded conformation to pass through the membrane-embedded translocation machinery (Driessen and Nouwen 2008; Rapoport 2007). About 90% of the soluble proteins exported by this system are translocated posttranslationally, resulting in the transient accumulation of substrates in the cytoplasm (Huber et al. 2005). Because folded proteins cannot be translocated across the membrane by the Sec system, SecB holds newly synthesized Sec substrates in an unfolded, translocation-competent conformation by wrapping them around the SecB tetramer (Crane et al. 2006). SecB then delivers its substrates to SecA, the secretion motor protein, for ATP-driven translocation through the SecY translocon of the inner membrane.
In summary, although they can substitute for one another to varying extents, each of these chaperone systems (TF, DnaK system, GroEL/ES, and SecB) plays a unique role in maintaining proteostasis in the cytoplasm under normal growth conditions by promoting (or in the case of SecB, inhibiting) folding of newly synthesized proteins.
ADJUSTING QUALITY CONTROL NETWORKS TO ENVIRONMENTAL STRESS: REGULATION OF STRESS RESPONSES IN BACTERIA
As a result of their lifestyle, unicellular prokaryotes (and to a lesser extent, those growing in biofilms) are often exposed to dramatic fluctuations in their environment, which can result in a perturbation of protein homeostasis. For example, the enteric commensals of mammals, such as E. coli, must survive a sudden shift from ambient to body temperature (∼36°C–40°C) at ingestion, acid shock in the stomach (∼pH 1–2), and a return to alkaline pH (∼pH 8–9) in the small intestine to successfully colonize a new host. Free-living organisms, which are more directly exposed to environmental fluctuations, must often survive even harsher folding stresses. These stresses not only disrupt the folding of newly synthesized proteins but can also cause misfolding of already folded proteins. Bacteria have therefore evolved sophisticated stress responses that can react to such threats to proteostasis through the induction of chaperones and proteases.
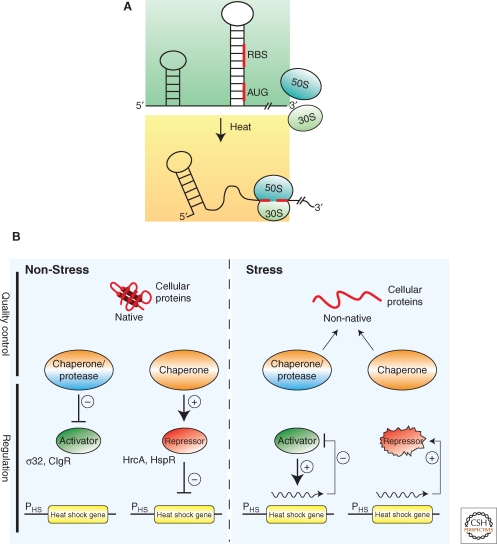
The best-studied folding stress, heat, induces the expression of chaperone and protease genes by various regulatory circuits, which work together to fine-tune the stress response. These circuits can be grouped into two general classes: (i) temperature-responsive mRNA and thermolabile transcriptional regulators that directly respond to temperature changes and (ii) transcriptional regulators that are controlled by chaperones or proteases, which indirectly monitor the general folding state of the cell (Fig. 2).
Figure 2.
Regulation of bacterial stress responses. (A) Principle of RNA thermometers. At low temperatures, the ribosomal binding site (RBS) and the AUG start codon of an mRNA encoding for a stress gene is base paired and not accessible. On heat shock, the structure around the RBS melts allowing for ribosome binding (30S and 50S) and translation. (B) Chaperones and proteases link the cellular folding state to stress gene expression. Under nonstress conditions, expression of stress genes is inhibited through (1) inhibition of transcriptional activators by chaperones or proteases that either sequester the regulators or degrade them or (2) repressor proteins that require chaperone assistance for activity. During environmental stress misfolded proteins accumulate, which titrate chaperones and proteases from their regulatory roles and resulting in the expression of stress genes through (1) release or stabilization of transcriptional activators or (2) inactivation of repressor proteins. Expression of stress genes initiates an inactivation feedback loop restoring nonstress gene expression.
Temperature-responsive RNAs (i.e., RNA thermometers) usually control translation of specific stress genes by occluding the translation initiation sequence and start codon in the mRNA in a hairpin structure (Narberhaus et al. 2006). Increased temperatures cause melting of the hairpin structure, which exposes the translation initiation sequence and permit translation of the stress gene (Fig. 2A). The first RNA thermometer identified in E. coli was the mRNA of rpoH encoding for the heat shock transcription factor σ32, which controls expression of approx. 90 ORFs (Morita et al. 1999; Nonaka et al. 2006). One abundant class of bacterial RNA thermometer, the ROSE element, can be found in the 5’ UTRs of many mRNAs encoding small heat shock proteins (sHsps) (Narberhaus et al. 1998; Waldminghaus et al. 2005), suggesting that the induction of their synthesis on stress treatment is particularly urgent. The ROSE RNA is a sensitive thermometer that gradually exposes the ribosome-binding site at the physiologically relevant temperature range. Its temperature-sensitivity is based on labile non–Watson-Crick base bairing in the melting RNA hairpin structure (Chowdhury et al. 2006). Alternatively, inactivation of temperature-sensitive transcriptional repressors, such as RheA of Streptomyces albus, by heat leads to the induction of heat-stress genes (Servant et al. 2000).
In the second class, chaperones and proteases inactivate transcriptional regulators of folding stress responses by binding or degrading them, respectively. During folding stresses misfolded proteins compete for binding to chaperones (or proteases), which releases or stabilizes these transcriptional regulators. These regulatory circuits allow the cell to respond to any stimulus that affects proteostasis and include a negative feedback loop through the induction of the same proteases and chaperones (Craig and Gross 1991) (Fig. 2B). In one well-characterized example, the DnaK and GroE chaperone systems inactivate σ32 by preventing it from associating with RNA polymerase (Gamer et al. 1992; Guisbert et al. 2004; Liberek et al. 1992). In addition, under physiological growth conditions, the membrane-bound protease FtsH rapidly degrades σ32 and degradation is transiently inhibited during heat stress (Tomoyasu et al. 1995). Inactivation by chaperones plays a particularly important role in the shut off of the heat shock response (Straus et al. 1990; Tilly et al. 1983). Notably, the DnaK binding site in σ32 resides is an exposed peptide stretch that is no longer accessible for chaperone interaction on complex formation with RNA polymerase (Rodriguez et al. 2008). Other examples of this kind of regulation are the repressors HrcA and HspR, which control chaperone gene expression in Gram-positive bacteria and require chaperone assistance for activity (Bucca et al. 2000; Mogk et al. 1997), and the transcriptional regulators CtsR from Bacillus subtilis and ClgR from Corynebacterium glutamicum, which are degraded by AAA+ proteases (Engels et al. 2005; Kirstein et al. 2007; Kruger et al. 2001). In all cases, an “unfolded protein titration model” permits the coupling of an imbalance in protein homeostasis to changes in stress gene expression (Bukau 1993; Craig and Gross 1991) (Fig. 2B). An even more direct sensing of misfolded proteins is key to the periplasmic stress response in E. coli, in which the protease DegS serves as a compartment-specific folding sensor. The proteolytic activity of DegS is allosterically activated on binding to the carboxy-terminal tails of outer membrane proteins, which are only accessible in a misassembled state (Hasselblatt et al. 2007; Sohn et al. 2007; Sohn et al. 2009). On activation, DegS degrades RseA, an integral inner membrane protein that functions as an anti-σ factor by sequestering the transcription factor σE (Walsh et al. 2003).
CHALLENGES TO THE QUALITY CONTROL SYSTEMS DURING STRESS CONDITIONS
Exposure of cells to physical and chemical denaturants, such as increased temperature, changes in ionic strength, oxidative stress or the presence of heavy metals, can disturb proteostasis and lead to the accumulation of misfolded proteins, which are at risk of aggregation. Here, we have concentrated on heat stress, because it is the best-characterized stress condition.
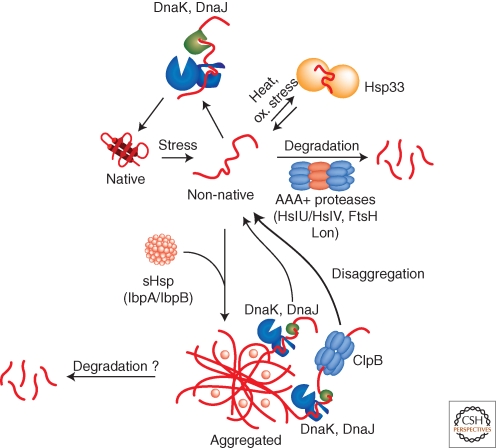
The primary strategy of cellular quality control systems during heat stress is to keep client proteins in a soluble, folding-competent state. Although most chaperone classes have been shown to prevent the aggregation of heat-labile reporters in vitro, they clearly have different functional roles during heat stress in vivo. In E. coli, the DnaK system is most important in preventing the aggregation of misfolded proteins during heat stress (Fig. 3) (Gragerov et al. 1992; Mogk et al. 1999). This role is based on its promiscuous and efficient substrate binding capacity and its high cellular concentration. Accordingly, dnaK mutants show a temperature-sensitive growth phenotype and strongly increased protein aggregation on heat shock (Georgopoulos et al. 1979; Mogk et al. 1999; Paek and Walker 1987). Heat shock temperatures also directly modulate the activity of the DnaK system by causing reversible inactivation of the nucleotide exchange factor GrpE (Groemping and Reinstein 2001). Inactivation of GrpE largely retards substrate release by slowing down ATP/ADP exchange, which freezes DnaK in the high affinity state for substrates. In this sense GrpE acts as a thermosensor and its activity control ensures that substrates remain bound to DnaK during stress conditions.
Figure 3.
Activities of bacterial quality control systems during environmental stress. Environmental stress like heat shock can cause protein-unfolding leading to the accumulation of misfolded protein species. Misfolded proteins are either refolded by the DnaK chaperone and its co-chaperone DnaJ or are removed by AAA+ proteases including e.g., Lon, ClpC/ClpP or HslU/HslV. The holding chaperone Hsp33 becomes important under oxidative and thermal stress and prevents protein aggregation. Severe stress conditions can overburden the protective capacity of quality control systems causing protein aggregation. sHsps coaggregate with misfolded protein species thereby changing the architecture (physical properties) of aggregates and allowing for more efficient protein disaggregation by chaperones. DnaK/DnaJ in cooperation with the AAA+ chaperone ClpB efficiently solubilize protein aggregates by extracting single unfolded protein species, whereas DnaK/DnaJ alone have limited disaggregation capacity. AAA+ proteases (ClpC/ClpP) might also act on aggregated protein species.
Heat stress is often accompanied by oxidative stress. This dual stress situation represents a further threat for cellular quality control as it causes inactivation of DnaK (Winter et al. 2005). The inactivation is caused by a rapid drop in intracellular ATP levels, leading to destabilization of the NBD of DnaK. In this situation Hsp33, a conserved and widespread chaperone in bacteria, takes over for DnaK function by protecting misfolded protein species from aggregation (Fig. 3). The chaperone activity of Hsp33 is directly controlled by environmental stress conditions. Activation of Hsp33 requires both, elevated temperatures (>40°C) and oxidative stress, which results in the formation of active oxidized Hsp33 dimers (Graf and Jakob 2002; Winter et al. 2008). Activated Hsp33 dimers can efficiently prevent the aggregation of misfolded proteins in an ATP-independent manner by forming stable complexes with substrates (Fig. 3). Hsp33 releases its substrates on return to reducing, nonstress conditions, ensuring that substrates stay protected by Hsp33 until permissive conditions for protein folding are restored (Hoffmann et al. 2004). Because DnaK inactivation is reversible, this mechanism also ensures that DnaK can take over for Hsp33 on return to nonstress conditions (Winter et al. 2005).
REMOVAL OF MISFOLDED PROTEINS BY AAA+ PROTEASES
Often, the cell cannot salvage misfolded proteins by the aforementioned chaperone systems. In such cases, prokaryotic cells protect themselves from the potentially detrimental effects of these terminally misfolded protein species by degrading them using AAA+ proteases (Fig. 3) (Goldberg et al. 1981). AAA+ proteases are compartmentalized proteases that consist of two functional units with separate activities: a barrel-shaped proteolytic subunit (e.g., heptameric ClpP or hexameric HslV) and a hexameric member of the AAA+ family of ATPases (e.g., ClpA, ClpC, HslU), which act as chaperones to unfold the protein substrate and feed it into the central proteolytic channel of the protease (Sauer et al. 2004). A number of AAA+ proteases are present in the cytoplasm of E. coli, in which the protease and AAA+ subunits may be encoded in one (e.g., Lon and FtsH), or two (e.g., ClpA/ClpP, ClpX/ClpP, and HslU/HslV) polypeptide chains.
AAA+ proteases act not only in general but also in regulatory proteolysis, in which they control signal transduction pathways by targeting regulators. The presence of multiple AAA+ proteases in the E. coli cytosol can be explained by their diverse functions in controlling many regulatory circuits, but also by overlapping functions in protein quality control. Thus, only E. coli cells deficient for multiple AAA+ proteases show strong growth defects and accumulate misfolded and aggregated proteins (Kanemori et al. 1997). In B. subtilis cells the ClpC/ClpP system plays a major role in the turnover of abnormal proteins, and cells lacking ClpC or ClpP show a temperature-sensitive growth phenotype and accumulate protein aggregates even under nonstress conditions (Kock et al. 2004; Kruger et al. 2000).
The mechanistic principle of substrate selection in general proteolysis by AAA+ proteases has been best characterized for Lon, which plays an important role in the removal of premature translational termination products and proteins containing nonnatural amino acids (Kowit and Goldberg 1977). The recognition of misfolded proteins seems to be largely mediated by the amino-terminal domain of Lon, as respective Lon deletion variants are impaired in the degradation of casein (Roudiak and Shrader 1998). Lon recognizes hydrophobic peptide stretches that are enriched in aromatic residues, which are typically buried in the structure of the folded protein and allows the selective identification of misfolded proteins (Gur and Sauer 2008).
Some misfolded model substrates are stabilized in cells lacking specific chaperone systems, which suggest that AAA+ proteases work together with chaperones in protein degradation (Huang et al. 2001; Kondror et al. 1995). However, the interplay between chaperones and proteases in determining the fate of a misfolded protein (i.e., refolding v. degradation) is not well understood. One possibility is that substrates are kinetically partitioned such that proteins that do not fold rapidly enough are degraded. Different misfolded protein substrates could display different affinities for proteases and chaperones. For example, although both DnaK and Lon recognize hydrophobic sequences, hydrophobic clusters lacking aromatic residues are poor Lon substrates (Gur and Sauer 2008) whereas the interaction of DnaK with a substrate is less dependent on the presence of aromatic amino acids (Rüdiger et al. 1997). It is also possible that chaperones alter the conformation of substrates, rendering them more accessible for proteolysis. One example is σ32, which requires the activity of the DnaK system for efficient degradation by the AAA+ protease FtsH (Tomoyasu et al. 1995). DnaK and DnaJ binding induces conformational changes within the σ32 polypeptide, which may be the prerequisite for recognition by FtsH (Rodriguez et al. 2008). However, in most cases, chaperones and proteases seem to compete for binding to misfolded protein species and their different activities–refolding vs. degradation—can at least partially compensate for the loss of each other. Thus, the AAA+ proteases ClpX/ClpP and Lon become essential for E. coli viability at high temperatures only if the levels of DnaK are reduced (Tomoyasu et al. 2001).
REVERSING PROTEIN AGGREGATION BY AAA+ CHAPERONES
Severe or persistent stress conditions lead to the accumulation of protein aggregates (e.g., approx. 1.5%–3% of the total cellular proteins aggregate after heat shock at 45°C) (Gragerov et al. 1992; Laskowska et al. 1996; Winkler et al. 2010). However, the concerted action of the DnaK system and the AAA+ chaperone ClpB can rescue the proteins contained in these aggregates by resolubilizing and refolding aggregated proteins (Fig. 3) (Goloubinoff et al. 1999; Motohashi et al. 1999; Zolkiewski 1999). The requirement for both of these systems for thermotolerance development (i.e., the ability to transiently survive normally lethal heat stress after acclimation by sublethal heat-treatment) highlights the importance of the disaggregation system in vivo (Squires et al. 1991; Weibezahn et al. 2004).
The mechanism of action of protein disaggregation requires the sequential activity of the DnaK system and ClpB (Weibezahn et al. 2004; Zietkiewicz et al. 2004). Initially, binding of DnaK simultaneously restricts the access of proteases and other chaperones to the aggregate and transfers aggregated polypeptides to ClpB (Haslberger et al. 2007; Haslberger et al. 2008). This transfer involves the ClpB M-domain, which is not present in other AAA+ family members (Haslberger et al. 2007). The mechanism of substrate transfer is unknown but likely involves a direct interaction between DnaK and ClpB as supported by their species-specific cooperation in aggregate solubilization (Glover and Lindquist 1998; Krzewska et al. 2001; Schlee et al. 2004). After substrate transfer, ClpB exerts a pulling force on individual polypeptides by threading them through its central pore, which leads to extraction of misfolded proteins from the aggregate one-by-one (Lum et al. 2004; Schlieker et al. 2004a; Schlieker et al. 2004b; Weibezahn et al. 2004). The pulling force generated by ClpB is coupled to ATP hydrolysis and is mediated by aromatic residues located in the pore of ClpB (Lum et al. 2004; Schlieker et al. 2004b; Weibezahn et al. 2004). Strikingly, ClpB activity is not restricted to accessible amino or carboxyl termini of substrates but can also act on internal substrate segments, involving the threading of looped polypeptide stretches (Haslberger et al. 2008). The processivity of ClpB is interrupted when it encounters a folded domain in a substrate protein, which suggests that ClpB acts by preventing the obstruction of refolding pathways by additional unfolded polypeptide segments (Haslberger et al. 2008).
Like ClpB, the AAA+ subunits of various AAA+ proteases (e.g., ClpA, ClpC) possess a disaggregation activity in vitro, illustrating the unique capacity of the AAA+ protein family to act on protein aggregates (Fig. 3) (Dougan et al. 2002; Schlothauer et al. 2003). In addition, several have suggested that the ClpC/ClpP and ClpE/ClpP AAA+ proteases contribute to protein disaggregation in Bacillus subtilis, which lacks a ClpB homolog. B. subtilis strains lacking ClpE, ClpC or ClpP show a significant delay in protein disaggregation (Kock et al. 2004; Miethke et al. 2006) and ClpC, ClpE and ClpP are recruited to stress-induced protein aggregates in vivo, supporting a role in aggregate clearance (Kirstein et al. 2008; Kruger et al. 2000). Finally, ClpL, another member of the AAA+ protein family, has been implicated in thermotolerance development in some Gram-positive bacteria (Frees et al. 2004; Suokko et al. 2008). ClpL does not harbor a “P-loop,” a short motif that mediates the interaction with ClpP in other AAA+ proteins, which suggests that ClpL could play a role in protein reactivation rather than in degradation.
SMALL HEAT SHOCK PROTEINS MODULATE PROTEIN AGGREGATION TO INCREASE PROTEIN DISAGGREGATION
The small heat shock proteins (sHsps) assist disaggregation by the DnaK/ClpB bi-chaperone system by binding to misfolded proteins and altering the process of aggregation (Haslbeck et al. 2005; van Montfort et al. 2002). The E. coli sHsps IbpA/B (inclusion body binding protein) were initially identified via their tight association with bacterial inclusion bodies and quantitatively associate with stress-induced protein aggregates (Fig. 3) (Allen et al. 1992; Laskowska et al. 2004; Mogk et al. 2003a).
sHsps consist of an amino-terminal domain, a conserved, ∼100 amino acids long α-crystallin domain and a carboxy-terminal domain. The α-crystallin domain is mainly responsible for dimerization of sHsps monomers, whereas the carboxy-terminal extensions promote oligomerization into 12 to >32-mers through contacts with adjacent α-crystallin domains (Kim et al. 1998; van Montfort et al. 2001b). The amino-terminal domains of sHsps have both, divergent sequences and variable lengths and appear to be mainly responsible for substrate binding (Jaya et al. 2009), although contributions of the other domains of sHSPs to substrate binding are not excluded.
sHsps function by binding tightly to misfolded protein species and sequestering them in a partially unfolded state into protein aggregates during stress conditions (Cheng et al. 2008; Friedrich et al. 2004; Mogk et al. 2003b). However, these aggregates are smaller and have an altered composition (Haslbeck et al. 1999; Mogk et al. 2003b), which allows them to be more efficiently resolved by the ClpB/DnaK machinery (Lee et al. 2000; Mogk et al. 2003a; Ratajczak et al. 2009). Moreover, aggregates containing sHsps can be disaggregated by the DnaK system without the assistance of other chaperone systems (Ehrnsperger et al. 1997; Lee et al. 1997; Mogk et al. 2003b). In E. coli the sHsps IbpA and IbpB cooperate to keep aggregated proteins in a disaggregation competent state (Matuszewska et al. 2005). However, IbpA and IbpB show distinct activities. Although IbpA efficiently changes the physical properties of protein aggregates, IbpB is required to promote the subsequent solubilization by disaggregating ClpB/KJE (Ratajczak et al. 2009).
The synthesis and activity of sHsps are tightly regulated by temperature (Haslbeck et al. 2005). Even compared to other chaperones, the synthesis of sHsps on heat stress is strongly induced. In addition, increased temperature causes an activation of sHsps by affecting their oligomeric state and increasing their affinity for substrates. (Haslbeck et al. 1999; Stengel et al. 2010; van Montfort et al. 2001a; van Montfort et al. 2001b). This rapid regulation could ensure that the sHSPs can participate in the organization of protein aggregates already during their formation.
ORGANIZING PROTEIN AGGREGATION: A SECOND LINE OF DEFENSE ALLOWING FOR AGGREGATE CLEARANCE BY ASYMMETRIC DAMAGE INHERITANCE
Occasionally, protein misfolding leads to the formation of aggregates that cannot be efficiently resolved by either the chaperone or proteolytic machineries, and even under physiological growth conditions, a small fraction of wild-type E. coli cells form stable protein aggregates (Lindner et al. 2008; Maisonneuve et al. 2008c). Such terminal aggregates are enriched for aberrant translation products and carbonylated proteins, which have been damaged irreversibly by oxidation (Maisonneuve et al. 2008b; Maisonneuve et al. 2008c). The presence of these aggregates is potentially dangerous for the cell because they could be a template for further aggregation or sequester the chaperones and proteases necessary for maintaining proteostasis, and cells that inherit protein aggregates have slower growth rates than those without (Lindner et al. 2008; Winkler et al. 2010). The finding that nonculturable E. coli cells contain increased amounts of aggregated proteins suggests that there might be a connection between bacterial senescence and protein aggregation (Maisonneuve et al. 2008a).
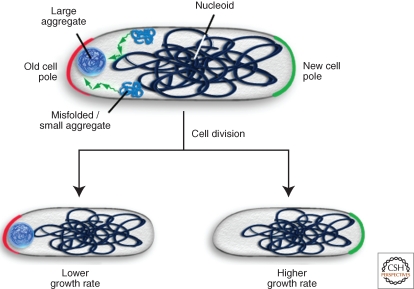
One strategy to limit the damage from terminal aggregates that is used by both prokaryotes and eukaryotes is the asymmetric inheritance of protein aggregates between the two daughter cells resulting from cell division (Aguilaniu et al. 2003; Rujano et al. 2006). For example, E. coli (and other rod-shaped prokaryotes) localizes terminal protein aggregates to insoluble compartments near the cell poles (Fig. 4A) (Lindner et al. 2008; Rokney et al. 2009; Winkler et al. 2010). After cell division in E. coli, each daughter cell contains an old pole and a new pole, which was generated during formation of the septum. E. coli cells that harbor a single aggregate preferentially localize it to the old cell pole (Fig. 4B) (Lindner et al. 2008; Winkler et al. 2010). The asymmetric deposition of aggregates at the old pole allows for the generation of aggregate-free new pole cells through a single division at the expense of old pole cells, which retain the main damage and show reduced growth rates (Fig. 4B). Accordingly, induction of the formation of randomly distributed aggregates using membrane-localized aggregation seeds results in the formation of two daughter cells that grow equally poorly (Winkler et al. 2010).
Figure 4.
Sequestration of protein aggregates at polar sites allows for aggregate clearance by cell division. In E. coli misfolded proteins are deposited during stress conditions as inclusion bodies at polar sites. The sequestration at these sites is driven by nucleoid occlusion resulting in the accumulation of misfolded protein species or small aggregates at the nucleoid-free space. The formation of a single protein aggregate, which is preferentially deposited at the old cell pole, allows for the asymmetric inheritance of aggregated proteins through cell division. New pole cells that are damage-free show higher growth rates at the expense of old pole cells, which retain the damage.
The polar deposition of aggregates in E. coli cells suggests that an active, energy-driven process could be responsible for such localization (Rokney et al. 2009). However, recent work showed that nucleoid occlusion is necessary and sufficient for controlling the polar localization of aggregated proteins, suggesting that E. coli uses a passive mechanism for aggregate sequestration (Fig. 4A) (Winkler et al. 2010).
PROTEOSTASIS IN EXTRACYTOPLASMIC COMPARTMENTS: QUALITY CONTROL IN THE PERIPLASM
In addition to the cytoplasm, bacteria contain a number of extracytoplasmic compartments known collectively as the cell envelope, which in E. coli and other Gram- negative bacteria consists of the cytoplasmic (or inner) membrane, the periplasm, and the outer membrane (Ruiz et al. 2006). The periplasm is a soluble compartment that is sandwiched between the inner and outer membranes and composes approximately 10% of the total cell volume. A number of important functions are performed in this compartment (e.g., maintenance of the peptidoglycan layer, energy metabolism, and assembly of flagella and pili), and as such Gram-negative bacteria export a large number of proteins to carry out these functions. Likewise, although Gram-positive bacteria lack an outer membrane, they do have an interstitial space between the cytoplasmic membrane and their thick cell walls, where many of the same functions are performed. One major difference between these compartments and the endoplasmic reticulum is that the periplasm and interstitial space are devoid of ATP (Ruiz et al. 2006). Thus, these compartments must maintain proteostasis without an obvious source of energy. Here, we focus on the periplasm of E. coli. However, the principles of maintaining proteostasis in the cell envelopes of other bacteria are likely very similar.
Most periplasmic proteins and outer membrane proteins (OMPs) are translocated across the cytoplasmic membrane by the Sec machinery (Driessen and Nouwen 2008; Rapoport 2007). Because this machinery requires that the substrate proteins pass through the SecYEG translocon channel in an unfolded conformation and folding proceeds from amino to carboxyl terminus, de novo folding of Sec-translocated proteins is similar in many respects to cotranslational folding in the cytoplasm. The chaperones with SurA, Skp, PpiD, FkpA, and PpiA interact with newly translocated proteins with broad specificity to assist de novo folding (Harms et al. 2001; Mogensen and Otzen 2005; Schafer et al. 1999). Many of these chaperones (particularly Skp, SurA, and PpiD) play an important role in the proper assembly of OMPs into the outer membrane, presumably by keeping them in an unfolded or semi-folded, insertion-competent state (Chen and Henning 1996; Dartigalongue and Raina 1998; Harms et al. 2001; Lazar and Kolter 1996; Rouviere and Gross 1996; Schafer et al. 1999) (Fig. 5), and SurA and FkpA have been suggested to play a role in the folding of soluble proteins (Arie et al. 2001; Bothmann and Pluckthun 2000; Ramm and Pluckthun 2000; Walton and Sousa 2004). In addition, the chaperones of chaperone-usher secretion pathways (e.g., PapD, FimC) assist the folding of pilus subunits by providing a folding template for their specific substrates (e.g., subunits of the P- or type 1 pili, respectively) and assist in their assembly into the pilus. In the absence of these chaperones, pilus subunits misfold and aggregate (Barnhart et al. 2000; Dodson et al. 1993; Thanassi et al. 1998; Waksman and Hultgren 2009). Finally, some periplasmic chaperones, such as HdeA, bind to and coaggregate with substrate proteins under conditions of folding stress. Interestingly, HdeA, which is activated by acid stress, is so far unique in that it can promote refolding of aggregated substrates after a return to neutral pH in the absence of ATP (Tapley et al. 2010).
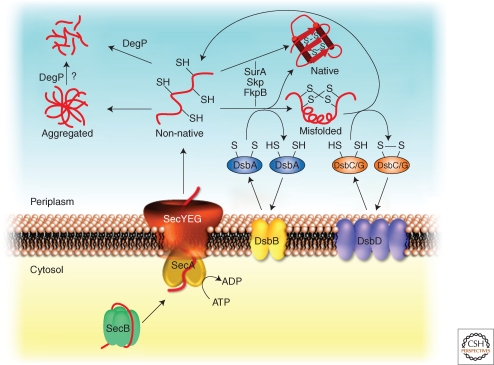
Figure 5.
Periplasmic quality control system. In the cytosol secretory proteins are kept in an export-competent conformation through association with the chaperone SecB. On delivery to SecA, the secretory proteins are translocated through the SecYEG translocon into the periplasm. Folding of newly imported proteins is supported by periplasmic chaperones including Skp, FkpB or SurA. Oxidative folding (formation of disulfide bonds) is catalyzed by DsbA, whereas incorrect disulfides are isomerized by DsbC/DsbG. Oxidized DsbA and DsbC/G are regenerated by DsbB and DsbD, respectively. Misfolded proteins are degraded by DegP. High levels of misfolded protein species can cause the generation of protein aggregates that might be removed by DegP.
A particularly important component of the periplasmic proteostasis network is DegP (HtrA). DegP has both protease and chaperone activities, and cells lacking DegP display a temperature-sensitive growth phenotype, which suggests that it plays an important role in maintaining proteostasis (Meltzer et al. 2009). This central role is also reflected by the dual control of degP gene expression, which can be activated by both periplasmic stress responses, the Cpx and the σE signaling systems (Ruiz and Silhavy 2005). In the presence of misfolded substrates or thermal stress, inactive hexameric DegP assembles into proteolytically active 12- and 24-mers (Krojer et al. 2008). These enormous structures are shaped like hollow spheres with an internal volume large enough to fit a folded or nearly-folded β-barrel OMP, and the active sites of the DegP subunits face the inside of this cavity. Thus, it has been proposed that DegP functions by a sink-or-swim type mechanism in which proteins that don’t fold rapidly enough are degraded (Huber and Bukau 2008; Krojer et al. 2008). Although it is clear that DegP plays an important role in degrading misfolded proteins and perhaps some protein aggregates, it remains a mystery how E. coli rids itself of terminally aggregated periplasmic proteins in the absence of an obvious energy source such as ATP (Fig. 5).
Many extracytoplasmic proteins contain disulfide bonds between cysteines within a polypeptide chain, which can serve to stabilize their tertiary structure. A dedicated oxidative machinery, consisting of DsbA and DsbB, catalyzes the formation of disulfide bonds and can act on a substrate polypeptide during or after translocation across the cytoplasmic membrane (Fig. 5) (Ito and Inaba 2008; Kadokura et al. 2003). DsbA (a member of the thioredoxin superfamily) oxidizes substrate proteins by transferring the disulfide bond at its active site and is then reoxidized by the integral cytoplasmic membrane protein DsbB, which in turn transfers its electrons to quinones that feed into the electron transport chain (Bardwell et al. 1993; Dailey and Berg 1993; Inaba et al. 2006a; Inaba et al. 2006b; Kamitani et al. 1992; Missiakas et al. 1993). Alternatively, many bacteria contain a homologue of VKOR (vitamin K epoxide reductase), which can substitute for the function of DsbB (Dutton et al. 2010; Li et al. 2010). Disulfide bonds can be formed either during or after translocation, and the mode of disulfide bond formation (i.e., co- or posttranslocational) appears to influence whether disulfide bonds are formed between consecutive cysteines in the sequence of a substrate protein or between cysteines that become contiguous only in the native structure (Kadokura and Beckwith 2009).
Often, the oxidative machinery forms disulfide bonds that are not compatible with the native structure of a protein. Such oxidative misfolding occurs more frequently in proteins whose native structure contains multiple disulfide bonds between cysteines that are not consecutive in the amino acid sequence (Berkmen et al. 2005). In such cases, the disulfide bond isomerases DsbC and DsbG catalyze isomerization of these disulfides to their native conformation (Fig. 5) (Ito and Inaba 2008; Kadokura et al. 2003). In addition, DsbC and DsbG have a role in periplasmic quality control by protecting proteins harboring a single cysteine residue from oxidation (Depuydt et al. 2009). Although it is possible that the isomerases could catalyze disulfide isomerization entirely through mixed-disulfide intermediates (Arredondo et al. 2009), DsbC is thought to function in vivo by binding to misfolded substrates through their dimerization domains and reducing them to allow another round of oxidation by DsbA (McCarthy et al. 2000; Shouldice et al. 2010). DsbC is maintained in a reduced state by the integral membrane protein DsbD, which is in turn reduced by cytoplasmic thioredoxin (Missiakas et al. 1995; Rietsch et al. 1996; Rietsch et al. 1997).
ACKNOWLEDGMENTS
We thank Yves Cully for help with figure preparation. This work was supported by grants from the Deutsche Forschungsgemeinschaft (Bu617/17-1, SFB638) to B.B. and A.M and a Humboldt fellowship to D.H.
Footnotes
Editors: Richard Morimoto, Jeffrey Kelly, and Dennis Selkoe
Additional Perspectives on Protein Homeostasis available at www.cshperspectives.org
REFERENCES
- Agashe VR, Guha S, Chang HC, Genevaux P, Hayer-Hartl M, Stemp M, Georgopoulos C, Hartl FU, Barral JM 2004. Function of trigger factor and DnaK in multidomain protein folding: increase in yield at the expense of folding speed. Cell 117: 199–209 [DOI] [PubMed] [Google Scholar]
- Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T 2003. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science 299: 1751–1753 [DOI] [PubMed] [Google Scholar]
- Allen SP, Polazzi JO, Gierse JK, Easton AM 1992. Two novel heat shock genes encoding proteins produced in response to heterologous protein expression in Escherichia coli. J Bacteriol 174: 6938–6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apetri AC, Horwich AL 2008. Chaperonin chamber accelerates protein folding through passive action of preventing aggregation. Proc Natl Acad Sci 105: 17351–17355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arie JP, Sassoon N, Betton JM 2001. Chaperone function of FkpA, a heat shock prolyl isomerase, in the periplasm of Escherichia coli. Mol Microbiol 39: 199–210 [DOI] [PubMed] [Google Scholar]
- Arredondo SA, Chen TF, Riggs AF, Gilbert HF, Georgiou G 2009. Role of dimerization in the catalytic properties of the Escherichia coli disulfide isomerase DsbC. J Biol Chem 284: 23972–23979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell JC, Lee JO, Jander G, Martin N, Belin D, Beckwith J 1993. A pathway for disulfide bond formation in vivo. Proc Natl Acad Sci 90: 1038–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MM, Pinkner JS, Soto GE, Sauer FG, Langermann S, Waksman G, Frieden C, Hultgren SJ 2000. PapD-like chaperones provide the missing information for folding of pilin proteins. Proc Natl Acad Sci 97: 7709–7714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Zvi A, De Los Rios P, Dietler G, Goloubinoff P 2004. Active solubilization and refolding of stable protein aggregates by cooperative unfolding action of individual hsp70 chaperones. J Biol Chem 279: 37298–37303 [DOI] [PubMed] [Google Scholar]
- Berkmen M, Boyd D, Beckwith J 2005. The non-consecutive disulfide bond of Escherichia coli phytase (AppA) renders it dependent on the protein-disulfide isomerase, DsbC. J Biol Chem 280: 11387–11394 [DOI] [PubMed] [Google Scholar]
- Bothmann H, Pluckthun A 2000. The periplasmic Escherichia coli peptidylprolyl cis,trans-isomerase FkpA. I. Increased functional expression of antibody fragments with and without cis-prolines. J Biol Chem 275: 17100–17105 [DOI] [PubMed] [Google Scholar]
- Bucca G, Brassington AM, Schonfeld HJ, Smith CP 2000. The HspR regulon of Streptomyces coelicolor: a role for the DnaK chaperone as a transcriptional co-repressordagger. Mol Microbiol 38: 1093–1103 [DOI] [PubMed] [Google Scholar]
- Bukau B 1993. Regulation of the E. coli heat shock response. Mol Microbiol 9: 671–680 [DOI] [PubMed] [Google Scholar]
- Chapman E, Farr GW, Usaite R, Furtak K, Fenton WA, Chaudhuri TK, Hondorp ER, Matthews RG, Wolf SG, Yates JR, et al. 2006. Global aggregation of newly translated proteins in an Escherichia coli strain deficient of the chaperonin GroEL. Proc Natl Acad Sci 103: 15800–15805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Henning U 1996. A periplasmic protein (Skp) of Escherichia coli selectively binds a class of outer membrane proteins. Mol Microbiol 19: 1287–1294 [DOI] [PubMed] [Google Scholar]
- Cheng G, Basha E, Wysocki VH, Vierling E 2008. Insights into small heat shock protein and substrate structure during chaperone action derived from hydrogen/deuterium exchange and mass spectrometry. J Biol Chem 283: 26634–26642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S, Maris C, Allain FH, Narberhaus F 2006. Molecular basis for temperature sensing by an RNA thermometer. EMBO J 25: 2487–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig EA, Gross CA 1991. Is Hsp70 the cellular thermometer? TiBS 16: 135–140 [DOI] [PubMed] [Google Scholar]
- Crane JM, Suo Y, Lilly AA, Mao C, Hubbell WL, Randall LL 2006. Sites of interaction of a precursor polypeptide on the export chaperone SecB mapped by site-directed spin labeling. J Mol Biol 363: 63–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey FE, Berg HC 1993. Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc Natl Acad Sci 90: 1043–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dartigalongue C, Raina S 1998. A new heat-shock gene, ppiD, encodes a peptidyl-prolyl isomerase required for folding of outer membrane proteins in Escherichia coli. EMBO J 17: 3968–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt M, Leonard SE, Vertommen D, Denoncin K, Morsomme P, Wahni K, Messens J, Carroll KS, Collet JF 2009. A periplasmic reducing system protects single cysteine residues from oxidation. Science 326: 1109–1111 [DOI] [PubMed] [Google Scholar]
- Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B 1999. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400: 693–696 [DOI] [PubMed] [Google Scholar]
- Dodson KW, Jacob-Dubuisson F, Striker RT, Hultgren SJ 1993. Outer-membrane PapC molecular usher discriminately recognizes periplasmic chaperone-pilus subunit complexes. Proc Natl Acad Sci 90: 3670–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan DA, Reid BG, Horwich AL, Bukau B 2002. ClpS, a substrate modulator of the ClpAP machine. Mol Cell 9: 673–683 [DOI] [PubMed] [Google Scholar]
- Driessen AJ, Nouwen N 2008. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem 77: 643–667 [DOI] [PubMed] [Google Scholar]
- Dutton RJ, Wayman A, Wei JR, Rubin EJ, Beckwith J, Boyd D 2010. Inhibition of bacterial disulfide bond formation by the anticoagulant warfarin. Proc Natl Acad Sci 107: 297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrnsperger M, Gräber S, Gaestel M, Buchner J 1997. Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J 16: 221–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels S, Ludwig C, Schweitzer JE, Mack C, Bott M, Schaffer S 2005. The transcriptional activator ClgR controls transcription of genes involved in proteolysis and DNA repair in Corynebacterium glutamicum. Mol Microbiol 57: 576–591 [DOI] [PubMed] [Google Scholar]
- Ewalt KL, Hendrick JP, Houry WA, Hartl FU 1997. In vivo observation of polypeptide flux through the bacterial chaperonin system. Cell 90: 491–500 [DOI] [PubMed] [Google Scholar]
- Farr GW, Furtak K, Rowland MB, Ranson NA, Saibil HR, Kirchhausen T, Horwich AL 2000. Multivalent binding of nonnative substrate proteins by the chaperonin GroEL. Cell 100: 561–573 [DOI] [PubMed] [Google Scholar]
- Fayet O, Ziegelhoffer T, Georgopoulos C 1989. The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J Bacteriol 171: 1379–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferbitz L, Maier T, Patzelt H, Bukau B, Deuerling E, Ban N 2004. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 431: 590–596 [DOI] [PubMed] [Google Scholar]
- Flaherty KM, Deluca-Flaherty C, McKay DB 1990. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature 346: 623–628 [DOI] [PubMed] [Google Scholar]
- Frees D, Chastanet A, Qazi S, Sorensen K, Hill P, Msadek T, Ingmer H 2004. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol Microbiol 54: 1445–1462 [DOI] [PubMed] [Google Scholar]
- Friedrich KL, Giese KC, Buan NR, Vierling E 2004. Interactions between small heat shock protein subunits and substrate in small heat shock protein-substrate complexes. J Biol Chem 279: 1080–1089 [DOI] [PubMed] [Google Scholar]
- Gamer J, Bujard H, Bukau B 1992. Physical interaction between heat shock proteins DnaK, DnaJ, GrpE and the bacterial heat shock transcription factor s32. Cell 69: 833–842 [DOI] [PubMed] [Google Scholar]
- Genevaux P, Keppel F, Schwager F, Langendijk-Genevaux PS, Hartl FU, Georgopoulos C 2004. In vivo analysis of the overlapping functions of DnaK and trigger factor. EMBO Rep 5: 195–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos CP, Lam B, Lundquist-Heil A, Rudolph CF, Yochem J, Feiss M 1979. Identification of the C. coli dnaK (groPC756) gene product. Mol Gen Genet 172: 143–149 [DOI] [PubMed] [Google Scholar]
- Glover JR, Lindquist S 1998. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell 94: 73–82 [DOI] [PubMed] [Google Scholar]
- Goldberg AL, Swamy KH, Chung CH, Larimore FS 1981. Proteases in Escherichia coli. Methods Enzymol 80: 680–702 [DOI] [PubMed] [Google Scholar]
- Goloubinoff P, Mogk A, Peres Ben Zvi A, Tomoyasu T, Bukau B 1999. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc Natl Acad Sci 96: 13732–13737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf PC, Jakob U 2002. Redox-regulated molecular chaperones. Cell Mol Life Sci 59: 1624–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gragerov A, Nudler E, Komissarova N, Gaitanaris G, Gottesman M, Nikiforov V 1992. Cooperation of GroEL/GroES and DnaK/DnaJ heat shock proteins in preventing protein misfolding in Eschrichia coli. Proc Natl Acad Sci 89: 10341–10344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groemping Y, Reinstein J 2001. Folding properties of the nucleotide exchange factor GrpE from Thermus thermophilus: GrpE is a thermosensor that mediates heat shock response. J Mol Biol 314: 167–178 [DOI] [PubMed] [Google Scholar]
- Guisbert E, Herman C, Lu CZ, Gross CA 2004. A chaperone network controls the heat shock response in E. coli. Genes Dev 18: 2812–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur E, Sauer RT 2008. Recognition of misfolded proteins by Lon, a AAA(+) protease. Genes Dev 22: 2267–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms N, Koningstein G, Dontje W, Muller M, Oudega B, Luirink J, de Cock H 2001. The early interaction of the outer membrane protein phoe with the periplasmic chaperone Skp occurs at the cytoplasmic membrane. J Biol Chem 276: 18804–18811 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M 2009. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol 16: 574–581 [DOI] [PubMed] [Google Scholar]
- Haslbeck M, Franzmann T, Weinfurtner D, Buchner J 2005. Some like it hot: the structure and function of small heat-shock proteins. Nat Struct Mol Biol 12: 842–846 [DOI] [PubMed] [Google Scholar]
- Haslbeck M, Walke S, Stromer T, Ehrnsperger M, White HE, Chen S, Saibil HR, Buchner J 1999. Hsp26: a temperature-regulated chaperone. EMBO J 18: 6744–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslberger T, Weibezahn J, Zahn R, Lee S, Tsai FT, Bukau B, Mogk A 2007. M domains couple the ClpB threading motor with the DnaK chaperone activity. Mol Cell 25: 247–260 [DOI] [PubMed] [Google Scholar]
- Haslberger T, Zdanowicz A, Brand I, Kirstein J, Turgay K, Mogk A, Bukau B 2008. Protein disaggregation by the AAA+ chaperone ClpB involves partial threading of looped polypeptide segments. Nat Struct Mol Biol 15: 641–650 [DOI] [PubMed] [Google Scholar]
- Hasselblatt H, Kurzbauer R, Wilken C, Krojer T, Sawa J, Kurt J, Kirk R, Hasenbein S, Ehrmann M, Clausen T 2007. Regulation of the sigmaE stress response by DegS: how the PDZ domain keeps the protease inactive in the resting state and allows integration of different OMP-derived stress signals on folding stress. Genes Dev 21: 2659–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesterkamp T, Hauser S, Lütcke H, Bukau B 1996. Escherichia coli trigger factor is a prolyl isomerase that associates with nascent polypeptide chains. Proc Natl Acad Sci 93: 4437–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Merz F, Rutkowska A, Zachmann-Brand B, Deuerling E, Bukau B 2006. Trigger factor forms a protective shield for nascent polypeptides at the ribosome. J Biol Chem 281: 6539–6545 [DOI] [PubMed] [Google Scholar]
- Hoffmann JH, Linke K, Graf PC, Lilie H, Jakob U 2004. Identification of a redox-regulated chaperone network. EMBO J 23: 160–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden JL, Nur-E-Kamal MSA, Fabri L, Nice E, Hammacher A, Maruta H 1991. Rsr1 and Rap1 GTPases are activated by the same GTPase-activating protein and require threonine 65 for their activation. J Biol Chem 266: 16992–16995 [PubMed] [Google Scholar]
- Horwich AL, Apetri AC, Fenton WA 2009. The GroEL/GroES cis cavity as a passive anti-aggregation device. FEBS Lett 583: 2654–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houry WA, Frishman D, Eckerskorn C, Lottspeich F, Hartl FU 1999. Identification of in vivo substrates of the chaperonin GroEL. Nature 402: 147–154 [DOI] [PubMed] [Google Scholar]
- Huang HC, Sherman MY, Kandror O, Goldberg AL 2001. The molecular chaperone DnaJ is required for the degradation of a soluble abnormal protein in Escherichia coli. J Biol Chem 276: 3920–3928 [DOI] [PubMed] [Google Scholar]
- Huber D, Bukau B 2008. DegP: a Protein “Death Star.” Structure 16: 989–990 [DOI] [PubMed] [Google Scholar]
- Huber D, Boyd D, Xia Y, Olma MH, Gerstein M, Beckwith J 2005. Use of thioredoxin as a reporter to identify a subset of Escherichia coli signal sequences that promote signal recognition particle-dependent translocation. J Bacteriol 187: 2983–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba K, Murakami S, Suzuki M, Nakagawa A, Yamashita E, Okada K, Ito K 2006a. Crystal structure of the DsbB-DsbA complex reveals a mechanism of disulfide bond generation. Cell 127: 789–801 [DOI] [PubMed] [Google Scholar]
- Inaba K, Takahashi YH, Ito K, Hayashi S 2006b. Critical role of a thiolate-quinone charge transfer complex and its adduct form in de novo disulfide bond generation by DsbB. Proc Natl Acad Sci 103: 287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Inaba K 2008. The disulfide bond formation (Dsb) system. Curr Opin Struct Biol 18: 450–458 [DOI] [PubMed] [Google Scholar]
- Jaya N, Garcia V, Vierling E 2009. Substrate binding site flexibility of the small heat shock protein molecular chaperones. Proc Natl Acad Sci 106: 15604–15609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadokura H, Beckwith J 2009. Detecting folding intermediates of a protein as it passes through the bacterial translocation channel. Cell 138: 1164–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadokura H, Katzen F, Beckwith J 2003. Protein disulfide bond formation in prokaryotes. Annu Rev Biochem 72: 111–135 [DOI] [PubMed] [Google Scholar]
- Kamitani S, Akiyama Y, Ito K 1992. Identification and characterization of an Escherichia coli gene required for the formation of correctly folded alkaline phosphatase, a periplasmic enzyme. Embo J 11: 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemori M, Nishihara K, Yanagi H, Yura T 1997. Synergistic Roles of HslVU and Other ATP-Dependent Proteases in Controlling In Vivo Turnover of s32 and Abnormal Proteins in Escherichia coli. J Bacteriol 179: 7219–7225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang HC, Stines AP, Georgopoulos C, Frishman D, Hayer-Hartl M, Mann M, et al. 2005. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122: 209–220 [DOI] [PubMed] [Google Scholar]
- Kim KK, Kim R, Kim S-H 1998. Crystal structure of a small heat shock protein. Nature 394: 595–599 [DOI] [PubMed] [Google Scholar]
- Kirstein J, Dougan DA, Gerth U, Hecker M, Turgay K 2007. The tyrosine kinase McsB is a regulated adaptor protein for ClpCP. EMBO J 26: 2061–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein J, Strahl H, Moliere N, Hamoen LW, Turgay K 2008. Localization of general and regulatory proteolysis in Bacillus subtilis cells. Mol Microbiol 70: 682–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kock H, Gerth U, Hecker M 2004. The ClpP peptidase is the major determinant of bulk protein turnover in Bacillus subtilis. J Bacteriol 186: 5856–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondror O, Sherman M, Rhode M, Goldberg AL 1995. Trigger factor is involved in GroEL-dependent protein degradation in Escherichia coli and promotes binding of GroEL to unfolded proteins. EMBO J 14: 6021–6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowit JD, Goldberg AL 1977. Intermediate steps in the degradation of a specific abnormal protein in Escherichia coli. J Biol Chem 252: 8350–8357 [PubMed] [Google Scholar]
- Kramer G, Boehringer D, Ban N, Bukau B 2009. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol 16: 589–597 [DOI] [PubMed] [Google Scholar]
- Kramer G, Patzelt H, Rauch T, Kurz TA, Vorderwulbecke S, Bukau B, Deuerling E 2004a. Trigger factor peptidyl-prolyl cis/trans isomerase activity is not essential for the folding of cytosolic proteins in Escherichia coli. J Biol Chem 279: 14165–14170 [DOI] [PubMed] [Google Scholar]
- Kramer G, Rauch T, Rist W, Vorderwulbecke S, Patzelt H, Schulze-Specking A, Ban N, Deuerling E, Bukau B 2002. L23 protein functions as a chaperone docking site on the ribosome. Nature 419: 171–174 [DOI] [PubMed] [Google Scholar]
- Kramer G, Rutkowska A, Wegrzyn RD, Patzelt H, Kurz TA, Merz F, Rauch T, Vorderwulbecke S, Deuerling E, Bukau B 2004b. Functional dissection of Escherichia coli trigger factor: unraveling the function of individual domains. J Bacteriol 186: 3777–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krojer T, Sawa J, Schafer E, Saibil HR, Ehrmann M, Clausen T 2008. Structural basis for the regulated protease and chaperone function of DegP. Nature 453: 885–890 [DOI] [PubMed] [Google Scholar]
- Kruger E, Witt E, Ohlmeier S, Hanschke R, Hecker M 2000. The clp proteases of Bacillus subtilis are directly involved in degradation of misfolded proteins. J Bacteriol 182: 3259–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger E, Zuhlke D, Witt E, Ludwig H, Hecker M 2001. Clp-mediated proteolysis in Gram-positive bacteria is autoregulated by the stability of a repressor. Embo J 20: 852–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzewska J, Langer T, Liberek K 2001. Mitochondrial Hsp78, a member of the Clp/Hsp100 family in Saccharomyces cerevisiae, cooperates with Hsp70 in protein refolding. FEBS Lett 489: 92–96 [DOI] [PubMed] [Google Scholar]
- Lakshmipathy SK, Tomic S, Kaiser CM, Chang HC, Genevaux P, Georgopoulos C, Barral JM, Johnson AE, Hartl FU, Etchells SA 2007. Identification of nascent chain interaction sites on trigger factor. J Biol Chem 282: 12186–12193 [DOI] [PubMed] [Google Scholar]
- Laskowska E, Bohdanowicz J, Kuczynska-Wisnik D, Matuszewska E, Kedzierska S, Taylor A 2004. Aggregation of heat-shock-denatured, endogenous proteins and distribution of the IbpA/B and Fda marker-proteins in Escherichia coli WT and grpE280 cells. Microbiology 150: 247–259 [DOI] [PubMed] [Google Scholar]
- Laskowska E, Wawrzynow A, Taylor A 1996. IbpA and IbpB, the new heat-shock proteins, bind to endogenous Escherichia coli proteins aggregated intracellularly by heat shock. Biochimie 78: 117–122 [DOI] [PubMed] [Google Scholar]
- Lazar SW, Kolter R 1996. SurA assists the folding of Escherichia coli outer membrane proteins. J Bacteriol 178: 1770–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GJ, Roseman AM, Saibil HR, Vierling E 1997. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. Embo J 16: 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Owen HA, Prochaska DJ, Barnum SR 2000. HSP16.6 is involved in the development of thermotolerance and thylakoid stability in the unicellular cyanobacterium, Synechocystis sp. PCC 6803. Curr Microbiol 40: 283–287 [DOI] [PubMed] [Google Scholar]
- Li W, Schulman S, Dutton RJ, Boyd D, Beckwith J, Rapoport TA 2010. Structure of a bacterial homologue of vitamin K epoxide reductase. Nature 463: 507–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K, Galitski TP, Zylicz M, Georgopoulos C 1992. The DnaK chaperone modulates the heat shock response of Escherichia coli by binding to the s32 transcription factor. Proc Natl Acad Sci 89: 3516–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Madan D, Rye HS 2008. GroEL stimulates protein folding through forced unfolding. Nat Struct Mol Biol 15: 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F 2008. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc Natl Acad Sci 105: 3076–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum R, Tkach JM, Vierling E, Glover JR 2004. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem 279: 29139–29146 [DOI] [PubMed] [Google Scholar]
- Maier T, Ferbitz L, Deuerling E, Ban N 2005. A cradle for new proteins: trigger factor at the ribosome. Curr Opin Struct Biol 15: 204–212 [DOI] [PubMed] [Google Scholar]
- Maisonneuve E, Ezraty B, Dukan S 2008a. Protein aggregates: an aging factor involved in cell death. J Bacteriol 190: 6070–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve E, Fraysse L, Lignon S, Capron L, Dukan S 2008b. Carbonylated proteins are detectable only in a degradation-resistant aggregate state in Escherichia coli. J Bacteriol 190: 6609–6614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve E, Fraysse L, Moinier D, Dukan S 2008c. Existence of abnormal protein aggregates in healthy Escherichia coli cells. J Bacteriol 190: 887–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Hackert E, Hendrickson WA 2009. Promiscuous substrate recognition in folding and assembly activities of the trigger factor chaperone. Cell 138: 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuszewska M, Kuczynska-Wisnik D, Laskowska E, Liberek K 2005. The small heat shock protein IbpA of Escherichia coli cooperates with IbpB in stabilization of thermally aggregated proteins in a disaggregation competent state. J Biol Chem 280: 12292–12298 [DOI] [PubMed] [Google Scholar]
- Mayer MP, Bukau B 2005. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 62: 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy AA, Haebel PW, Torronen A, Rybin V, Baker EN, Metcalf P 2000. Crystal structure of the protein disulfide bond isomerase, DsbC, from Escherichia coli. Nat Struct Biol 7: 196–199 [DOI] [PubMed] [Google Scholar]
- Meltzer M, Hasenbein S, Mamant N, Merdanovic M, Poepsel S, Hauske P, Kaiser M, Huber R, Krojer T, Clausen T, et al. 2009. Structure, function and regulation of the conserved serine proteases DegP and DegS of Escherichia coli. Res Microbiol 160: 660–666 [DOI] [PubMed] [Google Scholar]
- Merz F, Boehringer D, Schaffitzel C, Preissler S, Hoffmann A, Maier T, Rutkowska A, Lozza J, Ban N, Bukau B, et al. 2008. Molecular mechanism and structure of Trigger Factor bound to the translating ribosome. EMBO J 27: 1622–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miethke M, Hecker M, Gerth U 2006. Involvement of Bacillus subtilis ClpE in CtsR degradation and protein quality control. J Bacteriol 188: 4610–4619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missiakas D, Georgopoulos C, Raina S 1993. Identification and characterization of the Escherichia coli gene dsbB, whose product is involved in the formation of disulfide bonds in vivo. Proc Natl Acad Sci 90: 7084–7088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missiakas D, Schwager F, Raina S 1995. Identification and characterization of a new disulfide isomerase-like protein (DsbD) in Escherichia coli. Embo J 14: 3415–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen JE, Otzen DE 2005. Interactions between folding factors and bacterial outer membrane proteins. Mol Microbiol 57: 326–346 [DOI] [PubMed] [Google Scholar]
- Mogk A, Deuerling E, Vorderwulbecke S, Vierling E, Bukau B 2003a. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol Microbiol 50: 585–595 [DOI] [PubMed] [Google Scholar]
- Mogk A, Homuth G, Scholz C, Kim L, Schmid FX, Schumann W 1997. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO Journal 16: 4579–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Schlieker C, Friedrich KL, Schönfeld H-J, Vierling E, Bukau B 2003b. Refolding of substrates bound to small Hsps relies on a disaggregation reaction mediated most efficiently by ClpB/DnaK. J Biol Chem 278: 31033–31042 [DOI] [PubMed] [Google Scholar]
- Mogk A, Tomoyasu T, Goloubinoff P, Rüdiger S, Röder D, Langen H, Bukau B 1999. Identification of thermolabile E. coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J 18: 6934–6949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita MT, Tanaka Y, Kodama TS, Kyogoku Y, Yanagi H, Yura T 1999. Translational induction of heat shock transcription factor sigma32: evidence for a built-in RNA thermosensor. Genes Dev 13: 655–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K, Watanabe Y, Yohda M, Yoshida M 1999. Heat-inactivated proteins are rescued by the DnaK.J-GrpE set and ClpB chaperones. Proc Natl Acad Sci 96: 7184–7189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narberhaus F, Waldminghaus T, Chowdhury S 2006. RNA thermometers. FEMS Microbiol Rev 30: 3–16 [DOI] [PubMed] [Google Scholar]
- Narberhaus F, Käser R, Nocker A, Hennecke H 1998. A novel DNA element that controls bacterial heat shock gene expression. Mol Microbiol 28: 315–323 [DOI] [PubMed] [Google Scholar]
- Nonaka G, Blankschien M, Herman C, Gross CA, Rhodius VA 2006. Regulon and promoter analysis of the E. coli heat-shock factor, sigma32, reveals a multifaceted cellular response to heat stress. Genes Dev 20: 1776–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek KH, Walker GC 1987. Escherichia coli dnaK null mutants are inviable at high temperature. J Bacteriol 169: 283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramm K, Pluckthun A 2000. The periplasmic Escherichia coli peptidylprolyl cis,trans-isomerase FkpA. II. Isomerase-independent chaperone activity in vitro. J Biol Chem 275: 17106–17113 [DOI] [PubMed] [Google Scholar]
- Rapoport TA 2007. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450: 663–669 [DOI] [PubMed] [Google Scholar]
- Ratajczak E, Zietkiewicz S, Liberek K 2009. Distinct activities of Escherichia coli small heat shock proteins IbpA and IbpB promote efficient protein disaggregation. J Mol Biol 386: 178–189 [DOI] [PubMed] [Google Scholar]
- Rietsch A, Belin D, Martin N, Beckwith J 1996. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc Natl Acad Sci 93: 13048–13053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietsch A, Bessette P, Georgiou G, Beckwith J 1997. Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J Bacteriol 179: 6602–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez F, Arsene-Ploetze F, Rist W, Rudiger S, Schneider-Mergener J, Mayer MP, Bukau B 2008. Molecular basis for regulation of the heat shock transcription factor sigma32 by the DnaK and DnaJ chaperones. Mol Cell 32: 347–358 [DOI] [PubMed] [Google Scholar]
- Rokney A, Shagan M, Kessel M, Smith Y, Rosenshine I, Oppenheim AB 2009. E. coli transports aggregated proteins to the poles by a specific and energy-dependent process. J Mol Biol 392: 589–601 [DOI] [PubMed] [Google Scholar]
- Roudiak SG, Shrader TE 1998. Functional role of the N-terminal region of the Lon protease from Mycobacterium smegmatis. Biochemistry 37: 11255–11263 [DOI] [PubMed] [Google Scholar]
- Rouviere PE, Gross CA 1996. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev 10: 3170–3182 [DOI] [PubMed] [Google Scholar]
- Rüdiger S, Germeroth L, Schneider-Mergener J, Bukau B 1997. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J 16: 1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N, Silhavy TJ 2005. Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr Opin Microbiol 8: 122–126 [DOI] [PubMed] [Google Scholar]
- Ruiz N, Kahne D, Silhavy TJ 2006. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol 4: 57–66 [DOI] [PubMed] [Google Scholar]
- Rujano MA, Bosveld F, Salomons FA, Dijk F, van Waarde MA, van der Want JJ, de Vos RA, Brunt ER, Sibon OC, Kampinga HH 2006. Polarised asymmetric inheritance of accumulated protein damage in higher eukaryotes. PLoS Biol 4: e417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, et al. 2004. Sculpting the Proteome with AAA+ Proteases and Disassembly Machines. Cell 119: 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer U, Beck K, Muller M 1999. Skp, a molecular chaperone of gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J Biol Chem 274: 24567–24574 [DOI] [PubMed] [Google Scholar]
- Schlee S, Beinker P, Akhrymuk A, Reinstein J 2004. A chaperone network for the resolubilization of protein aggregates: direct interaction of ClpB and DnaK. J Mol Biol 336: 275–285 [DOI] [PubMed] [Google Scholar]
- Schlieker C, Tews I, Bukau B, Mogk A 2004a. Solubilization of aggregated proteins by ClpB/DnaK relies on the continuous extraction of unfolded polypeptides. FEBS Lett 578: 351–356 [DOI] [PubMed] [Google Scholar]
- Schlieker C, Weibezahn J, Patzelt H, Tessarz P, Strub C, Zeth K, Erbse A, Schneider-Mergener J, Chin JW, Schultz PG, et al. 2004b. Substrate recognition by the AAA+ chaperone ClpB. Nat Struct Mol Biol 11: 607–615 [DOI] [PubMed] [Google Scholar]
- Schlothauer T, Mogk A, Dougan DA, Bukau B, Turgay K 2003. MecA, an adaptor protein necessary for ClpC chaperone activity. Proc Natl Acad Sci 100: 2306–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant P, Grandvalet C, Mazodier P 2000. The RheA repressor is the thermosensor of the HSP18 heat shock response in Streptomyces albus. Proc Natl Acad Sci 97: 3538–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Chakraborty K, Muller BK, Astola N, Tang YC, Lamb DC, Hayer-Hartl M, Hartl FU 2008. Monitoring protein conformation along the pathway of chaperonin-assisted folding. Cell 133: 142–153 [DOI] [PubMed] [Google Scholar]
- Shouldice SR, Cho SH, Boyd D, Heras B, Eser M, Beckwith J, Riggs P, Martin JL, Berkmen M 2010. In vivo oxidative protein folding can be facilitated by oxidation-reduction cycling. Mol Microbiol 75: 13–28 [DOI] [PubMed] [Google Scholar]
- Skowyra D, Georgopoulos C, Zylicz M 1990. The E. coli dnaK gene product, the Hsp70 homolog, can reactivate heat-inactivated RNA polymerase in an ATP hydrolysis-dependent manner. Cell 62: 939–944 [DOI] [PubMed] [Google Scholar]
- Sohn J, Grant RA, Sauer RT 2007. Allosteric activation of DegS, a stress sensor PDZ protease. Cell 131: 572–583 [DOI] [PubMed] [Google Scholar]
- Sohn J, Grant RA, Sauer RT 2009. OMP peptides activate the DegS stress-sensor protease by a relief of inhibition mechanism. Structure 17: 1411–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires CL, Pedersen S, Ross BM, Squires C 1991. ClpB is the Escherichia coli heat shock protein F84.1. J Bacteriol 173: 4254–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel F, Baldwin AJ, Painter AJ, Jaya N, Basha E, Kay LE, Vierling E, Robinson CV, Benesch JL 2010. Quaternary dynamics and plasticity underlie small heat shock protein chaperone function. Proc Natl Acad Sci 107: 2007–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus D, Walter W, Gross CA 1990. DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of s32. Genes and Dev 4: 2202–2209 [DOI] [PubMed] [Google Scholar]
- Suokko A, Poutanen M, Savijoki K, Kalkkinen N, Varmanen P 2008. ClpL is essential for induction of thermotolerance and is potentially part of the HrcA regulon in Lactobacillus gasseri. Proteomics 8: 1029–1041 [DOI] [PubMed] [Google Scholar]
- Tang YC, Chang HC, Roeben A, Wischnewski D, Wischnewski N, Kerner MJ, Hartl FU, Hayer-Hartl M 2006. Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell 125: 903–914 [DOI] [PubMed] [Google Scholar]
- Tapley TL, Franzmann TM, Chakraborty S, Jakob U, Bardwell JC 2010. Protein refolding by pH-triggered chaperone binding and release. Proc Natl Acad Sci 107: 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teter SA, Houry WA, Ang D, Tradler T, Rockabrand D, Fischer G, Blum P, Georgopoulos C, Hartl FU 1999. Polypeptide flux through bacterial Hsp70: DnaK cooperates with Trigger Factor in chaperoning nascent chains. Cell 97: 755–765 [DOI] [PubMed] [Google Scholar]
- Thanassi DG, Saulino ET, Lombardo MJ, Roth R, Heuser J, Hultgren SJ 1998. The PapC usher forms an oligomeric channel: implications for pilus biogenesis across the outer membrane. Proc Natl Acad Sci 95: 3146–3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, McKittrick N, Zylicz M, Georgopoulos C 1983. The DnaK protein modulates the heat-shock response of Escherichia coli. Cell 34: 641–646 [DOI] [PubMed] [Google Scholar]
- Tomoyasu T, Gamer J, Bukau B, Kanemori M, Mori H, Rutman AJ, Oppenheim AB, Yura T, Yamanaka K, Niki H, et al. 1995. Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat shock transcription factor s32. EMBO J 14: 2551–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu T, Mogk A, Langen H, Goloubinoff P, Bukau B 2001. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol Microbiol 40: 397–413 [DOI] [PubMed] [Google Scholar]
- Tyagi NK, Fenton WA, Horwich AL 2009. GroEL/GroES cycling: ATP binds to an open ring before substrate protein favoring protein binding and production of the native state. Proc Natl Acad Sci 106: 20264–20269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullers RS, Luirink J, Harms N, Schwager F, Georgopoulos C, Genevaux P 2004. SecB is a bona fide generalized chaperone in Escherichia coli. Proc Natl Acad Sci 101: 7583–7588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent QA, Kendall DA, High S, Kusters R, Oudega B, Luirink J 1995. Early events in preprotein recognition in E. coli: interaction of SRP and trigger factor with nascent polypeptides. EMBO J 14: 5494–5505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Montfort R, Slingsby C, Vierling E 2001a. Structure and function of the small heat shock protein/α-crystallin family of molecular chaperones. Adv Protein Chem 59: 105–156 [DOI] [PubMed] [Google Scholar]
- van Montfort R, Slingsby C, Vierling E 2002. In Structure and Function of small heat shock protein/a-crystallin family of molecular chaperones, Vol. 59, New York: Academic press; [DOI] [PubMed] [Google Scholar]
- van Montfort RL, Basha E, Friedrich KL, Slingsby C, Vierling E 2001b. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol 8: 1025–1030 [DOI] [PubMed] [Google Scholar]
- Vorderwulbecke S, Kramer G, Merz F, Kurz TA, Rauch T, Zachmann-Brand B, Bukau B, Deuerling E 2004. Low temperature or GroEL/ES overproduction permits growth of Escherichia coli cells lacking trigger factor and DnaK. FEBS Lett 559: 181–187 [DOI] [PubMed] [Google Scholar]
- Waksman G, Hultgren SJ 2009. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat Rev Microbiol 7: 765–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldminghaus T, Fippinger A, Alfsmann J, Narberhaus F 2005. RNA thermometers are common in α- and γ-proteobacteria. Biol Chem 386: 1279–1286 [DOI] [PubMed] [Google Scholar]
- Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT 2003. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113: 61–71 [DOI] [PubMed] [Google Scholar]
- Walton TA, Sousa MC 2004. Crystal structure of Skp, a prefoldin-like chaperone that protects soluble and membrane proteins from aggregation. Mol Cell 15: 367–374 [DOI] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FT, et al. 2004. Thermotolerance Requires Refolding of Aggregated Proteins by Substrate Translocation through the Central Pore of ClpB. Cell 119: 653–665 [DOI] [PubMed] [Google Scholar]
- Winkler J, Seybert A, König L, Pruggnaller S, Haselmann U, Sourjik V, Weiss M, Frangakis AS, Mogk A, Bukau B 2010. Quantitative and spatio-temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO J 29: 910–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Ilbert M, Graf PC, Ozcelik D, Jakob U 2008. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell 135: 691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Linke K, Jatzek A, Jakob U 2005. Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol Cell 17: 381–392 [DOI] [PubMed] [Google Scholar]
- Xu Z, Horwich AL, Sigler PB 1997. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature 388: 741–750 [DOI] [PubMed] [Google Scholar]
- Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman M, Hendrickson WA 1996. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272: 1606–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietkiewicz S, Krzewska J, Liberek K 2004. Successive and synergistic action of the Hsp70 and Hsp100 chaperones in protein disaggregation. J Biol Chem 279: 44376–44383 [DOI] [PubMed] [Google Scholar]
- Zolkiewski M 1999. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli. J Biol Chem 274: 28083–28086 [DOI] [PubMed] [Google Scholar]